ABSTRACT
Cold acclimation is an important adaptive response of plants from temperate regions to increase their freezing tolerance after being exposed to low nonfreezing temperatures. The three CBF genes are well known to be involved in cold acclimation. As the 3 CBF genes are linked tandemly in the Arabidopsis genome, it is almost impossible to obtain cbf triple mutants using traditional genetic methods. Recently, using the CRISPR/Cas9 technology, we generated cbf single, double, and triple mutants. Our results showed that the cbf triple mutants are extremely sensitive to freezing stress. In addition, the cbf triple mutants are defective in early development and salt tolerance. Interestingly, the cbf1 cbf3 double mutants show increased expression of the CBF2 gene and some downstream cold-responsive genes and display increased freezing tolerance, compared to the wild type, revealing that CBF1 and CBF3 negatively regulate CBF2 expression.
KEYWORDS: CBF genes, cold acclimation, CRISPR/Cas9, freezing tolerance, non-coding RNA, transcriptional regulation
The three CBF genes have been well known to encode AP2/ERF (APETALA2/Ethylene-Responsive Factor) transcription factors involved in cold acclimation.1-3 When plants are exposed to low temperatures, CBF genes are rapidly up-regulated and the encoded proteins promote the transcription of downstream cold-responsive (COR) genes, which in turn increase the freezing tolerance of plants.4-6 The expression of CBF genes is tightly regulated, usually reaching maximal expression levels at 1–2 h, and beginning to decrease at 3 h after low temperature treatment.1 Experimental data have shown that the 3 CBF genes are regulated by 3 upstream transcription factors, ICE1, ICE2 and CAMTA3,7-9 and the CBF proteins also regulate the expression of a number of downstream transcription factors.4,10,11 These results suggest that transcriptional cascades are used by plants to magnify cold responses. Even though the role of CBF genes in cold acclimation had been supported by many studies,12,13 due to the lack of stable loss-of-function of cbf mutants, the extent of their contribution to freezing tolerance and cold acclimation was still not fully understood, and also little was known about the functions of CBF genes in development and other abiotic stress responses.
Recently, by using the newly developed CRISPR/Cas9 technology, we generated 2 independent cbf triple mutants, in each of which a large fragment, including the CDS regions of CBF1 and CBF3, the partial CDS region of CBF2 and the intergenic regions among these 3 genes, was deleted, which leads to the abolishment of the expression of all 3 CBF genes.4 Both electrolyte leakage and survival assays showed that the cbf triple mutants are extremely sensitive to freezing after cold acclimation, indicating that the 3 CBF genes are essential for cold acclimation. By analyzing the RNA-Seq data, we found a novel, transcriptionally active region between the CBF1 and CBF3 genes. Interestingly, the expression of this region was also induced by low temperature treatment (Fig. 1A). By analyzing this novel transcribed sequence using BLASTn, we found that this region has already been annotated as a putative long intergenic non-coding RNA (ncRNA),14 although the biological function of this ncRNA is still unknown. This region was deleted in both cbf123-1 and cbf123-2 mutants. To exclude the possibility that the freezing-hypersensitive phenotype of the cbf triple mutants is caused by the absence of this ncRNA, we generated a third line, named as cbf123-3, in which the whole CDS region of CBF1 was deleted, but only point mutations occurred in the CBF2 and CBF3 genes (Fig. 1B). So in the cbf123-3 mutant, the intergenic regions are still kept intact. Quantitative RT-PCR showed that the expression of this ncRNA was still induced in the cbf123-3 mutant after cold treatment (Fig. 1C), which confirms that this ncRNA was not disrupted in the cbf123-3 mutants. Freezing survival assays showed that, similar to cbf123-1, all seedlings of cbf123-3 were dead after freezing treatment, whereas many of the wild type seedlings survived (Fig. 1D), indicating that the 3 CBF genes, but not the ncRNA, are responsible for the observed difference in freezing tolerance between the triple mutants and wild type. In addition to the freezing-hypersensitive phenotype, our previous study also showed that the cbf triple mutants are defective in early development and salt stress tolerance.4 Further, our data showed unexpectedly that the cbf1 cbf3 double mutants had increased freezing tolerance compared to the wild type after cold acclimation, which was probably due to an increased expression of CBF2 and some of the downstream COR genes.4
Figure 1.
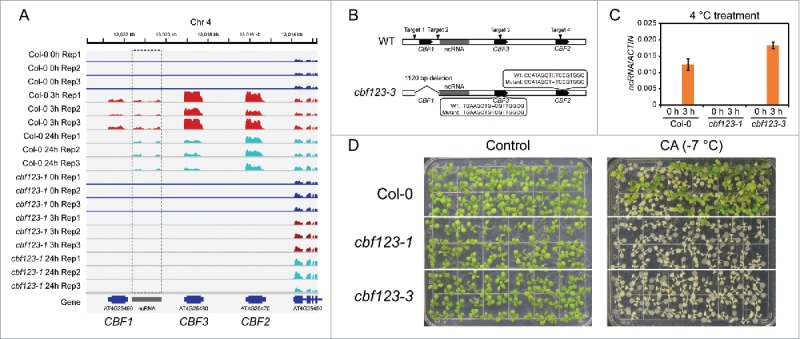
The cbf triple mutants are sensitive to freezing. (A) RNA-Seq data showing the expression of the 3 CBF genes in the wild type and cbf123-1 mutant after low temperature treatment for 0, 3, and 24 h. The dashed box indicates a long intergenic non-coding RNA that was also induced by low temperature treatment. (B) A diagram showing that 4 sgRNAs targets were used in the CRISPR/Cas9 system to generate the cbf123-3 mutant, in which the whole CDS region of CBF1 was deleted and point mutations occurred in both CBF2 and CBF3 genes. The ncRNA was not disrupted in the cbf123-3 mutant. (C) The expression of the ncRNA was examined in the wild type and cbf triple mutants that were treated with low temperature (4°C) for 3 h. Transcript accumulation was assessed by quantitative real-time RT-PCR, and ACTIN8 was used as the internal control. Error bars indicate the standard deviation of 3 biological replicates. (D) Freezing survival assay. Col-0, cbf123-1, and cbf123-3 seedlings were grown on MS medium for 12 d and then transferred to 4°C for cold acclimation. The acclimated plants were subjected to −7°C for 1 h before they were transferred to 23°C for recovery. The photographs show the seedlings before (left) and after (right) freezing treatment.
Analyzing the phenotypes of cbf single, double, and triple mutants in development, freezing tolerance, and other abiotic stresses enabled us to demonstrate the biological functions and the redundancy of the 3 CBF genes. However, there are important questions remain to be answered. Firstly, freezing assays showed that the 3 CBF genes are very important for cold acclimation, but it is also obvious that the cbf triple mutants do not completely lose the ability to acclimate to cold, implying that other components or pathways are also involved in cold acclimation. These other components most likely include the other “first-wave” transcription factors, such as HSFC1, ZAT12 and CZF1.4,10 To confirm this hypothesis in the future, an Arabidopsis line carrying mutations in all 3 CBF genes and other first-wave transcription factors need to be generated to determine whether the other first-wave transcription factors contribute to the remaining part of cold acclimation in the cbf triple mutant. Secondly, the cbf triple mutants were sensitive to salt stress and NaCl-induced expression of RD29A was down-regulated in the cbf triple mutant.4 How CBFs contribute to salt tolerance needs further investigation. Thirdly, a striking result of our earlier study is that the cbf1 cbf3 double mutants showed increased freezing tolerance after cold acclimation compared with the wild type plants. RNA-Seq data showed that CBF2 gene was upregulated in the cbf1 cbf3 double mutants,4 which could be one of the reasons for the increased freezing tolerance of cbf1 cbf3 double mutants. Alternatively, the CBF2 protein may have a higher transcriptional activity than CBF1 and CBF3, so in the cbf1 cbf3 double mutant, only CBF2 protein binds to the promoter regions of COR genes, which leads to the higher expression of some of the COR genes and increased freezing tolerance. Whether CBF2 has a higher transcriptional activity than CBF1 and CBF3 needs experimental evidence. Finally, whether and how the cold-inducible ncRNA may be involved in cold acclimation needs to be investigated.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Institutes of Health Grant R01 GM059138 and the Chinese Academy of Sciences (J.K.Z.).
References
- 1.Medina J, Catala R, Salinas J. The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 2011; 180:3-11; PMID:21421341; http://dx.doi.org/ 10.1016/j.plantsci.2010.06.019 [DOI] [PubMed] [Google Scholar]
- 2.Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 1999; 50:571-99; PMID:15012220; http://dx.doi.org/ 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- 3.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci 2007; 12:444-51; PMID:17855156; http://dx.doi.org/ 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 4. Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol 2016; 171:2744-59; PMID:27252305; http://dx.doi.org/15165189 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 2004; 38:982-93; PMID:15165189; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02100.x [DOI] [PubMed] [Google Scholar]
- 6.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002; 14:1675-90; PMID:12172015; http://dx.doi.org/ 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 2003; 17:1043-54; PMID:12672693; http://dx.doi.org/ 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009; 21:972-84; PMID:19270186; http://dx.doi.org/ 10.1105/tpc.108.063958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YS, Lee M, Lee JH, Lee HJ, Park CM. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol 2015; 89:187-201; PMID:26311645; http://dx.doi.org/ 10.1007/s11103-015-0365-3 [DOI] [PubMed] [Google Scholar]
- 10.Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 2015; 82:193-207; PMID:25736223; http://dx.doi.org/ 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- 11.Zhao C, Lang Z, Zhu JK. Cold responsive gene transcription becomes more complex. Trends Plant Sci 2015; 20:466-8; PMID:26072094; http://dx.doi.org/ 10.1016/j.tplants.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 2000; 124:1854-65; PMID:11115899; http://dx.doi.org/ 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998; 280:104-6; PMID:9525853; http://dx.doi.org/ 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012; 24:4333-45; PMID:23136377; http://dx.doi.org/ 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]


