Abstract
Locked nucleic acid (LNA) oligonucleotides have been successfully used to efficiently inhibit endogenous small noncoding RNAs in vitro and in vivo. We previously demonstrated that the direct miR-221 inhibition by the novel 13-mer LNA-i-miR-221 induces significant antimyeloma activity and upregulates canonical miR-221 targets in vitro and in vivo. To evaluate the LNA-i-miR-221 pharmacokinetics and pharmacodynamics, novel assays for oligonucleotides quantification in NOD.SCID mice and Cynomolgus monkeys (Macaca fascicularis) plasma, urine and tissues were developed. To this aim, a liquid chromatography/mass spectrometry method, after solid-phase extraction, was used for the detection of LNA-i-miR-221 in plasma and urine, while a specific in situ hybridization assay for tissue uptake analysis was designed. Our analysis revealed short half-life, optimal tissue biovailability and minimal urine excretion of LNA-i-miR-221 in mice and monkeys. Up to 3 weeks, LNA-i-miR-221 was still detectable in mice vital organs and in xenografted tumors, together with p27 target upregulation. Importantly, no toxicity in the pilot monkey study was observed. Overall, our findings indicate the suitability of LNA-i-miR-221 for clinical use and we provide here pilot data for safety analysis and further development of LNA-miRNA-based therapeutics for human cancer.
Keywords: Cynomolgus monkeys, LNA inhibitor, microRNA, miRNA therapeutics, multiple myeloma
Introduction
In the last decade noncoding RNAs have emerged as a novel landscape of regulatory networks and have been extensively studied in human cancer.1,2 Among noncoding RNAs, microRNAs (miRNAs) have been described as deeply dysregulated in cancer3,4,5,6 and have raised special interest for therapy.7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22 In fact, synthetic mimics of tumor suppressor miRNAs have been investigated for replacement strategies in several preclinical cancer models23,24,25,26,27,28,29 and the first cancer-targeted miRNA drug, MRX34, a liposome-based miR-34a mimic is currently under phase 1 investigation in cancer patients. By a different approach, the use of anti-oncopromoting miRNAs has been pursued for clinical translation, firstly by antisense oligonucleotides (ASOs) or short interference RNA30,31 and then, by a new generation of ASOs or Gapmers, which offer advantages in terms of pharmacokinetics (PK) and pharmacodynamics. It became evident that the real challenges for the therapeutic use of antisense molecules are their biological stability, target affinity, biodistribution as well as the ability to activate RNase H function, with reduced off-target toxicity. Among the modified oligonucleotides, the so called “second generation” phosphorothioate (PS) locked nucleic acid (LNA)-based oligonucleotides have demonstrated high affinity for the specific complementary strand (i.e., mature miRNA), with obvious advantages in term of target specificity.32,33 LNA-oligonucleotides have been successfully used in in vitro and in vivo studies and demonstrated great potential as inhibitors of small RNA targets.12,34,35,36 The high target affinity and specificity reduces nonhybridization-based interactions and related toxicities, making them suitable for systemic delivery.33 Moreover, LNA-oligonucleotides are stable for years under refrigeration, are highly soluble in water and can be diluted and injected in vivo in saline solution.12,34,35,36 However, at the present, poor information is available on the PK of LNA-oligonucleotides after systemic administration, either by intravenous (i.v.) infusion or intraperitoneal (i.p.) injection, due to the complex bioanalyticals that are needed for PK evaluation. The bioavailability of these compounds relies in their systemic distribution and retention by tissues: such processes involve surface protein interactions and endocytosis, thus finally lead to cell internalization and excretion.33
In a previous report, we demonstrated the therapeutic activity of a novel 13-mer LNA-miR-221 inhibitor (LNA-i-miR-221) in preclinical models of multiple myeloma (MM).12,20 Here we studied the LNA-i-miR-221 bioavailability profile following intravenous (i.v.) administration at antitumor dose in NOD.SCID mice and nonhuman primates. For the lack of standardized methodology, we developed a novel approach for the detection of LNA-i-miR-221 animal biofluids as well as in mouse tissues and tumor xenografts. Moreover, we studied the biological activity of LNA-i-miR-221 in animals and we searched for tissue and behavioral toxicity. Overall, our results provide novel information relevant for the design of oligonucleotide-based cancer therapeutics in the perspective of first-in-human regulatory studies.
Results
LNA-i-miR-221 is well tolerated in mouse
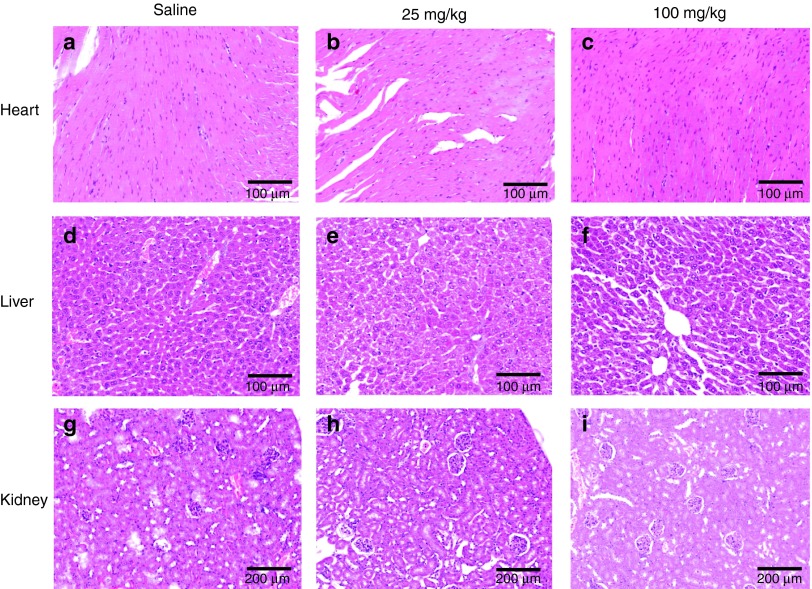
We first investigated the effect induced by LNA-i-miR-221 in mouse behavior or by pathologic analysis of vital organs including liver, kidney, and heart. Animals underwent one course of treatment, which includes injections on days 1, 4, 8, 15, and 22 with two different doses. We selected 25 mg/kg as investigation dose based on our previous findings.12 Thus we derived a fourfold higher dose of 100 mg/kg specifically selected to investigate tissue toxicity in mice. The animals did not show behavioral changes. One week following the completion of one treatment course, animal tissues were retrieved and hematoxylin and eosin stained. As shown in Figure 1, no morphological changes or abnormal findings were detected in retrieved tissues, confirming the absence of acute toxicity or inflammation. No toxicity even at highest dose was observed, indicating a good safety profile of LNA-i-miR-221 in mice.
Figure 1.
Toxicity evaluation of Locked Nucleic Acid (LNA) LNA-i-miR-221 in mouse organs. Hematoxylin and eosin (H&E) staining of heart, liver, and kidney retrieved from mouse 1 week after one cycle (day 1, 4, 8, 15, and 22) of treatment. a, b, c, show representative areas of heart retrieved after saline (a), 25 mg/kg LNA-i-miR-221 (b) or 100 mg/kg course of treatments. d, e, f show representative areas of liver, while g, h, i kidney areas, after saline (d,g), 25 mg/kg (e,h) or 100 mg/kg (f,i) LNA-i-miR-221 treatment. Frames a, b, c, d, e, f are 100× while g, h, i are 200× magnification.
LNA-i-miR-221 is detectable in xenografted tumors and mouse organs by in situ hybridization
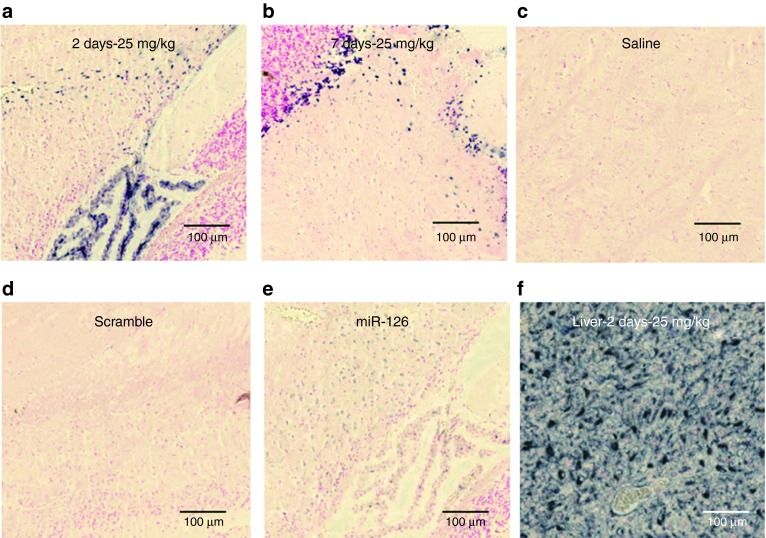
To evaluate the tissue bioavailability of LNA-i-miR-221, we assessed the localization of LNA-i-miR-221 by in situ hybridization (ISH) using a specific LNA antisense probe to LNA-i-miR-221 (miR-221i) in formalin-fixed paraffin-embedded tissue samples retrieved from single injection- (25 mg/kg of LNA-i-miR-221) treated animals after 2 or 7 days. Intense miR-221i ISH signal was detected in all organs analyzed including liver, kidney (Figure 2), heart, bone marrow, brain, and tumor samples retrieved from all treated mice (Figure 3, line I and II), whereas signal was not detectable from untreated animals (Figure 3 line III). ISH from LNA-i-miR-221 treated or untreated animals was validated by parallel staining with an LNA probe toward endogenous microRNA-126 (miR-126) (see Supplementary Figure S1). miR-126 was indeed detected in all samples and with staining intensity in endothelial cells independent from treatment, indicating that all tissue specimens had been prepared uniformly and therefore, the ISH signal detected with the miR-221i probe indeed reveals the genuine LNA-i-miR-221. A reduced miR-126 ISH signal was detected in tumor tissues probably due to a lower miR-126 expression by human MM cells. Intense LNA-i-miR-221 ISH signal was detected in liver samples, particularly in spindle-shaped cells, presumable liver macrophages/Kupfer cells and/or endothelial cells, but also epithelial liver cells had positive ISH signal (Figure 3a). In kidney tissue sections, LNA-i-miR-221 ISH signal was detected in tubules (Figure 3b). In heart samples, LNA-i-miR-221 ISH signal was detected in spindle-shaped cells presumably capillary endothelial cells (Figure 3c). In bone marrow sections, the ISH signal was detected in a subset of spherical cells (Figure 3d). In all organs retrieved from LNA-i-miR-221 treated animals, a LNA-i-miR-221 ISH specific signal was detected. An intense signal was detected also in xenograft MM tumor samples, demonstrating adequate tumor uptake (Figure 3e). Overall these findings demonstrated the detection of miR-221i/LNA-i-miR-221 heteroduplex within cells, with prevalent perinuclear localization in most tissues. No obvious differences in ISH signal were seen in tissues from animals sacrificed 2 or 7 days after a single 25 mg/kg dose of LNA-i-miR-221. Our findings indicate that ISH is a suitable technique for detection of LNA-i-miR-221 in normal and tumor tissues.
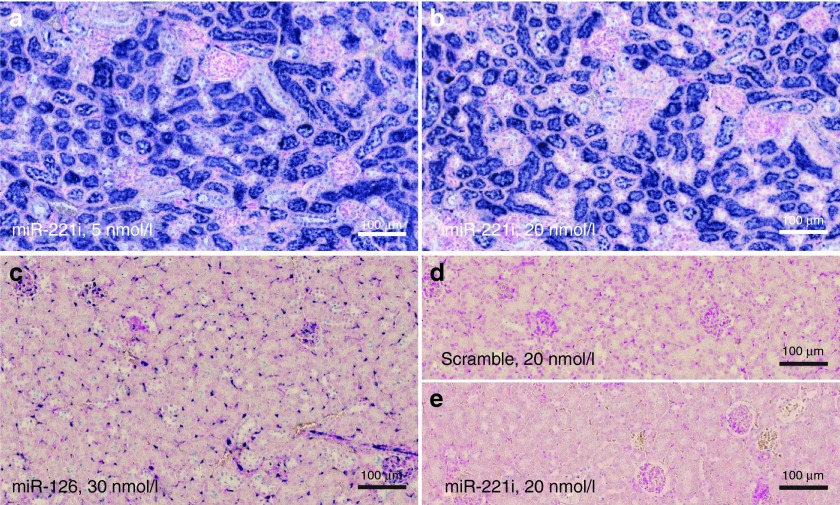
Figure 2.
In situ hybridization (ISH) analysis of LNA-i-miR-221. Serial sections from formalin-fixed paraffin-embedded (FFPE) tissue samples of NOD.SCID mouse kidney after a single i.p. (intraperitoneal) injection of 25 mg/kg LNA-i-miR-221 (a–d) or saline (e), processed by ISH with Locked Nucleic Acid (LNA) probes for LNA-i-miR-221 (a, b, e), miR-126 (c), and miR-221 scramble (d). Intense LNA-i-miR-221 ISH signal is detectable in tubules at both low (5 nmol/l) and high (20 nmol/l) probe concentrations (a, b), whereas the probe results in no ISH signal in saline-treated kidney. The positive control probe, miR-126, stains endothelial cells including capillaries in the glomeruli (c), and only background staining is seen by the use of the scramble negative control probe (d).
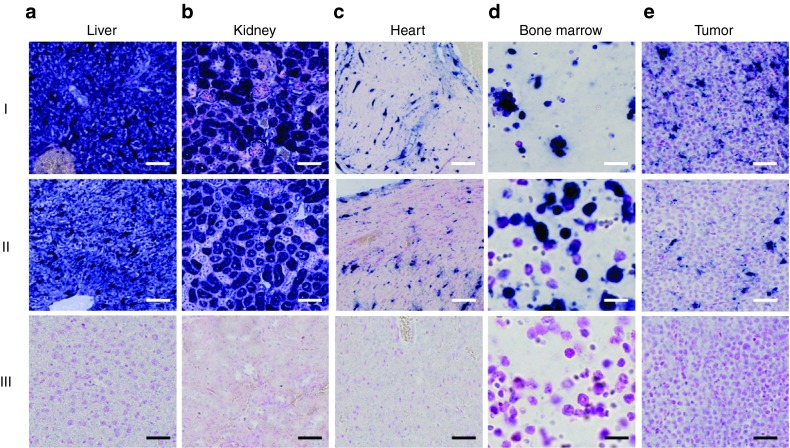
Figure 3.
In situ hybridization (ISH) detection of LNA-i-miR-221 in mouse tissues. LNA-i-miR-221 ISH on sections from organs retrieved from mice after 2 (I) and 7 (II) days of a single intraperitoneal (i.p.) injection of the LNA-i-miR-221 at therapeutic dose of 25 mg/kg or saline (III). Tissue sections from LNA-i-miR-221-treated (I and II) or saline-treated (III) liver (a), kidney (b), heart (c), bone marrow (d), and NCI-H929 xenograft tumor (e) samples after miR-221i probe hybridization. LNA-i-miR-221 staining is detectable in macrophages and/or vessels (liver and tumor), tubules (kidney), probably vascular cells (heart), subpopulation of hematopoietic cells (bone marrow). Bars: Liver, kidney, and heart: 100 µm; bone marrow: 12 µm; tumor: 50 µm.
Long-lasting LNA-i-miR-221 detection in mouse tissues
In order to evaluate bioavailability of LNA-i-miR-221, we performed a mouse study to analyze the inhibitor in different tissues retrieved from animals which had undergone to one course of treatments (days 1, 4, 8, 15, 22) with LNA-i-miR-221 at doses of 25 mg/kg or fourfold higher dose (100 mg/kg). Animals were then sacrificed 1 week or 3 weeks after last injection, and vital organs including liver, kidney, heart, bone marrow, and xenografted tumors were collected. ISH showed variations in staining intensity comparing various organs at different time points, with a signal reduction within 3 weeks from last treatment. Specifically, the ISH signal was weaker in the heart and xenograft tumors after 3 weeks from last injection (25 mg/kg), while liver, kidney, and bone marrow maintained an intense signal up to 3 weeks from last treatment at both doses (Figure 4, line II and S2). No obvious difference in the LNA-i-miR-221 staining intensity was found among animals receiving single treatment (Figure 3) or all scheduled treatments (Figure 4).
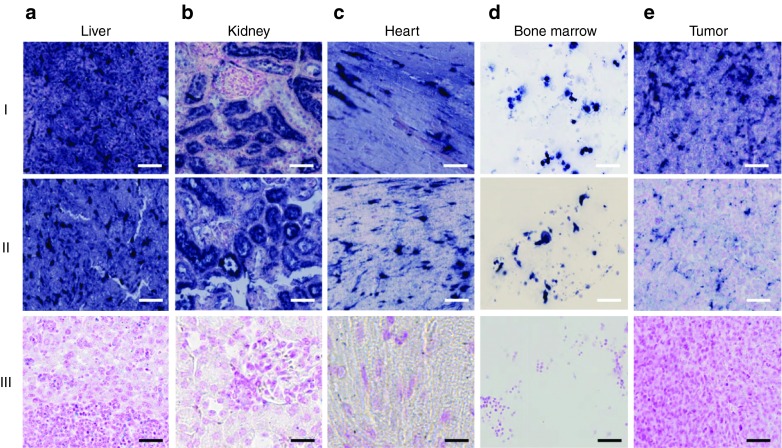
Figure 4.
Long-lasting LNA-i-miR-221 signal in mouse tissues. LNA-i-miR-221 in situ hybridization (ISH) in NOD.SCID mouse organs after injection of therapeutic doses of LNA-i-miR-221 at 25 mg/kg at days 1, 4, 8, 15, and 22. The organs were harvested at 1 week (I) or 3 weeks (II) after last injection. The staining intensity is high both after 1 and 3 weeks: (a) liver and (b) kidney maintain an intense signal throughout the period, while a slightly weaker signal is detectable in heart (c), bone marrow (d) and tumor tissue (e) 3 weeks after last dose. No ISH signal is seen in the organs from saline-treated animals (III). Bars: row I, II, and III (bone marrow and tumor) 50 µm, row III (liver, kidney, and heart) 25 µm.
LNA-i-miR-221 ISH analysis in brain
To evaluate if LNA-i-miR-221 is detectable in the brain tissues, we performed ISH on samples retrieved from six mice, 2 and 7 days after a single injection at the dose of 25 mg/kg. Positive ISH signal was confined to the capillary glial cells and endothelial cells of the venous capillaries in arachnoid meninges, whereas all neurons were negative (Figure 5a,b). No ISH signal was seen in brain tissue from untreated animals with miR-221i probe assay (Figure 5c) or in treated animals with miR-221i scramble probe assay (Figure 5d). Liver samples from the same animals were collected and analyzed to demonstrate the successful delivery of LNA-i-miR-221 in these animals (Figure 5f). Additional images of brain ISH are included in Supplementary Figure S2. Our results indicate that LNA-i-miR-221 does not cross the blood-brain barrier (BBB).
Figure 5.
LNA-i-miR-221 in situ hybridization (ISH) signal in mouse brain. LNA-i-miR-221 ISH in brain samples retrieved from mice after 2 (a) or 7 days (b) of a single intraperitoneal (i.p.) injection of the LNA-i-miR-221 at the therapeutic dose of 25 mg/kg or saline (c). Brain sections from LNA-i-miR-221 (a, b) or saline treated (c) animals using miR-221i probe (a, b, c), scramble negative control probe, (d) or positive control probe against miR-126 (e). Intense staining is seen in the liver (f) from the same mouse as in a. No nervous parenchyma showed detectable LNA-i-miR-221 ISH signal.
P27 is upregulated in mouse tissues after LNA-i-miR-221 treatment
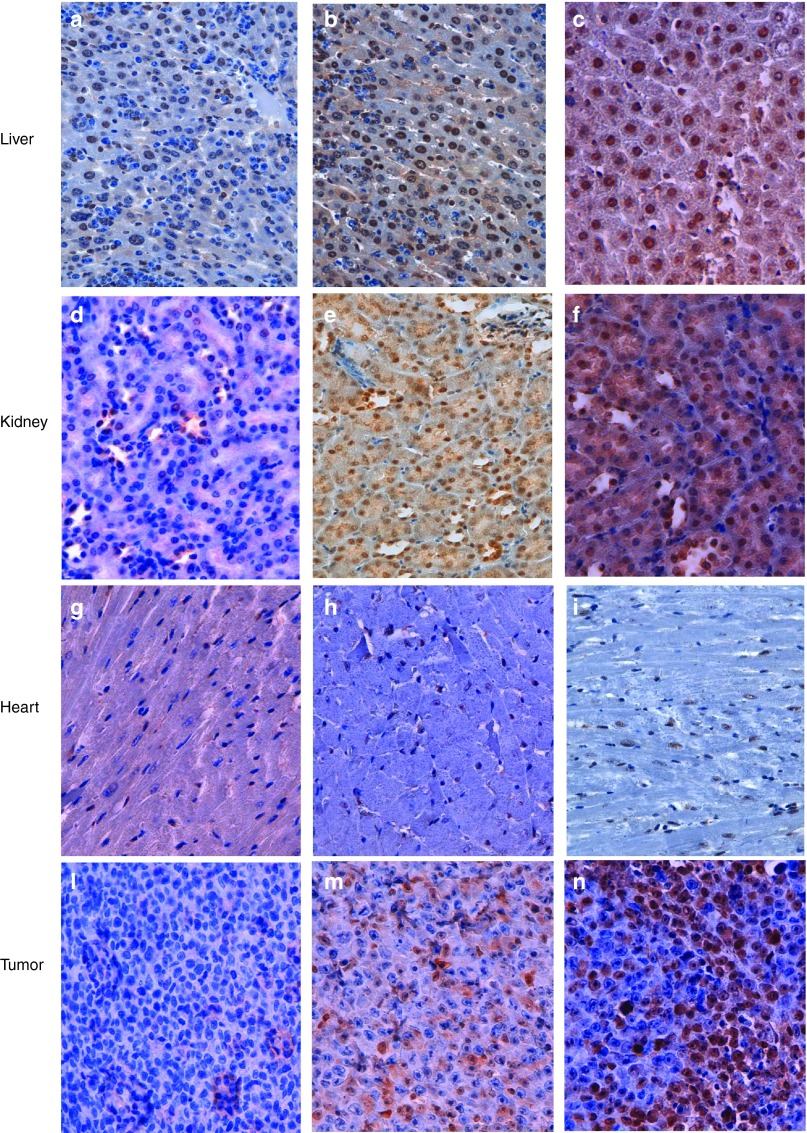
A major goal in the monitoring of an oligonucleotide-based therapy is the demonstration of its biological activity in vivo. At this aim, we investigated whether LNA-i-miR-221 efficiently inhibits miR-221 in tissues by modulation of p27 expression, the major miR-221 canonical target. We analyzed by immunohistochemistry, normal tissues, and xenografted tumors retrieved from mice. We confirmed the effectiveness of our oligonucleotide in functional inhibition of miR-221. Interestingly, this effect was observed till 3 weeks after the last injection at dose of 25 mg/kg, as shown by increased p27 expression in liver and kidney (90% of positive cells in both tissues after 1 week; 80 and 90%, respectively, after 3 weeks) (Figure 6b,c,e,f), as compared with control animals (60 and 50%, respectively) (Figure 6a,d). In the heart, we did not detect any change in p27 expression, as assessed by 30% of positive cells in all animal groups (Figure 6g,i), while xenografted tumors showed an increase of positive cells from 20% in the control group (Figure 6l) to 40% after 1 week and 60% after 3 weeks from the last injection (Figure 6m,n). The p27 signal intensity was strongly homogeneous in all analyzed specimens. Interestingly, the increase of p27 did not result in toxicity on normal liver where Ki67 was never detected (see Supplementary Figure S9). Conversely, the increase of p27 in xenografted tumors was associated with reduction of Ki67 positive cells 1 week after the last treatment (15% of positive cells) as compared with untreated tumors (30%, Supplementary Figure S9d,e). However, the percentage of Ki67-positive cells increased again 3 weeks after last treatment (30%; Supplementary Figure S9f), probably for resumed cell proliferation even though the persistence of the inhibitor in tumor cells, assessed by ISH (Figure 4). In these same specimens, we also evaluated apoptosis by Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) assay. Normal liver sections from all animals showed the absence of apoptotic events (see Supplementary Figure S10a–c), while MM xenografts showed 0.1% of apoptotic cells in untreated mice, 15% and 5%, respectively 1 and 3 weeks after last injection in treated animals (see Supplementary Figure S10 d–f). Overall, our data indicate that LNA-i-miR-221 efficiently antagonizes miR-221 activity in tumors, as also demonstrated by the inverse correlation of miR-221 and p27 mRNA expression assessed by quantitative real time polymerase chain reaction (see Supplementary Figure S11). In addition, 3 weeks after ending the treatment, an increase of miR-221 and a reduction of p27 expression was detected, indicating release from the inhibitory activity, as also demonstrated by Ki67 increase. Interestingly, the increased expression of p27 in vital organs did not result in abnormal morphology, as assessed by hematoxylin and eosin staining (Figure 1), nor resulted in apoptosis, as evaluated by TUNEL (see Supplementary Figure S10).
Figure 6.
miR-221 inhibition by LNA-i-miR-221 induces p27 upregulation in mouse tissues. Immunohistochemistry (IHC) of p27 in liver and kidney mouse tissue from untreated mice (a, d) and from mice that underwent a course of 25 mg/kg LNA-i-miR-221 treatment retrieved after 1 (b,e) or 3 (c,f) weeks from last treatment. Increased p27 expression is showed in representative image of liver and kidney (80 and 90% of positive cells, respectively) from treated animals, compared with the same control tissue (60 and 50%) in liver and kidney, respectively. Heart tissue of untreated (g) and treated animals showing the same low expression of p27 (30%) at both time-points evaluated (h,i). MM xenograft tumors showed an increase of positive cells form 20% in the saline treated animals (l) to 40% after 1 week (m) and 60% after 3 weeks (n) from last injection. Positive cells were counted in 10 high-magnification random fields of the most representative areas; the degree of immunostaining for each antibody was expressed as the percentage of positive cells among the total number of cells. Representative image of different tissues are shown as 400× magnification.
LNA-i-miR-221 is detectable in animal plasma and urine
Short oligonucleotides are not suitable for quantification with quantitative real time polymerase chain reaction. Moreover, ASOs, such as LNA-PS derivatives, require a high degree of quality control and identification of minor impurities. Until recently, analytical tools have been limited to high-performance liquid chromatography and gel electrophoresis, but the qualitative conclusions were based on retention or migration times and not sufficient for absolute identification. A number of high-resolution separation systems have been developed for oligonucleotides detection in biological fluids, including electrospray ionization mass spectrometry. In our feasibility study, we first used plasma from treated mouse to set-up a LNA-i-miR-221 quantification method, based on mass spectrometry. To this aim aliquots of 120 µl of plasma from mouse blood samples, treated with a single dose of 25 mg/kg with LNA-i-miR-221, were used for preparation of LC-MS/MS injection solution. LNA-i-miR-221 concentration were calculated in the range of 25–2,500 ng/ml, based on calibration curve fitted by weighted linear regression function (l/x2), with at least 75% of back-calculated concentrations within 15% accuracy. In these conditions, in mouse plasma the limit of quantification was fixed at 25 ng/ml. The retention time for LNA-i-miR-221 was 0.45 minutes (see Supplementary Figure S3a) and we drown a linear (r = 0.9943) calibration curve (see Supplementary Figure S3b) that we used for quantification of LNA-i-miR-221 in mouse and monkey studies. Likewise, 10 µl of monkey urine were used for preparation of LC-MS/MS injection solution. In these conditions LNA-i-miR-221 concentration was calculated in the range of 75–5,000 ng/ml, based on calibration curve fitted by weighted linear regression function (l/x2), with at least 75% of back-calculated concentrations within 20% accuracy. Limit of quantification in monkey urine was fixed at 75 ng/ml. In Supplementary Figure S3 is reported typical LC-MS/MS chromatograms obtained for urine samples and an example of calibration curve.
Pharmacokinetic profile of LNA-i-miR-221 in mouse
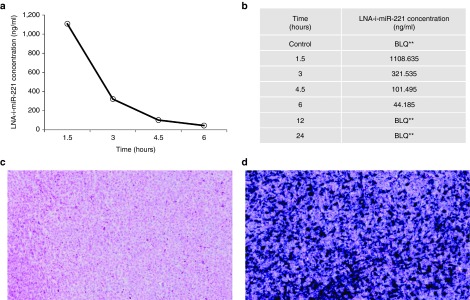
Analysis of the mouse PK following a single injection of 25 mg/kg of LNA-i-miR-221 indicated a plasma concentration peak at a Tmax of 1.5 hours with a Cmax of 1,110 ng/ml (Figure 7a). After 3 hours from i.v. injection the plasma concentration was reduced to ~ 60% and after 6–24 hours the molecule was no more detectable (Figure 7b ). The t1/2 of the molecule was calculated as 1.05 hours. The plasma exposure of the molecules shown an AUC last of 1,730 ng*hour/ml. The PK study in mice demonstrated a rapid serum clearance of LNA-i-miR-221 and a rapid uptake from cells, as also demonstrated by the strong signal detected by ISH in xenografted tumors after 12 hours of single LNA-i-miR-221 injection (Figure 7c,d).
Figure 7.
Pharmacokinetics (PK) profile of LNA-i-miR-221 in mice. (a) Plot of plasma concentrations at different time-points for LNA-i-miR-221 indicates a rapid plasmatic clearance from 1.5 to 6 hours after single dose injection (25 mg/kg), followed by a distribution phase, till to almost no detectability 6 hours after injection; (b) plasma LNA-i-miR-221 concentration (ng/ml) at each time point from 1.5 to 24 hours; (c–d) ISH for LNA-i-miR-221 in xenograft tumors of untreated (c) and treated animals (d) retrieved after 12 hours of single injection of LNA-i-miR-221 (25 mg/kg). **: BLQ ≤ LLQ (LLQ = 25 ng/ml).
LNA-i-miR-221 is nontoxic in Cynomolgus monkey
To further evaluate the safety profile of LNA-i-miR-221, we designed a pilot toxicity study in nonhuman primates. To this aim five non-naive Cynomolgus monkeys were used for the study and represented five treatment “groups” including, control, low, mid, high dose, and PK study group. Animals underwent five injections (on days 1, 4, 8, 15, and 22) with 0, 2.5, 6.25, and 8.75 mg/kg of LNA-i-miR-221, respectively. The animal of the PK group received a single dose of 8.75 mg/kg, the highest dose evaluated, defined as maximum tolerated dose in our study. Specifically, the mid dose (6.25 mg/kg) tested in monkey was selected, because it was equivalent to the therapeutic dose in xenografted mice (25 mg/kg) (Preclinical Safety Evaluation of Biotechnology-derived Pharmaceuticals S6 (R1), Parent Guideline, 1997). Observations were done daily (food and water consumption; behavior; eyes, and skin signs) or weekly (body weight) or prior to the treatment (hematology or serum biochemistry; for detailed evaluation, see supplemental material). After completion of the treatment period in the first four animals, no changes were observed on behavior of treated animals, including body weight and food consumption, as compared with the control (see Supplementary Figure S4). Moreover, no changes in heart rate were recorded in all animals in the time frame between the first and the last injection, when compared with the predose values (data not shown). Blood evaluations included in the study are listed in Supplementary Table S1. All the recorded values confirmed the safety profile of LNA-i-miR-221 in monkeys, as shown by hematology, coagulation, liver and kidney parameters (see Supplementary Figure S5–S7). All data before and after treatments for each animal are listed in Supplementary Table S2. The findings indicated normal liver and kidney function in all treated animals. Moreover, activation of the immune response was investigated by flow cytometer evaluation of the plasma cytokines mostly involved in inflammation. LNA-i-miR-221 treatment progressively reduced plasma levels of IL-8, MCP-1, and IL-6, suggesting anti-inflammatory activity. No changes in plasma levels of TNF-α, IFN-γ, and IL-4 could be detected. These cytokines remained below the detection level (< 10 pg/ml, Supplementary Figure S8) during treatments.
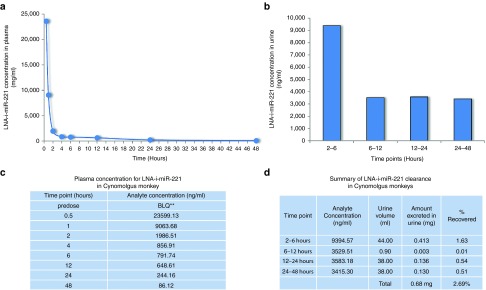
The animal included in the PK study (#5) received a single maximum tolerated dose of LNA-i-miR-221. Plasma and urines were collected from this animal. Figure 8 shows the plasma PK profile. LNA-i-miR-221 plasma concentration of a single high dose (8.75 mg/kg/day) peaked at a Tmax of half an hour with a Cmax of 23,600 ng/ml (Figure 8a,c). After 1 hour from i.v. injection, LNA-i-miR-221 plasma concentration was reduced to ~ 60% and after 6 hours, it was no more detectable. The plasma exposure of LNA-i-miR-221 showed an AUC-last of 49,300 hour*ng/ml and C0 of 61,400 ng/ml. The t1/2 of the molecule in monkey study was 12.83 hours. The monkey urine PK evaluation was performed on samples collected in a four time-range from 2–48 hours. For each interval the urine volumes and LNA-i-miR-221 concentrations were recorded. LNA-i-miR-221 excreted in urine was quantified =0.68 mg in 48 hours, mostly in the first 6 hours (0.413 mg), with a total urinary recovery of ~2.68% of the total amount injected (25,375 mg). Specifically, the LNA-i-miR-221 urinary concentration/time-points plot showed the excretion of 9,394.57 ng/ml (1.63% of recovery) in the first range-time (2–6 hours) in 44 ml of urine. During the other 3 time-points, LNA-i-miR-221 concentrations were lower (Figure 8b,d). Overall these PK findings demonstrated a rapid serum clearance of LNA-i-miR-221, with maximum tissues uptake and minimum urinary excretion.
Figure 8.
Pharmacokinetics (PK) profile of LNA-i-miR-221 in monkey study. (a) Plot of plasma concentration at different time-points for LNA-i-miR-221 indicates a rapid plasmatic clearance from 0.5 to 48 hours after single dose injection (875 mg/kg), followed by distribution phase, till to almost no detectablility after 48 hours; (b) plot of urine concentration versus four range of urine collection spanning from 2 hours to 48 hours, showing that most LNA-i-miR-221 excretion occurred within 6 hours; (c) plasma LNA-i-miR-221 concentration (ng/ml) at each time points (hours); (d) urine LNA-i-miR-221 concentration (ng/ml) at each time point. **: BLQ ≤ LLQ (LLQ = 50 ng/ml).
Discussion
The basic strategy of miRNA therapeutics relies on the ability to efficiently replace or sequester endogenous miRNAs dysregulated in human cancer. Different chemical modifications have been recently introduced in oligonucleotides synthesis to improve stability and specificity which, in turn, strengthen their biomodulatory activity. It is now well established that LNA-based ASOs ensure high potency and affinity as inhibitors of upregulated onco-miRNA, so that this class of compounds are presently approaching cancer treatment in the clinics.34,36,37,38,39,40 However, one of the major issues in the aim of clinical translation has been the lack of quantitative assays for detection of LNA-miRNA inhibitors in biological fluids and tissues, which is relevant for PK and pharmacodynamics investigations and therefore for the design of clinical studies. Among available technologies for qualitative and quantitative evaluation, LC combined with MS is the method-of-choice for the detection of oligonucleotides in body fluids.41 Insufficient PK and poor cellular uptake are the main hurdles for successful therapeutic development of oligonucleotide-based therapeutics. Second generation ASOs, including LNA-mixmers, following systemic administration, have rapid distribution, long-terminal half-life and plasma clearance, primarily due to extensive distribution in tissues.33 However, few data are presently available on the use of LC/MS for the study of LNA-miRNA inhibitors. To shed light on this important point, we used an ultra-high pressure LC-MS/MS protocol following solid-phase extraction, which allows quantification of very low amounts of LNA-miR-221 inhibitor by the use of small volumes of mouse and monkey plasma as well as of monkey urine. In our mouse study, we observed that the 13-mer LNA-i-miR-221 is rapidly distributed into systemic circulation. Following injection at therapeutic doses, a peak plasma concentration (Cmax) at 1.5 hour was detected. After reaching Cmax, plasma concentration declines after the fast initial distribution phase (distribution half-life of about 3–4.5 hours), then a terminal elimination phase starting from ~ 4.5 hours with very low plasma concentration. Six hours after the treatment LNA-i-miR-221 became almost undetectable in the mouse plasma. Similarly, in Cynomolgus monkeys, following i.v. infusion of a single dose of 8.75 mg/kg of LNA-i-miR-221, we observed again the peak of Cmax after 0.5 hour and its rapid decline in the plasma, where the concentration was reduced more than 10-folds in 1.5 hours from the peak. Consistent with the plasma distribution profile dominating plasma exposure and clearance of the second generation ASOs, partial area plasma AUC(0.5–6 hours) values of LNA-i-miR-221 represent 73.8% of AUC-last (0–48 hours) values. We could conclude that LNA-i-miR-221, evaluated in two different species by LC-MS/MS showed a similar PK profile.
It has been demonstrated that second generation ASOs in plasma are mostly bound to plasma proteins which reduce the glomerular filtration and urine excretion.42 According to these findings, only a minimal amount (0.68 mg, 2,679%) of the intact LNA-i-miR-221, after a single active dose (25,375 mg), was recovered in monkey urines. On the other hand, the low affinity binding of ASOs to plasma proteins favors the binding to cell surface proteins with consequent extensive cellular uptake.43 This is confirmed by our ISH analyses in mouse tissues where high LNA-i-miR-221 content 48 hours after the single active dose was demonstrated. The prolonged presence of LNA-i-miR-221 in tissues several weeks after the oligonucleotide have been cleared from the circulation indicates cell specific uptake. We detected strong LNA-i-miR-221 ISH signal in a variety of cell types. In the liver, stained cells were spindle-shaped cells which can be identified as vascular cells or macrophages. In the kidney, many tubular cells were positive and in the heart also spindle-shaped cells were positive. These observations indicate that different cell populations indeed bind and retain the LNA-i-miR-221. Further studies are needed to better characterize LNA-i-miR-221 retaining cells. Interestingly, xenografted MM tumors accumulate the LNA-i-miR-221 rapidly and the single 25 mg/kg i.p. dose reaches the tumor after 12 hours, even if these tumors are not well vascularized as compared to normal mouse vital organs.
Oligonucleotides are distinguished from other classes of pharmaceutical agents by both the mechanism of action and physical–chemical or pharmaceutical properties. PK studies of PS-oligonucleotides demonstrate that they are well absorbed from injection sites, rapidly distributed broadly to all peripheral tissues, and do not cross the BBB.44 However, PS-oligonucleotides have been used in some studies to knock-down central nervous system (CNS) proteins, for their characteristics to be transported across the BBB45,46 indicating that the PS-oligonucleotides injected peripherally can accumulate in the central nervous system in sufficient amounts to knockdown proteins in brain. Many reports showed that PS-oligonucleotides are transported across the BBB by saturable transporter even if at moderate rate to enter both the parenchymal space of the brain and the cerebrospinal fluid, and particularly high rate in hippocampus in comparison to the cerebral cortex or whole brain.47 In other studies, however the delivery of ASOs to the CNS has demonstrated important limitations since the BBB prevented blood-borne oligonucleotides from leaving the brain's vascular space to enter the CNS.48,49 The substitution of one nonbridging oxygen with the more hydrophobic sulfur atom, as in the case of PS nucleotides, increases both plasma stability and plasma protein binding and thus, tissue bioavailability and broad biodistribution. Our ISH analyses suggest that the 13-mer LNA-i-miR-221 does not cross the normal BBB.
This observation further supports the framework for the clinical translation of the LNA-i-miR-221 molecule because long-term CNS toxicity is avoided. The safety toxicity profile has been evaluated in our mouse and monkey studies. In fact, it has been described that ASOs with PS backbone exhibit a significant hybridization-independent toxicity profile. The observed effects include increase of coagulation time, pro-inflammatory activity, activation of the complement pathway, and at higher concentrations, PS-ASOs lead to renal tubule changes and thrombocytopenia and induce a strong immunostimulatory response through their interactions with Toll-like receptors.50 Data from our mouse study excluded damage of vital organs, since no changes in liver, kidney, and heart cellular morphology were observed after postmortem pathology. Furthermore, even p27 increase has been observed in liver, no apoptosis was detected by TUNEL assay. In monkey study, biochemical and hematologic analysis did not demonstrate acute toxicity induced by LNA-i-miR-221 treatment, nor liver and renal failure, even at 10-folds increase of therapeutic dose. No immune stimulation was induced in monkeys after systemic administration even at 10-folds increase. All findings from pathology and blood and serum evaluations, demonstrating absence of acute toxicity, were parallel by the lack of weight loss or behavioral changes by close monitoring of treated animals at all the investigated oligonucleotide doses.
In conclusion, our work firstly reports the PK and tissue bioavailability of LNA-i-miR-221 in two different animal models after i.v. or i.p. injection, which appears highly promising for translation in clinics. LNA-i-miR-221 is rapidly absorbed and completely distributed and excreted, with a short half-life. We speculate that our PK results support the hypothesis of high plasma protein binding of LNA-i-miR-221, which may explain the reduced renal excretion and, therefore, facilitate uptake into peripheral tissues via high affinity receptors, resulting in increase of therapeutic efficacy. The well tolerated dose of 25 mg/kg in mouse, equivalent to 8.75 mg/kg in monkey, is recommended for the dosage calculation for animal treatment in regulatory toxicity studies and for the design of an early clinical trial. On the basis of our findings it will be reasonable to consider intensive loading schedule, for instance by consecutive daily administration, in order to maximize the tissue distribution up to tolerated doses in dose finding early clinical trials.
Materials and methods
Animal studies
Mouse study. Different doses of LNA-i-miR-221 were evaluated in NCI-H929 xenografted NOD.SCID mice. Two different in vivo experiments were performed for LNA-i-miR-221 detection by ISH. In the first experiment, mice were randomized into two groups and treated i.p. with a single dose of 25 mg/kg LNA-i-miR-221, defined as therapeutic dose, or scramble control. Tumors and organs including liver, kidney, heart, bone marrow, brain, and tumor were collected after 2 and 7 days from treatment. In a second in vivo experiment animals received a cycle (days 1, 4, 8, 15, and 22) of LNA-i-miR-221 i.p. treatment at two different doses (25 and 100 mg/kg) including saline control group. MM xenografted tumors and organs were collected after 1 and 3 weeks from the last dose. All specimens were immediately formalin-fixed and paraffin embedded in 24–36 hours. For PK study mice were treated i.p. with a single dose of 25 mg/kg LNA-i-miR-221, or saline as control and blood sampling were performed 90 minutes and 3, 4.5, 6, 12, and 24 hours following injection into a K2EDTA tube and immediately placed on ice. Thereafter, plasma was obtained from the blood sample by centrifugation at 3,000×g for 10 minutes at +4 °C. The resulting plasma was stored at approximately −80°C until analysis.
Tissue samples. Mouse tissues and organs were collected after treatment at different time points. In brief, after removal from mice, the tissues/organs were dissected into appropriate specimens, formalin fixed for 24–36 hours and then embedded in paraffin at the Pathology Unit (Hospital Lamezia Terme, Calabria, Italy). Five micrometer-thick serial sections were obtained from all paraffin samples for standard hematoxylin and eosin, and immunohistochemistry staining, for immunofluorescence assay and for ISH analysis. This was performed at Bioneer (Horsholm, Denmark).
LNA probes. Double FAM-labeled LNA:DNA chimeric antisense probes (hereafter referred to LNA probes) were all obtained from Exiqon (Exiqon, Vedbaek, Denmark). LNA probes included the antisense probe to the therapeutic LNA-i-miR-221 named miR-221i (sequence 3′-AGCTACATTGTCTGTA-5′, DNA/LNA probe, Tm = 79 °C, LNA content = 31%), miR-126-3p (3′-CATTATTACTCACGGTACGA-5′, DNA/LNA probe, Tm = 84 °C, LNA content = 40%), used as positive control, and a scramble probe (3′-tgtaacacgtctatacgccca-5′, Tm = 87 °C, LNA content = 33%), as negative control.51 The melting temperatures for all the above indicated LNA probes are RNA-Tm, and are predicted (according to Exiqon's software tools) toward complementary RNA oligonucleotides using spectrophotometer. The Tm for the interaction between the LNA molecule, LNA-i-miR-221, and the LNA probe, miR-221i, may well be higher than predicted by the software because of the LNA content in both sequences.
In situ hybridization. Five µm-thick paraffin sections were obtained and processed for LNA-based ISH essentially as previously described.52,53 In brief, the sections were deparaffinized in xylene, and then the following steps were performed in a Tecan Freedom Evo automated hybridization instrument (Tecan, Mannedorf, Switzerland): pre-digestion with 3 µg/ml proteinase-K (for 8 minutes at 37 °C), prehybridization and ISH in Exiqon hybridization buffer (Exiqon) at 55 °C for 15 minutes and 60 minutes, respectively, stringent washes with SSC buffers, probe detection using alkaline phosphatase-conjugated anti-FAM sheep immunoglobulin (Roche Diagnostics, Manheim, Germany) for 30 minutes, 4-nitro-blue tetrazolium, and 5-bromo-4-chloro-30-indolyl-phosphate substrate incubation for 60 minutes. Finally, sections were counterstained with nuclear fast red (Vector Laboratories, Burlingname, CA) and mounted using Eukitt mounting medium. The three LNA probes, LNA-i-miR-221, miR-126, and scramble, were incubated on tissue sections during the ISH step at 10, 30 and 20 nmol/l.
Immunohistochemistry and fluorescence assays. For each case 4 µm-thick serial sections were cut from a representative block of formalin fixed, paraffin-embedded tissue, mounted on acid-cleaned glass slides, and heated at 55 °C for 60 minutes. Slides were, dewaxed with xylene, and processed for hematoxylin and eosin and immunohistochemistry. All the procedures were performed at room temperature. Immunohistochemical evaluation of anti-p27 was performed using the LSAB+ System HRP (DakoEnvision System, CA), followed by the addition of 3,3′-diaminobenzidine as a chromogen. Endogenous peroxidase activity was quenched for 5 minutes in 3% hydrogen peroxidase, and the slides were rinsed in wash solution (TBST, 0.05 mol/l Tris Buffered Saline with Tween20). Antigen retrieval was performed with EDTA buffer pH 9 for 30 minutes at 98 °C. Slides were washed three times in phosphate-buffered saline (PBS; pH 7.4) for 5 minutes. Immunostaining was performed using a purified mouse monoclonal antibody anti-p27 (clone 57/Kip-1/27, diluted 1:150, BD Biosciences, CA) and a purified mouse monoclonal antibody ki67 (clone MIB-1, 1:150 dilution, DakoEnvision System) for 1 hour at 25 °C. Sections were finally counterstained with hematoxylin. Negative controls were performed in each run by substituting primary antibodies with antibodies with irrelevant specificity but of the same isotype of the primary antibodies. The apoptosis evaluation was performed by TUNEL Assay, designed for the specific detection and quantitation of apoptotic cells (DeadEnd Fluorometric Tunel System, Promega Corporation, Madison, WI). Deparaffined slides were washed twice in PBS for 5 minutes each time, permeabilized and equilibrated with an Equilibration buffer at room temperature for 10 minutes. On the Slides were added a Terminal Deoxynucleotidyl Transferase reaction mix, covered with plastic coverslips to ensure even distribution of the mix and incubate for 60 minutes at 37°C in a humidified chamber avoid exposure to light. The reaction was stopped immerging slides in 2× SSC for 15 minutes. Then slides were washed three times in PBS for 5 minutes each time. Finally all nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI). The fragmented DNA of apoptotic cells by catalytically incorporating fluorescein-12-dUTP at 3′-OH DNA ends using Terminal Deoxynucleotidyl Transferase were analyzed by fluorescence microscopy.
Determination of LNA-i-miR-221 in plasma mouse samples by LC-MS/MS. The samples were analyzed in Aptuit (Verona, Italy). Ultra-high pressure liquid chromatography (UHPLC) after solid-phase extraction was used. To obtain LNA-miR-221 inhibitor from mouse plasma, an aliquot of 120 µl of these samples were diluted with 120 µl of 0.1 mol/l triethylammonium acetate (TEAAC) buffer pH = 7 and gently mixed. After, 200 µl of samples were loaded onto a µ-elution HLB SPE 96-well plate, which were preactivated and preconditioned using 200 µl of acetonitrile followed by 200 µl of 0.1 mol/l TEAAC buffer pH = 7. Then, samples were drawn through using a low vacuum. SPE wells were first washed using 200 µl 0.1 mol/l TEAAC buffer and then 200µl of water. The wells were eluted two times using 200 µl 0.1 mol/l TEAAC buffer pH 7/acetonitrile (30:70, v/v). The eluates after evaporation to dryness, at ~25 °C, under nitrogen were reconstituted with 75µl of mobile phase A. An aliquot of 10 µl were injected into LC-MS/MS system. Then a tandem mass spectrometry (MS/MS) by a triple quadripole Turbo Ion Spray ionization source permitted the detection of very low quantity (25 ng/ml) of LNA-miR-221 inhibitor in mouse plasma. For calibration curve, a weighed linear regression function (1/x2) was used to fit calibration lines, and consequently, to calculate LNA-miR-221 inhibitor concentrations in the calibration range 25–2,500 ng/ml, with at least 75% of back-calculated concentrations within 15% accuracy. Mouse plasma concentrations obtained at various sampling time-points were used to calculate the time to achieve maximum plasma concentration at first experimental observation (Tmax), the maximum plasma concentration at the first observation (Cmax), the area under the curve up to the last (Tlast 6 hours) measurable concentration (AUClast), and the elimination half-life (t1/2).
Nonhuman primates study. Number seven male purpose-bred Cynomolgus monkeys (Macaca fascicularis) were selected for entry into this study. The animals were allowed to acclimatize to the CiToxLab (Evreux, France) primate toxicology accommodation for a period of 4 weeks before the commencement of the study. All animals were dewormed by CiToxLab. The animals were selected for entry into the study based on the results of a satisfactory preliminary health screening. The route of administration was i.v. bolus injection into the saphenous vein. The control group was treated with saline (sodium chloride 0.9%) as well the dilution of the molecule. The quantity of dose formulation administered to each animal was adjusted according to the most recently recorded body weight. A constant dosage-volume of 1 ml/kg was used. The administration system was weighed before and after dosing in order to determine the precise quantity of test item administered. At the beginning of the study, the animals were 33–58 months old and they had a mean body weight of 4.1 kg (range: 3.2–4.8 kg) (groups 1–4) and 3.1 kg on Day 7 (group 5). After allocation to the study, the animals were acclimated to the study conditions for a period of 7 days before the beginning of the treatment period. During this period, pretreatment investigations were performed. Two supernumerary animals were allocated to the study and acclimated, in order to permit the selection and/or replacement of individuals. During the pretreatment period, five male animals were selected according to the results of clinical and laboratory examinations. The animals were individually identified by a subcutaneous microchip for automated identification at the breeder's facility. At the beginning of the study, each animal received a unique CiToxLab France identity number and were housed in a dedicated primate unit. The animal room conditions were set with a temperature range 22 ± 3 °C, relative humidity of 50 ± 30%, light/dark cycle of 12 hours/12 hours, and ventilation range between 8 and 15 cycles/hour of filtered, nonrecycled air.
Toxicity profile of LNA-i-miR-221 in nonhuman primates. Five male Cynomolgus monkeys were allocated in the study. The first four animals (#1–#4) received LNA-i-miR-221 on days 1, 4, 8, 15, and 22, respectively at 0, 2.5, 6.25 or 8.75 mg/kg/day by i.v. infusion (bolus) in order to determine the maximum tolerated dose. On day 22, the last animal (#5) received the highest dose (8.75 mg/kg) to perform PKs investigations in plasma and urine. Animals were checked at least twice a day for mortality and clinical signs, including weekends. Special attention was paid to skin, eyes, and mucous membranes as well as injection sites. Heart rate was evaluated using a stethoscope in a calm environment over 10 seconds and then multiply by six in order to determine approximate heart rate of the animals in beats per minute. Body weight was recorded once before the beginning of the treatment period (once in pretest, day 5), on the first day of treatment and then, on each day of the first two treatments for the animals of groups 1–4 and on the eve of the day of treatment for the following administration including the animal of the group 5. Blood samples for hematology and blood biochemistry investigations were collected as following: in pretest (day 5), predose prior the first administration, on days 1, 4, 8, 15, and 22 from animals of groups 1–4. Blood and urine were collected from animal #5 only in order to establish PKs profile of the LNA-i-miR-221, accordingly: (i) for blood sampling, before treatment 0, 30, and 60 minutes, 2, 4, 6, 12, 24, and 48 hours following i.v. injection of LNA-i-miR-221 and (ii) for urine sampling, intervals (0–2 hours), (2—6 hours), (6—12 hours), (12 —24 hours), and (24—48 hours) after i.v. administration. Before blood collection, the animals were deprived of food for an overnight period of at least 14 hours. Blood samples were collected without anesthetic from an appropriate vein into BD Microtainer® (K2EDTA) tubes for the ADVIA 120 Hematology analyzer; into sodium citrate tubes for ACL Elite Pro blood coagulation analyzer; into lithium heparin tubes for ADVIA 1,800 blood biochemistry analyzer. For PK analysis, venous blood (~0.5 ml) was taken from an appropriate vein of animal #5 into a K2EDTA tube and immediately placed on ice. Thereafter, plasma was obtained from the blood sample by centrifugation at 3,000×g for 10 minutes at +4 °C (start of centrifugation within 30 minutes after blood sampling). The resulting plasma was stored at ~−80 °C until analysis. Urine collection was performed for the determination of LNA-i-miR-221 levels from animal #5. To increase diuresis, drinking water (~15 ml) was administered (gavage) 2 hours after treatment and prior to the beginning of urine collection. All samples were collected on ice. Urine samples were weighed and the volumes recorded, then stored at −80 °C until analysis. Plasma collected at time 0, 24, and 48 hours after single highest dose were used for TNF-α, IFN-γ, IL-4, IL-6, IL-8 and MCP-1 inflammatory cytokines measurement, by using the flex-set BD cytometric bead array (BD CBA flex set; BD Biosciences, Franklin Lakes, NJ) assay, according to producer guidelines.
Pharmacokinetic profile of LNA-i-miR-221 in nonhuman primates. The determination of LNA-i-miR-221 in plasma monkey samples were performed according to LC-MS/MS protocols before described for mouse plasma. The determination of LNA-i-miR-221 in urine monkey samples was performed by ultra-high pressure liquid chromatography (UHPLC) after solid-phase extraction. To obtain LNA-miR-221 inhibitor from urine, an aliquots of 220 µl of the urine samples were diluted with 220 µl of 0.1 mol/l Triethylammonium acetate (TEAAC) buffer pH = 6 with acetic acid (10%) and gently mixed. 400 µl of mixed samples were loaded onto a conditioned µ-elution HLB SPE 96-well plate. The SPE wells were activated and conditioned using 200 µl of acetonitrile followed by 200 µl of 0.1 mol/l TEAAC buffer pH = 6. After loading of the mixed samples and drawn through using a low vacuum, the SPE wells were washed using first 200 µl 0.1 mol/l TEAAC buffer pH = 6 and then 200 µl of water. The compound was eluted two times using 200 µl of 0.1 mol/l TEAAC buffer pH 6/acetonitrile (30:70, v/v). The eluates were leave to dry by evaporation approximately at 25 °C under nitrogen, then were reconstituted with 50 µl of mobile phase A. Aliquots of 10 µl were injected into the LC-MS/MS system. A tandem mass spectrometry (MS/MS) equipped with a triple quadripole API4,000 (Sciex) and Turbo Ion Spray ionization source in positive ion mode, allowed the detection of very low quantity (25 ng/ml) of LNA-miR-221 inhibitor in mouse plasma. For calibration curve, a weighed linear regression function (1/x2) was used to fit calibration lines and consequently to calculate LNA-miR-221 inhibitor concentration in the calibration range 75–5,000 ng/ml, with at least 75% of back-calculated concentrations within 20% accuracy. The limit of quantification in Cynomolgus monkey urine was 75 ng/ml. The monkey's plasma concentrations obtained at various sampling time points were used to calculate the time to achieve the maximum plasma concentration (Tmax), the initial concentration, extrapolated at dosing time (C0), the maximum plasma concentration (Cmax) and the area under the curve up to the last (Tlast 48 hours) measurable concentration (AUClast).
SUPPLEMENTARY MATERIAL Figure S1. Pilot ISH assay with miR-221i or miR-126 or scramble probes in different organs retrieved from LNA-i-miR-221 treated animals. Figure S2. ISH assay of brain and liver tissues retrieved from mice treated with a single dose of LNA-i-miR-221 after 2 or 7 days from treatment. Figure S3. Feasibility of an Analytical Method for the Determination of LNA-i-miR-221 in plasma and urine by LC-MS/MS. Figure S4. Body weight evaluation of each Cynomolgus monkey included in the study. Figure S5. Hematology evaluation in monkey. Figure S6. Hematology evaluation in monkey. In the y-axis is reported the specific parameters. Figure S7. Hematology evaluation in monkey. In the y-axis is reported the specific parameters. Figure S8. Serum levels of IL-4, IL-6, IL-8, TNF-α, IFN-γ and MCP-1 in cynomolgus monkey at baseline and 24 hours and 48 hours after the administration of LNA-i-miR-221. Figure S9. Ki67 proliferation index assay. Figure S10. Apoptosis evaluation by TUNEL assay in normal liver and in tumor tissues of treated and untreated after 1 and 3 weeks from a course of treatment with LNA-i-miR-221. Figure S11. Quantitative real time polymerase chain reaction (qRT-PCR) of miR-221 and p27 expression levels in MM xenograft. Table S1. List of parameters evaluated in blood of Cynomolgus monkey. Table S2. Blood evaluation in Cynomolgus monkey study.
Acknowledgments
This work has been supported by the Italian Association for Cancer Research (AIRC), PI: PT. “Special Program Molecular Clinical Oncology - 5 per mille” no. 9980, 2010/15. M.E.G.C and M.T.D. designed and made the experimental work. P. T., P T., and M.T.D. wrote the manuscript. B.S.N. performed the in situ hybridization. M.A. and C.R. contribute to PK data interpretation, C.M. contributed to histological analysis, C.B. performed cytokines evaluations. We thank CiToxLab for monkey study, Aptuit for MS/MS analysis, Caterina Camastra and Maria Cerra for the valuable technical assistance in histological analysis.
Supplementary Material
References
- Amodio, N, Di Martino, MT, Neri, A, Tagliaferri, P and Tassone, P (2013). Non-coding RNA: a novel opportunity for the personalized treatment of multiple myeloma. Expert Opin Biol Ther 13 (Suppl. 1): S125–S137. [DOI] [PubMed] [Google Scholar]
- Esteller, M (2011). Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874. [DOI] [PubMed] [Google Scholar]
- Calin, GA, Sevignani, C, Dumitru, CD, Hyslop, T, Noch, E, Yendamuri, S et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C, Li, X, Zhang, Y, Guo, Y, Zhou, J, Gao, K et al. (2015). The microRNA feedback regulation of p63 in cancer progression. Oncotarget 6: 8434–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Guzzi, PH, Caracciolo, D, Agnelli, L, Neri, A, Walker, BA et al. (2015). Integrated analysis of microRNAs, transcription factors and target genes expression discloses a specific molecular architecture of hyperdiploid multiple myeloma. Oncotarget 6: 19132–19147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti, M, Musto, P, Di Martino, MT, Fabris, S, Agnelli, L, Todoerti, K et al. (2013). Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res 19: 3130–3142. [DOI] [PubMed] [Google Scholar]
- Tagliaferri, P, Rossi, M, Di Martino, MT, Amodio, N, Leone, E, Gulla, A et al. (2012). Promises and challenges of MicroRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets 12: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio, N, Rossi, M, Raimondi, L, Pitari, MR, Botta, C, Tagliaferri, P et al. (2015). miR-29s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 6: 12837–12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, M, Amodio, N, Di Martino, MT, Caracciolo, D, Tagliaferri, P and Tassone, P (2013). From target therapy to miRNA therapeutics of human multiple myeloma: theoretical and technological issues in the evolving scenario. Curr Drug Targets 14: 1144–1149. [DOI] [PubMed] [Google Scholar]
- Thorsen, SB, Obad, S, Jensen, NF, Stenvang, J and Kauppinen, S (2012). The therapeutic potential of microRNAs in cancer. Cancer J 18: 275–284. [DOI] [PubMed] [Google Scholar]
- Amodio, N, Bellizzi, D, Leotta, M, Raimondi, L, Biamonte, L, D'Aquila, P et al. (2013). miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle 12: 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Gullà, A, Gallo Cantafio, ME, Altomare, E, Amodio, N, Leone, E et al. (2014). In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One 9: e89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi, L, Amodio, N, Di Martino, MT, Altomare, E, Leotta, M, Caracciolo, D et al. (2014). Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget 5: 3039–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, M, Amodio, N, Di Martino, MT, Tagliaferri, P, Tassone, P and Cho, WC (2014). MicroRNA and multiple myeloma: from laboratory findings to translational therapeutic approaches. Curr Pharm Biotechnol 15: 459–467. [DOI] [PubMed] [Google Scholar]
- Leotta, M, Biamonte, L, Raimondi, L, Ronchetti, D, Di Martino, MT, Botta, C et al. (2014). A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol 229: 2106–2116. [DOI] [PubMed] [Google Scholar]
- Leone, E, Morelli, E, Di Martino, MT, Amodio, N, Foresta, U, Gullà, A et al. (2013). Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res 19: 2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio, I, Di Martino, MT, Campani, V, Virgilio, A, Galeone, A, Gullà, A et al. (2014). Transferrin-conjugated SNALPs encapsulating 2'-O-methylated miR-34a for the treatment of multiple myeloma. Biomed Res Int 2014: 217–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio, N, Leotta, M, Bellizzi, D, Di Martino, MT, D'Aquila, P, Lionetti, M et al. (2012). DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget 3: 1246–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Campani, V, Misso, G, Gallo Cantafio, ME, Gullà, A, Foresta, U et al. (2014). In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS One 9: e90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Gullà, A, Cantafio, ME, Lionetti, M, Leone, E, Amodio, N et al. (2013). In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 4: 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone, P, Neri, P, Burger, R, Di Martino, MT, Leone, E, Amodio, N et al. (2012). Mouse models as a translational platform for the development of new therapeutic agents in multiple myeloma. Curr Cancer Drug Targets 12: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, AG (2012). miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 3: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio, N, Di Martino, MT, Foresta, U, Leone, E, Lionetti, M, Leotta, M et al. (2012). miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis 3: e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Leone, E, Amodio, N, Foresta, U, Lionetti, M, Pitari, MR et al. (2012). Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res 18: 6260–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli, E, Leone, E, Cantafio, ME, Di Martino, MT, Amodio, N, Biamonte, L et al. (2015). Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia 29: 2173–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Z, Zhang, Y, Yang, Q, Liu, B, Wu, J, Zhang, Y et al. (2015). miR-497 and miR-34a retard lung cancer growth by co-inhibiting cyclin E1 (CCNE1). Oncotarget 6: 13149–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y, Li, X, Zhu, S, Zhang, JG, Yang, M, Qin, Q et al. (2015). Ectopic expression of miR-494 inhibited the proliferation, invasion and chemoresistance of pancreatic cancer by regulating SIRT1 and c-Myc. Gene Ther 22: 729–738. [DOI] [PubMed] [Google Scholar]
- Tréhoux, S, Lahdaoui, F, Delpu, Y, Renaud, F, Leteurtre, E, Torrisani, J et al. (2015). Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta 1853(10 Pt A): 2392–2403. [DOI] [PubMed] [Google Scholar]
- Zheng, F, Liao, YJ, Cai, MY, Liu, TH, Chen, SP, Wu, PH et al. (2015). Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet 11: e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Aranguez, A, Loma, P and Pintor, J (2013). Small-interfering RNAs (siRNAs) as a promising tool for ocular therapy. Br J Pharmacol 170: 730–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, PM and Pêgo, AP (2014). Therapeutic antisense oligonucleotides against cancer: hurdling to the clinic. Front Chem 2: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin, KE, Højland, T, Hansen, BR, Persson, R, Bramsen, JB, Kjems, J et al. (2013). Biological activity and biotechnological aspects of locked nucleic acids. Adv Genet 82: 47–107. [DOI] [PubMed] [Google Scholar]
- Yu, RZ, Grundy, JS and Geary, RS (2013). Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin Drug Metab Toxicol 9: 169–182. [DOI] [PubMed] [Google Scholar]
- Elmén, J, Lindow, M, Schütz, S, Lawrence, M, Petri, A, Obad, S et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899. [DOI] [PubMed] [Google Scholar]
- Straarup, EM, Fisker, N, Hedtjärn, M, Lindholm, MW, Rosenbohm, C, Aarup, V et al. (2010). Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res 38: 7100–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford, RE, Hildebrandt-Eriksen, ES, Petri, A, Persson, R, Lindow, M, Munk, ME et al. (2010). Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, JB, Fisker, N, Westergaard, M, Kjaerulff, LS, Hansen, HF, Thrue, CA et al. (2008). SPC3042: a proapoptotic survivin inhibitor. Mol Cancer Ther 7: 2736–2745. [DOI] [PubMed] [Google Scholar]
- Obad, S, dos Santos, CO, Petri, A, Heidenblad, M, Broom, O, Ruse, C et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43: 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini, D, Omlin, A, Pezaro, C, Lorente, D, Ferraldeschi, R, Mukherji, D et al. (2013). First-in-human phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br J Cancer 109: 2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, HL, Reesink, HW, Lawitz, EJ, Zeuzem, S, Rodriguez-Torres, M, Patel, K et al. (2013). Treatment of HCV infection by targeting microRNA. N Engl J Med 368: 1685–1694. [DOI] [PubMed] [Google Scholar]
- van Dongen, WD and Niessen, WM (2011). Bioanalytical LC-MS of therapeutic oligonucleotides. Bioanalysis 3: 541–564. [DOI] [PubMed] [Google Scholar]
- Yu, RZ, Kim, TW, Hong, A, Watanabe, TA, Gaus, HJ and Geary, RS (2007). Cross-species pharmacokinetic comparison from mouse to man of a second-generation antisense oligonucleotide, ISIS 301012, targeting human apolipoprotein B-100. Drug Metab Dispos 35: 460–468. [DOI] [PubMed] [Google Scholar]
- Koller, E, Vincent, TM, Chappell, A, De, S, Manoharan, M and Bennett, CF (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39: 4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary, RS, Yu, RZ and Levin, AA (2001). Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr Opin Investig Drugs 2: 562–573. [PubMed] [Google Scholar]
- Erickson, MA, Niehoff, ML, Farr, SA, Morley, JE, Dillman, LA, Lynch, KM et al. (2012). Peripheral administration of antisense oligonucleotides targeting the amyloid-β protein precursor reverses AβPP and LRP-1 overexpression in the aged SAMP8 mouse brain. J Alzheimers Dis 28: 951–960. [DOI] [PubMed] [Google Scholar]
- Banks, WA (2011). Measurement of phosphorothioate oligodeoxynucleotide antisense transport across the blood-brain barrier. Methods Mol Biol 789: 337–342. [DOI] [PubMed] [Google Scholar]
- Banks, WA, Farr, SA, Butt, W, Kumar, VB, Franko, MW and Morley, JE (2001). Delivery across the blood-brain barrier of antisense directed against amyloid beta: reversal of learning and memory deficits in mice overexpressing amyloid precursor protein. J Pharmacol Exp Ther 297: 1113–1121. [PubMed] [Google Scholar]
- Evers, MM, Toonen, LJ and van Roon-Mom, WM (2015). Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev 87: 90–103. [DOI] [PubMed] [Google Scholar]
- Sands, H, Gorey-Feret, LJ, Cocuzza, AJ, Hobbs, FW, Chidester, D and Trainor, GL (1994). Biodistribution and metabolism of internally 3H-labeled oligonucleotides. I. Comparison of a phosphodiester and a phosphorothioate. Mol Pharmacol 45: 932–943. [PubMed] [Google Scholar]
- Kole, R, Krainer, AR and Altman, S (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, S, Baker, A, Møller, S and Nielsen, BS (2010). Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 52: 375–381. [DOI] [PubMed] [Google Scholar]
- Nielsen, BS, Jørgensen, S, Fog, JU, Søkilde, R, Christensen, IJ, Hansen, U et al. (2011). High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 28: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, BS, Møller, T and Holmstrøm, K (2014). Chromogen detection of microRNA in frozen clinical tissue samples using LNA™ probe technology. Methods Mol Biol 1211: 77–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.