Abstract
Rapid detection of the foodborne pathogen Escherichia coli O157:H7 is of vital importance for public health worldwide. Among detection methods, reporter phages represent unique and sensitive tools for the detection of E. coli O157:H7 from food as they are host-specific and able to differentiate live cells from dead ones. Upon infection, target bacteria become identifiable since reporter genes are expressed from the engineered phage genome. The E. coli O157:H7 bacteriophage ΦV10 was modified to express NanoLuc luciferase (Nluc) derived from the deep-sea shrimp Oplophorus gracilirostris. Once infected by the ΦV10 reporter phage, E. coli O157:H7 produces a strong bioluminescent signal upon addition of commercial luciferin (Nano-Glo®). Enrichment assays using E. coli O157:H7 grown in LB broth with a reporter phage concentration of 1.76 × 102 pfu ml−1 are capable of detecting approximately 5 CFU in 7 hours. Comparable detection was achieved within 9 hours using 9.23 × 103 pfu ml−1 of phage in selective culture enrichments of ground beef as a representative food matrix. Therefore we conclude that this NanoLuc reporter phage assay shows promise for detection of E. coli O157:H7 from food in a simple, fast and sensitive manner.
The prevalence of Shiga toxin-producing bacterium Escherichia coli O157:H7 in the food supply has accounted for hundreds of the reported foodborne outbreaks resulting in thousands of illness in the United States in the past decade1,2. Outbreak-related food vehicles that are frequently associated with E. coli O157:H7 are meat products (especially ground beef)3,4,5,6 and fresh produce (such as lettuce, spinach and sprouts)7,8,9. For instance, an outbreak caused by E. coli O157:H7 EDL933 in 1982 afflicted approximately 47 people with bloody diarrhea from undercooked meat10. In 2006, a large E. coli O157:H7 outbreak was linked to contaminated pre-package spinach and 205 individuals were affected across 26 US states with 29% of infected individuals developing severe Hemolytic Uremic Syndrome (HUS)7. Rapid and sensitive detection of E. coli O157:H7 from food is vital for the prevention of foodborne illness. The extant detection methods for zero-tolerance pathogens of immunological separation and PCR rely on a culture enrichment step. The strength of bacteriophage-based detection methods is that they may exploit this enrichment period to create a reporter signal indicating the presence of the sought-after pathogen. Furthermore, bacteriophages are a useful approach for detection due to their extreme specificity for their hosts and capability of distinguishing between live and dead target cells11,12.
Bacteriophages have been engineered in the past to include reporter genes so that a fluorescent or bioluminescent signal can be produced only after viable cells are infected and propagated12. To date, the reporters used for detection of foodborne pathogens typically include green fluorescent protein (gfp), the bacterial luciferase (luxAB or luxCDABE coupled with luxI and luxR) and β-galactosidase (lacZ)13,14,15,16,17,18,19. Recently, a new luciferase, NanoLuc luciferase (Nluc) was engineered from the deep-sea shrimp Oplophorus gracilirostris by Promega20. Nluc is a small 19 kDa protein and produces bright luminescence with an imidazopyrazinone substrate (furimazine) in a reaction that is independent of ATP. When introduced into group A Streptococcus, Nluc, in spite of its eukaryotic origin, has shown a superior sensitivity to firefly luciferase (FFluc) and bacterial luciferase (Lux) in terms of signal strength21. Furthermore, it is reported that E. coli O157:H7 strains expressing NanoLuc were able to generate a readily detectable signal over two days22. While the 6 kb luxCDABE cassette is difficult to incorporate into phage genomes, the size of nluc gene is small enough (516 base pairs) to have a potential to be inserted into a phage genome with a proper headful packaging23. The genetic manipulation of a phage genome using eukaryotic luc has so far been limited to the assessment of drug-resistance in the respiratory pathogen Mycobacterium tuberculosis24.
In this study, NanoLuc for the first time was introduced into the lysogenic bacteriophage ΦV10 for the detection of E. coli O157:H7. Previously, Waddell and Poppe15 chose transposon mutagenesis to construct a luxAB-based ΦV10 reporter phage due to the lack of knowledge of the genetic makeup of this phage. The resultant mutant phage was capable of transducing bioluminescence but failed to propagate on E. coli O157:H7 cells. Now the genetic analysis of the ΦV10 phage allows identification of non-essential genes to facilitate the construction of a recombinant phage25,26,27. Therefore in the present report, the previously identified putative gene 37 (recET) of phage ΦV1026 was replaced by nluc via homologous recombination. This novel recombinant phage was used to detect E. coli O157:H7 in pure culture and in E. coli O157:H7-inoculated ground beef enrichment.
Results
Construction of NanoLuc reporter phage
The 536 bp DNA fragment encoding the NanoLuc luciferase (nluc) along with a translation initiation region was cloned downstream of a constitutive kanamycin resistance determinant. The resultant kanR-nluc cassette is approximately 1.7 kb, the size of which is very close to that of the recET locus of approximately 1.8 kb. A pair of short homologous sequences flanked by the kanR-nluc cassette successfully replaced the complementary regions that are adjacent to recET, and the knockout of recET was confirmed by sequencing of the DNA fragments around the recombination junctions (Fig. 1). Nluc typically produces a blue light with a maximum emission of 460 nm20. Three recombinant isolates emitted blue light when mixed with the Nanoluc-Glo substrate. One isolate B-C2 produced the highest bioluminescence and was chosen to be used in the subsequent experiments. The mutant phage (ΦV10nluc) showed spontaneous induction from the B-C2 lysogen. However, when mitomycin C (at a final concentration of 0.5 μg ml−1) was added to the culture of strain B-C2 in the exponential phase, the titer of the phage increased by approximately 10-fold. Furthermore the propagation of phage ΦV10nluc from the overlay top agar was easily achieved. That phage ΦV10nluc was produced from either spontaneously induced lysogens or wild type E. coli O157:H7 suggests that the insertion of nluc into the phage genome does not compromise phage lytic growth. However, the ΦV10nluc plaques appeared to be small pinpointed dimples, as opposed to the large plaques of wild type ΦV10. A similar morphology change was observed in a previous study involving a defective ΦV10 particle15. Nonetheless, this defective phage particle was still able to transduce bioluminescence to E. coli O157:H715. These observations indicate that the modification of ΦV10 genome can lead to certain phenotypic changes. The visibility of the plaques was increased by the addition of Coomassie Brilliant blue G-250 dye to the top agar (Supplementary Figure S1) without interfering with the phage titer (data not shown).
Figure 1. Scheme of the recombination event.
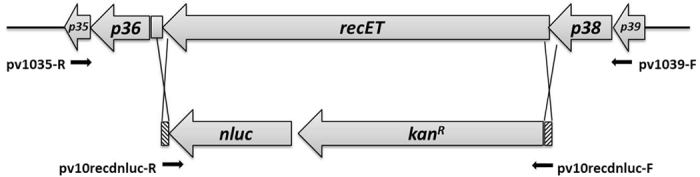
The Xs (double crossover) represent the gene exchange sites between the ΦV10 genome and the PCR amplicon containing the kanR-nluc cassette amplified by primers pv10recdnluc-F and pv10recdnluc-R. The absence of recET and the presence of kanR-nluc cassette flanked by the homologous sequences 5′-GTGGCGCCGCGGTGGTGGTTACATAGATGTTTCGTTT-3′ and 5′-TCGCCCGCTGGGATCTGGCGGATCAGCTTGATGGAC-3′ (shown in shaded bars) in the ΦV10 genome was confirmed by sequencing using primers pv1035-R and pv1039-F. The map was drawn based on the sequence of the Enterobacteria phage PhiV10 (GenBank accession NO. DQ126339.2).
Host specificity of bacteriophage ΦV10
Perry et al.25 previously reported that the O157 antigen was the receptor for ΦV10 as the acetylation of the O antigen by Oac during lysogeny. To examine the host specificity, 281 E. coli O157:H7 isolates from various sources including: FDA, USDA-ARS and Indiana State Department of Health, were tested for plaque formation with ΦV10. All of the E. coli O157:H7 isolates examined formed plaques. Additionally, 39 non-O157:H7 E. coli isolates were screened with ΦV10 and no plaques were detected. However 4 of these strains contained the O157 antigen but did not have the H7 antigen. Although this is a limited number of isolates, it suggests specificity for E. coli O157:H7.
Detection of E. coli O157:H7 in pure culture by NanoLuc phage
LB broth was used to evaluate the ability of phage ΦV10nluc to detect E. coli O157:H7. Phage were incubated statically with E. coli O157:H7 C7927 cells at dilutions ranging from 5.4 × 108 CFU to 5.4 × 100 CFU in 40 ml broth. Kinetic analysis showed time to detectable luminescence (2x background) increased with decreasing cell populations (Fig. 2). The NanoLuc-induced bioluminescence was readily detected from an initial calculated inoculum of 5.4 cells per assay (in 40 ml) within approximately 7 hours without a pre-incubation period (Table 1). When the initial inoculum was within the range of 10–100 cells per assay, that is 54 cells per assay in this study, it only took approximately 6 hours to detect a positive signal (Table 1).
Figure 2. Profile of phage ΦV10nluc induced bioluminescence from E. coli O157:H7 grown in LB broth.
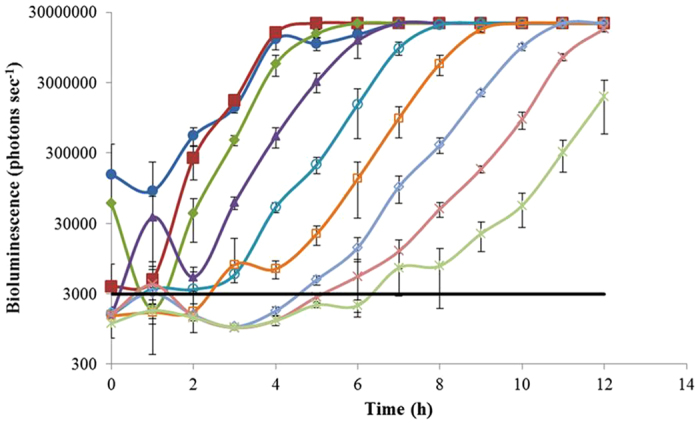
Wild type E. coli O157:H7 C7927 was inoculated at 5.40 × 108 ( ), 5.40 × 107 (
), 5.40 × 107 ( ), 5.40 × 106 (
), 5.40 × 106 ( ), 5.40 × 105 (
), 5.40 × 105 ( ), 5.40 × 104 (
), 5.40 × 104 ( ), 5.40 × 103 (
), 5.40 × 103 ( ), 5.40 × 102 (
), 5.40 × 102 ( ), 5.40 × 101 (
), 5.40 × 101 ( ), 5.40 × 100 (
), 5.40 × 100 ( ) CFU per assay (in a total of 40 ml), and the threshold (—) was set at double the background value. The phage added per assay was 1.76 × 102 pfu ml−1. At 1 hour intervals, individual light readings were recorded for triplicate samples. The cutoff of the bioluminescence reading was set up at 2.1 × 107 photons per second which is the upper detection limit of the Sirius luminometer used in this study.
) CFU per assay (in a total of 40 ml), and the threshold (—) was set at double the background value. The phage added per assay was 1.76 × 102 pfu ml−1. At 1 hour intervals, individual light readings were recorded for triplicate samples. The cutoff of the bioluminescence reading was set up at 2.1 × 107 photons per second which is the upper detection limit of the Sirius luminometer used in this study.
Table 1. Time to detection using the ΦV10nluc based assay.
| Enrichment medium | Initial inoculum of E. coli O157:H7 (CFU/Assay) | Time to detection* (h) |
|---|---|---|
| LB brotha | 5.4 × 104 | 2.30 ± 0.62 |
| 5.4 × 103 | 3.07 ± 0.76 | |
| 5.4 × 102 | 4.60 ± 0.10 | |
| 5.4 × 101 | 5.57 ± 0.60 | |
| 5.4 × 100 | 6.70 ± 0.61 | |
| Ground beefb | 4.68 × 108 | 0.00 ± 0.00 |
| 4.68 × 107 | 0.50 ± 0.32 | |
| 4.68 × 106 | 1.13 ± 0.12 | |
| 4.68 × 105 | 1.43 ± 0.23 | |
| 4.68 × 104 | 2.23 ± 0.06 | |
| 4.68 × 103 | 2.80 ± 0.30 | |
| 4.68 × 102 | 5.13 ± 0.06 | |
| 4.68 × 101 | 6.47 ± 0.81 | |
| 4.68 × 100 | 8.67 ± 0.29 |
*A positive detection corresponds to 2x background.
aLinear regression of initial CFU (log) versus time to detection: y = −0.8781x + 6.6372, R2 = 0.9923. The higher initial inocula (5.4 × 108 to 5.4 × 105 CFU/Assay) were excluded due to high variability.
bLinear regression of initial CFU (log) versus time to detection: y = −0.8811x + 7.4474, R2 = 0.9095.
Detection of E. coli O157:H7 in ground beef enrichment by NanoLuc phage
Ground beef has been identified as a major food source that is associated with most recent outbreaks of E. coli O157:H71. Therefore ground beef was used as an example to test the ability of our NanoLuc phage (ΦV10nluc) for detection of E. coli O157:H7 in a complex food matrix during selective enrichment. Ground beef was homogenized in mTSB + n medium and the resultant beef broth was artificially contaminated with E. coli O157:H7 C7927 dilutions with a range from 4.68 × 108 CFU to 4.68 × 100 CFU in a total of 40 ml. A similar kinetic response of bioluminescence production was observed from ground beef enrichment as well (Fig. 3). The NanoLuc reporter phage was able to detect E. coli O157:H7 C7927 with an initial calculated inoculum as low as 4.68 CFU per assay (in 40 ml) in approximately 9 hours (Table 1). At the level of fewer than 100 cells per assay, this method detects an inoculum of 46.8 cells in a period of approximately 7 hours (Table 1).
Figure 3. Profile of phage ΦV10nluc induced bioluminescence from ground beef artificially contaminated with E. coli O157:H7.
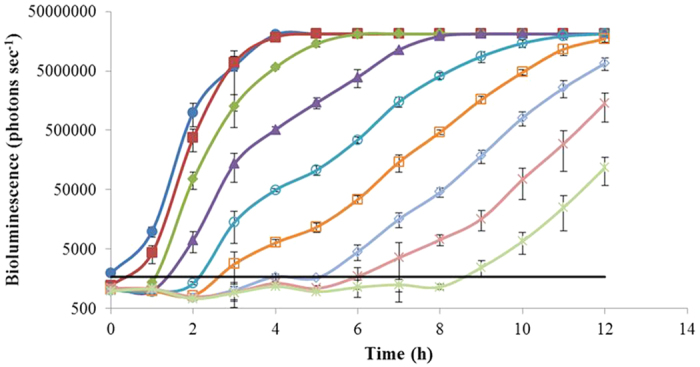
Wild type E. coli O157:H7 C7927 was originally inoculated at 4.68 × 108 ( ), 4.68 × 107 (
), 4.68 × 107 ( ), 4.68 × 106 (
), 4.68 × 106 ( ), 4.68 × 105 (
), 4.68 × 105 ( ), 4.68 × 104 (
), 4.68 × 104 ( ), 4.68 × 103 (
), 4.68 × 103 ( ), 4.68 × 102 (
), 4.68 × 102 ( ), 4.68 × 101 (
), 4.68 × 101 ( ), 4.68 × 100 (
), 4.68 × 100 ( ) CFU per assay (in a total of 40 ml), and the threshold (—) was set at double the background value. The concentration of phage added was 9.23 × 103 pfu ml−1 per assay. At 1 hour intervals, individual light readings were recorded for triplicate samples. The cutoff of the bioluminescence reading was at 2.1 × 107 photons per second which is the upper detection limit of the Sirius luminometer used in this study.
) CFU per assay (in a total of 40 ml), and the threshold (—) was set at double the background value. The concentration of phage added was 9.23 × 103 pfu ml−1 per assay. At 1 hour intervals, individual light readings were recorded for triplicate samples. The cutoff of the bioluminescence reading was at 2.1 × 107 photons per second which is the upper detection limit of the Sirius luminometer used in this study.
Discussion
Food matrices that are contaminated with Shiga toxin-producing E. coli O157:H7 even at the low infectious dose of around 10–100 cells can cause severe outbreaks28. A zero-tolerance policy adopted by USDA-FSIS has been used for regulatory-based analysis of samples after E. coli O157:H7 was declared as an adulterant in ground beef2. In order to detect pathogens at low levels from food, very few of the current detection methods available can bypass enrichment and the subsequent extraction of potential bacteria due to the intrinsic complex nature of food matrices. However, with the use of a bioluminescence reporter phage, the detection signal that is generated during the culture enrichment can be easily non-destructively interrogated in real-time, greatly reducing the time-to-result with minimal labor. In this study, we have constructed a bioluminescent reporter phage from ΦV10. ΦV10 has demonstrated the ability to infect a large number of E. coli O157:H7 isolates from a variety of sources and no verified false positives or negatives (with the exception of ΦV10 lysogens) were detected (Supplementary Table S1).
The luciferase gene nluc was introduced into ΦV10 genome via the λ Red recombination system using linear PCR amplicons resulting in the reporter phage ΦV10nluc. It has been reported that DNA substrates with longer regions of homology have higher recombination efficiency than shorter substrates29,30. However our attempt to use long homologous substrates was not successful, probably due to low yield from PCR (data not shown). Instead, all the positive recombinant isolates we obtained were generated from short homologous substrates (Table 2). Based on our observation, use of short homologous substrates is adequate to promote the gene exchange in phage ΦV10. While the specific modification of the ΦV10 genome in this study does not affect phage propagation or its ability to transduce bioluminescence to E. coli O157:H7, the plaque size was reduced, which made the phage counting difficult. Coomassie Brilliant blue dye is commonly used for protein staining and has been added to agar to detect the fish pathogen Aeromonas salmonicida31. Here we report adding Coomassie Brilliant blue dye into top agar to improve the visualization of the small plaques with no significant change in titer (data not shown). It may be possible that this staining method can be applicable to other phages as well.
Table 2. The oligonucleotide primers used for this study.
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| nluc-F | ATGGTCTTCACACTCGAAGATTTCGTTGG |
| nluc-R | CCGACTCTAGAGTCGCGGCCTTACGCCAGAATGCG |
| nluc-F′a | ATTAACTTTATAAGGAGGAAAAACATATGGTCTTCACACTCGAAGATTTCG |
| pv10recdnluc-Fb | TCGCCCGCTGGGATCTGGCGGATCAGCTTGATGGACCCATCATCGATGAATTGTGTC |
| pv10recdnluc-Rc | GTGGCGCCGCGGTGGTGGTTACATAGATGTTTCGTTTCCGACTCTAGAGTCGCGGCCTTACGCCAGAATGCG |
| pv1039-F | ATGCAGTGGAAAATCATC |
| pv1035-R | GTCGGCTTAACTTTCTCAC |
aThe sequence of the translation initiation region was shown in bold letters.
bThe sequence underlined was homologous to the upstream of the kanamycin resistant determinant.
cThe sequence underlined was homologous to the downstream of the nluc gene.
Initial studies to test the ability of phage ΦV10nluc to transduce bioluminescence to E. coli O157:H7 for detection were conducted using LB broth in a pure culture system. When 1.76 × 102 pfu ml−1 of phage ΦV10nluc was applied, we detected 5.4 cells per assay (in 40 ml) within 7 hours (Fig. 2). There were luminescent fluctuations at the high inoculation levels (108–105 CFU per assay) in the first two hours. However, such a large load of pathogen in food matrices does not normally occur naturally therefore the analysis of such fluctuations was not included in this report. Ground beef was used as a complex matrix to demonstrate the capability of this phage system to detect E. coli O157:H7 in food during selective enrichment. The very low dose of approximately 5 cells per assay (in 40 ml) was detected within 9 hours when using 9.23 × 103 pfu ml−1 of phage (Fig. 3). The detection period was further reduced to 7 hours if approximately 50 cells per assay were inoculated. Other phage-based luminescence assays used high concentrations of phage for detection, such as 108 pfu ml−1 16,17, however the phage concentration used in this study was less than 104 pfu ml−1, demonstrating the potential to deliver a cost-effective platform which could be easily integrated into current E. coli O157:H7 detection regimens. On the other hand, since the developed assay exploits the ability of ΦV10nluc to form lysogens, the addition of higher phage concentrations could potentially shorten the time to detection. However it is important to note that ΦV10nluc is a temperate phage and upon infection there are two outcomes: either chromosomal integration or lytic propagation. Therefore the initial multiplicity of infection on time to detection needs to be further studied. The resistance marker also allows selective isolation of the resultant lysogens on plates supplemented with kanamycin (Supplementary Figure S2). Isolates can then be screened with luciferin for further verification. The genomic sequence and attachment site of ΦV10 in the host chromosome are known25, therefore the ability to trace the origin of the pathogen is not compromised when using sequencing or PFGE for identification.
In summary, a novel NanoLuc-labeled reporter phage was constructed in this study. This recombinant phage was able to stably transduce NanoLuc-induced bioluminescence into E. coli O157:H7 to function as a sensitive signaling system. The detection of a very low quantity of E. coli O157:H7 (5–6 cells) was achieved in 7–9 hours in pure culture and ground beef enrichment when a small amount of reporter phage (102–104 pfu ml−1) was used. In addition, because of the strong signal produced by this NanoLuc luciferase, this reporter phage system could be applied to test other food matrices such as vegetables and dairy products. It is also interesting that the kinetic data plotted in Figs 2 and 3 using this lysogen based method during selective enrichment resemble graphs routinely obtained from using real time PCR for pathogen detection. This suggests the assay could be further developed into a (semi) quantitative method in which time to detection of a specific threshold value could approximate initial levels of contamination. Previously developed luxCDABE based reporter phages17,18 have an advantage over the assay described here as they do not require addition of an exogenous substrate for signal detection. The substrate addition adds complexity to the assay although the brighter signal improves sensitivity. Cost of the reagent has to be considered but at approximately $0.20 per assay in presence/absence format is not prohibitive. In conclusion, this reporter phage can offer a fast and sensitive detection for E. coli O157:H7 from food matrices in a simple low cost detection platform.
Materials and Methods
Bacterial strains and culture media
Bacterial strains, bacteriophages and plasmids used in this study are listed in Table 3. The E. coli O157:H7 C7927 bacterial strain was used for the cultivation of phage ΦV1032. Homologous recombination was carried out in a lysogenic strain E. coli O157:H7 C7927 (ΦV10) bearing the lambda Red expression plasmid pKD46. For the construction of ΦV10 reporter phage, bacterial strains were grown in Luria-Bertani (LB) broth (Difco Laboratories, MI) or LB agar plates supplemented with antibiotics as needed [ampicillin (Ap), 100 μg ml−1; kanamycin (Kan), 50 μg ml−1 (Sigma-Aldrich, MO)]. Salt-Optimized Carbon broth medium (SOC) (Clontech Laboratories, Inc, CA) was used for recovery of cells after electroporation. Modified tryptone soya broth (Oxoid Ltd., UK) containing 1% casamino acids (VWR International, PA) with novobiocin (Sigma-Aldrich, MO) of 8 mg l−1 (mTSB + n) was used for ground beef enrichment as specified by the USDA-FSIS protocol33,34. Cell dilutions were done in phosphate buffered saline (PBS) (8 mM Na2HPO4, 6 mM NaH2PO4, 145 mM NaCl, pH7.6). Phage buffer (50 mM Tris, 100 mM MgCl2, pH7.6) was used to dilute and preserve the phage stock. LB Top agar (1% (wt/vol) tryptone (Becton Dickinson, NJ), 1% (wt/vol) NaCl (Macron, PA), 0.5% (wt/vol) yeast extract (Hardy Diagnostics, CA) and 0.6% (wt/vol) agar (Alfa Aesar, MA)) was used for the overlay plaque assay.
Table 3. Bacterial strains, phages and plasmids used and constructed in this study.
| Bacteria/Phage/Plasmid | Description | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli O157:H7 C7927 | Human isolate, host cell for propagation of ΦV10 and its mutant | 32 |
| E. coli O157:H7 C7927 (ΦV10) | A ΦV10 lysogenic strain | Bruce Applegate |
| E. coli O157:H7 C7927 (ΦV10nluc) | A ΦV10nluc lysogenic strain | This study |
| OneShot TOP10 | General E. coli cloning host | Invitrogen |
| Phage | ||
| ΦV10 | Non-virulent phage, E. coli O157:H7 specific | 15 |
| ΦV10nluc | ΔrecET::kanRnluc mutant strain of ΦV10 | This study |
| Plasmids | ||
| pGEM-T Easy | General cloning vector | Promega |
| pNL1.1 | NanoLuc luciferase reporter vector | Promega |
| pNluc | The nluc gene with a translation initiation region cloned into pGEM-T Easy vector | This study |
| pFSP138 | The kanamycin resistance determinant of Tn5 cloned into pCR 2.1 TOPO vector | Bruce Applegate |
| pNluc-kan | pFSP138 with insertion of kanR determinant upstream of nluc | This study |
| pKD46 | A lambda Red recombinase plasmid | 30 |
Construction of plasmids
The complete coding sequence of the nluc gene (GenBank accession no. JQ437370.1) was amplified by touch down PCR using primers nluc-F and nluc-R (Table 2). The NanoLuc luciferase reporter vector pNL1.1 was used as the template. Ready-to-go PCR beads (GE Healthcare, WI) were used as instructed by the manufacturer and the concentration of each primer per reaction was 0.4 μM. The touch down PCR cycling program was performed in two phases35. Phase one started with 95 °C for 15 s for initial denaturation, followed by cycles at 94 °C for 15 s per cycle, annealing at successively decreasing temperatures from 68 °C to 56 °C for 15 s (decreasing in increments of 1 °C with 2 cycles per temperature), and then a 72 °C extension step for 1 min per cycle. Phase two consisted of 14 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 15 s, and extension at 72 °C for 1 min with a final extension at 72 °C for 2 min. In order to introduce a translation initiation region (Table 2)36 upstream of the nluc ORF, a second forward primer nluc-F′ paired with nluc-R was used to amplify the nluc-F and nluc-R amplicon. The same PCR cycling program was used as above, except that the annealing temperature of phase one was decreased from 68 °C to 61 °C and in phase two, the annealing temperature was held at 60 °C for 22 cycles. The amplicon consisting of the translation initiation region was cloned into pGEM-T Easy Vector system I (Promega, WI) to create pNluc. A kanamycin-resistance determinant isolated from EZ-Tn5TM < R6Kγori/KAN-2 > Tnp TransposomeTM kit (Epicentre Biotechonologies, WI) was inserted into pCR2.1 TOPO vector (Invitrogen, CA) to generate pFSP138. Both pNluc and pFSP138 were digested with NotI. Then pFSP138 was further dephosphorylated by shrimp alkaline phosphatase (New England biolab, MA) and ligated with pNluc at the NotI site to produce pNluc-Kan.
Homologous recombination
For constructing a NanoLuc-labeled ΦV10 reporter phage, a gene replacement between recET and nluc was promoted by using lambda Red recombineering technique for the enterohemorrhagic E. coli29. Briefly, the forward primer pv10recdnluc-F was designed to contain 36 bases at 5′ end that are homologous to the upstream of recET and 21 bases at 3′ end to amplify the kanamycin resistance determinant (Table 2). The reverse primer pv10recdnluc-R was composed of 36 bases at 5′ end to complement the downstream region of recET and with 35 bases at 3′ end to amplify the nluc sequence (Table 2). The PCR cycling program was 95 °C for 15 s, followed by 38 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 90 s. DNA prepared from a loopful of a single colony of TOP10 (pNluc-kan) was used as the template. A previously constructed ΦV10 lysogenic strain E. coli O157:H7 C7927 (ΦV10) containing the Red expression vector pKD46 was used to promote the gene exchange. The protocol recommended by Sawitzke et al.37 for making electro-competent cells for lambda Red-driven recombineering was modified accordingly. Briefly, an overnight culture of E. coli O157:H7 C7927 (ΦV10) was sub-cultured into 25 ml of LB broth supplemented with ampicillin at 30 °C until the OD600 was 0.6. Then L-arabinose solution was added to reach a final concentration of 1 M. After 30 min of incubation at 30 °C with shaking (120 rpm), the cells were made electro-competent at 4 °C. Four microliters of the PCR amplicon digested with DpnI was mixed with fresh competent cells and electroporated. These transformed cells were recovered in SOC medium for 3 hours at 37 °C and spread onto LB-Kan agar plates. All the plates were incubated at 37 °C overnight. The positive colonies, which can emit light detected by the Nano-Glo® Luciferase Assay System (Promega, WI), were isolated and grown in LB broth with kanamycin overnight. After centrifugation of the overnight culture at 10,000 g for 10 min, the remaining bacterial cells in the supernatant were removed by a sterile syringe filter with a pore size of 0.2 μm (VWR International, PA). A standard plaque assay38 described below was performed to confirm the presence of the presumptive recombinant phage in the culture supernatant. Primers pv1039-F and pv1035-R were designed to amplify either upstream or downstream of the recombination sites. A DNA fragment amplified from a single colony of the presumptive E. coli O157:H7 C7927 (ΦV10nluc) using primers pv1039-F and pv1035-R was sequenced (the Purdue Genomics Core Facility, Purdue University,West Lafayette, IN) to confirm the deletion of recET and the integration of kanR-nluc cassette to the ΦV10 phage genome. All the primers used for PCR and sequencing are shown in Table 2.
Host specificity of bacteriophage ΦV10
Previously obtained E. coli strains from Dr. Arun Bhunia’s laboratory collection were screened against bacteriophage ΦV10 by a modified standard plaque assay38. Briefly, after LB top agar was melted and tempered to 42 °C in a water bath, 200 μl of the bacteria overnight culture mixed with 100 μl of the appropriate dilution phage was added. The top agar mix was vortexed for two seconds and immediately poured onto the LB base plate to form a top layer. All the plates were incubated at 37 °C overnight after they were solidified.
Measurement of bioluminescence and propagation of NanoLuc phage
Based on the instructions of the Nano-Glo® Luciferase Assay System, 10 μl of the reconstituted reagent, which was composed of 20 μl of substrate mixed with 1 ml of lysis buffer, was added into a 1 ml aliquot of cell culture or food samples. The light reading was recorded in photons per second by a Sirius luminometer (Berthold Detection Systems GmbH, Germany).
For propagation of this phage, a single colony of the confirmed lysogenic strain E. coli O157:H7 C7927 (ΦV10nluc) was grown overnight in LB-kan broth at 37 °C with shaking (120 rpm), and then centrifuged to remove cell debris. The supernatant was sterile-filtered to create a crude phage stock. The standard plaque assay described above with a slight modification was performed to enumerate the phage count. Coomassie Brilliant blue G-250 dye (Bio-Rad laboratories, CA) at 1% (wt/vol) in water was added to the original top agar formula to make a final concentration of 0.01% (wt/vol) in the agar medium, to increase visibility of phage plaques.
Detection of E. coli O157:H7 in pure LB broth by using NanoLuc phage
The recombinant phage lysate (ΦV10nluc) was purified via dialysis to remove NanoLuc luciferase protein before use for detection. Wild type E. coli O157:H7 C7927 was grown in LB broth at 37 °C overnight. Then 10-fold serial dilutions of this stationary phase culture were prepared in sterile PBS. Four ml of each dilution was added to triplicate 36 ml volumes of LB broth where phage ΦV10nluc with a final concentration of 1.76 × 102 pfu ml−1 was pre-added. LB broth that contained only phage ΦV10nluc was included as a background control. All the samples were incubated at 37 °C without agitation. The initial inoculum counts were verified in triplicate by spread plating of the serial-diluted stationary phase culture on LB agar plates. Bioluminescent measurement of a 1 ml sample was taken at 1 hour intervals over a period of 12 hours by using a Sirius luminometer. The detection threshold was set at twice the background value.
Application of NanoLuc phage to detect E. coli O157:H7 in ground beef enrichment
Ground beef was purchased from a local store and processed upon arrival. The USDA-FSIS protocol for preparation of ground beef enrichment broth was followed accordingly34. Briefly, one portion of raw ground beef was mixed with 3 portions of modified tryptone soya broth with novobiocin (mTSB + n) in a sterile Seward Ltd. Classic 400 Stomacher strainer bag (Davie, FL) and pummeled for 2 min in a Stomacher LabBlender 400 apparatus (Cooke Laboratory Products, VA). The homogenized beef juice was divided into aliquots of 32 ml in 50 ml sterile polypropylene conical tubes (Corning Inc, NY). An overnight culture of wild type E. coli O157:H7 C7927 was diluted as described previously and 4 ml of each dilution in triplicate was added to the tubes containing beef homogenate. Also 4 ml of purified phage lysate was added to each tube to make a final concentration of 9.23 × 103 pfu ml−1. Beef homogenate containing only phage lysate was used as a background control. The sampling of culture, the bioluminescence measurement, the incubation time and the set-up of the detection threshold are the same as described previously.
Additional Information
How to cite this article: Zhang, D. et al. The Use of a Novel NanoLuc-Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 6, 33235; doi: 10.1038/srep33235 (2016).
Supplementary Material
Acknowledgments
This research was supported through a cooperative agreement with the Agricultural Research Service of the US Department of Agriculture project number 1935-42000-035. Dandan Zhang was also supported by a Purdue Research Foundation Fellowship. Claudia Coronel-Aguilera acknowledges the financial support from National Council for Science and Technology of Mexico (CONACYT, Post-doctoral program). Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Bruce Applegate is the founder of Phicrobe an LLC located in West Lafayette Indiana which has licensed technology related to using ΦV10 and its derivatives for the detection of E. coli O157:H7. Authors Bruce Applegate, Dandan Zhang, Claudia P Coronel-Aguilera, Patricia L Romero, Lynda Perry, Udit Minocha and Carla Rosenfield are coinventors on recent disclosures of IP related to ΦV10 and its derivatives.
Author Contributions B.A. conceived the idea and supervised this effort. D.Z., C.P.C.-A., C.R. and U.M. performed the experiments and analyzed the data. P.R. and L.P. generated the lysogenic strain of E. coli O157:H7 C7927 (ΦV10) containing the Red expression vector pKD46 and identified recET as an optimum location for modifications. A.G.G. and G.C.P. provided input on experimental design. A.B. provided direction on ΦV10 specificity assays. D.Z. wrote the main manuscript text. D.Z. and C.P.C.-A. prepared the figures. D.Z., C.P.C.-A., C.R., A.G.G., G.C.P., A.B. and B.A. reviewed the manuscript.
References
- Heiman K. E., Mody R. K., Johnson S. D., Griffin P. M. & Gould L. H. Escherichia coli O157 Outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 21, 1293–1301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli G. C., Wijey C. & Uhlich G. A. Genetically Marked Strains of Shiga Toxin–Producing O157:H7 and Non-O157 Escherichia coli: Tools for Detection and Modeling. J. Food Prot. 78, 888–901 (2015). [DOI] [PubMed] [Google Scholar]
- Rangel J. M., Sparling P. H., Crowe C., Griffin P. M. & Swerdlow D. L. Epidemiology of Escherichia coli O157:H7 Outbreaks,United States,1982–2002. Emerg. Infect. Dis. 11, 603–609 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torso L. M. et al. Escherichia coli O157:H7 Outbreak Associated with Restaurant Beef Grinding. J. Food Prot. 78, 1272–1279 (2015). [DOI] [PubMed] [Google Scholar]
- Bell B. P. et al. A multistate outbreak of Escherichia coli O157:H7—associated bloody diarrhea and hemolytic uremic syndrome from hamburgers: the washington experience. JAMA 272, 1349–1353 (1994). [PubMed] [Google Scholar]
- Conedera G. et al. A family outbreak of Escherichia coli O157 haemorrhagic colitis caused by pork meat salami. Epidemiol. Infect. 135, 311–314 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J. et al. Spinach-associated Escherichia coli O157:H7 Outbreak, Utah and New Mexico, 2006. Emerg. Infect. Dis. 14, 1633–1636 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers M. L. et al. An Outbreak of Escherichia coli O157:H7 Infections Associated with Leaf Lettuce Consumption. J. Infect. Dis. 177, 1588–1593 (1998). [DOI] [PubMed] [Google Scholar]
- Hilborn E. D. et al. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159, 1758–1764 (1999). [DOI] [PubMed] [Google Scholar]
- Riley L. W. et al. Hemorrhagic Colitis Associated with a Rare Escherichia coli Serotype. N Engl J Med 308, 681–685 (1983). [DOI] [PubMed] [Google Scholar]
- Hagens S. & Loessner M. J. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76, 513–519 (2007). [DOI] [PubMed] [Google Scholar]
- Smartt A. E. & Ripp S. Bacteriophage reporter technology for sensing and detecting microbial targets. Anal. Bioanal. Chem. 400, 991–1007 (2010). [DOI] [PubMed] [Google Scholar]
- Oda M., Morita M., Unno H. & Tanji Y. Rapid Detection of Escherichia coli O157:H7 by Using Green Fluorescent Protein-Labeled PP01 Bacteriophage. Appl. Environ. Microbiol. 70, 527–534 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awais R., Fukudomi H., Miyanaga K., Unno H. & Tanji Y. A Recombinant Bacteriophage-Based Assay for the Discriminative Detection of Culturable and Viable but Nonculturable Escherichia coli O157:H7. Biotechnol. Prog. 22, 853–859 (2006). [DOI] [PubMed] [Google Scholar]
- Waddell T. E. & Poppe C. Construction of mini-Tn10luxABcam/Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol. Lett. 182, 285–289 (2000). [DOI] [PubMed] [Google Scholar]
- Brigati J. R. et al. Bacteriophage-Based Bioluminescent Bioreporter for the Detection of Escherichia coli O157:H7. J. Food Prot. 70, 1386–1392 (2007). [DOI] [PubMed] [Google Scholar]
- Kim S., Kim M. & Ryu S. Development of an Engineered Bioluminescent Reporter Phage for the Sensitive Detection of Viable Salmonella Typhimurium. Anal. Chem. 86, 5858–5864 (2014). [DOI] [PubMed] [Google Scholar]
- Franche N., Vinay M. & Ansaldi M. Substrate-independent luminescent phage-based biosensor to specifically detect enteric bacteria such as E. coli. Environ. Sci. Pollut. Res. 1–10 (2016). [DOI] [PubMed] [Google Scholar]
- Willford J. & Goodridge L. D. Integrated assay for rapid detection of Escherichia coli OI57:H7 on beef samples. Food Prot. Trends. 28, 468–472 (2008). [Google Scholar]
- Hall M. P. et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 7, 1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J. M. S. & Proft T. Comparison of firefly luciferase and NanoLuc luciferase for biophotonic labeling of group A Streptococcus. Biotechnol. Lett. 36, 829–834 (2013). [DOI] [PubMed] [Google Scholar]
- Lee J. & Mittar D. Evaluation of NanoLuc and GFP reporter-labeled control strains for Shiga toxin-producing Escherichia coli (STEC) assays Available at: http://www.atcc.org/~/media/PDFs/Presentations/2014/IAFP%202014%20Poster%20Evaluation%20of%20NanoLuc%20and%20GFP%20reporter-labeled%20control%20strains.ashx. (Accessed: 6th April 2016) (2014)
- Smartt A. E. et al. Pathogen detection using engineered bacteriophages. Anal. Bioanal. Chem. 402, 3127–3146 (2012). [DOI] [PubMed] [Google Scholar]
- Jacobs W. R. et al. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260, 819–822 (1993). [DOI] [PubMed] [Google Scholar]
- Perry L. L. et al. Sequence analysis of Escherichia coli O157:H7 bacteriophage ΦV10 and identification of a phage-encoded immunity protein that modifies the O157 antigen. FEMS Microbiol. Lett. 292, 182–186 (2009). [DOI] [PubMed] [Google Scholar]
- Romero P. L. A cobA-based reporter bacteriophage for the detection of E. coli O157:H7. Theses Diss. Available at ProQuest. 1–79 (2008).
- Ulitzur S. & Kuhn J. In Methods In Enzymology, Vol. 305 (eds. Ziegler M.M. & Baldwin T.O.) 543–557 (Academic Press, 2000). [DOI] [PubMed] [Google Scholar]
- Teunis P., Takumi K. & Shinagawa K. Dose response for infection by Escherichia coli O157:H7 from outbreak data. Risk Anal. 24, 401–407 (2004). [DOI] [PubMed] [Google Scholar]
- Murphy K. C. & Campellone K. G. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4, 11 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R., Acosta S., Hernalsteens J. P., Jofre J. & Muniesa M. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 7, 1–12 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano R. C. & Bertolini J. Selection for virulence in the fish pathogen Aeromonas salmonicida, using coomassie brilliant blue agar. J. Wildl. Dis. 24, 672–678 (1988). [DOI] [PubMed] [Google Scholar]
- Uljas H. E. & Ingham S. C. Combinations of Intervention Treatments Resulting in 5-Log10-Unit Reductions in Numbers of Escherichia coli O157:H7 and Salmonella typhimurium DT104 Organisms in Apple Cider. Appl. Environ. Microbiol. 65, 1924–1929 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Agriculture-Food Safety and Inspection Service. MLG Appendix 1.08. Media and Reagents Available at: http://www.fsis.usda.gov/wps/wcm/connect/b089fb25-5d69-4b01-a470-da1d0182a5d7/MLG-Appendix-1-Media-Reagents.pdf?MOD=AJPERES. (Accessed: 11th November 2015) (2013).
- U.S. Department of Agriculture-Food Safety and Inspection Service. MLG 5.09. Detection, Isolation and Identification of Escherichia coli O157:H7 from Meat Products and Carcass and Environmental Sponges Available at: http://www.fsis.usda.gov/wps/wcm/connect/51507fdb-dded-47f7-862d-ad80c3ee1738/MLG-5.pdf?MOD=AJPERES. (Accessed: 11th November 2015) (2015).
- Don R. H., Cox P. T., Wainwright B. J., Baker K. & Mattick J. S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. G. & Lindow S. E. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191, 149–153 (1997). [DOI] [PubMed] [Google Scholar]
- Sawitzke J. A. et al. In Methods In Enzymology, Vol. 421 (eds Hughes K. T. & Maloy S. R.) 171–199 (Academic Press, 2007).17352923 [Google Scholar]
- Sambrook J., Fritsch E. F. & Maniatis T. In Molecular Cloning: A Laboratory Manual 2nd edn, Vol. 1, Ch. 2, 2.61 (Cold Spring Harbor Laboratory Press, 1989). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


