ABSTRACT
A/J and 129P3/J mice strains have been widely studied over the last few years because they respond quite differently to fluoride (F) exposure. 129P3/J mice are remarkably resistant to the development of dental fluorosis, despite excreting less F in urine and having higher circulating F levels. These two strains also present different characteristics regardless of F exposure.
Objective
In this study, we investigated the differential pattern of protein expression in the liver of these mice to provide insights on why they have different responses to F.
Material and Methods
Weanling male A/J and 129P3/J mice (n=10 from each strain) were pared and housed in metabolic cages with ad libitum access to low-F food and deionized water for 42 days. Liver proteome profiles were examined using nLC-MS/MS. Protein function was classified by GO biological process (Cluego v2.0.7 + Clupedia v1.0.8) and protein-protein interaction network was constructed (PSICQUIC, Cytoscape).
Results
Most proteins with fold change were increased in A/J mice. The functional category with the highest percentage of altered genes was oxidation-reduction process (20%). Subnetwork analysis revealed that proteins with fold change interacted with Disks large homolog 4 and Calcium-activated potassium channel subunit alpha-1. A/J mice had an increase in proteins related to energy flux and oxidative stress.
Conclusion
This could be a possible explanation for the high susceptibility of these mice to the effects of F, since the exposure also induces oxidative stress.
Keywords: Proteomics, Fluorides, Liver, Oxidative stress
INTRODUCTION
A/J and 129P3/J mice strains have been widely studied over the last few years because they respond quite differently to fluoride (F) exposure. When given the same dose of F, the A/J strain responds with a rapid onset and severe development of dental fluorosis, while the 129P3/J strain develops minimal fluorosis 8 . This was believed to be a consequence of the faster excretion of F by the 129P3/J strain. Surprisingly, a metabolic study showed that the 129P3/J mice excrete less F in urine, have higher circulating F levels and, consequently, higher bone F levels, however, they still are remarkably resistant to the development of dental fluorosis 5 .
Some differences between these strains are intrinsic to themselves and do not depend on the F exposure. For example, the A/J mice drink significantly higher volumes of water than their 129P3/J counterparts 4 , which can be explained by the increased expression of Alpha-aminoadipic semialdehyde dehydrogenase in the kidney of 129P3/J mice, regardless of F exposure. This enzyme metabolyzes irreversibly betaine aldehyde to betaine that is the most effective osmoprotectant accumulated by eukaryotic organisms to cope with osmotic stress 4 . In addition, exclusive proteins expressed in the kidney of A/J or 129P3/J mice exhibited the same profile, regardless of F exposure. This suggests that the genetic background per se accounts for such differences between these two strains of mice.
Liver represents the main detoxifying tissue in the body by processing, neutralizing, and eliminating toxins from the digestive tract through hepatocyte-mediated enzymatic detoxification systems. Due to these important functions, liver is one of the body’s organs most subject to injury. Thus, it is believed that the differential pattern of protein expression in the liver of A/J and 129P3/J mice can provide new insights that could explain why they respond differently when exposed to F. To achieve this, state-of-the-art shotgun proteomics combined to bioinformatics approaches were used.
MATERIAL AND METHODS
Animals and samples collection
Weanling male mice from the A/J and 129P3/J inbred strains (3-week-old; n=10 from each strain) were pared and housed in metabolic cages with ad libitum access to low-F food (AIN76A, PMI Nutrition, Richmond, IN, USA, 0.95 mg/Kg F) and deionized water for 42 days. The temperature and humidity in the climate-controlled room, which had a 12 h light/dark cycle, were 23±1°C and 40%-80%, respectively. All experimental protocols were approved by the Ethics Committee for Animal Experiments of Bauru School of Dentistry, University of São Paulo (Protocol # 031/2013). At the end of the study, the mice were anesthetized with ketamine/xylazine and livers were collected. Samples designated for proteomic analysis were stored at -80°C, while those designated for F analysis were stored at -20°C.
Fluoride analysis in liver
Fluoride analysis was done with the ion-sensitive electrode, after hexamethyldisiloxane-facilitated diffusion 22 , exactly as previously described 20 .
Statistical analysis
For liver F concentration, the GraphPad InStat software version 4.0 for Windows (GraphPad software Inc., La Jolla, California USA) was used. Data were analyzed by unpaired t test (p<0.05).
Sample preparation for proteomic analysis
Samples were prepared for analysis as previously described 17 . The frozen tissue was homogenized in a cryogenic mill (model 6770, Spex, Metuchen, NJ, EUA). For protein extraction, liver homogenate was incubated in lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 1% IPG buffer pH 3-10, 40 mM DTT for 1 h at 4°C with occasional shaking. After this period, the homogenate was centrifuged at 15,000 rpm for 30 min at 4°C and the supernatant containing soluble proteins was recovered. The proteins were precipitated using the kit PlusOne 2D Cleanup (GE Healthcare, Uppsala, Sweden), as recommended by the manufacturer. Pellets were resuspended in rehydration buffer (7 M urea, 2 M thiourea, 0.5% CHAPS, 0.5% IPG buffer pH 3–10, 18 mM DTT, 0.002% bromophenol blue). Twenty-five μL of liver proteins from each animal of the same group were combined to constitute a pool that was centrifuged for clarification. To each pool, 50 mM AMBIC, containing 3 M urea, were added. Each sample was filtered twice in 3 kDa AMICON (Millipore, St Charles, MO, USA). Protein quantification was measured in the pooled samples by Bradford protein assay 3 . To each sample (50 µg of total protein for each pool in a volume of 50 µL), 10 µL of 50 mM AMBIC were added. In sequence, 25 µL of 0.2% RapiGEST™ (Waters Co., Manchester, UK) were added and incubated at 80°C for 15 min. Following, 2.5 µL of 100 mM DTT were added and incubated at 60°C for 30 min. Also, 2.5 µL of 300 mM IAA were added and incubated for 30 min at room temperature (under dark). Then, 10 µL of trypsin (100 ng; Trypsin Gold Mass Spectrometry, Promega, Madison, USA) were added and digestion occurred for 14 h at 37°C. After digestion, 10 µl of 5 % TFA were added, incubated for 90 min at 37°C and the sample was centrifuged (14,000 rpm for 30 min). The supernatant was collected and 5 µL of ADH (1 pmol/µL) plus 85 µL 3% ACN were added.
LC-MS/MS and bioinformatics analyses
Separation and identification of peptides were performed on a nanoAcquity UPLC-Xevo QTof MS system (Waters, Manchester, UK), exactly as previously described 15 . Difference in expression among the groups was obtained using PLGS software and expressed as p<0.05 for down-regulated proteins 1-p>0.95 for up-regulated proteins (Table 1). Bioinformatics analysis was performed, as reported earlier 1 , 15 , 17 - 19 . Briefly, Uniprot protein ID accession numbers were mapped back to their associated encoding Uniprot gene entries for the comparison A/J X 129P3/J. Gene Ontology annotation of Broad Biological Process was performed using Cluego v2.0.7 + Clupedia v1.0.8, a Cytoscape plugin. Uniprot IDs were uploaded to Table 1 and analyzed with default parameters, which specify a Enrichment (right-sided hypergeometric test) correction method using Bonferroni step down, analysis mode “Function” and load gene cluster list for Mus musculus, Evidence Codes “All”, set networking specificity “medium” (GO levels 3 to 8) and KappaScoreThreshold 0.03. The protein-protein interaction network was downloaded from PSICQUIC, built in Cytoscape version 3.0.2 and constructed as proposed by Millan 18 (2013). A network was then created, providing global view of potentially relevant interacting partners of proteins whose abundances change.
Table 1. Identified proteins with expression significantly altered in the liver of mice of group A/J control vs. 129 control (0 ppm F).
| Foldchange | |||||
|---|---|---|---|---|---|
| aAccess | Gene | Protein name description | PLGS score | A/J 0 ppm | 129P3/ J 0 ppm |
| Number | name | ||||
| Q921H8 | Acaa1a | 3-ketoacyl-CoA thiolase A, peroxisomal | 195.3 | 1.65 | -1.65 |
| Q8VCH0 | Acaa1b | 3-ketoacyl-CoA thiolase B, peroxisomal | 195.3 | 1.70 | -1.70 |
| Q8BWT1 | Acaa2 | 3-ketoacyl-CoA thiolase, mitochondrial | 189.2 | 1,42 | -1,42 |
| P63038 | Hspd1 | 60 kDa heat shock protein, mitochondrial | 153.6 | 1.55 | -1.55 |
| P20029 | Hspa5 | 78 kDa glucose-regulatedprotein | 254.4 | 1.43 | -1.43 |
| P68033 | Actc1 | Actin, alpha cardiacmuscle 1 | 630.1 | 1.28 | -1.28 |
| P68134 | Acta1 | Actin, alpha skeletalmuscle | 630.1 | 1.28 | -1.28 |
| P62737 | Acta2 | Actin, aorticsmoothmuscle | 60.2 | 1.35 | -1.35 |
| P60710 | Actb | Actin, cytoplasmic 1 | 62.4 | 1.25 | -1.25 |
| P63260 | Actg1 | Actin, cytoplasmic 2 | 62.4 | 1.26 | -1.26 |
| P63268 | Actg2 | Actin, gamma-enteric smooth muscle | 60.2 | 1.34 | -1.34 |
| P47738 | Aldh2 | Aldehydedehydrogenase, mitochondrial | 72.6 | 1.67 | -1.67 |
| P17182 | Eno1 | Alpha-enolase OS=Mus musculus | 129.4 | 1.46 | -1.46 |
| P16460 | Ass1 | Argininosuccinatesynthase | 58.6 | 1.28 | -1.28 |
| P05202 | Got2 | Aspartateaminotransferase, mitochondrial | 79.3 | 1.34 | -1.34 |
| Q03265 | Atp5a1 | ATP synthase subunit alpha, mitochondrial | 74.7 | 1.43 | -1.43 |
| P56480 | Atp5b | ATP synthasesubunit beta, mitochondrial | 138.6 | 1.35 | -1.35 |
| O35490 | Bhmt | Betaine--homocysteine S-methyltransferase 1 | 40.6 | 1.23 | -1.23 |
| Q8C196 | Cps1 | Carbamoyl-phosphate synthase [ammonia], mitochondrial | 269.2 | 1.39 | -1.39 |
| Q63880 | Ces3a | Carboxylesterase 3A | 336.9 | 1.46 | -1.46 |
| Q8VCU1 | Ces3b | Carboxylesterase 3B | 139.1 | 1.65 | -1.65 |
| P24270 | Cat | Catalase | 260.8 | 1.62 | -1.62 |
| Q8R0Y6 | Aldh1l1 | Cytosolic 10-formyltetrahydrofolate dehydrogenase | 53.1 | 1.55 | -1.55 |
| Q9DCW4 | Etfb | Electron transfer flavoprotein subunit beta | 174.4 | 1.48 | -1.48 |
| P10126 | Eef1a1 | Elongationfactor 1-alpha 1 | 245.5 | 1.39 | -1.39 |
| P70694 | Akr1c6 | Estradiol 17 beta-dehydrogenase 5 | 207.5 | 1.48 | -1.48 |
| Q91XD4 | Ftcd | Formimidoyltransferase-cyclodeaminase | 121.1 | 3.82 | -3.82 |
| Q91Y97 | Aldob | Fructose-bisphosphatealdolase B | 96.1 | 1.62 | -1.62 |
| P35505 | Fah | Fumarylacetoacetase | 136.0 | 1.46 | -1.46 |
| P26443 | Glud1 | Glutamatedehydrogenase 1, mitochondrial | 467.9 | 1.84 | -1.84 |
| P10649 | Gstm1 | Glutathione S-transferase Mu 1 | 129.1 | 1.26 | -1.26 |
| P15626 | Gstm2 | Glutathione S-transferase Mu 2 | 109.8 | 1.32 | -1.32 |
| P48774 | Gstm5 | Glutathione S-transferase Mu 5 | 109.8 | 1.32 | -1.32 |
| P19157 | Gstp1 | Glutathione S-transferase P 1 | 317.2 | -0.66 | 0.66 |
| P63017 | Hspa8 | Heat shock cognate 71 kDa protein | 275.2 | 1.36 | -1.36 |
| P01942 | Hba | Hemoglobinsubunit alpha | 1252.1 | -0.85 | 0.85 |
| P02104 | Hbb-y | Hemoglobinsubunit epsilon-Y2 | 854.2 | -0.48 | 0.48 |
| Q8CGP6 | Hist1h2ah | Histone H2A type 1-H | 193.0 | 1.22 | -1.22 |
| Q64522 | Hist2h2ab | Histone H2A type 2-B | 241.3 | 1.51 | -1.51 |
| P62806 | Hist1h4a | Histone H4 | 88.1 | 1.54 | -1.54 |
| P54869 | Hmgcs2 | Hydroxymethylglutaryl-CoAsynthase, mitochondrial | 292.1 | 1.22 | -1.22 |
| P11588 | Mup1 | Major urinaryprotein 1 | 815.0 | -0.53 | 0.53 |
| B5X0G2 | Mup17 | Major urinaryprotein 17 | 824.6 | -0.54 | 0.54 |
| P11589 | Mup2 | Major urinaryprotein 2 | 815.0 | -0.54 | 0.54 |
| P11591 | Mup5 | Major urinaryprotein 5 | 389.7 | -0.57 | 0.57 |
| P02762 | Mup6 | Major urinaryprotein 6 | 815.0 | -0.53 | 0.53 |
| P04938 | Mup8 | Major urinary proteins 11 and 8 (Fragment) | 815.0 | -0.54 | 0.54 |
| P08249 | Mdh2 | Malatedehydrogenase, mitochondrial | 247.9 | 1.45 | -1.45 |
| Q64374 | Rgn | Regucalcin | 107.2 | 1.36 | -1.36 |
| P24549 | Aldh1a1 | Retinaldehydrogenase 1 | 208.9 | 1.49 | -1.49 |
| P07724 | Alb | Serumalbumin | 108.5 | 1.34 | -1.34 |
| P00329 | Adh1 | Alcoholdehydrogenase 1 | 163.3 | + | - |
| Q61234 | Snta1 | Alpha-1-syntrophin | 77.6 | + | - |
| Q8VCT3 | Rnpep | Aminopeptidase B | 73.8 | + | - |
| Q9D3D9 | Atp5d | ATP synthasesubunit delta, mitochondrial | 183.6 | + | - |
| Q62210 | Birc2 | Baculoviral IAP repeat-containing protein 2 | 65.9 | + | - |
| Bad | Q61337 | Bcl2 antagonist of cell death | 116.2 | - | + |
| P21550 | Eno3 | Beta-enolase | 161.0 | + | - |
| P34914 | Ephx2 | Bifunctionalepoxidehydrolase 2 | 441.9 | + | - |
| Q8R1G2 | Cmbl | Carboxymethylenebutenolidasehomolog | 73.2 | + | - |
| Q61686 | Cbx5 | Chromoboxproteinhomolog 5 | 96.9 | + | - |
| Q3V079 | Ccdc176 | Coiled-coil domain-containing protein 176 | 66.5 | + | - |
| P50172 | Hsd11b1 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | 100.4 | + | - |
| Cth | Q8VCN5 | Cystathioninegamma-lyase | 100.5 | - | + |
| P48771 | Cox7a2 | Cytochrome c oxidase subunit 7A2, mitochondrial | 185.6 | + | - |
| P10518 | Alad | Delta-aminolevulinicaciddehydratase | 316.8 | + | - |
| Q9DBT9 | Dmgdh | Dimethylglycinedehydrogenase, mitochondrial | 89.4 | + | - |
| Q99LC5 | Etfa | Electron transfer flavoprotein subunit alpha, mitochondrial | 77.6 | + | - |
| Q9ER73 | Elp4 | Elongatorcomplexprotein 4 | 103.4 | + | - |
| P63242 | Eif5a | Eukaryotic translation initiation factor 5A-1 | 104.8 | + | - |
| Q9QXD6 | Fbp1 | Fructose-1,6-bisphosphatase 1 | 154.4 | + | - |
| P17183 | Eno2 | Gamma-enolase | 159.3 | + | - |
| Q3UHD2 | Gfod1 | Glucose-fructose oxidoreductase domain-containing protein 1 | 83.6 | + | - |
| P11352 | Gpx1 | Glutathioneperoxidase 1 | 419.0 | + | - |
| P24472 | Gsta4 | Glutathione S-transferase A4 | 127.0 | + | - |
| Q9QYE6 | Golga5 | Golginsubfamily A member 5 | 103.4 | + | - |
| P07901 | Hsp90aa1 | Heat shock protein HSP 90-alpha | 67.4 | + | - |
| P11499 | Hsp90ab1 | Heat shock protein HSP 90-beta | 107.9 | + | - |
| P68433 | Hist1h3a | Histone H3.1 | 163.6 | + | - |
| P84228 | Hist1h3b | Histone H3.2 | 163.6 | + | - |
| P84244 | H3f3a | Histone H3.3 | 163.6 | + | - |
| P02301 | H3f3c | Histone H3.3C | 163.6 | + | - |
| Hgd | O09173 | Homogentisate 1,2-dioxygenase | 95.6 | - | + |
| Hadh | Q61425 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 183.9 | - | + |
| Q5U5V2 | Hykk | Hydroxylysinekinase | 78.0 | + | - |
| Q8BLR9 | Hif1an | Hypoxia-induciblefactor 1-alpha inhibitor | 96.3 | + | - |
| O88844 | Idh1 | Isocitratedehydrogenase [NADP] cytoplasmic | 69.5 | + | - |
| Q9CPU0 | Glo1 | Lactoylglutathionelyase | 203.5 | + | - |
| P06151 | Ldha | L-lactatedehydrogenase A chain | 153.0 | + | - |
| Acsl1 | P41216 | Long-chain-fatty-acid--CoA ligase 1 | 48.0 | - | + |
| Q9DB40 | Med27 | Mediator of RNA polymerase II transcription subunit 27 | 68.9 | + | - |
| Q8BPT6 | Immp2l | Mitochondrial inner membrane protease subunit 2 | 65.7 | + | - |
| Myef2 | Q8C854 | Myelinexpressionfactor 2 | 44.9 | - | + |
| Q9DC69 | Ndufa9 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplexsubunit 9, mitochondrial | 79.2 | + | - |
| Ncoa5 | Q91W39 | Nuclear receptor coactivator 5 | 67.7 | - | + |
| P11725 | Otc | Ornithinecarbamoyltransferase, mitochondrial | 217.0 | + | - |
| O08807 | Prdx4 | Peroxiredoxin-4 | 391.3 | + | - |
| Prdx5 | P99029 | Peroxiredoxin-5, mitochondrial | 174.7 | - | + |
| O08709 | Prdx6 | Peroxiredoxin-6 | 321.1 | + | - |
| P09411 | Pgk1 | Phosphoglyceratekinase 1 | 106.8 | + | - |
| Pgap2 | Q3TQR0 | Post-GPI attachment to proteins factor 2 | 60.0 | - | + |
| Prdm12 | A2AJ77 | PR domainzincfingerprotein 12 | 43.7 | - | + |
| Q80U40 | Rimbp2 | RIMS-bindingprotein 2 | 74.3 | + | - |
| B2RY56 | Rbm25 | RNA-bindingprotein 25 | 80.8 | + | - |
| Q91X83 | Mat1a | S-adenosylmethionine synthase isoform type-1 | 177.4 | + | - |
| Q99J08 | Sec14l2 | SEC14-like protein 2 | 106.4 | + | - |
| P47758 | Srprb | Signal recognition particle receptor subunit beta | 68.7 | + | - |
| Hspa9 | P38647 | Stress-70 protein, mitochondrial | 119.8 | - | + |
| Q8K2B3 | Sdha | Succinatedehydrogenase [ubiquinone] flavoproteinsubunit, mitochondrial | 74.3 | + | - |
| Q62264 | Thrsp | Thyroid hormone-inducible hepatic protein | 180.0 | + | - |
| P97360 | Etv6 | Transcriptionfactor ETV6 | 64.7 | + | - |
| Tmem42 | Q9CR22 | Transmembraneprotein 42 | 110.6 | - | + |
| Tpi1 | P17751 | Triosephosphateisomerase | 149.7 | - | + |
| Q9D6F9 | Tubb4a | Tubulin beta-4A chain | 101.3 | + | - |
| P68372 | Tubb4b | Tubulin beta-4B chain | 109.0 | + | - |
| Ube2w | Q8VDW4 | Ubiquitin-conjugatingenzyme E2 W | 102.0 | - | + |
| Q5QNV8 | Heatr9 | Uncharacterizedprotein C17orf66 homolog | 91.1 | + | - |
| N/A | Q8C4X7 | UPF0258 protein KIAA1024-like homolog | 38.4 | - | + |
| P25688 | Uox | Uricase | 92.7 | + | - |
The identified proteins are organized according to alphabetical order. Relative differential is indicated by + sign, when the protein is up-regulated and by - sign, when the protein is down-regulated in the respective comparison. aIdentification is based on protein ID from UniProt protein database (http://www.uniprot.org/)
RESULTS
Liver F analysis
Mean±SD liver F concentrations found in 129P3/J mice (0.022±0.003 µg/g) were significantly higher than those found in A/J mice (0.015±0.002 µg/g) (t=4.929, p=0.0006).
Liver proteome profile and identification of differentially expressed proteins
Table 1 shows proteins with expression changes in A/J and 129P3/J mice. In general, most proteins with fold change were increased in A/J mice.
Gene ontology annotation
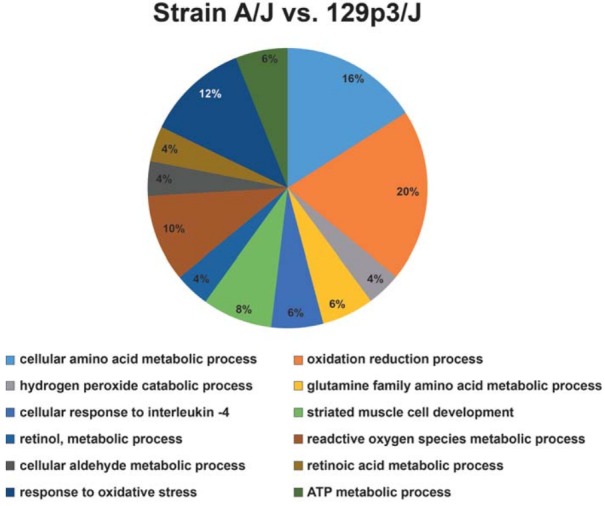
Figure 1 shows the functional classification according to the biological process with the most significant term. Twelve categories were observed. Among them, the category with the highest percentage of genes was oxidation-reduction process (20%), followed by cellular amino acid metabolic process (16%) and response to oxidative stress (12%).
Figure 1. Functional distribution of proteins identified with differential expression in liver of mice belonging to A/J vs. 129p3/J strains. Categories of proteins based on GO annotation Biological Process. Terms significant (Kappa=0.03) and distribution according to percentage of number of genes association.
Protein-protein interaction network
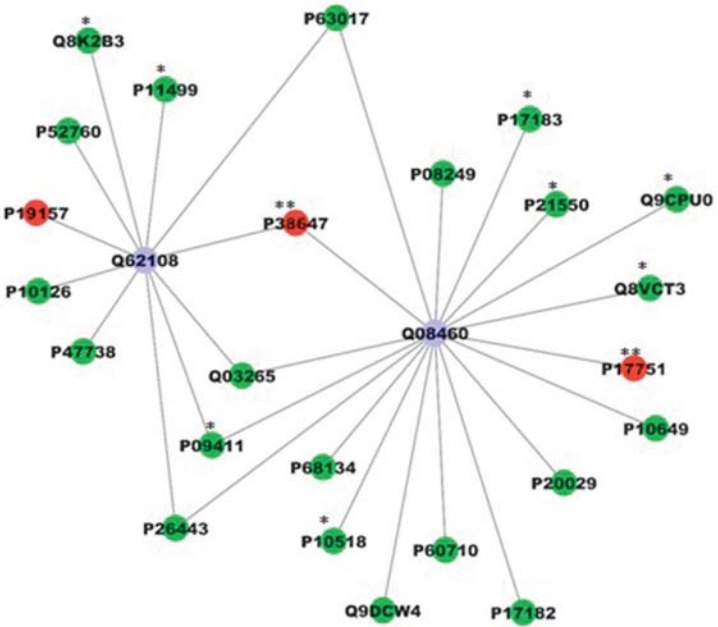
For the comparison displayed above, a network was created; employing all the interactions found in the search conducted using PSICQUIC. After the global network was created, nodes and edges were filtered using the specification for Mus musculus taxonomy (10090). The value of fold change and also the p-value were added in new columns. The ActiveModules 1.8 plug-in to Cytoscape was used to make active modules connected subnetworks within the molecular interaction network whose genes presented significant coordinated changes in fold changes and p-value, as shown in the original proteomic analysis. Figure 2 shows the subnetwork generated by VizMapper. As can be seen, most proteins with fold change present interaction with Disks large homolog 4 (Q62108; 11 proteins) and Calcium-activated potassium channel subunit alpha-1 (Q08460; 18 proteins).
Figure 2. Subnetworks generated by VizMapper for each comparison – A Group A/J vs. 129p3/J. Color of node and * indicate the differential expression of the respective protein, for each comparison. Red and green nodes indicate protein down-regulation and up-regulation, respectively, while * and ** indicate presence and absence of protein, respectively, in the respective group. Purple node indicates proteins presenting interaction but that were not identified in the present study. The access numbers in nodes correspond to: P68134- (Acta1)Actin, alpha skeletal muscle; P10518- (Alad) Delta-aminolevulinic acid dehydratase; Q9DCW4- (Etfb) Electron transfer flavoprotein subunit beta; P60710- (Actb) Actin, cytoplasmic 1; P17182- (Eno1) Alpha-enolase; P20029- (Hspa5) 78 kDa glucose-regulated protein; P10649- (Gstm1) Glutathione S-transferase Mu 1; P17751- (Tpi1) Triosephosphate isomerase; Q8VCT3- (Rnpep) Aminopeptidase B; Q9CPU0- (Glo1) Lactoylglutathionelyase; P21550- (Eno3) Beta-enolase; P17183- (Eno2) Gamma-enolase; P08249- (Mdh2) Malate dehydrogenase; P63017- (Hspa8) Heat shock cognate; P38647- (Hspa9) Stress-70 protein; Q03265- (Atp5a1) ATP synthase subunit alpha; P09411- (Pgk1) Phosphoglycerate kinase 1; P26443- (Glud1) Glutamate dehydrogenase 1; P47738- (Aldh2) Aldehyde dehydrogenase; P10126- (Eef1a1) Elongation factor 1-alpha 1; P19157- (Gstp1) Glutathione S-transferase P 1; P52760- (Hrsp12) Ribonuclease; Q8K2B3- (Sdha) Succinate dehydrogenase; P11499- (Hsp90ab1) Heat shock protein; Q62108- (Dlg4) Disks large homolog 4; Q08460- (Kcnma1) Calcium-activated potassium channel subunit alpha-1.
DISCUSSION
129P3/J mice interestingly have been reported to excrete less F and as consequence to have higher circulating F levels, bone and enamel F levels and they still are remarkably resistant to the development of dental bfluorosis 5 , 7 - 8 , 12 . In this study, even without administration of F through the drinking water and with consumption of a low-F diet, 129P3/J mice had significantly higher liver F concentrations, which might have been due to the residual amounts of F present in their diets and is in-line with the metabolic characteristics of this strain regarding F 4 - 5 .
In this study, proteomic analysis of liver of 129P3/J and A/J mice was employed to provide insights into the possible mechanisms that could explain the differential metabolic handling and effects of F in these two strains. It has been shown that even without exposure to F, A/J mice present a higher retention of proteins in the maturing enamel 9 . For this reason, the mice were not treated with F, because we wanted to see differences in the liver proteome profile that were intrinsic to the strains. Most proteins with fold change were increased in the A/J mice (Table 1), with fold changes ranging between 1 and 2. Formimidoyltransferase-cyclodeaminase, however, was increased 3.82 times in A/J mice. This enzyme is a liver-specific antigen recognized by sera of patients with autoimmune hepatitis 14 and is found down-regulated in hepatocellular carcinoma 16 . Formimidoyltransferase-cyclodeaminase has two enzymatic functions. In one of them, formiminotetrahydrofolate and glutamate are produced. Through its cyclodeaminase function, the enzyme breaks down formiminotetrahydrofolate, involved in the synthesis of purines and pyrimidines, and amino acids (UNIPROT). Thus, the increase in this enzyme might explain the increased expression of other liver proteins in A/J mice due to higher supply of nucleotides and amino acids.
Remarkably, the functional category with the highest percentage of altered genes was oxidation-reduction process. The increase of proteins such as ATP synthase subunit alpha, mitochondrial, Heat shock cognate 71 kDa protein, Electron transfer flavoprotein subunit beta, Alpha-enolase, Beta-enolase, Gamma-enolase and, Malate dehydrogenase in the A/J mice indicate an increased energy flux in this strain, which might generate oxidative stress. This can be confirmed by the concomitant increase in GRP78, which suggests endoplasmic reticulum (ER) stress 20 . ER stress occurs when nascent proteins are misfolded or not folded properly, leading to the initiation of the unfolded protein response, as the unfolded proteins accumulate in the ER 13 . It has been demonstrated that F is able to induce an ER stress response in the LS8 ameloblast-derived cell line, which could be implicated in the pathogenesis of dental fluorosis 13 . In addition, administration of F through the drinking water is able to increase the expression of GRP78 in the liver of rats 20 . Thus, considering that A/J mice present an increased energy flux and tendency to oxidative stress even without exposure to F, this exposure has been shown to worsen oxidative stress 20 , which can implicate in the pathogenesis of dental fluorosis 8 , this can be a hypothesis for the high susceptibility of the A/J to the effects of F.
The proteins in the center of the protein-protein interaction network are related to potassium channels. One of them (calcium-activated potassium channel subunit alpha-1) is a potassium channel activated either by membrane depolarization or increase in cytosolic Ca2+ that mediates export of K+. It is also activated by the concentration of cytosolic Mg2+. Its activation dampens the excitatory events that elevate the cytosolic Ca2+ concentration and/or depolarize the cell membrane. Therefore, it contributes to the repolarization of the membrane potential and plays a key role in controlling excitability in a number of systems, such as regulation of the contraction of smooth muscle 21 , the tuning of hair cells in the cochlea 6 , regulation of transmitter release 6 and innate immunity 2 . The other one is Disks large homolog 4 that is required for synaptic plasticity associated with NMDA (N-methyl-D-aspartate) receptor signaling 11 . It interacts with shaker-type potassium channels and the cytoplasmic tail of NMDA receptor subunits. At first glance, it may seem odd the presence of a protein associated with the nervous system in the center of the network in this study. However, we must consider that liver failure leaves to the accumulation of ammonia, which affects the cerebral function 10 . As mentioned above, A/J mice presented several proteins related to the energy flux increased in the liver, which might have caused oxidative stress and contributed to liver damage, which in turn might have provoked cerebral alterations. Since this was a preliminary exploratory work, future studies comparing the proteomic profile of the brain of these mice strains should be conducted to add new light into this topic. Also, additional studies should be done to quantify, by other techniques, the proteins with changing expression in this study. Despite being an exploratory study, the lack of additional techniques to confirm the proteins with altered expression identified by nLC-MS/MS might be considered a limitation of this study.
CONCLUSIONS
In conclusion, A/J mice had an increase in proteins related to energy flux and oxidative stress. This could be a possible explanation for the high susceptibility of these mice to the effects of F, since F exposure also induces oxidative stress.
ACKNOWLEDGMENTS
The authors thank CNPq/TWAS for granting the scholarship to the first author.
REFERENCES
- 1.Bauer-Mehren A. Integration of genomic information with biological networks using Cytoscape. Methods Mol Biol. 2013;1021:37–61. doi: 10.1007/978-1-62703-450-0_3. [DOI] [PubMed] [Google Scholar]
- 2.Butler A, Tsunoda S, McCobb DP, Wei A, Salkoff L. mSlo, a complex mouse gene encoding "maxi" calcium-activated potassium channels. Science. 1993;261(5118):221–224. doi: 10.1126/science.7687074. [DOI] [PubMed] [Google Scholar]
- 3.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho JG, Leite AL, Peres-Buzalaf C, Salvato F, Labate CA, Everett ET, et al. Renal proteome in mice with different susceptibilities to fluorosis. e53261PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho JG, Leite AL, Yan D, Everett ET, Whitford GM, Buzalaf MA. Influence of genetic background on fluoride metabolism in mice. J Dent Res. 2009;88(11):1054–1058. doi: 10.1177/0022034509347249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabo R, Zichichi R, Viña E, Guerrera MC, Vázquez G, García-Suárez O, et al. Calcium-activated potassium channel SK1 is widely expressed in the peripheral nervous system and sensory organs of adult zebrafish. Neurosci Lett. 2013;555:62–67. doi: 10.1016/j.neulet.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 7.Charone S, Leite AL, Peres-Buzalaf C, Fernandes MS, Almeida LF, Graeff MS, et al. Proteomics of secretory and maturation stage enamel of genetically distinct mice. Caries Res. 2016;50:24–31. doi: 10.1159/000442301. [DOI] [PubMed] [Google Scholar]
- 8.Everett ET, McHenry MA, Reynolds N, Eggertsson H, Sullivan J, Kantmann C, et al. Dental fluorosis: variability among different inbred mouse strains. J Dent Res. 2002;81(11):794–798. doi: 10.1177/0810794. [DOI] [PubMed] [Google Scholar]
- 9.Everett ET, Yan D, Weaver M, Liu L, Foroud T, Martinez-Mier EA. Detection of dental fluorosis-associated quantitative trait Loci on mouse chromosomes 2 and 11. Cells Tissues Organs. 2009;189(1-4):212–218. doi: 10.1159/000151383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felipo V. Hepatic encephalopathy: effects of liver failure on brain function. Nat Rev Neurosci. 2013;14(12):851–858. doi: 10.1038/nrn3587. [DOI] [PubMed] [Google Scholar]
- 11.Halff AW, Gómez-Varela D, John D, Berg DK. A novel mechanism for nicotinic potentiation of glutamatergic synapses. J Neurosci. 2014;34(6):2051–2064. doi: 10.1523/JNEUROSCI.2795-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi CA, Leite AL, Peres-Buzalaf C, Carvalho JG, Whitford GM, Everett ET, et al. Bone response to fluoride exposure is influenced by genetics. e114343PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota K, Lee DH, Tsuchiya M, Young CS, Everett ET, Martinez-Mier EA, et al. Fluoride induces endoplasmic reticulum stress in ameloblasts responsible for dental enamel formation. J Biol Chem. 2005;280(24):23194–23202. doi: 10.1074/jbc.M503288200. [DOI] [PubMed] [Google Scholar]
- 14.Lapierre P, Hajoui O, Homberg JC, Alvarez F. Formiminotransferase cyclodeaminase is an organ-specific autoantigen recognized by sera of patients with autoimmune hepatitis. Gastroenterology. 1999;116:643–649. doi: 10.1016/s0016-5085(99)70186-1. [DOI] [PubMed] [Google Scholar]
- 15.Leite AL, Lobo GV, Pereira HA, Fernandes MS, Martini T, Zucki F, et al. Proteomic analysis of gastrocnemius muscle in rats with streptozotocin-induced diabetes and chronically exposed to fluoride. e106646PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang CR, Leow CK, Neo JC, Tan GS, Lo SL, Lim JW, et al. Proteome analysis of human hepatocellular carcinoma tissues by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5(8):2258–2271. doi: 10.1002/pmic.200401256. [DOI] [PubMed] [Google Scholar]
- 17.Lobo JG, Leite AL, Pereira HA, Fernandes MS, Peres-Buzalaf C, Sumida DH, et al. Low-level fluoride exposure increases insulin sensitivity in experimental diabetes. J Dent Res. 2015;94(7):990–997. doi: 10.1177/0022034515581186. [DOI] [PubMed] [Google Scholar]
- 18.Millan PP. Visualization and analysis of biological networks. Methods Mol Biol. 2013;1021:63–88. doi: 10.1007/978-1-62703-450-0_4. [DOI] [PubMed] [Google Scholar]
- 19.Orchard S. Molecular interaction databases. Proteomics. 2012;12(10):1656–1662. doi: 10.1002/pmic.201100484. [DOI] [PubMed] [Google Scholar]
- 20.Pereira HA, Leite AL, Charone S, Lobo JG, Cestari TM, Peres-Buzalaf C, et al. Proteomic analysis of liver in rats chronically exposed to fluoride. e75343PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0075343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-Pastor E, Andrade F, Sánchez-Pastor JM, Elizalde A, Huerta M, Virgen-Ortiz A, et al. Cannabinoid receptor type 1 activation by arachidonylcyclopropylamide in rat aortic rings causes vasorelaxation involving calcium-activated potassium channel subunit alpha-1 and calcium channel, voltage-dependent, L type, alpha 1C subunit. Eur J Pharmacol. 2014;15(729):100–106. doi: 10.1016/j.ejphar.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Taves DR. Separation of fluoride by rapid diffusion using hexamethyldisiloxane. Talanta. 1968;15(9):969–974. doi: 10.1016/0039-9140(68)80097-9. [DOI] [PubMed] [Google Scholar]