Abstract
There is a growing emphasis in the field of psychiatry on the need to identify candidate biomarkers to aid in diagnosis and clinical management of depression, particularly with respect to predicting response to specific therapeutic strategies. MicroRNAs are small nucleotide sequences with the ability to regulate gene expression at the transcriptomic level and emerging evidence from a range of studies has highlighted their biomarker potential. Here we compared healthy controls (n=20) with patients diagnosed with major depression (n=40) and who were treatment-resistant to identify peripheral microRNA biomarkers, which could be used for diagnosis and to predict response to electroconvulsive therapy (ECT) and ketamine (KET) infusions, treatments that have previously shown to be effective in treatment-resistant depression (TRD). At baseline and after treatment, blood samples were taken and symptom severity scores rated using the Hamilton Depression Rating Scale (HDRS). Samples were analyzed for microRNA expression using microarray and validated using quantitative PCR. As expected, both treatments reduced HDRS scores. Compared with controls, the baseline expression of the microRNA let-7b was less by ~40% in TRD patients compared with controls. The baseline expression of let-7c was also lower by ~50% in TRD patients who received ECT. Bioinformatic analysis revealed that let-7b and let-7c regulates the expression of 27 genes in the PI3k-Akt-mTOR signaling pathway, which has previously been reported to be dysfunctional in depression. The expression of miR-16, miR-182, miR-451 and miR-223 were similar to that in controls. Baseline microRNA expression could not predict treatment response and microRNAs were unaffected by treatment. Taken together, we have identified let-7b and let-7c as candidate biomarkers of major depression.
Introduction
Depression is the most prevalent psychiatric disorder and current projections indicate that it will be the leading cause of disability by the year 2030.1 Subjective diagnostic schemes such as DSM-IV, ICD-10 are constrained in their ability to accurately diagnose depression2 and its subtypes such as treatment-resistant depression (TRD), which affects a significant proportion of patients.3, 4 As such, the potential benefits of using biomarkers to improve diagnostic precision and refine therapeutic strategies are significant.5 In this regard, emerging evidence from a range of clinical studies has reported the potential utility of microRNAs as biomarkers for a range of psychiatric disorders including depression.6, 7
MicroRNAs are small non-coding nucleotide sequences (18–24 nt), which regulate the expression of ~60% of the mammalian genome.8 Each microRNA can alter the translation of multiple messenger RNAs (mRNA) into proteins and each mRNA is the target of multiple microRNAs. This has led microRNAs to being known as ‘master-regulators' of cellular processes.9 The existence of microRNAs in bodily fluids such as blood and saliva10 has provided the impetus to evaluate their potential as biomarkers of illnesses and to predict response to different therapeutic strategies. In the context of depression, several microRNAs have shown biomarker potential including miR-16,11 miR-182,12 miR-223 and miR-451.13 There is also emerging evidence that basal microRNA expression can predict therapeutic response to antidepressants,14 consistent with their putative role as mediators of antidepressant effects.15 Moreover, several preclinical studies have shown that microRNAs mediate the antidepressant effects of electroconvulsive therapy (ECT, electroconvulsive stimulation in rodents)16 and the NMDA-receptor antagonist, ketamine (KET),16, 17, 18 therapeutic strategies, which have previously shown efficacy in TRD patients.19, 20, 21
Against this background, the aims of our study were to identify microRNA biomarkers that could be used for diagnosis of major depression as well as to predict response to treatments with ECT or KET.
Materials and methods
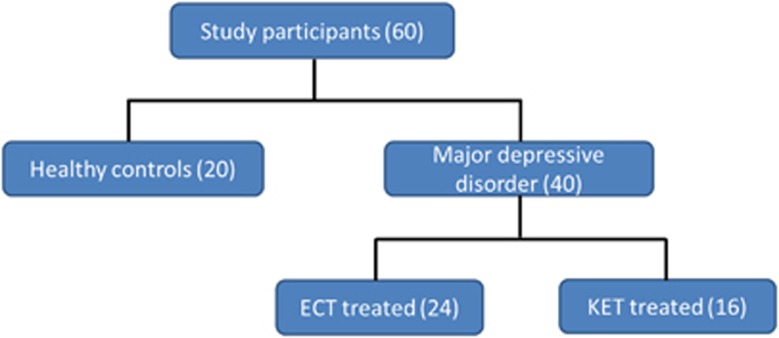
The study design (Figure 1), recruitment process and treatment procedures have been described previously.21 Briefly, patients who received KET treatment (n=16) were recruited from a mental health service in Cork, Ireland. Patients who received ECT (n=24) were inpatients at St Patrick's University Hospital. All patients had a diagnosis of major depressive disorder and had failed at least two adequate trials of antidepressant medication. Patients in the ECT group were older than healthy controls and patients in the KET group (Table 1). The majority of patients were receiving antidepressant therapy. Controls were recruited from Cork University Hospital staff and were screened for a personal or family history (1st degree relative) of a mental disorder and excluded if positive. Informed consent was obtained from all participants. There was no difference in gender profile between groups.
Figure 1.
Breakdown and number of study participants according to treatment received. ECT, electroconvulsive therapy; KET, ketamine.
Table 1. Characteristics of healthy controls and patients.
| Controls (n=20) | ECT (n=24)a | Ketamine (n=16)b | P-value | |
|---|---|---|---|---|
| Age | 42.85±2.22 | 56.88±2.75 | 44.63±3.40 | Control vs ECT: P<0.01 ECT vs KET: P<0.01 |
| Gender | 11M, 9F | 9M, 15F | 10M, 6F | NS |
| Baseline HDRS | — | 22.42±0.90 | 21.38±0.75 | NS |
| Post-treatment HDRS | — | 9.26±1.26 | 7.15±0.84 | NS |
| Responder | 5.67±0.78 (15/23) | 6.36±0.76 (11/13) 5.2±0.58 (5/11 long-term responders) | NS | |
| Non-responder | 16.00±1.49 (8/23) | 11.00±4.00 (2/13) | NS | |
| Medication profilec | ||||
| Selective serotonin reuptake inhibitor | 3 | 5 | ||
| Selective noradrenaline reuptake inhibitor | 8 | 4 | ||
| Serotonin agonist and reuptake inhibitor | 1 | — | ||
| Tricyclic antidepressant | 7 | — | ||
| Noradrenergic and specific serotonergic antidepressant | 4 | 1 | ||
| Monoamine oxidase inhibitor | 1 | — | ||
| Buproprion | 1 | 2 | ||
| Other | 7 | 4 | ||
Abbreviations: ECT, electroconvulsive therapy; F, female; HDRS, Hamilton Depression Rating Scale; KET, ketamine; M, male; NS, not significant.
One sample from a patient post-ECT session was unavailable.
Three samples from patient post-KET infusion were unavailable.
Eight patients in the ECT group were on more than one of antidepressants listed in this table.
The ketamine component of the study was approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals (EMC (3nn) 08/11/11) and the Irish Medicines Board (IMB: EudraCT number: 2011-003654-40). The ECT component of the study was approved by the Ethics Committee of St Patrick's University Hospital (Protocol No. 21/12).
Patients received either KET intravenously (0.5 mg kg−1) once a week for up to three sessions, or bi-weekly brief-pulse bitemporal ECT based on published protocols. Blood samples were collected from all study participants at baseline between 0800 and 1100 hours on the morning of the first visit, prior to the first treatment session. Blood samples were obtained from KET-treated patients for microRNA analysis 24 h after the first infusion. For patients who received ECT treatment, samples were collected 4–7 days after the final session. For all patients, depression was assessed using the Hamilton Depression Rating Scale (HDRS) at the same time points at which blood was collected. Baseline HDRS scores were similar between the two groups of patients (Table 1). For patients who received KET treatment, additional HDRS scores were obtained 1 week after the first infusion. Treatment response was classified as at least a 50% reduction in HDRS scores relative to baseline. All blood samples were collected in PAXgene blood RNA tubes (PreAnalytix, Hombrechtikon, Switzerland) and stored at −80 °C until processing.
Blood microRNA extraction
Total mRNA was isolated from blood samples using PAXgene Blood miRNA Kit (PreAnalytix). Samples were left at room temperature overnight prior to RNA extraction. Isolation was performed according to the protocol provided with the kit. Samples were eluted in 80 μl of buffer solution and stored in aliquots at −80 °C. Total RNA yield and quality were verified using the Nanodrop2000 spectrophotometer (ThermoScientific, Waltham, MA, USA) and RNA Integrity Number (RIN value) was assessed using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Only samples with a RIN value ⩾7 were used for microarray analysis.
Microarray analysis
MicroRNA Array analysis was completed by Exiqon Services (Exiqon, Vedbæk, Denmark). The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. Four hundred nanograms of total RNA from both sample and reference was labeled with Hy3TM and Hy5TM fluorescent label, respectively, using the miRCURY LNA microRNA Hi-Power Labeling Kit, Hy3TM/Hy5TM (Exiqon) following the procedure described by the manufacturer. The quantified signals were background corrected (Normexp with offset value 10) and normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm. Array data have been deposited in the NCBI's Gene Expression Omnibus (GSE81152).
MicroRNA quantification
Quantitative real-time PCR (qRT-PCR) was carried out on available samples using probes (6 carboxy fluorescein-FAM) designed by Applied Biosystems (Carlsbad, CA, USA): miR-16 (assay ID:000391), miR-182 (assay ID:002334), miR-451 (assay ID:464419_mat), miR-223 (assay ID:002098), let-7b (assay ID:002619) and let-7c (assay ID:000379). RNA was reverse-transcribed to complementary DNA using hairpin primers specific to each microRNA gene of interest on Applied Biosystem's GeneAmp PCR System 9700. qRT-PCR was carried out on the StepOnePlus PCR machine (Applied Biosystems). Samples were heated to 95 °C for 10 min, and then subjected to 40 cycles of amplification by melting at 95 °C and annealing at 60 °C for 1 min. Experimental samples were run in technical replicates with 1.33 μl complementary DNA per reaction. To check for amplicon contamination, each run also contained template free controls for each probe used.
Bioinformatic analysis
Bioinformatic analysis was performed using MicroT-CDS (v5.0) and the DIANA miRPath server (v3.0). MicroT-CDS is a database of over 11 million predicted in silico interactions between microRNAs and the 3′-UTR of their target mRNAs across a range of different species.22 miRPath uses a statistical algorithm which combines the Fisher's exact test, EASE Scores and false discovery rates to assign miRNA targets to biological pathways provided by the Kyoto Encyclopaedia of Genes and Genomes.23, 24 All identified pathways are arranged according to enrichment statistical scores (P-values) in addition to the number and names of miRNA target genes implicated in each Kyoto Encyclopaedia of Genes and Genomes pathway. For multiple microRNAs, the miRPath server calculates significance levels between each microRNA and every pathway. A merged P-value is then calculated and signifies if a particular pathway is targeted by at least one or more of the selected microRNAs. Pathways with a P-value <0.05 were considered. Gene ontology analysis of identified miRNA targets was performed with a focus on molecular function25 and interactions between targets using string analysis were examined.26
Statistical analysis
Relationships between the various categorical variables were evaluated using the χ2-test. Age and HDRS scores (baseline and post treatment) between groups were analyzed using Student's t-test for parametric data and Mann–Whitney tests for the non-parametric data.
For microarray data, statistical analyses were performed using a two-way analysis of variance. P-values were corrected for multiple testing by the Benjamini and Hochberg adjustment method. Genes found to be significant by the one-way analysis of variance test have been subjected to the Tukey's ‘Honest Significant Difference' test to determine which groups contribute most to the significant difference. All calculations have been done in the software R/bioconductor using the limma package. To enable quick visual identification of those microRNAs that display large-magnitude changes that are also statistically significant, the expression data have been plotted in a Volcano plot (−log10(P-value) versus log2(fold-change)). We only quantified with qRT-PCR the expression of microRNAs that were were found to differentially expressed between groups (P<0.05).
PCR data were analyzed using the 2−ΔCt method and outliers were defined by Grubb's method as previously described.27 The microRNA miR-25, which was stably expressed in all samples, was used as the endogenous control. Differences in microRNA expression between groups were determined using Student's t-test for parametric data and Mann–Whitney's test for the non-parametric data. Effects of treatments were determined using Student's t-test for parametric data and Mann–Whitney tests for the non-parametric data. To explore the associations between baseline microRNA expression and other clinical factors (for example, age and gender), Pearson's test was used for normally distributed data and Spearman's test was used for the non-normally distributed data. All statistics were calculated using SPSS-18 (IBM, Chicago, IL, USA). A P-value of 0.05 was selected as the threshold of statistical significance.
Results
Clinical effects of ketamine and ECT
Both ECT and KET treatments reduced the HDRS in the majority of patients (P<0.001; Table 1). There was no difference in post-treatment HDRS scores between the groups of TRD patients. There were no correlations between age, gender or medication profile and the ECT/KET-induced reduction in HDRS scores. Overall, there were more responders than non-responders. Five out of 11 patients demonstrated a sustained reduction 1 week after the first ketamine session.
Microarray analysis
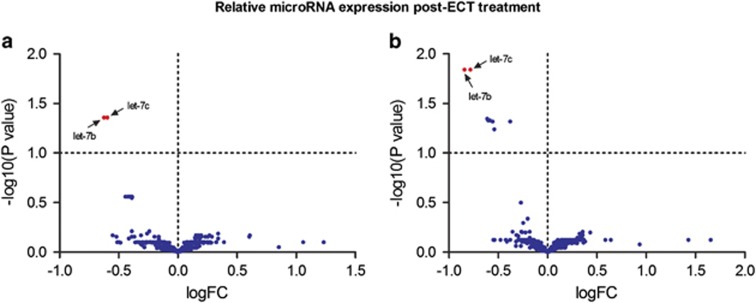
A total of 502 out of 2087 possible microRNAs were detected in the blood samples. After controlling for false discovery rates with the Benjamini–Hochberg correction (see Materials and Methods), the only significant finding was a reduction in the expression of let-7b and let-7c in all TRD patients who received ECT (responders and non-responders; Figure 2, Supplementary Table 1). No microRNAs identified at baseline in TRD patients were predictive of response to ECT or KET and there were no other microRNAs that were affected by treatments.
Figure 2.
Microarray volcano plot showing differential expression of let-7b and let-7c (red) in (a) responders and (b) non-responders to ECT compared with healthy controls. ECT, electroconvulsive therapy.
qRT-PCR analysis
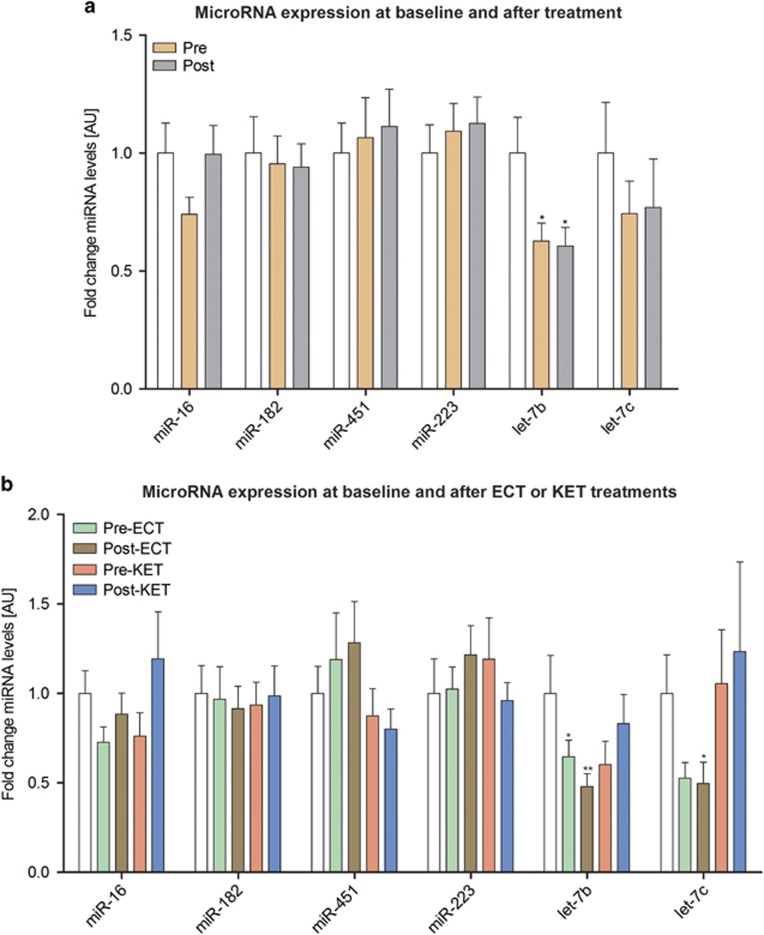
On the basis of microarray analysis, we first quantified baseline differences in the expression of let-7b and let-7c in all TRD patients compared with controls. The expression of let-7b was less in TRD patients compared with that in controls; there was no difference in let-7c expression between patients and controls (Figure 3a, Supplementary Table 2).
Figure 3.
qRT-PCR data of microRNA expression (mean±s.e.m.) in controls (white column) and in (a) all patients at baseline (Pre) and after treatment (Post) and (b) according to type of treatment received (ECT or KET). *P<0.05, **P<0.01 compared with controls. ECT, electroconvulsive therapy; KET, ketamine; qRT-PCR, quantitative real-time PCR.
When patient groups were split according to the treatments they subsequently received, baseline expression let-7b was lower in patients who progressed to receive ECT treatment (Figure 3b, Supplementary Table 3). In patients who received KET treatment, there was a trend towards a lower baseline expression of let-7b. There was no difference in baseline let-7b expression between these two patient groups. Given that the age of patients in the ECT group were older than controls and patients in the KET group, we assessed whether there was a correlation between age and baseline expression of let-7b, but this was found to be not statistically significant. Patients in the ECT group were also receiving more medication, but there was no significant correlation between medication profile and let-7b expression. Compared with controls the baseline expression of let-7c tended to be lower in patients in the ECT group. In addition to let-7b and let-7c we also quantified the expression of miR-16, miR-182, miR-451 and miR-223, all of which have been implicated in depression7 and found that their baseline expression was similar to those of controls (Figures 3a and b).
Overall, there were no microRNAs affected by KET or ECT treatments. However, there was a trend toward higher post-treatment expression of let-7b in patients who had received KET than ECT. This increase in let-7b expression was subsumed into the post-ECT group, which could explain the group significance seen in Figure 3a.
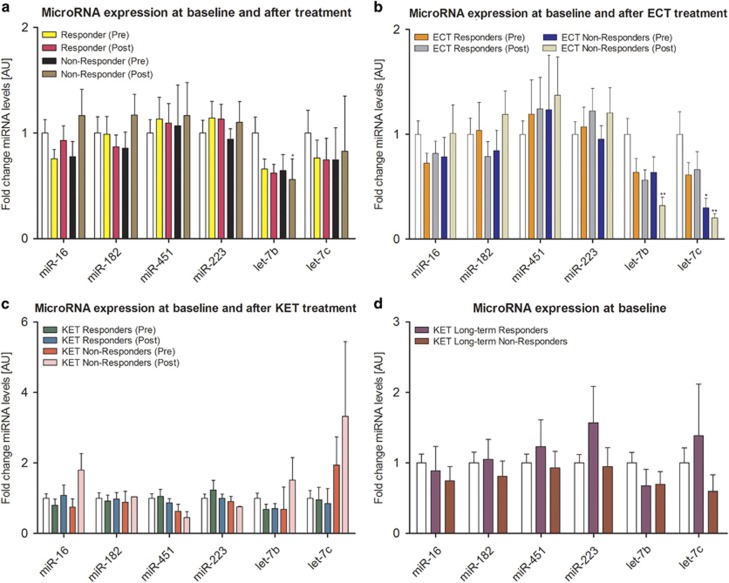
We next determined whether baseline microRNA expression could predict response to treatment in TRD patients. Relative to controls, there was a trend towards lower expression of let-7b in responders but overall, the baseline expression of both let-7b and the other microRNAs did not differ significantly between responders and non-responders. In both responders and non-responders, there was no effect of treatment, but post-treatment expression of let-7b was reduced compared with controls (Figure 4a, Supplementary Table 4).
Figure 4.
qRT-PCR data of microRNA expression (mean±s.e.m.) in controls (white column) and in all (a) patients, (b) ECT patients and (c) KET patients at baseline and after treatment according to response. (d) Baseline microRNA expression in long-term responders and non-responders to KET treatment. *P<0.05, **P<0.01 compared with controls. ECT, electroconvulsive therapy; KET, ketamine; qRT-PCR, quantitative real-time PCR.
When we stratified the analysis according to the types of treatment, in patients who received ECT, baseline expression of let-7c was low in non-responders relative to controls (Figure 4b, Supplementary Table 5). However, the overall baseline expression of all microRNAs was similar in responders and non-responders. In responders, ECT treatment had no effect on microRNA expression. In non-responders, post-treatment expression of let-7b and let-7c was less compared with that of controls, but pre- and post-treatment levels were similar.
For patients who received KET treatment, there were no differences in the baseline expression of all microRNAs between responders and non-responders (Figure 4c, Supplementary Table 6). Similarly, there was no effect of treatment on microRNA expression in responders or in non-responders. Last, we examined whether baseline microRNA expression could predict response to KET at 1 week and found there were no differences between those who exhibited a sustained reponse and non-responders at this later time point (Figure 4d, Supplementary Table 7).
Taken together, these results would suggest that microRNAs let-7b and let-7c are potential biomarkers of TRD. Baseline expression of let-7b, let-7c or any other microRNAs in TRD did not predict treatment response. Interestingly, both the direction and the magnitude of the changes in the microarray were significantly different from qRT-PCR analyses. In particular, whereas microarray results showed a significant reduction in let-7b and let-7c expression following ECT treatment, qRT-PCR analysis did not detect this difference. We have addressed this discrepancy in detail in our discussion.
Bioinformatics analysis of let-7b and let-7c gene targets and pathways
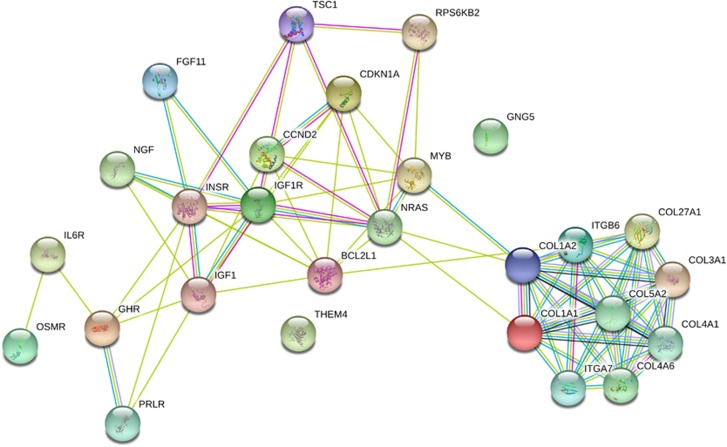
Bioinformatics analysis revealed that there are 1343 genes targeted by let-7b (682) and let-7c (661). Pathway union analysis revealed a significant over-representation of 27 genes (Table 2) in the PI3k-Akt signaling pathway. Gene ontology analysis revealed that the 12 out of the 27 genes (Table 3) were involved in receptor activity and protein binding. String analysis showed that genes involved in structural molecular activity and composition of the extracellular matrix, such as collagen and integrin, clustered together in terms of their predicted interactions. A separate and larger cluster included genes for receptors implicated in other molecular functions including those for hormones and growth factors such as the growth hormone, insulin and insulin-like growth factor (Figure 5).
Table 2. Genes in the PI3K-Akt pathway under the regulation of let-7b and let-7c.
| Gene ID | Description |
|---|---|
| COL1A1 | Collagen, type I, alpha 1 |
| GHR | Growth hormone receptor |
| PRLR | Prolactin receptor |
| ITGB6 | Integrin, beta 6 |
| MYB | V-myb avian myeloblastosis viral oncogene homolog |
| CCND2 | Cyclin D2 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| NGF | Nerve growth factor |
| ITGA7 | Integrin, alpha 7 |
| IGF1R | Insulin-like growth factor 1 receptor |
| OSMR | Oncostatin M receptor |
| THEM4 | Thioesterase superfamily member 4 |
| IL6R | Interleukin 6 receptor |
| COL1A2 | Collagen, type I, alpha 2 |
| TSC1 | Tuberous sclerosis 1 |
| COL3A1 | Collagen, type III, alpha 1 |
| INSR | Insulin receptor |
| BCL2L1 | BCL2-like 1 |
| GNG5 | Guanine nucleotide binding protein (G protein), gamma 5 |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 |
| COL4A1 | Collagen, type IV, alpha 1 |
| COL27A1 | Collagen, type XXVII, alpha 1 |
| COL4A6 | Collagen, type IV, alpha 6 |
| COL5A2 | Collagen, type V, alpha 2 |
| NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog |
| FGF11 | Fibroblast growth factor 11 |
| IGF1 | Insulin-like growth factor 1 |
Table 3. Gene ontology analysis of molecular function of identified targets of let-7b and let-7c.
| Molecular function | Frequency | P-value |
|---|---|---|
| Receptor activity (COL5A2, OSMR, IL6R, PRLR, COL4A6, COL4A1, COL3A1,COL27A1, ITGB6, COL1A2, COL1A1, GHR) | 12/1636 | 5.30e−5 |
| Protein binding (FGF11, BCL2L1, NRAS, NGF, OSMR, IL6R, GNG5, PRLR, CCND2, CDKN1A, IGF1, GHR) | 12/2819 | 1.53e−2 |
| Extracellular matrix structural constituent (COL5A2, COL4A6, COL4A1, COL3A1, COL27A1, COL1A2, COL1A1) | 7/76 | 1.22e−9 |
| Transmembrane transporter activity (COL5A2, COL4A6, COL4A1, COL3A1, COL27A1, COL1A2, COL1A1) | 7/1010 | 4.02e−2 |
| Receptor binding (FGF11, NGF, OSMR, IL6R, PRLR, IGF1, GHR) | 7/980 | 3.34e2 |
| Structural molecular activity (COL5A2, COL4A6, COL4A1, COL3A1, COL27A1, COL1A2, COL1A1) | 7/1034 | 4.64e−2 |
| Cytokine activity (NGF, OSMR, IL6R, PRLR, GHR) | 5/230 | 1.84e−3 |
Frequency is based on number of genes identified relative to total number of reference genes in humans for each molecular function.
Figure 5.
String analysis showing interactions between identified genes under the regulation of let-7b and let-7c. Darker lines indicate stronger network connections between genes.
Discussion
In this study, we sought to identify microRNAs that were differentially expressed in patients with major depression and TRD compared with controls and which were predictive of response to either ECT or KET treatments. We observed that baseline expression of let-7b was significantly lower in all patients compared with healthy controls. When analysis was split according to the type of treatments received, let-7b expression was comparable between these two groups. This was significant in the ECT group and displayed a strong trend toward significance in the KET group. The baseline expression of let-7c was also lower in patients who subsequently received ECT compared with controls and was unaffected by this treatment. These findings suggest that let-7b and let-7c are candidate biomarkers of major depression. We also speculate as to whether they may also be biomarkers for TRD. However, our conclusions come with several limitations discussed below.
The let-7 family of microRNAs were the first to be discovered in humans with roles in neurogenesis and synapse formation.28, 29 One previous study has shown dysregulation in another member of the let-7 family, let-7p-5p, in patients with schizophrenia,30 but to our knowledge, there have been no investigations into whether this family of microRNAs is dysregulated in depression. However, they have been shown to be responsive to a variety of mood stabilizers and antidepressants.31 There are ~1343 predicted experimental targets of let-7b and let-7c, but pathway analysis revealed that there is a significant overexpression of 27 genes in the intracellular PI3K-Akt signaling pathway. Moreover, one of the downstream targets of activated Akt is the mTOR signaling pathway, which has been previously implicated in the pathophysiology of depression32, 33 and the rapid antidepressant effects of KET.34
Gene ontology analysis revealed that the 12 out of the 27 genes were involved in binding of growth factors such as insulin and growth hormone (GH). Evidence suggests that the insulin-like growth factor (IGF1) can promote the signaling effects of the brain-derived neurotrophic factor and also act synergistically with brain-derived neurotrophic factor to induce antidepressant-like effects.35 Both IGF1 and insulin can bind to IGF1 and insulin receptors (IGF1R and INSR) to activate downstream signaling cascades including the PI3k-Akt signaling pathway.36, 37 Dysregulation of GH release from the anterior pituitary is associated with atypical depression.38 In depressed patients, secretion is reportedly abnormal, but this appears to depend on the age of the patients; studies show significantly reduced secretion of GH in depressed children and adolescents,39 but no change in adults.40, 41 The activation of the PI3K-Akt signaling pathway by GH is indirect via JAK2 tyrosine kinase phosphorylation which in turn phosphorylates insulin-like receptor substrates.42 String analysis showed that genes for several collagen factors clustered together. Collagen is a structural protein found in the basement membranes of blood vessels and given that the current study utilized blood samples for microRNA analysis, this finding is expected.
The ability to predict treatment response based on expression of biomarkers confers clinical advantage in terms of choosing the right therapeutic strategy for the right patient. We explored this possibility in our study, but identified no microRNAs at baseline that predicted response to ECT or KET treatments. To our knowledge, there has been only one study that studied microRNAs as biomarkers to predict antidepressant response. Lopez et al.14 showed that baseline expression of miR-1202 was lower in patients with depression who subsequently responded to an 8-week regimen of the selective serotonin reuptake inhibitor, citalopram. However, patients were drug-naive at study commencement. They were classified as responders and non-responders based on changes to HDRS scores following treatment, but it is unclear what their cutoff criteria were for response and non-response. Previous studies have used cutoffs ranging from 30 to 60% improvement in HDRS scores43, 44 to distinguish responders from non-responders. For our study, our classification was based on at least a 50% reduction in HDRS scores.21
Our results with miR-16 are consistent with recent work, which reported that expression of miR-16 in the blood was not different between controls and patients with major depressive disorder.11 The fact that peripheral miR-16 expression is unchanged relative to controls in major depression and TRD argues against its involvement in the pathophysiology. However, its expression was found to be reduced in the cerebrospinal fluid of patients with major depression.11 Overlaps in the transcriptome profiles between the periphery and the brain is reportedly high (81.9%),45 but this does not imply that alterations in the profile in one compartment will be mirrored in the other, either in the context of health or during periods of illness. Although its role as a diagnostic biomarker remains to be validated, previous work has shown that miR-16 is a molecular mediator of the antidepressant effect, particularly for selective serotonin reuptake inhibitors.46 Our results suggest that peripheral miR-16 levels do not reflect a potential role in the therapeutic mechanisms of ECT or KET treatment.
In contrast to previous studies, we observed no differences in expression of miR-182 and miR-223 between patients and controls.12, 13 Potential causes for the discrepancy include medication status at study commencement (drug-naive versus medicated), sampling source (for example, serum and plasma) and the fact that patients in our study were approximately 10–20 years older. Another important factor which has been discussed in other disease contexts, but not in the depression-microRNA literature is ethnicity.47, 48, 49 This study was conducted in Ireland, whereas the previous studies were conducted in China and Turkey.
We have previously shown that the maternal separation paradigm in rats can induce significant down-regulation of miR-451 expression in the hippocampus,16 a brain region that is consistently implicated in depression.50, 51 In contrast, a recent study has shown increased peripheral expression of miR-451 in patients with depression.13 We found no differential expression of miR-451 between patients and controls, but factors already described (for example, age and sampling source) could explain the discrepancy in clinical findings. The lack of consistency between clinical and preclinical studies suggests poor correlation between central and peripheral readouts of miR-451 expression. The expression of miR-451 was not altered following treatment with ketamine, a result which was consistent with our preclinical study, suggesting that it is unlikely to be involved in the therapeutic mechanism of this novel antidepressant.
The results of this study should be interpreted in the light of its limitations. The patients were on medication at the time of study commencement and throughout the study itself. This was owing to ethical considerations clinically but could have confounded the results. Furthermore, our patient group was heterogeneous with melancholic and non-melancholic patients included. Moreover, patients in the ECT group were older than all other study participants and were thus more likely to be on multiple medications for a variety of health conditions. They were also institutionalized. Also, extraction of microRNAs for patients in the KET group was only 24 h after the first infusion whereas for the ECT group it was several days after treatment. These differences could explain the divergence in baseline and post-treatment microRNA expression levels for let-7b and let-7c. Moreover, we did not include patients who were responsive to conventional antidepressants in our study to support our hypothesis that let-7b and let-7c were also potential biomarkers for TRD. In addition, some factors inherent to the ECT procedure may also hinder a clear interpretation of the studies, that is, electrode placement, stimulus intensity and frequency, and seizure duration (which all may have different effects on microRNAs), as well as the use of general anaesthesia. We also did not assess patient insulin, IGF or GH levels, which could have supported the findings of the bioinformatics analyses. However, our approach of using only an in silico methodology to draw conclusions about the involvement of microRNAs in pathological processes has precedent.13, 52, 53 Given the role of stress as a major risk factor for the development of psychopathologies, we did not determine whether the patients had early-life or current stress experiences. To our knowledge, there have been no microRNA studies in clinical depression that have taken this variable into account. Last, we stress that these findings are preliminary and needs replication.
It is also important to note that the PCR results were in contrast to what was observed from microarray analysis. Fidelity between the two techniques is an ongoing issue due to a range of factors including the sensitivity of microarray probes to differentiate between mature and precursor microRNA sequences.54 It is also possible that pathology and medication could have had effects on microRNAs, which could only be detected by more accurate PCR that is considered a ‘gold-standard'.55
Although it is clear that microRNAs function as a mechanism for post-transcriptional regulation, it has not been conclusively proven whether, under conditions of homeostasis or pathology, their presence in body fluids is simply a by-product of cell degradation or whether are they actively secreted into the body fluids to mediate intercellular gene regulation. Nevertheless, the correlation between circulating microRNAs and peripheral tissue microRNAs suggests that in human fluids they might serve as biomarkers for various diseases.56 However, in the context of depression few studies have supported such correlations between circulating and central readouts of microRNA expression.14, 57 Indeed, of the microRNAs that we analyzed in this study, a correlation between changes in peripheral blood and in brain tissue remains to be established. Thus, in the case of let-7b and let-7c future clinical studies could focus on post-mortem samples from patients who had suffered from major depression to determine whether this microRNA is a valid diagnostic biomarker. In vivo and in vitro models could also be used to further investigate the functional expression of these microRNAs.
In conclusion, we provide preliminary evidence that let-7b and let-7c are candidate diagnostic biomarkers of major depression. Future studies utilizing larger patient samples with more detailed medical histories and the extraction of both peripheral blood samples and cerebrospinal fluid samples will allow us to validate let-7b, let-7c and other potential microRNAs that could be used for diagnosis, predict response to various therapeutic strategies and provide novel insights into the neuromolecular pathophysiology of depression.
Acknowledgments
This research was funded by the Health Research Board (HRB: HRA_POR/2012/32) and conducted in the APC Microbiome Institute, which is funded by Science Foundation Ireland (SFI; Grant nos. SFI/12/RC/2273, 02/CE/B124 and 07/CE/B1368). JFC is also funded by the European Community's Seventh Framework Programme (Grant no. FP7/2007–2013, Grant agreement 201714). GC is supported by a NARSAD Young Investigator Grant from the Brain and Behaviour Research Foundation (Grant no. 20771). We thank all the patients and volunteers that took part in the study.
Author contributions
GC, GS, DMM, JFC and TGD designed and managed the study. MN, JD, AW, FI and LS conducted the clinical research. AG and KAS performed microRNA analyses. AG and GM analyzed the data. AG and MN wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Lancet T. Depression and the global economic crisis: is there hope? Lancet 2012; 380: 1203. [DOI] [PubMed] [Google Scholar]
- Schmidt HD, Shelton RC, Duman RS. Functional biomarkers of depression: diagnosis, treatment, and pathophysiology. Neuropsychopharmacology 2011; 36: 2375–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR. Beyond Efficacy: The STAR*D Trial. Am J Psychiatry 2006; 163: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden D, Rush AJ, Trivedi M, Fava M, Wisniewski S. The STAR*D project results: A comprehensive review of findings. Curr Psychiatry Rep 2007; 9: 449–459. [DOI] [PubMed] [Google Scholar]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69: 89–95. [DOI] [PubMed] [Google Scholar]
- Geaghan M, Cairns MJ. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol Psychiatry 2014; 78: 231–239. [DOI] [PubMed] [Google Scholar]
- Gururajan A, Clarke G, Dinan TG, Cryan JF. Molecular biomarkers of depression. Neurosci Biobehav Rev 2016; 64: 101–133. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng 2010; 12: 1–27. [DOI] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010; 56: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M-F, Dong J-Z, Wang Y-W, He J, Ju X, Zhang L et al. CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J Affect Disord 2015; 178: 25–31. [DOI] [PubMed] [Google Scholar]
- Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang YX et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS ONE 2013; 8: e63648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camkurt MA, Acar Ş, Coşkun S, Güneş M, Güneş S, Yılmaz MF et al. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res 2015; 69: 67–71. [DOI] [PubMed] [Google Scholar]
- Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 2014; 20: 764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S, Baudry A, Launay J-M, Kellermann O. MicroRNAs and depression. Neurobiol Dis 2012; 46: 272–278. [DOI] [PubMed] [Google Scholar]
- O'Connor RM, Grenham S, Dinan TG, Cryan JF. microRNAs as novel antidepressant targets: converging effects of ketamine and electroconvulsive shock therapy in the rat hippocampus. Int J Neuropsychopharmacol 2013; 16: 1885–1892. [DOI] [PubMed] [Google Scholar]
- Yang X, Yang Q, Wang X, Luo C, Wan Y, Li J et al. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. Neuromolecular Med 2014; 16: 594–605. [DOI] [PubMed] [Google Scholar]
- Naughton M, Clarke G, O'Leary OF, Cryan JF, Dinan TG. A review of ketamine in affective disorders: current evidence of clinical efficacy, limitations of use and pre-clinical evidence on proposed mechanisms of action. J Affect Disord 2014; 156: 24–35. [DOI] [PubMed] [Google Scholar]
- Wijkstra J, Nolen WA, Algra A, van Vliet IM, Kahn RS. Relapse prevention in major depressive disorder after successful ECT: a literature review and a naturalistic case series. Acta Psychiatr Scand 2000; 102: 454–460. [DOI] [PubMed] [Google Scholar]
- DeWilde KE, Levitch CF, Murrough JW, Mathew SJ, Iosifescu DV. The promise of ketamine for treatment-resistant depression: current evidence and future directions. Ann N Y Acad Sci 2015; 1345: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AP, Naughton M, Dowling J, Walsh A, Ismail F, Shorten G et al. Serum BDNF as a peripheral biomarker of treatment-resistant depression and the rapid antidepressant response: A comparison of ketamine and ECT. J Affect Disord 2015; 186: 306–311. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013; 41: W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015; 43: W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput Biol 2009; 5: e1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ et al. Microbes & neurodevelopment – Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun 2015; 50: 209–220. [DOI] [PubMed] [Google Scholar]
- Rehfeld F, Rohde AM, Nguyen DT, Wulczyn FG. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res 2015; 359: 145–160. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci 2009; 10: 842–849. [DOI] [PubMed] [Google Scholar]
- Rizos E, Siafakas N, Katsantoni E, Skourti E, Salpeas V, Rizos I et al. Let-7, Mir-98 and Mir-181 as biomarkers for cancer and schizophrenia. PLoS ONE 2015; 10: e0123522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KA, Hoban AE, Clarke G, Moloney GM, Dinan TG, Cryan JF. Thinking small: towards microRNA-based therapeutics for anxiety disorders. Expert Opin Invest Drugs 2015; 24: 529–542. [DOI] [PubMed] [Google Scholar]
- Jernigan CS, Goswami DB, Austin MC, Iyo AH, Chandran A, Stockmeier CA et al. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry 2011; 35: 1774–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran A, Iyo AH, Jernigan CS, Legutko B, Austin MC, Karolewicz B. Reduced phosphorylation of the mTOR signaling pathway components in the amygdala of rats exposed to chronic stress. Prog Neuropsychopharmacol Biol Psychiatry 2013; 40: 240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 2010; 329: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paslakis G, Blum WF, Deuschle M. Intranasal insulin-like growth factor I (IGF-I) as a plausible future treatment of depression. Med Hypotheses 2012; 79: 222–225. [DOI] [PubMed] [Google Scholar]
- Plum L, Schubert M, Brüning JC. The role of insulin receptor signaling in the brain. Trends Endocrinol Metab 2005; 16: 59–65. [DOI] [PubMed] [Google Scholar]
- Bondy CA, Cheng CM. Signaling by insulin-like growth factor 1 in brain. Eur J Pharmacol 2004; 490: 25–31. [DOI] [PubMed] [Google Scholar]
- Mahajan T, Crown A, Checkley S, Farmer A, Lightman S. Atypical depression in growth hormone deficient adults, and the beneficial effects of growth hormone treatment on depression and quality of life. Eur J Endocrinol 2004; 151: 325–332. [DOI] [PubMed] [Google Scholar]
- Birmaher B, Dahl RE, Williamson DE, Perel JM, Brent DA, Axelson DA et al. Growth hormone secretion in children and adolescents at high risk for major depressive disorder. Arch Gen Psychiatry 2000; 57: 867–872. [DOI] [PubMed] [Google Scholar]
- Koenigsberg HW, Teicher MH, Mitropoulou V, Navalta C, New AS, Trestman R et al. 24-h Monitoring of plasma norepinephrine, MHPG, cortisol, growth hormone and prolactin in depression. J Psychiatr Res 2004; 38: 503–511. [DOI] [PubMed] [Google Scholar]
- Franz B, Buysse DJ, Cherry CR, Gray NS, Grochocinski VJ, Frank E et al. Insulin-like growth factor 1 and growth hormone binding protein in depression: a preliminary communication. J Psychiatr Res 1999; 33: 121–127. [DOI] [PubMed] [Google Scholar]
- Herrington J, Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab 2001; 12: 252–257. [DOI] [PubMed] [Google Scholar]
- Mulder RT, Joyce PR, Frampton C. Relationships among measures of treatment outcome in depressed patients. J Affect Disord 2003; 76: 127–135. [DOI] [PubMed] [Google Scholar]
- Clark CP, Golshan S. Polysomnography and criteria for the antidepressant response to sleep deprivation. J Affect Disord 2007; 101: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew C-C, Ma J, Tang H-C, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med 2006; 147: 126–132. [DOI] [PubMed] [Google Scholar]
- Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 2010; 329: 1537–1541. [DOI] [PubMed] [Google Scholar]
- Wang X, Sundquist J, Zöller B, Memon AA, Palmér K, Sundquist K et al. Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PLoS ONE 2014; 9: e86792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Li S, Li J, Yin L, Zhou T, Zhang C et al. Ethnic differences in microRNA-375 expression level and DNA methylation status in type 2 diabetes of Han and Kazak populations. J Diabetes Res 2014; 2014: 761938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings-Goss RA, Campbell MC, Tishkoff SA. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics 2014; 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malykhin NV, Coupland NJ. Hippocampal neuroplasticity in major depressive disorder. Neuroscience 2015; 309: 200–213. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature 2008; 455: 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacology 2013; 23: 602–611. [DOI] [PubMed] [Google Scholar]
- Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J et al. Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS ONE 2015; 10: e0121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand S, Weber J, Lemoine F, Consales F, Paillusson A, Dupasquier M et al. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques 2010; 48: 219–222. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 2009; 10: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 2014; 83: 344–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.