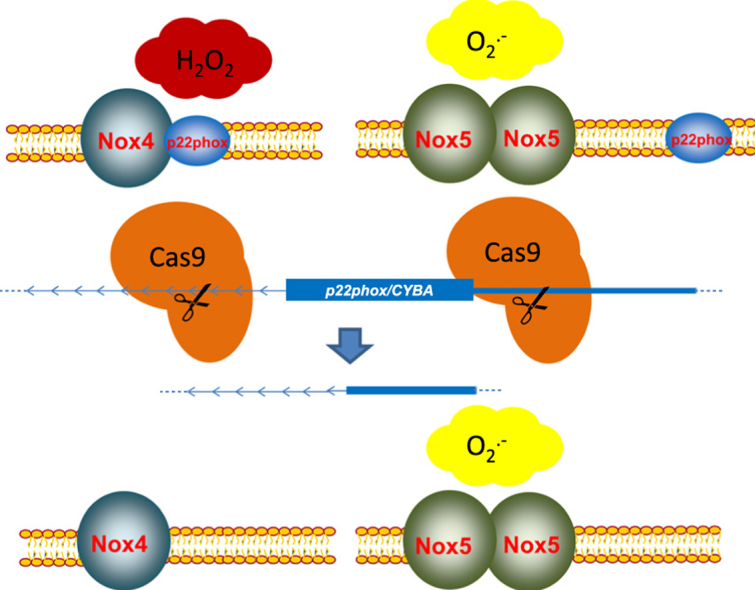
Abstract
The NADPH oxidases are important transmembrane proteins producing reactive oxygen species (ROS). Within the Nox family, different modes of activation can be discriminated. Nox1-3 are dependent on different cytosolic subunits, Nox4 seems to be constitutively active and Nox5 is directly activated by calcium. With the exception of Nox5, all Nox family members are thought to depend on the small transmembrane protein p22phox. With the discovery of the CRISPR/Cas9-system, a tool to alter genomic DNA sequences has become available. So far, this method has not been widely used in the redox community. On such basis, we decided to study the requirement of p22phox in the Nox complex using CRISPR/Cas9-mediated knockout. Knockout of the gene of p22phox, CYBA, led to an ablation of activity of Nox4 and Nox1 but not of Nox5. Production of hydrogen peroxide or superoxide after knockout could be rescued with either human or rat p22phox, but not with the DUOX-maturation factors DUOXA1/A2. Furthermore, different mutations of p22phox were studied regarding the influence on Nox4-dependent H2O2 production. P22phox Q130* and Y121H affected maturation and activity of Nox4. Hence, Nox5-dependent O2•− production is independent of p22phox, but native p22phox is needed for maturation of Nox4 and production of H2O2.
Abbreviations: CRISPR, Clustered regulatory interspaced short palindromic repeats; Cas9, CRISPR-associated protein 9; HEK293, human embryonic kidney 293 cells; HRP, horseradish peroxidase; Nox4-HEK293, HEK293 cells stably expressing Nox4; PMA, phorbol 12-myristate 13-acetate; tetNox4-HEK293, HEK293 cells expressing Nox4 in tetracycline-inducible tet-on vector
Keywords: NADPH oxidase, CYBA/p22phox, CRISPR/Cas9
Graphical abstract
1. Introduction
The Nox family of NADPH oxidases is an important source of reactive oxygen species (ROS). Through ROS, Nox enzymes play diverse roles in cellular signaling, regulation of gene expression, cell differentiation and posttranslational protein modifications [4]. In mammals, seven members are expressed and grouped according to their mode of activation. Nox1, Nox2 and Nox3 are activated by an assembly with specific cytosolic proteins, which translocate to the plasma membrane. On the other hand, Nox5 and DUOX1 as well as DUOX2 (Dual oxidase 1 and 2) are directly activated by calcium in an EF-hand motif dependent manner. To our current knowledge, Nox4, in contrast, is constitutively active and ROS production of the enzyme is mainly controlled by its expression level [5].
For DUOX1 or DUOX2, maturating factors are needed to achieve proper expression and activity. The small transmembrane protein p22phox/CYBA (Cytochrome B-245 Alpha Chain) was described to be essential for Nox1, Nox2 and Nox3 maturation, stabilization and activation [2], [20], [21], [31]. P22phox has been found to interact with Nox4, indicating importance for Nox4 activity and localization [18], [19], [2]. The exact site where p22phox interacts and activates Nox4 appears non-canonical with respect to Nox1, Nox2 or Nox3 [17], [31]. P22phox consists of two (or four) predicted transmembrane domains and an extended C-terminus with a prolin-rich region, which is required for the assembly with p47phox or NoxO1 to membrane-bound Nox1, Nox2 and Nox3 [31], [5]. This feature is dispensable for Nox4 indicating a different role for the p22phox-Nox4 activation [16], [17]. Mutations in p22phox are associated with a loss of function leading to severe immunodeficiency called chronic granulomatous disease (CGD), with inability to clear some pathogens and subsequent life threatening infections [10], [6].
Further analysis of the structural and functional role of p22phox are necessary not just for understanding the Nox2-dependent pathogen defense, but also for the analysis of Nox4, as a protective cardiovascular enzyme.
In the kidney, the endothelium and fibroblasts, Nox4 is the predominantly expressed Nox homolog [1], [11], [7]. Nox4 is fundamentally different to the other Nox enzymes and exhibits a different dependency on p22phox. Approaches to study p22phox-Nox4 interaction like the p22phox mutant mouse nmf333 could only partially reveal the mechanisms underlying the relevant protein interaction [17], [22]. There is, however, no doubt that p22phox is important for Nox4 and changing the expression of this ubiquitously expressed protein alters H2O2 formation of most cells overexpressing Nox4 [16], [2], [26]. In previous work, Löhneysen et al. inserted mutations in p22phox which reduced Nox4-dependent H2O2 formation. A dual tryptophan motif in the N-terminal amino acids (aa) 6-11 was found necessary for Nox4 activity and localization. P22phox mutants of the first five -but not ten- aa did not impact on activity and are thus tolerated as well as deletion of aa after residue 130, which are involved in binding of p47phox or NoxO1 and activation of Nox1-3. In line with this, p22phox mutations in the C-terminal prolin-rich region (P156Q-P160Q) are tolerated for Nox4-derived H2O2 formation [16], [17]. Point mutations in the putative membrane spanning domain are less well tolerated, e.g. non-conservative exchange of arginine 90 (R90Q) abolished Nox4-dependent H2O2 production [17], [27].
Given that protein abundance of p22phox is largely controlled by posttranscriptional mechanisms and that a few studies on complete knockout of p22phox have been performed so far, we set out to study the requirement of p22phox in the Nox complex using Clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated knockout. CRISPR/Cas9-mediated genome editing offers a fast and easy way to manipulate genomes in cell culture requiring just a 20-nucleotide targeting sequence, called guide RNA (gRNA). In one variant of RNA-guided Cas9 nuclease, the BbsI enzyme-mediated knockout strategy, a pair of gRNAs have been successfully used to mediate microdeletions e. g. in the Homeobox protein EMX1 [25].
The p22phox gene (CYBA) is located on the long arm of chromosome 16 and consists of six exons with a total length of approx. 8500 nucleotides [10]. As the first exon is found in all predicted variants of p22phox, gRNAs targeting or flanking the exon 1 were chosen for microdeletions. In this study CRISPR/Cas9-mediated p22phox HEK (human embryonic kidney cells) knockout cell lines were used to study the role of endogenous p22phox on the expression, maturation and activity of Nox4, but also Nox1 and Nox5. Moreover, truncated and modified p22phox variants with essential N-terminal and membrane domain mutations were analyzed in a constitutive and inducible Nox4-overexpressing and p22phox-deficient cellular system.
2. Materials and methods
2.1. Mutagenesis of p22phox
The QuikChange II XL Site-Directed Mutagenesis Kit (#200521, Agilent) was used to mutate p22phox. Primers were designed with the web-based QuikChange Primer Design Program. Briefly, the plasmid coding for human p22phox (kindly provided by Leto, NIH) was used for mutant strand synthesis reaction with PfuUltra HF DNA polymerase (2.5 U/µl) according to the kit, followed by a DpnI (10 U/µl) digestion of the original strand. The mutated plasmid was transformed into XL10-Gold Ultracompetent cells, the generated plasmid was purified with the GenJet Plasmid Miniprep Kit (K0503, Thermo Scientific). Mutations were verified by sequencing. The following primers were used for the introduction of single amino acid mutation: CYBA A91stop 5′-GAG AGC AGG AGA TGC AGG ACC TAC CGA ACA TAG TAA TTC CTG G-3′ and 5′-CCA GGA ATT ACT ATG TTC GGT AGG TCC TGC ATC TCC TGC TCT C-3′; CYBA E5A 5′-CCA CAT GGC CCA CGC GAT CTG CCC CAT-3′ and 5′ATG GGG CAG ATC GCG TGG GCC ATG TGG-3; CYBA E12A 5′CCA GCG CCT GCG CGT TGG CCC AC-3′2 and 5′-GTG GGC CAA CGC GCA GGC GCT GG-3′; CYBA G110stop 5′-CTG CTG GCC ACC ATC CTT TAG ACC GCC TGC-3′and 5′-GCA GGC GGT CTA AAG GAT GGT GGC CAG CAG-3′; CYBA I110C 5′-CCG CCA GTA GGT AGC AGC CGC TCG CAA TGG-3′ and 5′-CCA TTG CGA GCG GCT GCT ACC TAC TGG CGG-3; CYBA Q130stop 5′-TGG GCG TCC ACT ACT CGC CAC GCA C-3′ and 5′-GTG CGT GGC GAG TAG TGG ACG CCC A-3′; CYBA R90Q 5′-GCC CTT TAC CAG GAA TTA CTA TGT TGA GGC CGT CCT GCA T-3′and 5′-ATG CAG GAC GGC CTC AAC ATA GTA ATT CCT GGT AAA GGG C-3′; CYBA R90stop 5′-ATG CAG GAC GGC CTA AAC ATA GTA ATT CCT GGT AAA GGG C-3; CYBA r90stop 5′-GCC CTT TAC CAG GAA TTA CTA TGT TTA GGC CGT CCT GCA T-3; CYBA Y121H 5′-CCG CCA GTA GGT GGA TGC CGC TCG C-3′ and 5′-GCG AGC GGC ATC CAC CTA CTG GCG G-3′ as well Nox5 T493K 5′-CAG GAG CAC TGC TGA TCT TGA AGG GGT GCC ACTC-3′ and 5′-GAG TGG CAC CCC TTC AAG ATC AGC AGT GCT CCT G-3′.
2.2. Cell culture
Human embryonic kidney 293 cells (HEK293) cells were obtained from ATCC (Manassas, VA, USA) and cultured in Modified Eagle's Medium (MEM, Gibco) supplemented with fetal calf serum (FCS; 8%), non-essential amino acids (0.1 mM), sodium pyruvate (1 mM) and gentamycin (50 μg/mL) in a humidified atmosphere (5% CO2, 37 °C). HEK293 cells expressing human Nox4 with a tetracycline-inducible tet-on operator (Nox4-pDEST30) were kindly provided by K.H. Krause (University of Geneva, Switzerland) [29]. Induction with 1 µg/mL tetracycline was performed for 24 h in HEK medium. HEK293 cells stably expressing human Nox4 (Nox4-HEK293) were generated as described before using lentiviral transfection and selection [12].
2.3. Transfection of Nox constructs
Transient transfection of HEK293 cells was performed using Lipofectamine 2000 (#11668027, ThermoFisher) and 0.75 µg/mL plasmid DNA according to the manufacturer's instructions. Cells were kept in culture for 48 h. DUOXA1 and DUOXA2 plasmids were kindly provided by H. Grasberger (University of Michigan Medical School, Michigan).
2.4. Cloning of pHAGE2-NoxO1-NoxA1-Nox1 for transient overexpression
To clone NoxO1, NoxA1 and Nox1 as lentiviral triple overexpression plasmid under control of the human EF1a promoter, in which NoxO1 and NoxA1 are divided by a 2A peptide (F2A) and NoxA1 and Nox1 separated by an internal ribosome entry site (IRES), PCR amplifications of NoxO1, NoxA1 and Nox1 were performed from pCMV6-NoxO1 (NM_172167, #RC214973, Origene), pCMV6-NoxA1 (NM_001256067, #RC234211, Origene) and pCMV6-Nox1 (NM007052, #RC210426, Origene) Human cDNA ORF clones with the following primers: NoxO1 5′-ATA GCG GCC GCC ATG GCA GGC CCC CGA TAC CCA GTT TCA GTG-3′ and 5′-ATT TGC TTA GCC CCT GCT CCG TCG TGG GGT GCG GC-3′, NoxA1 5′-ATT AGC TAG CAT GGC CTC TCT GGG GGA CCT GGT GCG-3′ and 5′-ATT TGG ATC CTT AGG GCT GAT CTC CCT GCT GGG ATC GGG-3′ and 5′-ATA TTA CAT ATG ATG GGA AAC TGG GTG GTT AAC CAC-3′ and 5′-ATT GAT TAG GAT CTA TCG ACT CAA AAA TTT TCT TTG TTG AAG TAG-3′. In parallel, a F2A region for placing between NoxO1 and NoxA1 was amplified with 5′-ATT TGC TAA GCA CGG GTT CGG GTG CGC CAG TAA AGC AGA C-3′ and 5′-ATT TGC TAG CTG GAC CTG GAT TTG ACT CTA CAT CAC-3′ from pHAGE2, which was a gift from G. Mostoslavsky (Boston University, MA, USA) [30]. PCR products were cut in the following way to achieve a specific order of integration into pHAGE2: NoxO1 (NotI/BlpI (#ER0591, #ER0091, ThermoFisher)), F2A (BlpI/NheI (#ER0091, #ER0972, ThermoFisher)), NoxA1 (NheI/BamHI (#ER0972, #ER0055, ThermoFisher)) and Nox1 (NdeI/BsaBI (#ER0581, #ER1711, ThermoFisher)). In a first ligation reaction, pHAGE2 cut by BamHI/NotI (#ER0055, #ER0591, ThermoFisher) was used to be ligated with restricted NoxO1, F2A and NoxA1. This intermediate product was purified and further cut by NdeI/BsaBI (#ER0581, #ER1711, ThermoFisher) to be ligated in a second reaction with restricted Nox1. The final plasmid (pHAGE2-NoxO1-NoxA1-Nox1) was purified and sequenced.
2.5. Cloning of pHAGE2-Nox5 for stable overexpression
To clone pHAGE2-Nox5 for lentiviral overexpression under the control of the human EF1a promoter, Nox5 was amplified from pCMV6-Nox5 (NM_001184779) Human cDNA ORF Clone (#RC230471, Origene) with the following primers: 5′-ATT GCG GCC GCC ATG AAC ACA TCT GGA GAC CCA GCC CAG AC-3′ and 5′-ATT TGA TTA GGA TCT ATC GAC CTA GAA ATT CTC TTG GAA AAA TCT G-3′. pHAGE2 was a gift from Gustavo Mostoslavsky [30]. Nox5 PCR product and pHAGE2 were cut with NotI/BsaBI (#ER0591, #ER1711, ThermoFisher) followed by ligation. Plasmid was purified and sequenced.
2.6. Lentiviral transfection of HEK293 cells with pHAGE2-Nox5
Vesicular stomatitis virus (VSV) pseudotyped lentivirus was produced by transfection of HEK293T cells with pHAGE2-Nox5 in combination with HGPM2, Tat1B, Rev1B and VSV-G, which were a gift from Bianling Liu. Viral supernatants were collected, concentrated and snap-frozen after four days. Transfection of 70% subconfluent HEK293 with Nox5 viral particles was performed for 12 days, followed by clonal expansion.
2.7. CRISPR/Cas9
The web interface of CRISPR design (http://crispr.mit.edu/) was used to develop gRNAs. Off-target activity was evaluated additionally with Blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The pSpCas9(BB)-2A-Puro (PX459) v2.0 was a gift from Feng Zhang (Addgene plasmid # 62988) and used as cloning backbone. CRISPR/Cas9 was carried out similar as in [14], [25]. Briefly, phosphorylation and annealing was performed with the following oligonucleotides harboring a BbsI overhang: For (A) CYBA_1_Exon1 with 5′-CAC CGG CCG GGT TCG TGT CGC CAT G-3′ and 5′-AAA CCA TGG CGA CAC GAA CCC GGC C-3′, for (B) CYBA_2_Exon1 with 5′-CAC CGG GAC GCC AGC GCC TGT TCG T-3′ and 5′-AAA CAC GAA CAG GCG CTG GCG TCC C-3′, for (C) CYBA_3_Exon1 with 5′-CAC CGG GCC ATG TGG GCC AAC GAA C-3′ and 5′-AAA CGT TCG TTG GCC CAC ATG GCC C-3′, for (D) CYBA_1_Intron1 with 5′-CAC CGG TGC ACG TCA GGG ACG GTG G-3′ and 5′-AAA CCC ACC GTC CCT GAC GTG CAC C-3′, for (E) CYBA_2_Intron1 with 5′-CAC CGC GGG ACC TCG GGC TAC TTA C-3′ and 5′-AAA CGT AAG TAG CCC GAG GTC CCG C-3′, for (F) CYBA_3_Intron1 with 5′-CAC CGC CCA CCC TGT AAG TAG CCC G-3′ and 5′-AAA CCG GGC TAC TTA CAG GGT GGG C-3′ and for (G) CYBA_2_5′UTR with 5′-CAC CGC GGG GTT CGG CCG GGA GCG C-3′ and 5′-AAA CGC GCT CCC GGC CGA ACC CCG C-3′. Afterwards, BbsI (#FD1014, ThermoFisher) mediated digestion and T7 DNA ligase (#M0318L, NEB) directed ligation was performed followed by PlasmidSafe exonuclease treatment (#E3101K, Epicenter/Biozym) according to the manufacturers protocol. Plasmids were purified and sequenced. After transfection of the indicated combinations of pSpCas9(BB)-2A-Puro-gRNAs (Fig. 1B), positive cells were selected using puromycin (2 μg mL−1) for five days prior to clonal expansion. Empty pSpCas9(BB)-2A-Puro was used as negative control.
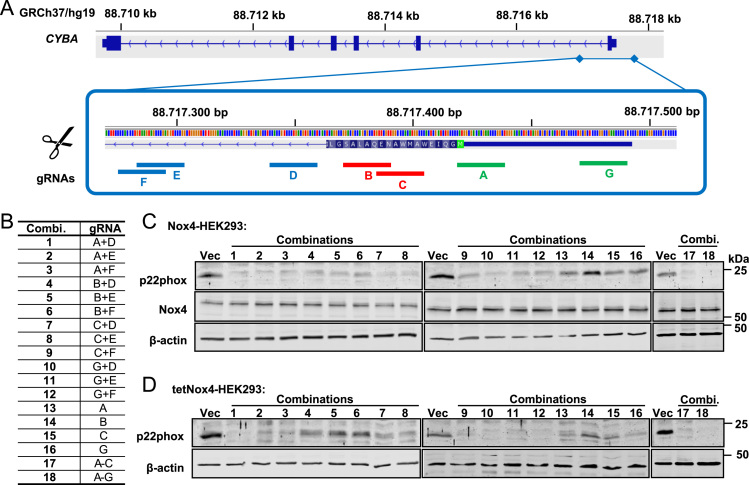
Fig. 1.
CRISPR/Cas9-mediated knockout of p22phox in Nox4/tetNox4-HEK293 cells. A, Schematic representation of the designed guide RNAs (gRNA) targeting 5′UTR (untranslated region; 1&7, green), exon 1 (2&3, red) or intron 1 (4–6, blue) of the p22phox gene (CYBA; on chromosome 16). Image taken from UCSC genome browser (http://genome-euro.ucsc.edu/index.html) is modified. B, Used combinations (Combi.) of different gRNAs A-G for transfection of HEK293 cells. C, D, Representative Western blot for p22phox, Nox4 and β-Actin protein expression in empty vector control cells (Vec) and after CRISPR/Cas9-mediated p22phox knockout with different combinations or single gRNAs as indicated and defined in B. Every combination (combi.) was tested in Nox4-HEK293 (C) or tetNox4-HEK293 cells (D). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
2.8. Determination of ROS production
ROS production was measured as described before [24], [26]. As probe, luminol (100 µM, #123072, Sigma) catalyzed by horseradish peroxidase (HRP, 1 U/mL, #P6782, Sigma) or L012 (250 µM, #120-04891, Wako) was used and measured in a TriStar2 Multimode Reader (LB942, Berthold, Wildbad, Germany).
2.9. Western blot analysis
Triton-lyzed samples were substituted with sample buffer (8.5% glycerin, 2% SDS, 6.25% TRIS/HCl pH 6.8, 20 mM DTT, 0.013% bromphenol blue) but not heat-denatured. As a reducing agent TCEP (tris(2-carboxyethyl)phosphine, Thermo Scientific) was used as described before [28]. Protein amounts were determined by Bradford protein assay. Proteins were separated by SDS-PAGE, substituted to Western analysis and detected by primary antibodies. Infrared-fluorescent-labeled secondary antibodies were used and visualized in the Odyssey system (Licor, Bad Homburg, Germany). Nox4 was detected by an anti-Nox4 antibody (1:2000) reported by Anilkumar et al. [3]. Primary antibodies against Nox1 (Mox1, #sc-5821, 1:500), Nox5 (#sc-67006, 1:500) and p22phox (#sc-20781, 1:500) were purchased from Santa Cruz, β-Actin (#A1978, 1:2000) was obtained from Sigma.
2.10. RNA isolation and RT-qPCR
RNA was isolated with the RNA Mini-Kit (#BS67.311, Bio&Sell) according to the manufacturer's instructions. Random hexamer primers (#C1181, Promega) and SuperScript III Reverse Transcriptase (RT, #18080044 ThermoFisher) were used for reverse transcription. A non-RT control was used. CDNA was used for semi-quantitative real-time PCR using iQ™ SYBR® Green Supermix (#1708880, BioRad) in a Mx3000P qPCR cycler (Agilent Technologies). Relative mRNA expression was normalized to RNA Polymerase IIa (POLR2A) and analyzed by the delta-delta-Ct method. Results are shown as relative to the corresponding control. Primer sequences are summarized in Table 1.
Table 1.
RT-qPCR Primer.
| Gen | Sequence (5′–3′) | |
|---|---|---|
| h POLR2A | forward | GCACCACGTCCAATGACAT |
| reverse | GTGCGGCTGCTTCCATAA | |
| h NOX4 | forward | TCCGGAGCAATAAGCCAGTC |
| reverse | CCATTCGGATTTCCATGACAT | |
| h CYBA | forward | TACTATGTTCGGGCCGTCCT |
| reverse | CACAGCCGCCAGTAGGTA |
2.11. Statistical analysis
Values are mean±SEM (standard error of mean) if not otherwise indicated. Statistical analysis was carried out using Prism 3.02. For multiple testing ANOVA followed by post hoc testing (Bonferroni's Multiple Comparison Test) was used, unless otherwise indicated; n indicates the number of individual experiments. Densitometry was performed using the Odyssey-software (Image studio lite 5.0) and normalized to corresponding reference protein β-actin. A p-value of less than 0.05 was considered statistically significant (*p<0.05; **p<0.01; ***p<0.001).
3. Results
3.1. CRISPR/Cas9-mediated knockout of CYBA/p22phox
To study the role of endogenous p22phox on Nox-dependent ROS formation, different guide RNAs (gRNAs) against the p22phox gene CYBA were designed (Fig. 1A, B). Seven different gRNAs were transfected into stable Nox4- (Fig. 1C; Nox4-HEK293) or tetracycline-inducible Nox4-HEK293 cells (Fig. 1D; tetNox4-HEK293) and CRISPR/Cas9-mediated knockout was validated by Western Blot. Whereas single gRNA usage resulted in p22phox protein reduction, most of the gRNA combinations displayed loss of p22phox in both HEK293 cell types (Fig. 1B, C, D). Complete loss of p22phox was achieved with a combination of three gRNAs (Fig. 1B and C; combination 17) or all seven gRNAs (Fig. 1B and C; combination 18). For the subsequent experiments, the most effective combinations were used with cells targeted with two or three gRNAs with the combinations 1, 4, 7, 8, 10 and 17.
3.2. Knockout of endogenous p22phox reduces Nox4-dependent H2O2 production, but not Nox4 expression
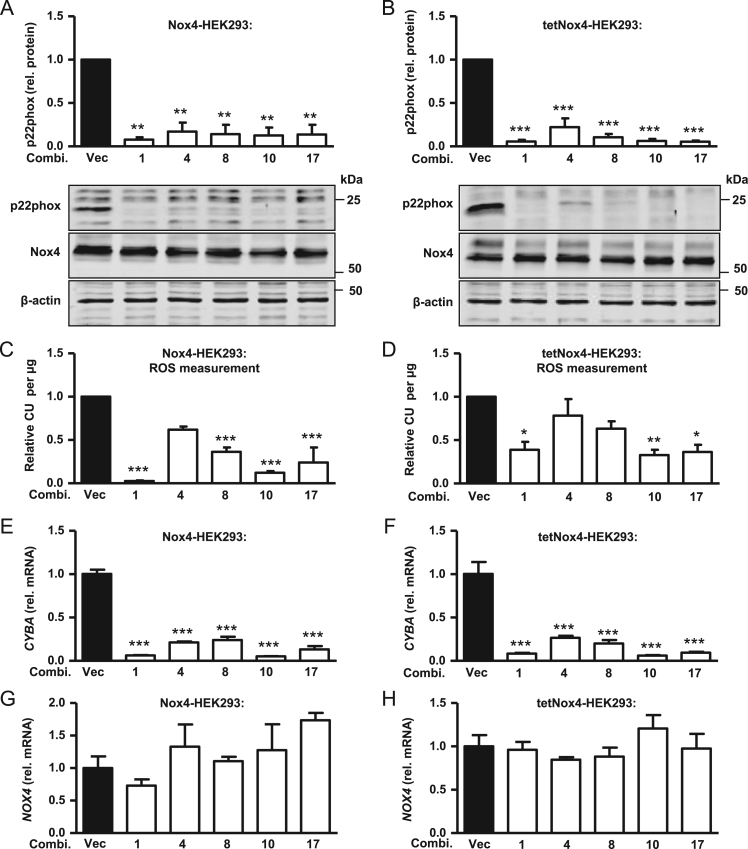
To test the influence of CRISPR/Cas9-mediated p22phox knockout on Nox4-dependent ROS production, Nox4 constitutively overexpressing (Fig. 2 left) and induced tetNox4-overexpressing HEK293 cells (Fig. 2 right, tetracycline 1 µg/mL, 24 h) were studied in the luminol/HRP assay. P22phox and Nox4 mRNA as well as protein expression were studied in the cells subjected to ROS measurements (Fig. 2A, B, E, F, G, H). Deletion of p22phox resulted in a strong reduction of ROS production, and the extent of the effect correlated with the p22phox mRNA and protein expression but not with Nox4 mRNA or protein expression. Interestingly, the inhibitory effect on ROS formation appeared more prominent in the constitutively overexpressing than in the tet-inducible cells (Fig. 2C and D). Although deletion of p22phox lowered ROS formation, it certainly did not abolish it. This effect could potentially be a consequence of insufficient transfection efficiency.
Fig. 2.
Nox4-dependent H2O2 production and expression in p22phox-CRISPR/Cas9 knockout Nox4/tetNox4-HEK293 cells. A, B, Representative Western blot with densitometry for p22phox protein expression normalized to β-Actin in empty vector (Vec) control cells and after CRISPR/Cas9-mediated p22phox knockout (combinations of gRNAs: 1, 4, 8, 10, 17) in Nox4-HEK293 (A) or induced tetNox4-HEK293 cells (B). C, D, Relative luminol/HRP assay in controls (Vec) and after CRISPR/Cas9-mediated p22phox knockout (combi. 1, 4, 8, 10, 17) in Nox4-HEK293 (C) or tetNox4-HEK293 cells (D). CU (chemiluminescence unit) was normalized to protein amount (μg) and control (Vec). E–H, Normalized, relative mRNA level for CYBA (E, F) and NOX4 (G, H) in control (Vec) and CRISPR/Cas9-mediated p22phox knockout (combi. 1, 4, 8, 10, 17) in Nox4-HEK293 (E, G) or induced tetNox4-HEK293 cells (F, H). n≥3, mean±SEM,*p<0.05; **p<0.01; ***p<0.001 relative to the corresponding vector treated cells (Vec).
3.3. Knockout of endogenous p22phox completely abolishes Nox4-dependent H2O2 production
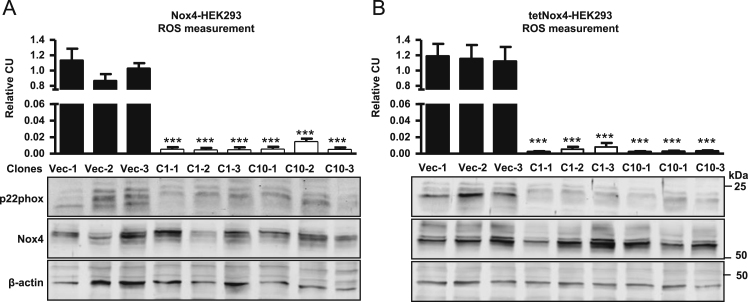
To generate defined p22phox-deficient cells, Nox4- and tetNox4-HEK293 cells, harboring the most effectively reduced p22phox expression (gRNA combination 1 and combination 10), were clonally expanded. Multiple clones were tested on p22phox expression and H2O2 production and three of those were chosen for the subsequent studies. The three tested clones of either constitutive expressing Nox4-HEK293 cells (Fig. 3A) or tetracycline-induced tetNox4-HEK293 cells (Fig. 3B) displayed a complete loss of H2O2 production measured by luminol/HRP assay, in comparison to the empty vector control. Despite some unspecific bands in the Western blot, p22phox could not be detected in all three clonally expanded CRISPR/Cas9-knockout cell lines (C1 or C10). As the starting population of the clonal expansion was variable regarding Nox4 expression, Nox4 expression level also varied in the different p22phox-knockout cell lines. Hence, the H2O2 production is not dependent on Nox4 expression as seen in the Western blot, but on p22phox expression only. The clones C1-1 and C10-1 were used for all following experiments.
Fig. 3.
Nox4-dependent H2O2 production in subclonal expanded p22phox-CRISPR/Cas9 knockout Nox4/tetNox4-HEK293 cells. Either Nox4-HEK293 (A) or induced tetNox4-HEK293 cells (B) were transfected with empty vector (Vec) or gRNA combination 1 or 10, selected with puromycin and subclonally expanded. Each clone is named according to the vector control (Vec) or gRNA combination (1 or 10) and ongoing number (e.g. C1-1). Relative luminol/HRP assay was performed in subclonally expanded control cells (Vec) or CRISPR/Cas9-mediated p22phox knockout (combinations 1 or 10) Nox4-HEK293 (A) or tetNox4-HEK293 cell lines (B). Also shown are representative Western blots for p22phox, Nox4 and β-Actin. n≥3, mean±SEM, ***p<0.001 relative to corresponding vector clones.
3.4. ROS production after p22phox knockout can be restored with p22phox, but not with DUOXA1 or DUOXA2
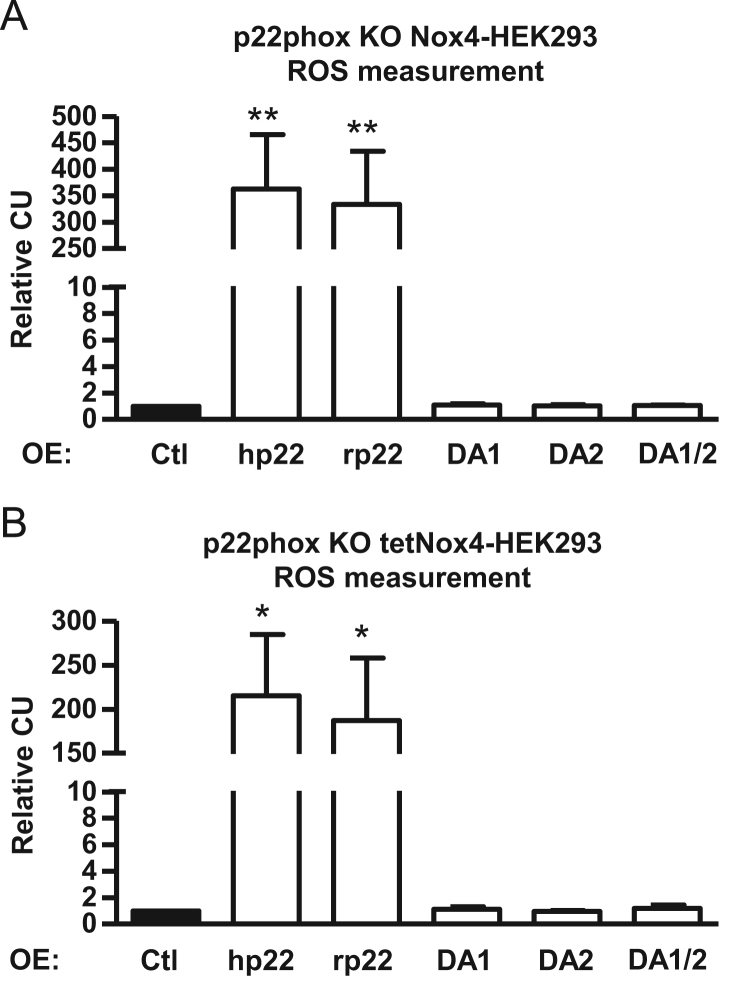
The clonally expanded p22phox knockout cells are an ideal tool for reconstitution experiments. Transfection of the cells with plasmids coding for human p22phox (hp22) as well as rat p22phox (rp22) restored the ROS production in Nox4-HEK293 cells as well as tetNox4-HEK293 cells (Fig. 4A and B). To test the hypothesis that DUOX1/2 maturation factors may substitute p22phox, DUOXA1 (DA1) and/or DUOXA2 (DA2) were also transfected. Importantly, the DUOX maturation factors did not affect ROS formation (Fig. 4A and B) and therefore cannot substitute p22phox.
Fig. 4.
Nox4-dependent H2O2 production in p22phox-CRISPR/Cas9 knockout Nox4/tetNox4-HEK293 cells. Either subclonally expanded p22phox knockout Nox4-HEK293 (C1-1 and C10-1; A) or tetNox4-HEK293 cells (C1-1 and C10-1; B) were transiently transfected (overexpression: OE) with human (hp22), rat p22phox (rp22) or DUOXA1 (DA1), DUOXA2 (DA2) or a combination of both (DA1/2). Relative luminol/HRP assay in Nox4-HEK293 (A) or induced tetNox4-HEK293 cells (B) was performed. n=6, mean±SEM, *p<0.05; **p<0.01; relative to the corresponding control cells.
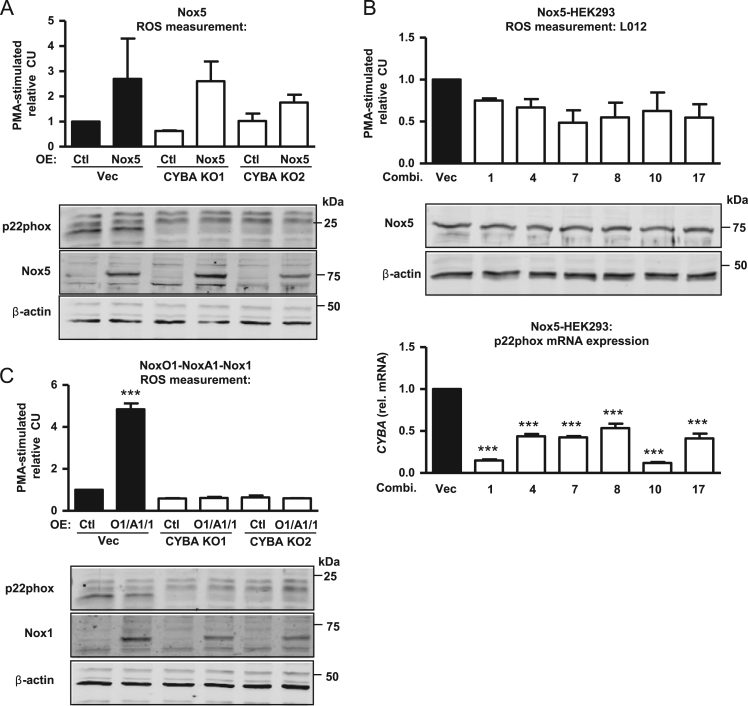
3.5. P22phox is required for Nox1-, but not Nox5-dependent O2•− production
To test whether other Nox isoforms are also influenced by the knockout of endogenous p22phox, p22phox deficient cells were transiently transfected with either a NoxO1-NoxA1-Nox1 expressing plasmid or with a Nox5 plasmid. Despite knockout of p22phox, Nox5 transfected cells exhibited PMA (phorbol 12-myristate 13-acetate; 100 nM)-stimulated ROS production as detected by L012 chemiluminescence (Fig. 5A, CYBA KO1&2). A similar effect was observed in stably transduced Nox5-HEK293 cells subjected to CRISPR/Cas9 p22phox knockout with different combinations of gRNAs (1, 4, 7, 8, 10, 17) (Fig. 5B). In contrast, p22phox-deficiency completely abolished Nox1-dependent O2•− production (Fig. 5C, CYBA KO1&2), whereas Nox1 expression was not changed (Fig. 5C). Thus, Nox5 but not Nox1-mediated ROS is independent of p22phox in contrast to Nox1 or Nox4.
Fig. 5.
Nox1- or Nox5-dependent O2•− production in p22phox-CRISPR/Cas9 knockout HEK293 cells. A, p22phox-knockout HEK293 cells (KO) or control cells (Vec) were transiently transfected with control (Ctl) or human Nox5 overexpression (OE) plasmid and treated with PMA (100 nM, 30 min). Relative L012 assay of the transiently transfected cells was performed. Also shown are representative Western blots for p22phox, Nox5 and β-Actin. B, Stably transduced Nox5-HEK293 cells were treated with empty vector (Vec) or different gRNAs combinations targeting p22phox (combi. 1, 4, 7, 8, 10, 17) and treated with PMA (100 nM, 30 min). Relative L012 assay was performed. Normalized, relative Western Blot for Nox5 and mRNA expression of p22phox (CYBA) are shown. C, p22phox-deficient HEK293 cells (KO) or control cells (Vec) were transient transfected with human NoxO1-NoxA1-Nox1 triple overexpression plasmid (OE) and treated with PMA (100 nM, 30 min). Relative L012 assay was performed. n≥3, mean±SEM, ***p<0.001.
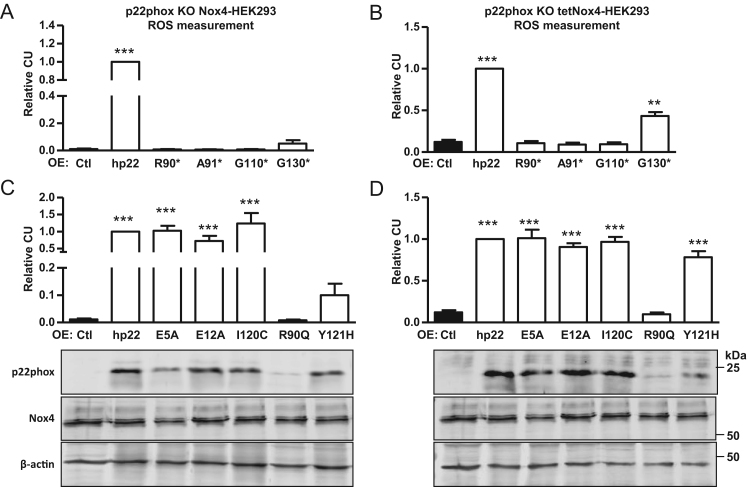
3.6. Analysis of p22phox mutants and their effect on Nox4-dependent ROS production
To gain insights into the function of p22phox in Nox4-dependent ROS production, mutations were studied. First, C-terminal truncations were analyzed (Fig. 6A and B). Truncation beyond Q130 resulted in a complete loss of enzyme activity, whereas the protein truncated at Q130 still showed some ROS activity. Interestingly, this effect was more pronounced in the tetNox4-HEK293 cells as compared to the Nox4 stably expressing cells, potentially suggesting that the dynamic of Nox4 protein formation is relevant for this mutation. Importantly, these observations are in stark contrast to a previous publication, which reported that a p22phox-Q130 truncation was fully active [17]. Truncations at aa90, aa91 or aa110 were indispensable for Nox4 activity (Fig. 6A and B).
Fig. 6.
Nox4-dependent H2O2 production with p22phox mutants in p22phox-CRISPR/Cas9 knockout HEK293 cells. Either subclonally expanded p22phox knockout (KO) Nox4-HEK293 (C1-1 and C10-1; A and C) or induced tetNox4-HEK293 cells (C1-1 and C10-1; B and D) were transiently transfected (overexpression: OE) with full-length human (hp22), truncated (*; stop (A and B)) or point mutated versions of p22phox (C and D). Relative luminol/HRP assay in Nox4-HEK293 (A and C) or induced tetNox4-HEK293 cells (B and D) was performed. n=6, mean±SEM, **p<0.01; ***p<0.001, relative to the corresponding p22phox (hp22) transfected cells.
As several CGD patients carry the p22phox missense mutation R90, we also tested a R90Q mutant [27] on its effect on Nox4 (Fig. 6C and D). The mutation not only leads to a loss of Nox4-dependent H2O2 formation, it also exhibited reduced p22phox protein expression. Mutations of negatively charged amino acids at the beginning of the N-terminus (E5A, E12A) or aliphatic amino acid close to the predicted Nox1/2-p22phox interaction site (I120C) had no influence on Nox4-dependent ROS production (Fig. 6C and D). Interestingly, the Y121H mutation, previously described by Banfi et al. in Cyba-deficient mice [22] only resulted in ROS formation in the tetracycline-induced Nox4-HEK293 cells, whereas in the constitutive Nox4-HEK293 cells only a minimal effect on ROS production was achieved. None of the mutants had a dominant negative effect. I.e. transfection of the p22phox mutants into HEK cells overexpressing Nox4 but expressing endogenous p22phox had only a very minor effect on ROS formation (Sup. Fig. 1).
These results highlight the importance of R90, the transmembrane region aa90-130 and also indicate importance for Y121 and aa130-195 for Nox4 activity.
4. Discussion
In this study using CRISPR/Cas9-mediated knockout cell lines we demonstrate that p22phox deficit completely abolishes Nox1 and Nox4 but not Nox5-dependent ROS production. In contrast, expression levels of these Nox enzymes were independent of p22phox. The effect of the knockouts could be rescued with either human or rat p22phox, but not with the DUOX-maturation factors DUOXA1/A2. Truncation of p22phox till the amino acid 130 was without a functional consequence, while further truncation leads to a reduced function of the protein. Exchange of conserved negatively charged amino acids at the very N-terminus did not impact on Nox4-dependent ROS formation, whereas the R90Q mutant was not able to produce significant amounts of ROS. Surprisingly, the C-Terminus of p22phox was just dispensable in tetracycline-induced HEK293 cells. In constitutive Nox4-expressing cells, truncation of p22phox with Q130stop could not rescue the p22phox knockout. The same was observed for Y121H p22phox, which just restored the ROS producing capacity of tetracycline-induced Nox4. This might suggest that these parts of the protein are important during the de novo formation of the Nox-p22phox complex.
The functional interaction of the catalytically active Nox enzyme with p22phox on ROS formation is in line with previous work employing p22phox shRNA, siRNA or overexpression of Nox and p22phox [16], [17], [19]. The rescue with either truncated Q130stop or Y121H p22phox was described for co-overexpression of p22phox and Nox4 [17]. This could be confirmed with the tetracycline-inducible Nox4 expression 24 h after the transfection with p22phox. Therefore, Nox4 is transcribed, translated and matured after the p22phox Q130stop or Y121H p22phox overexpression. In the constitutively expressing Nox4-HEK293 cells, Nox4 is already expressed in the absence of p22phox. Our data therefore suggest an involvement of the C-terminal part of p22phox in the maturation of the p22phox-Nox4 interaction. Since Nox4 is not dependent on cytosolic subunits to form a functional Nox complex the role of p22phox is rather stabilization then activation [23], [29], [9]. Nox1, Nox2 and Nox3 maturation and glycosylation have also been described to be p22phox-dependent, but these isoforms are furthermore dependent on the direct activation by p22phox by the prolin-rich region in the C-terminus of p22phox [20], [21]. This region is dispensable for Nox4 activation as shown in the tetNox4-HEKs and co-expression studies by others [16], [17].
Zhu et al. described the dependence of Nox2 maturation on amino acids 6-142 [31]. Therefore we propose that also Nox4 maturation is dependent on the amino acids 6-130. In line with this, deletion of the first five amino acids, but not 11 amino acids, is tolerated by Nox4 as shown in a previous study [17]. Controversially, a peptide targeting the amino acids 6-11 (WAMWAN) did not inhibit Nox4-derived ROS production [8]. Furthermore, it was also reported that the D-Loop of Nox4 is responsible for the Nox4-p22phox activation suggesting an ionic interaction [18]. We tested if substitution of inter-species conserved negatively charged glutamic acid in the N-terminal region influence Nox4 activity. Neither E5A nor E12 A reduced Nox4 activity in p22phox-deficient HEK293 cells. Further studies on the structural relevance of the N-Terminus for Nox4 maturation and activity are thus needed to understand the mechanism underlying p22phox function.
For Nox1-3 a 1:1 p22phox heterocomplex formation is proposed for the activation and assembly of functional Nox complexes at the plasma membrane, whereas Nox5 forms homooligomers and does not require p22phox [15]. In this and in other studies Nox4 overexpression yielded high ROS production with endogenous p22phox [12], [16], [2], [26]. In HEK293 cells, co-overexpression of p22phox or its mutants did not further increase ROS production. This effect, however, is cell-type specific and in other cells, like COS-7 or PLB cells, basal p22phox mRNA expression might be insufficient to provide sufficient protein to the Nox enzyme. Maturation of Nox enzymes in general is a complex process and other proteins, like protein disulfide isomerase [13] or Calnexin [24] also contribute to the process. Further studies are needed to understand the process in detail to dissect the precise mode of p22phox – Nox interaction.
In summary, we used CRISPR/Cas9-technology to reliably and rapidly alter genomic CYBA DNA sequences and induced a knockout of p22phox in two Nox4-overexpressing cell lines. We demonstrated that rapid deletion of p22phox is possible and that the activity of Nox1 and Nox4 but not Nox5 exclusively depends on p22phox.
Conflict of interest
The authors declare that they have no relevant financial, personal or professional relationships to disclose which could be perceived as a conflict of interest or as potentially influencing or biasing the authors' work.
Acknowledgment
The study was supported by the DFG Excellence Cluster “Cardiopulmonary System – ECCPS”, SFB 815 (TPA1 to KS and SFB 834 (TPA2 to RPB)) and the Faculty of Medicine, Goethe-Universität, Frankfurt am Main, Germany. AMS is supported by the British Heart Foundation.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.redox.2016.08.013.
Appendix A. Supplementary materials
Supplementary Material
.
References
- 1.Ago Tetsuro, Kitazono Takanari, Kuroda Junya, Kumai Yasuhiro, Kamouchi Masahiro, Ooboshi Hiroaki. NAD(P)H oxidases in rat basilar arterial endothelial cells. Stroke: J. Cereb. Circ. 2005;36:1040–1046. doi: 10.1161/01.STR.0000163111.05825.0b. [DOI] [PubMed] [Google Scholar]
- 2.Ambasta Rashmi K., Kumar Pravir, Griendling Kathy K., Schmidt Harald H.H.W., Busse Rudi, Brandes Ralf P. Direct interaction of the novel Nox proteins with p22phox is required for the formation of a functionally active NADPH oxidase. J. Biol. Chem. 2004;279:45935–45941. doi: 10.1074/jbc.M406486200. [DOI] [PubMed] [Google Scholar]
- 3.Anilkumar Narayana, Weber Roberta, Zhang Min, Brewer Alison, Shah Ajay M. Nox4 and nox2 NADPH oxidases mediate distinct cellular redox signaling responses to agonist stimulation. Arterioscler. Thromb. Vasc. Biol. 2008;28(7):1347–1354. doi: 10.1161/ATVBAHA.108.164277. [DOI] [PubMed] [Google Scholar]
- 4.Bedard Karen, Krause Karl-Heinz. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 5.Brandes Ralf P., Weissmann Norbert, Schröder Katrin. Nox family NADPH oxidases: molecular mechanisms of activation. Free Radic. Biol. Med. 2014;76:208–226. doi: 10.1016/j.freeradbiomed.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 6.Bu-Ghanim H.N., Casimir C.M., Povey S., Segal A.W. The alpha subunit of cytochrome b-245 mapped to chromosome 16. Genomics. 1990;8(3):568–570. doi: 10.1016/0888-7543(90)90045-v. [DOI] [PubMed] [Google Scholar]
- 7.Clempus Roza E., Sorescu Dan, Dikalova Anna E., Pounkova Lily, Jo Patricia, Sorescu George P. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 2007;27(1):42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csányi Gábor, Pagano Patrick J. Strategies aimed at Nox4 oxidase inhibition employing peptides from Nox4 B-loop and C-terminus and p22 (phox) N-terminus: an elusive target. Int J. Hypertens. 2013;2013:842827. doi: 10.1155/2013/842827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai Leena P., Zhou Yong, Estrada Aida V., Ding Qiang, Cheng Guangjie, Collawn James F., Thannickal Victor J. Negative regulation of NADPH oxidase 4 by hydrogen peroxide-inducible clone 5 (Hic-5) protein. J. Biol. Chem. 2014;289(26):18270–18278. doi: 10.1074/jbc.M114.562249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinauer M.C., Pierce E.A., Bruns G.A., Curnutte J.T., Orkin S.H. Human neutrophil cytochrome b light chain (p22-phox). Gene structure, chromosomal location, and mutations in cytochrome-negative autosomal recessive chronic granulomatous disease. J. Clin. Investig. 1990;86(5):1729–1737. doi: 10.1172/JCI114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiszt M., Kopp J.B., Varnai P., Leto T.L. Identification of renox, an NAD(P)H oxidase in kidney. Proc. Natl. Acad. Sci. USA. 2000;97(14):8010–8014. doi: 10.1073/pnas.130135897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmcke Ina, Heumüller Sabine, Tikkanen Ritva, Schröder Katrin, Brandes Ralf P. Identification of structural elements in Nox1 and Nox4 controlling localization and activity. Antioxid. Redox Signal. 2009;11(6):1279–1287. doi: 10.1089/ars.2008.2383. [DOI] [PubMed] [Google Scholar]
- 13.Janiszewski Mariano, Lopes Lucia Rossetti, Carmo Alípio O., Pedro Marcelo A., Brandes Ralf P., Santos C.élio X.C., Laurindo Francisco R.M. Regulation of NAD(P)H oxidase by associated protein disulfide isomerase in vascular smooth muscle cells. J. Biol. Chem. 2005;280(49):40813–40819. doi: 10.1074/jbc.M509255200. [DOI] [PubMed] [Google Scholar]
- 14.Josipovic I., Fork C., Preussner J., Prior K.K., Iloska D., Vasconez A.E. Acta physiologica; Oxford, England: 2016. PAFAH1B1 and the lncRNA NONHSAT073641 maintain an angiogenic phenotype in human endothelial cells. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara Tsukasa, Jackson, Heather M., Smith, Susan M.E., Simpson, Paul D., Lambeth J., David Nox5 forms a functional oligomer mediated by self-association of its dehydrogenase domain. Biochemistry. 2011;50(12):2013–2025. doi: 10.1021/bi1020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawahara Tsukasa, Ritsick Darren, Cheng Guangjie, Lambeth J. David. Mutations in the proline-rich region of p22phox are dominant inhibitors of Nox1- and Nox2-dependent reactive oxygen generation. J. Biol. Che.m. 2005;280:31859–31869. doi: 10.1074/jbc.M501882200. [DOI] [PubMed] [Google Scholar]
- 17.Löhneysen Katharina von, Noack Deborah, Jesaitis Algirdas J., Dinauer Mary C., Knaus Ulla G. Mutational analysis reveals distinct features of the Nox4-p22 phox complex. J. Biol. Chem. 2008;283(50):35273–35282. doi: 10.1074/jbc.M804200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löhneysen Katharina von, Noack Deborah, Wood Malcolm R., Friedman Jeffrey S., Knaus Ulla G. Structural insights into Nox4 and Nox2: motifs involved in function and cellular localization. Mol. Cell. Biol. 2010;30(4):961–975. doi: 10.1128/MCB.01393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martyn Kendra D., Frederick Linda M., Loehneysen Katharina von, Dinauer Mary C., Knaus Ulla G. Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cell. Signal. 2006;18(1):69–82. doi: 10.1016/j.cellsig.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 20.Miyano K., Sumimoto H. N-Linked glycosylation of the superoxide-producing NADPH oxidase Nox1. Biochem. Biophys. Res. Commun. 2014;443(3):1060–1065. doi: 10.1016/j.bbrc.2013.12.086. [DOI] [PubMed] [Google Scholar]
- 21.Nakano Y., Banfi B., Jesaitis A., Dinauer M., Allen La, Nauseef W. Critical roles for p22phox in the structural maturation and subcellular targeting of Nox3. Biochem. J. 2007;403(Pt 1):97–108. doi: 10.1042/BJ20060819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano Yoko, Longo-Guess, Chantal M., Bergstrom, David E., Nauseef, William M., Jones, Sherri M., Bánfi Botond. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J. Clin. Invest. 2008;118(3):1176–1185. doi: 10.1172/JCI33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peshavariya Hitesh, Jiang Fan, Taylor Caroline J., Selemidis Stavros, Chang Catherine W.T., Dusting Gregory J. Translation-linked mRNA destabilization accompanying serum-induced Nox4 expression in human endothelial cells. Antioxid. Redox Signal. 2009;11(10):2399–2408. doi: 10.1089/ars.2009.2579. [DOI] [PubMed] [Google Scholar]
- 24.Prior Kim-Kristin, Wittig Ilka, Leisegang Matthias S., Groenendyk Jody, Weissmann Norbert, Michalak Marek. The endoplasmic reticulum chaperone calnexin is a NADPH oxidase NOX4 interacting protein. J. Biol. Chem. 2016;291(13):7045–7059. doi: 10.1074/jbc.M115.710772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran F. Ann, Hsu Patrick D., Wright Jason, Agarwala Vineeta, Scott David A., Zhang Feng. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezende Flavia, Löwe Oliver, Helfinger Valeska, Prior Kim-Kristin, Walter Maria, Zukunft Sven. Unchanged NADPH oxidase activity in Nox1-Nox2-Nox4 triple knockout mice: what Do NADPH-stimulated chemiluminescence assays really detect? Antioxid. Redox Signal. 2016;24(7):392–399. doi: 10.1089/ars.2015.6314. [DOI] [PubMed] [Google Scholar]
- 27.Roos D., Boer M. de, Kuribayashi F., Meischl C., Weening R.S., Segal A.W. Mutations in the X-linked and autosomal recessive forms of chronic granulomatous disease. Blood. 1996;87(5):1663–1681. [PubMed] [Google Scholar]
- 28.Schröder Katrin, Zhang Min, Benkhoff Sebastian, Mieth Anja, Pliquett Rainer, Kosowski Judith. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110(9):1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 29.Serrander Lena, Cartier Laetitia, Bedard Karen, Banfi Botond, Lardy Bernard, Plastre Olivier. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406(41):105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somers Aba, Jean Jyh-Chang, Sommer, Cesar A., Omari Amel, Ford Christopher C., Mills Jason A. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28(10):1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Yanmin, Marchal, Christophe C., Casbon Amy-Jo, Stull Natalie, Löhneysen Katharina von, Knaus Ulla G. Deletion mutagenesis of p22phox subunit of flavocytochrome b558: identification of regions critical for gp91phox maturation and NADPH oxidase activity. J. Biol. Chem. 2006;281(41):30336–30346. doi: 10.1074/jbc.M607191200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material