Abstract
The first step of lipid A biosynthesis in Escherichia coli (E. coli) is catalyzed by LpxA (EcLpxA), an acyltransferase selective for UDP-N-acetylglucosamine (UDP-GlcNAc) and R-3-hydroxymyristoyl-acyl carrier protein (3-OH-C14-ACP), and is an essential step in majority of Gram-negative bacteria. Since the majority of lipid A species isolated from F. novicida contains 3-OH-C16 or 3-OH-C18 at its C3 and C3' positions, FnLpxA was thought to be selective for longer acyl chain (3-OH-C16 and 3-OH-C18) over short acyl chain (3-OH-C14, 3-OH-C12, and 3-OH-C10). Here we demonstrate that Francisella novicida (F. novicida) lpxA functionally complements an E. coli lpxA knockout mutant and efficiently transfers 3-OH-C14 as well as 3-OH-C16 in E. coli. Our results implicate that the acyl chain length of lipid A is determined by several factors including acyl chain selectivity of LpxA and downstream enzymes, as well as the composition of the acyl-ACP pool in vivo. We also report the crystal structure of F. novicida LpxA (FnLpxA) at 2.06 Å. The N-terminal parallel beta-helix (LβH) and C-terminal alpha-helical domain are similar to other reported structures of LpxAs. However, our structure indicates that the supposed ruler residues for hydrocarbon length, 171L in one monomer and 168H in the adjacent monomer in a functional trimer of FnLpxA, are located just 3.8 Å apart that renders not enough space for binding of 3-OH-C12 or longer acyl chains. This implicates that FnLpxA may have an alternative hydrophobic pocket, or the acyl chain may bend while binding to FnLpxA. In addition, the FnLpxA structure suggests a potential inhibitor binding site for development of antibiotics.
Keywords: F. novicida lipid A, LpxA crystal structure, acyl chain ruler, LpxA, acyl chain selectivity, inhibitor binding site
1. Introduction
Francisella tularensis (F. tularensis) is a highly infectious Gram-negative pathogen and is responsible for tularemia, a fatal infectious disease in mammals. It is a potential bioweapon, as it takes as few as 10 bacteria of F. tularensis subspecies tularensis to cause tularemia. Francisella novicida (F. novicida) is an environmental isolate, and can be an excellent model system for studying F. tularensis biochemistry as it is infectious not to humans but to mice, and shares 97.7% average nucleotide identities [1].
Lipid A, the hydrophobic moiety of lipopolysaccharide (LPS), is an essential component of the outer membrane of Gram-negative bacteria, and is recognized by mammalian innate immune system [2, 3]. While the structure of lipid A portion of LPS is relatively conserved, there exist some variations among species. The lipid A in F. novicida and other strains of Francisella has several interesting features. First, the length of R-3-hydroxyacyl chains is longer than that of other bacteria, such as Escherichia coli (E. coli), and we can observe micro-heterogeneity in the lengths of acyl chains (Fig. 1A). In addition, over 90% of their lipid A is in free form, not attached to the core and O-antigen sugars, even though Francisella species contain LPS [4].
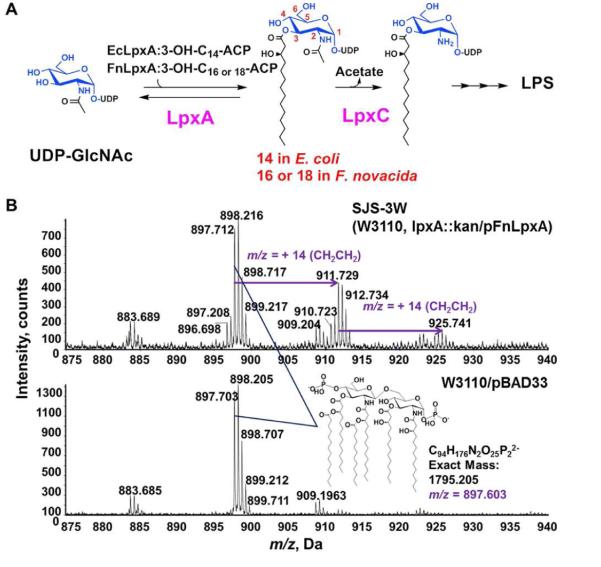
Figure 1. F. novicida lpxA complements E. coli lpxA knockout.
A. LPS biosynthetic pathway B. ESI-MS spectra of Lipid A species expressing FnLpxA in E. coli SJS-3W and wild type E. coli W3110/pBAD33. Cells expressing FnLpxA accumulate lipid A derivatives (predicted [M-2H]2− at m/z 897.603) similar to wild type E. coli W3110/pBAD33 (lower panel) as well as 28 and 56 amu (14 m/z and 28 m/z, respectively in the upper panel) larger. Lipids were extracted after acid hydrolysis of LPS.
LpxA catalyzes the first reaction of lipid A biosynthesis by transferring an acyl chain from acyl-ACP to UDP-N-acetylglucosamine (UDP-GlcNAc). Even though E. coli LpxA (EcLpxA) and F. novicida LpxA (FnLpxA) share significant sequence homology (44% identity and 74% similarity) as analyzed by Clustal omega [5], FnLpxA transfers longer hydroxyacyl chains, R-3-hydroxypalmitoyl group (3-OH-C16) and R-3-hydroxystearoyl group (3-OH-C18), compared to EcLpxA (R-3-hydroxymyristoylgroup, 3-OH-C14). To understand acyl chain length selectivity, the species-specificities of FnLpxA, and establish a structural basis for molecular mechanism and inhibitor design, we investigated the biochemical properties of FnLpxA in vitro, and in vivo, and determined its crystal structure at 2.06 Å resolution.
2. Materials and Methods
2.1 Materials
Chloroform, methanol, and silica gel 60 (0.25 mm) thin layer chromatography (TLC) plates were from EMD Millipore (Germany). QIAprep Miniprep kit for plasmid purification and Ni-NTA superflow resin were from Qiagen (Germany). Easy-DNA kit for genomic DNA purification was from Invitrogen (USA). A HiLoad 26/60 Superdex 200 prep grade column and Akta protein purification system, and PhosphorImager were from GE Healthcare (United Kingdom). Tryptone, yeast extract, and agar were from BD Sciences (USA). [α-32P]UTP was purchased from PerkinElmer (USA). Oligonucleotide primers were obtained from Integrated DNA Technologies (USA). Enzymes for cloning were from New England Biolabs (USA). The crystallography reagents and Crystal Screen HT were from Hampton Research (USA). Other chemical reagents were from Sigma-Aldrich (USA).
2.2 Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are described in Table S1. Protocols for handling DNA and preparing E. coli cells for electroporation were those of Sambrook and Russell [6]. A C-terminally His6-tagged SJD-02 was constructed using primers SJP-015F (5'- GAGATATACATATGATACATAGTTTGGCAGTAGTACATGAG -3') and SJP-015His (5'- CGAATTCGGATCCCTTAGTATACCTCTTCGCGAAGTACC -3'), pET-21b(+) vector, and a cDNA library of F. novicida, a gift from Jinshi Zhao (Duke University, USA). The resulting plasmid was confirmed with primers T7P (5'- TAATACGACTCACTATAGGG -3) and T7T (5'- GCTAGTTATTGCTCAGCGG -3'), and transformed into C41(DE3) [7] for protein expression. The native FnlpxA gene was amplified from SJD-02 with primers SJP-022 (5'- CTAGCGAGCTCAAGGAGATATACCATGATACATAGTTTG GCAGTAGTACATGAG -3') and SJP-022B (5'- CTGCAGGTCGACTTATCTTAGTATACCTCTTC GCGAAGTACC -3') and inserted into pBAD33 [8] using the SacI and SalI sites to construct pFnLpxA. Plasmids were isolated with QIAprep Miniprep kit, and the bacterial genomic DNAs were purified with the Easy-DNA kit. All DNA constructs were confirmed by dideoxy sequencing.
2.3 Construction of a conditional lpxA chromosomal knockout strain
A conditional lpxA chromosomal knockout strain, SJS-3W, was constructed by transducing lpxA::kan into the E. coli W3110 cells harboring pFnLpxA as previously described [9]. Briefly, the lpxA::kan cassette was moved into W3110 using P1 transduction in the presence of 0.2% L-arabinose. Consistent with lpxA being required for growth, transductants were only viable when cells carried pFnLpxA but not the control vector pBAD33. The chromosomal lpxA deletion was confirmed by sequencing the lpxA region on the chromosome of W3110 lpxA::kan/pFnLpxA strain, and the knockout strain was named as SJS-3W. To check in vivo complementation phenotype, SJS-3W and W3110 harboring pBAD33 (W3110/pBAD33) as a control were streaked on the LB agar plates containing 25 μg/mL chloramphenicol and 0%, 0.02%, or 0.2% L-arabinose.
2.4 FnLpxA expression and purification from E. coli
Full-length (residues 1-259) FnLpxA, tagged with 6 histidine residues at its C-terminus, was over-expressed in E. coli C41(DE3) cells [7], and purified by a Ni-NTA superflow column and a HiLoad 26/60 Superdex 200 prep grade column.
2.5 LpxA enzymatic activity assays
R,S-3-hydroxypalmitoyl-ACP was obtained as previously described [10, 11], and the LpxA assay conditions were similar as reported previously [12, 13]. A 40 mM HEPES (pH 7.4) reaction buffer containing 20 μM R,S-3-hydroxypalmitoyl-ACP, 100 μM [α-32P]UDP-GlcNAc (4.5 × 104 μCi/mmol) and 100 mM NaCl was mixed with the enzyme and incubated at 30 °C. The reactions were terminated by spotting the reaction mixture onto a silica gel TLC plate. The plates were developed in CHCl3/CH3OH/H2O/CH3COOH (25:1:4:2, v/v), and exposed to a PhosphorImager screen for measuring the extent of acylation of [α-32P] UDP-GlcNAc.
2.6 Isolation and characterization of lipid A species
Lipid A species were prepared from the overnight cultures of E. coli SJS-3W cells and W3110/pBAD33 by acid hydrolysis of LPS [14] followed by two-phase Bligh-Dyer extraction [15], as described previously [16]. The isolated lipid A samples were subjected to mass spectrometry analysis as described elsewhere [17, 18].
2.7 Crystallization, data collection, structure determination, and refinement
The initial crystallization screening was done with Hampton crystal screen HT using hanging-drop vapor diffusion method at 17 °C. The reservoir solution containing 0.2 M lithium sulfate, 0.1 M Tris HCl pH 8.5, and 30% PEG 4,000 mixed with the protein at ~10 mg/mL concentration (2 μL each) yielded crystals that were rhomboid-shaped (~80 μm each side) in 1-2 days. For data collection, the crystals were frozen in liquid N2. X-ray diffraction data were collected with R-AXIS IV++ image-plate detector using Rigaku RU-200 rotating anode generator. The diffraction images were indexed and scaled with XDS [19].
The FnLpxA structure was solved by the molecular replacement method in PHASER using E. coli LpxA (PDB id: 1LXA) as the search model. Regions outside the hexapeptide repeats were removed from the initial molecular replacement solution, and manually rebuilt using COOT [20]. The structure refinement was carried out with REFMAC5 [21] within the CCP4 software suite [22]. The final model was validated using MOLPROBITY [23]. Structural alignments and figures were prepared using PyMol [24], and the atomic coordinates and structure factor have been deposited in the Protein Data Bank (PDB id: 5F42). The crystallographic information is summarized in Table S2.
3. Results and discussion
3.1 In vitro activity of the FnLpxA enzyme
FnLpxA was expressed as a His6-tagged protein in E. coli and purified to homogeneity. In vitro assays showed that purified FnLpxA transfers a 3-hydroxypalmitoyl group from 3-hydroxypalmitoyl-ACP to the 3-OH of UDP-GlcNAc with a specific activity of ~2.1 nmol min−1 mg−1 (see Fig S1, Table S3). Even though the acyltransferase activity of FnLpxA was about 20 times lower than that of purified EcLpxA, it is comparable to the enzymatic activity (~5 nmol min−1 mg−1) of other LpxA ortholog from Arabidopsis thaliana [9], which were purified from E. coli expression system.
3.2 Complementation of E. coli lpxA knockout mutant by F. novicida lpxA
Since the lipid A of F. novicida majorly contains longer hydroxyacyl chains (3-OH-C16/3-OH-C18) at its C3 and C3′positions, FnLpxA should have been evolved to transfer 3-OH-C16 or 3-OH-C18 effectively. Therefore, it was questioned whether FnLpxA could functionally replace chromosomal EclpxA that transfers 3-OH-C14 in E. coli. To answer this, we constructed a strain SJS-3W, a conditional mutant of E. coli chromosomal lpxA knockout containing a plasmid harboring FnlpxA under control of an arabinose inducible promoter, pFnLpxA. Interestingly, FnLpxA supported the survival and growth of E. coli lpxA knockout mutant in the presence of L-arabinose (Fig. S2).
3.3 Characterization of lipid A species
As FnLpxA can functionally replace EcLpxA despite the difference in the acyl-chain selectivity between the two orthologs, we wondered about its effects on lipid A structure in E. coli under the conditions that all other enzymes and substrates are the same as the wild type E. coli. LPS samples isolated from SJS-3W cells and wild type W3110/pBAD33 were subjected to mild acid hydrolysis to release lipid A for structural characterizations using negative-ion electrospray ionization quadrupole time-of-flight mass spectrometry. The major negative-ions observed are doubly charged [M–2H+]2− ions, and the prominent ion in both spectra was the [M–2H+]2− at m/z 897.7, corresponding to the major hexa-acylated lipid A species with 3-OH-C14 in its 3 and 3' positions, usually found in wild-type E. coli (Fig. 1C). In addition, we could observe the presence of [M–2H+]2− at m/z 911.7 and m/z 925.7 in the spectrum of lipid A species of SJS-3W. These ions correspond to the lipid A in which one or two of the acyl chains are hydroxypalmitoyl group (3-OH-C16). Considering the fact that the major species founded in F. novicida lipid A are 3-OH-C16/3-OH-C18 at 3 and 3' positions, the observed major lipid A species containing 3-OH-C14 was unexpected. However, other factors such as the relative abundance of 3-OH-C14 acyl ACP to 3-OH-C16 acyl ACP in E. coli and/or the selective pressure from the downstream enzymes of the lipid A pathway, could explain the efficient incorporation of 3-OH-C14 into UDP-GlcNAc by FnLpxA to support E. coli growth.
3.4 Crystal structure of FnLpxA
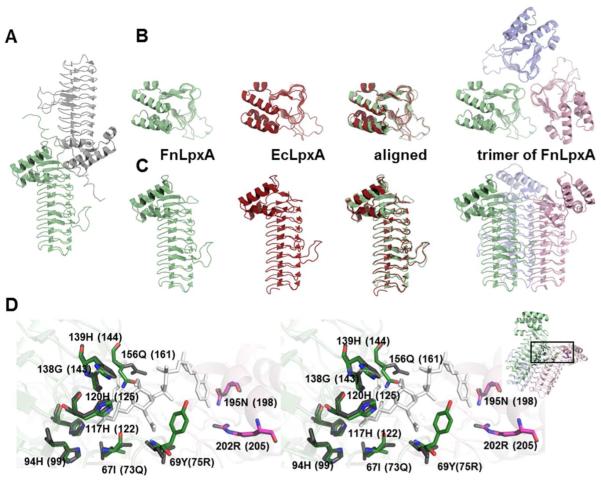
To gain a better insight into the FnLpxA catalytic mechanism and the species-specificities, we crystallized a His6-tagged FnLpxA as described in section 2.7. A single crystal diffracted to 2.06 Å, and the FnLpxA structure was solved by molecular replacement, using a poly-Ala model derived from EcLpxA (PDB id: 1LXA) [25]. The FnLpxA crystal belongs to the space group H32, with cell parameters of a = b = 105.4 Å, c = 285.8 Å (Table S2). The FnLpxA asymmetric unit contains two monomers (Fig. 2A), and each monomer is in parallel with the crystallographic threefold axis. The homotrimeric structure of FnLpxA, which can be generated by a 120° rotation about the axis of symmetry, agrees well with the trimeric structure of EcLpxA (Fig. 2B, and 2C). The Rwork and Rfree values for FnLpxA converged at 21%, and 26%, respectively. Among 280 residues of the protein including a linker and a His6-tag, 4 histidine residues in the carboxyl tag region were not included in the model due to insufficient electron density.
Figure 2. Structure of FnLpxA.
A. Two chains of the FnLpxA in the asymmetric unit. B. Top-down views of the monomers of FnLpxA (pale green), EcLpxA (red), and the biological trimer of FnLpxA (pale green, light pink, and light blue). C. Side views as indicated in B. D. Overlay of the conserved residues within the active site of FnLpxA and EcLpxA (stereo view). Residues in the active site of FnLpxA superimposed with the structure of EcLpxA complexed with its product, UDP-3-O-[(R)-hydroxymyristoyl]-GlcNAc (PDB id: 2QIA [26]). EcLpxA residues and the product are colored grey and white, respectively, while FnLpxA residues are colored by element. The residue names and numbers of FnLpxA are shown, with the corresponding residues from EcLpxA in parentheses. In the inset, the box on the FnLpxA trimer indicates the active site area enlarged in the figure.
The structure of FnLpxA contains the characteristic N-terminal left-handed parallel β helix (LβH) domain, in a shape of triangular prism, followed by a C-terminal portion composed mainly of α-helices. The N-terminal LβH domain consists of 10 beta-helical coils (C1 through C10). Each helical coil consists of 3 hexapeptide repeats, with the exception of coil C1 and C10 that lack the first and the last hexapeptide repeat, respectively. There are two loops, loop 1 and loop2, inserted at coils C4-C5 and C5-C6, respectively. The overall structure of FnLpxA is very similar to those structures of LpxA orthologs reported thus far, and superposition of FnLpxA and EcLpxA yielded an RMS deviation of 0.5 Å for 208 Cα atoms pairs, as analyzed by PyMol [24] (Fig. 2B and 2C). Most of key active site residues are similarly positioned when the FnLpxA is compared with EcLpxA bound with UDP-3-O-[(R)-3-hydroxymyristoyl]-GlcNAc (PDB id: 2QIA [26]). His120 (His125 in EcLpxA) is proposed to be a catalytic base, activating GlcNAc 3-OH group to transfer the acyl chain from acyl-ACP, and His139 (His144 in EcLpxA) is proposed to form a hydrogen bond with the GlcNAc 6-OH group of the substrate. Although 73Q and 75L of EcLpxA are replaced with 67I and 69Y in FnLpxA, theses substitutions would not affect the binding of substrate as the backbone amide nitrogens, not the side chains, are involved with interaction (Fig. 2D).
3.5 Acyl-chain binding site of FnLpxA
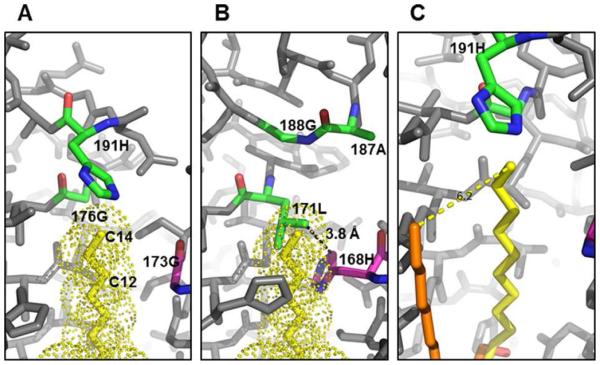
To explain selectivity of hydrocarbon length of LpxAs, certain residues have been suggested to act as ‘hydrocarbon rulers’ for other LpxA orthologs. For example, 168M of Pseudomonas aeruginosa LpxA (PaLpxA) dictates 3-OH-C10 acyl chain selectivity [27], whereas 191H of EcLpxA dictates binding of 3-OH-C14 acyl chain [26]. Based on these suggested rulers, we searched the corresponding residues in FnLpxA structure. For 3-OH-C14 acyl chain ruler, a non-bulky residue 188G, located in the same position as 191H of EcLpxA, enables to accommodate the longer acyl chain such as 3-OH-C16 or 3-OH-C18. However, 176G of EcLpxA that allows the accommodation of 3-OH-C12 acyl group is replaced with a bulky residue 171L in FnLpxA. Moreover, the equivalent residue to 168M of PaLpxA, reportedly a 3-OH-C10 acyl chain ruler, is replaced with a glycine in EcLpxA but with a bulky residue (168H) in FnLpxA. In EcLpxA, the lack of side chains in 173G and 176G provides a passage for the acyl chain to extend through the binding groove between two neighboring monomers of a functional homotrimer (Fig 3A). However, in FnLpxA, the distance between the 168H of FnLpxA monomer and the 171L from the adjacent monomer becomes 3.8 Å due to bulky side chains (Fig 3B). Therefore, the space corresponding to the binding grooves for 3-OH-C14 in EcLpxA and 3-OH-C-12 in Leptospira interrogans LpxA, respectively [26, 28], is practically sealed where the 12th carbon of the acyl chain would be located in FnLpxA. While it remains to be determined how FnLpxA binds acyl chain, it stands to reason that there might be an alternative acyl chain binding groove in the trimeric structure of LpxA. In fact, the structure of Helicobacter pylori LpxA (PDB id: 1J2Z) [29], crystalized with a 1-s-octyl-β-D-thioglucoside, suggests a possible acyl chain binding groove: the octyl group of 1-s-octyl-β-D-thioglucoside runs in parallel, but ~ 6 Å apart from the acyl binding groove known for EcLpxA (Fig. 3C). This alternative hydrocarbon binding pocket might explain how FnLpxA can bind longer hydrocarbon acyl chain. Instead, the acyl chain might start bend before it reaches the ceiling formed by 168H and 171L pair, rather than extend straight as observed in 2QIA [26] (Fig. 3A).
Figure 3. Hydrocarbon binding groove of FnLpxA.
A. The hydrocarbon binding pocket and R-OH-C14 rulers of EcLpxA. The carbon atoms of the ligand (dotted) are shown in yellow, and three ruler residues, 173G (C10 ruler), 176G (C12 ruler), and 191H (C14 ruler), reported are colored by element based on monomer colors while others are grey. 191H is near the end of the terminal methyl group (C14). 176G and 173G allow enough space for alkyl group in the binding groove. B. FnLpxA cannot accommodate 3-OH-C14 if acyl chain binds to FnLpxA in the same manner observed in 2QIA [26]. The structure of FnLpxA trimer is overlaid to the EcLpxA. While the top roof area contains the small residue 188G, the other two ruler residues 171L and 168H are too bulky to accommodate alkyl chain in the same binding groove of EcLpxA. C. A possible, alternative binding groove for 3-OH-C14, 16, or 18. The structure of 1-s-octyl-β-D-thioglucoside complexed with HpLpxA (PDB id: 1J2Z [29]) is overlaid to panel A. The octyl group extends in parallel, ~ 6 Å apart from hydroxymyristoyl group of UDP-3-O-[(R)-hydroxymyristoyl]-GlcNAc in 2QIA. Figures are drawn from PDB ids 2QIA and 1J2Z.
3.6 A potential inhibitor binding site of FnLpxA
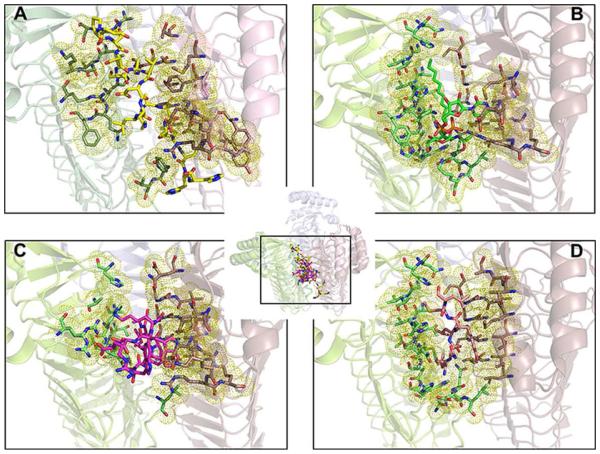
The FnLpxA protein used for crystallization contains a long expression tag of 21 residues (residues 260~280; DPNSSSVDKLAAALEHHHHHH). Out of those residues, 17 except the last 4 histidine residues could be built into the structure. In aqueous solution, the His6-tagged FnLpxA is a homotrimer, as judged by size-exclusion chromatography, and it is enzymatically active. In the crystal packing, the expression tag extends to the other monomer in the asymmetric unit (Fig. 2A), located at the interface of two adjacent monomers in another functional trimer: the presence of the tag sequence keeps two separate trimers intertwined. The tag sequence contains a helical region (262N~272A), and the residues 268K through 272N correspond to the site that accommodates the UDP-GlcNAc in EcLpxA (Fig. 4A, 4B) [26]. Looking at interactions between the tag region and the other FnLpxA monomer in the asymmetric unit, we identified a salt bridge between 267D and 255R and hydrogen bonds between backbone carbonyl of 274E and side chain of 202R. As the part of the tag sequence is localized to the suggested substrate binding site, we overlaid the structure of FnLpxA and the complex structures of inhibitors and EcLpxA (Fig. 4). Notably, the position of the tag coincides very well with the reported binding location of peptide inhibitors of EcLpxA, Peptide 920 [30] (Fig. 4C) and RJPXD33 [31] (Fig. 4D). Those residues of FnLpxA in close contact (within 5 Å) with the tag sequence agree very well with those residues of EcLpxA close to the known inhibitors and UDP-GlcNAc of the product of EcLpxA (Fig. 4 and Fig. S3). These results suggest the possibility of development of broad spectrum inhibitors targeting these binding sites against LpxA, regardless of acyl chain length selectivity.
Figure 4. A potential inhibitor binding site in FnLpxA.
A. The expression tag of FnLpxA (yellow sticks) extends to the other monomer in the asymmetric unit, located at the interface of two adjacent monomers in another functional trimer. B. The product of EcLpxA, uridine-5'-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-D-glucosamine (UDP-3-O-(R-3-OH-C14)-GlcNAc (green sticks) is located at the interface of two adjacent monomers in the functional trimer (PDB id: 2QIA [26]). C. P920, a peptide inhibitor (pink sticks) of EcLpxA, is located at the interface of two adjacent monomers of EcLpxA in the functional trimer. (PDB id: 2AQ9 [30]). D. RJPXD33, another peptide inhibitor (salmon sticks) of EcLpxA, is located at the interface of two adjacent monomers of EcLpxA in the functional trimer (PDB id: 4J09 [31]). In the inset, the FnLpxA trimer and EcLpxE trimers are overlaid and the binding sites of individual structures are enlarged (boxed region) in Fig. 4A, 4B, 4C, and 4D. Amino acids within 5 Å are highlighted with yellow dots.
Supplementary Material
Highlights.
F. novacida lpxA functionally complements an E. coli lpxA knockout mutant.
• FnLpxA efficiently transfers 3-OH-C14 as well as 3-OH-C16 in E. coli
Crystal structure of FnLpxA is determined at 2.06 Å resolution
FnLpxA does not follow “hydrocarbon ruler” suggested for other LpxA orthologs
FnLpxA structure implicates a potential inhibitor binding site
Acknowledgements
We thank the late professor Christian R. H. Raetz for his insightful contributions to the foundations of this work and his mentorship. We specially thank Dr. Ali Masoudi for critical reading of the manuscript.
Funding sources
This work was supported by National Institutes of Health Grant GM-51310 to C.R.H.R, by the Intramural Research Program of KIST, and by the Multi-omics program of the Korean Ministry of Science, ICT, and Future Planning.
Abbreviations
- ACP
acyl carrier protein
- EcLpxA
Escherichia coli LpxA
- FnLpxA
Francisella novicida LpxA
- HpLpxA
Helicobacter pylori LpxA
- LβH
left-handed parallel β helix
- LPS
lipopolysaccharide
- LB
Luria Bertani
- ESI
electrospray ionization
- MS
mass spectrometry
- amu
atomic mass unit
- PBS
phosphate-buffered saline
- GlcNAc
N-acetylglucosamine
- RMS
root mean square
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- acyl-ACP
acylated-acyl carrier protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A. References
- 1.Siddaramappa S, Challacombe JF, Petersen JM, Pillai S, Hogg G, Kuske CR. Common Ancestry and Novel Genetic Traits of Francisella novicida-Like Isolates from North America and Australia as Revealed by Comparative Genomic Analyses. Appl Environ Microb. 2011;77:5110–5122. doi: 10.1128/AEM.00337-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raetz CR, Guan Z, Ingram BO, Six DA, Song F, Wang X, Zhao J. Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res. 2009;50(Suppl):S103–108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambrook JG, Russel DW. Plainview, NY. 3rd 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 7.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 8.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joo SH, Chung HS, Raetz CR, Garrett TA. Activity and Crystal Structure of Arabidopsis thalianaUDP-N-Acetylglucosamine Acyltransferase. Biochemistry. 2012;51:4322–4330. doi: 10.1021/bi3002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartling CM, Raetz CR. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–5302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartling CM, Raetz CR. Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2009;48:8672–8683. doi: 10.1021/bi901025v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyckoff T, Lin S, Cotter R, Dotson G, Raetz C. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J Biol Chem. 1998;273:32369–32372. doi: 10.1074/jbc.273.49.32369. [DOI] [PubMed] [Google Scholar]
- 13.Wyckoff T, Raetz C. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J Biol Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, White K, Polissi A, Georgopoulos C, Raetz C. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 15.BLIGH EG, DYER WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 16.Chung HS, Raetz CR. Interchangeable domains in the Kdo transferases of Escherichia coli and Haemophilus influenzae. Biochemistry. 2010;49:4126–4137. doi: 10.1021/bi100343e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Ribeiro A, Guan Z, Abraham S, Raetz C. Attenuated virulence of a Francisella mutant lacking the lipid A 4'-phosphatase. Proc Natl Acad Sci U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W. Xds, Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.N. Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrodinger, LLC The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 25.Raetz C, Roderick S. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 26.Williams AH, Raetz CR. Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc Natl Acad Sci U S A. 2007;104:13543–13550. doi: 10.1073/pnas.0705833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith EW, Zhang XJ, Behzadi C, Andrews LD, Cohen F, Chen Y. Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity. Biochemistry. 2015;54:5937–5948. doi: 10.1021/acs.biochem.5b00720. [DOI] [PubMed] [Google Scholar]
- 28.Robins LI, Williams AH, Raetz CR. Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA. Biochemistry. 2009;48:6191–6201. doi: 10.1021/bi900629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BI, Suh SW. Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori. Proteins. 2003;53:772–774. doi: 10.1002/prot.10436. [DOI] [PubMed] [Google Scholar]
- 30.Williams AH, Immormino RM, Gewirth DT, Raetz CRH. Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. P Natl Acad Sci USA. 2006;103:10877–10882. doi: 10.1073/pnas.0604465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins RJ, Heslip KA, Meagher JL, Stuckey JA, Dotson GD. Structural basis for the recognition of peptide RJPXD33 by acyltransferases in lipid A biosynthesis. J Biol Chem. 2014;289:15527–15535. doi: 10.1074/jbc.M114.564278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.