ABSTRACT
Characterizing the molecular mechanisms governing the response of plant roots and shoots to drought stress could aid the development of strategies aiming to ameliorate drought stress. Small open reading frames (sORFs), putatively encoding small peptides, may play a significant role in the response to different abiotic stresses. Microarray analyses revealed that after 5, 7 and 9 d of a drought treatment, 2, 77, and 104 sORFs were up-regulated in roots, respectively; while the number of upregulated sORFs in shoots was 12, 45, and 158, respectively. RT-qPCR analysis confirmed the up-regulated expression of ATRIKEN29196 and ATRIKEN32280 specifically in roots. The identified upregulated sORFs, particularly those in roots, may contribute to drought stress tolerance.
KEYWORDS: Abiotic stresses, Arabidopsis thaliana, drought, microarray, small open reading frames
Abiotic stresses severely limit crop production and reduce the yield and quality of crops. Drought stress is a serious agricultural constraint that has a significant impact on plant growth and crop production. Water scarcity is predicted to increase in future due to global warming and climate change. Thus, understanding the molecular response of plants to drought stress and utilizing this knowledge to develop various molecular strategies to ameliorate the harmful effects of water deficit is essential to meet the demand for increases in food production.1-6 Plants differentially regulate the expression of specific genes in roots and shoots in response to drought stress, which results in adjustments of osmotic potential and metabolism.7,8 During a progressive drought stress, the transcriptional changes in roots during the early phase of drought stress (3–5 d after the onset of drought stress) are more significant than those that occur in shoots; indicating that roots sense water scarcity much earlier than shoots.8 The genes that are up-regulated in roots during the initial stages of drought stress are related to ABA synthesis and transport, as well as solute transport. Moreover, genes belonging to major facilitator superfamily transporters, multidrug and toxin extrusion efflux transporters, microRNA genes, suberin, pectin and secondary cell wall biosynthesis/modification-related genes, pre-tRNA genes, and various S-adenosyl-L-methionine (SAM) dependent transferases are also upregulated in roots.8
Despite recent advances in genome and RNA sequencing, it is still a challenge to identify genes encoding small proteins (less than 100 amino acids). As a result, the contribution of small open reading frames (sORF) to the development and response to abiotic and biotic stresses has not been investigated in much detail.9,10 Greater than 8,000 sORFs have been reported in Arabidopsis11,12 and many of these have been reported to be associated with morphogenesis.12 Although some of the sORFs are present as antisense to the Arabidopsis Genome Initiative (AGI)-annotated genes (4.78%), the majority of these sORFs lie in intergenic regions. Several small peptides putatively encoded by sORFs have also been identified through ribosomal sequencing.13,14 Although transcriptional changes in response to drought stress have been well documented,8,15,16 changes in the expression of sORFs in roots and shoots of Arabidopsis plants have not been investigated. We recently developed a system in which plants are grown in a ceramics-based granular soil. The characteristics of this substrate allows root and shoot samples to be easily sampled independently, which in turn enables a separate dissection of the root and shoot transcriptome. Here we report that, similar to other reported genes, Arabidopsis sORFs are also differentially regulated in roots and shoots of Arabidopsis plants grown in a ceramics-based granular soil in response to a progressive drought stress. It is plausible that an increased understanding of the role of these sORFs could provide the opportunity to develop novel approaches for the development of drought-stress tolerant crop plants.
A customized microarray analysis was used to monitor the expression of sORFs that respond to a drought stress.8 The experimental conditions and the microarray data have been previously described, and are available through GenBank under accession number GSE76827.8 Briefly, seeds of Arabidopsis thaliana (Col-0 ecotype) were grown on MS medium for 9 d and were then transferred to a ceramics-based, granular soil (size 2.5 L, Sakatanotane, Japan) and grown for 8 additional days at 22°C (16 h light/8 h dark cycle, 60 µmol m−2 s−1 photon flux density). The plants were then subjected to drought stress by removing excess water and subsequently ceasing to water the plants. Roots and shoots were separately harvested at 0, 1, 3, 5, 7, and 9 d after the onset of the drought treatment. Three biological replicates were collected at each time point. RNA was extracted with a mirVana™ miRNA Isolation Kit (Ambion, USA) as previously described.8 This kit is specifically designed to extract small RNAs from tissue and cells using a glass fiber filter (GFF)-based method, and is therefore also suitable for analyzing the expression of sORFs. A student's t-test (p-value) was performed on normalized microarray data and the Benjamini and Hochberg False Discovery Rate (FDR; q-value) procedure was used to control the certainty level.17 sORFs with at least a 2-fold change in expression and having a q-value <0.1 were considered to be differentially expressed.
Due to its large pore size, the water holding capacity of the ceramics-based, granular soil is poor as compared to normal soil; thus the drought stress progressed more rapidly than it would in normal soil. The water retention capacity of the soil was 119% at day 0, after which it dropped rapidly during the early stages of the drought stress. By day 3, the water content had decreased to 73% (Fig. 1A). The water content of ceramics-based, granular soil dropped by 46% during the first 3 d of drought stress. By day 5 of drought stress, the water content continued to drop and reached 44%. By day 7 and 9, the soil water content of the ceramics-based, granular soil was 16% and 4%, respectively. These data clearly indicate that the drought progressed very rapidly in the ceramics-based, granular soil, as compared to normal soil conditions which have a greater water retention capacity. A water content around 38% and 15% has been reported on the 8th and 13th day of drought stress in normal soil, respectively.18 Root and shoot fresh weight increased up to the fifth day of the drought stress, and then began to drop by the seventh day (Fig. 1B). The initial increase in root and shoot fresh weight indicated that plants were actively growing, despite sensing the change in water availability during the early stages of the drought stress, up to the point where soil water content was approximately 44%.
Figure 1.
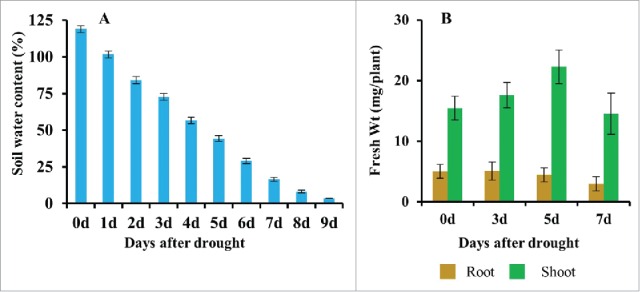
Soil water content and root and shoot fresh weight during a progressive drought stress treatment. (A): Percent soil water content, (B): Root and shoot fresh weight. Data represent the mean ± sd. n = 8.
Microarray data indicated that the expression of many sORFs was impacted in roots and shoots in response to a progressive drought stress (Fig. 2). The effect on the expression of these sORFs was slightly later than in AGI annotated genes. By day 3 of the drought stress, several drought-stress related genes, including RD29A, RD29B, PP2C1, PP2C2, PP2C3, and ABA biosynthesis-related genes, were up-regulated,8 while no change in the expression of the sORFs was observed in roots or shoots on day 1 or day 3 of the drought stress treatment. By day 5, 7, and 9 of the drought stress treatment, the number of upregulated sORFs in roots progressed from 2 to 77, and 104, respectively (Fig. 2). The two sORFs that were up-regulated on day 5 of the drought stress were also upregulated on day 7 and 9. In total, 61 sORFs were up-regulated in roots in response to the drought stress treatment at day 7 and day 9 (Fig. 2B). A total of 12, 45, and 158 sORFs were upregulated in shoots in response to the drought stress treatment on day 5, 7, and 9, respectively (Fig. 2). Among the 12 sORFs that were up-regulated at day 5, the expression of 11 of these sORFs was also upregulated at day 7 and 9 (Fig. 2B). A total of 41 sORFs were up-regulated in shoots at day 7 and day 9 of the drought stress treatment. Collectively, 120 and 162 sORFs were upregulated in roots and shoots, respectively (Fig. S1). Among these, 64 sORFs were up-regulated in both roots and shoots.
Figure 2.
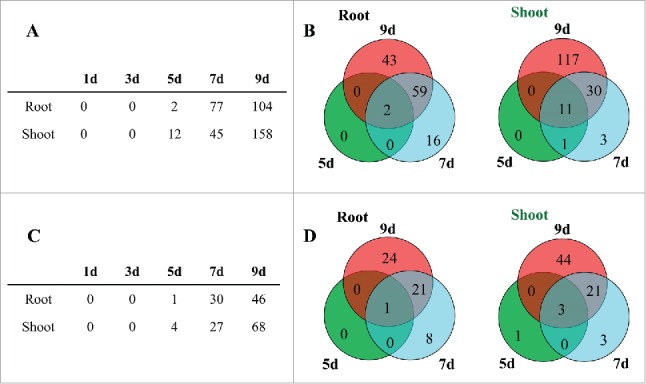
Arabidopsis sORFs up or downregulated in response to a progressive drought stress treatment. (A-B): sORFs up-regulated by drought stress in roots and shoots. A): Number of sORFs upregulated in response to a progressive drought stress. (B): Venn diagram of up-regulated sORFs at day 5, 7, and 9 of a drought stress treatment (C-D): sORFs down-regulated in roots and shoots in response to a drought stress treatment. (C): Number of sORFs downregulated in response to progressive drought stress treatment. (D): Venn diagram of down-regulated sORFs at day 5, 7, and 9 of a drought stress treatment.
None of the sORFs whose expression was impacted by the drought stress treatment was down-regulated at day 1 or 3 (Fig. 2C). At day 5, 7, and 9, however, the number of downregulated sORFs in roots was 1, 30, and 46 respectively (Fig. 2C), while the number of down-regulated sORFs in shoots was 4, 27 and 68, respectively. The down-regulation of the majority of sORFs in roots and shoots occurred at day 9 of the drought stress treatment (Fig. 2D). In total, 54 and 72 sORFs were downregulated in roots and shoots, respectively, in response to the imposed drought stress (Fig. S2). The expression of 14 of the sORFs was down-regulated in both roots and shoots. Many of the sORFs identified in the current study may be involved in drought stress response through signal transduction or some other unknown molecular mechanism. Some of these sORFs, however, may not be related to drought stress response and are potentially involved in other physiological processes. For example, since the plants started flowering on the fifth day of the drought stress treatment as previously reported,8 many of the sORFs, particularly those that were upregulated in shoots, may be related to flowering.
The sequence and expression profiles of the sORFs identified in the present study can be acquired from the HanaDB-AT19 database maintained at http://evolver.psc.riken.jp/seiken/search.html. The sORFs are predicted to encode peptides of varying lengths (Table S1). For example, ATRIKEN27513 is predicted to encode a peptide of 53 amino acids (a.a.), ATRIKEN28546 is predicted to encode a 35 a.a. peptide, while ATRIKEN32280 is predicted to encode a 72 a.a peptide. ATRIKEN27811 and ATRIKEN28145 are predicted to encode peptides of 71 and 49 a.a., respectively, while ATRIKEN29195 and ATRIKEN29358 are each predicted to encode a polypeptide of 70 a.a. (Table S1). The longer peptides may be representing pre-peptides and may be truncated to encode functional peptides, e.g. Arabidopsis CLE like (CLEL) genes encode precursors of peptide hormones.20-22
While analyzing the microarray data, we focused on sORFs that were up-regulated in roots at day 7 of the drought stress treatment (q-value <0.1, fold change ≥2) and divided them into 2 categories; sORFs upregulated only in roots (Fig. 3), and sORFs up-regulated in both roots and shoots (Fig. 4). It is plausible that the peptides encoded by sORFs that were specifically upregulated in roots may be involved in root to shoot signaling. In order to validate the data obtained in the microarray analysis, RT-qPCR analysis was performed for sORFs that were specifically up-regulated in roots only (ATRIKEN 29196 and ATRIKEN32280) or in both roots and shoots (ATRIKEN27811, ATRIKEN28145 and ATRIKEN29195). The sequences of the primers used to confirm the expression of different sORFs are shown in Table-S2. Actin 2 was used as an internal control to normalize the data. RT-qPCR analysis confirmed that ATRIKEN29196 and ATRIKEN32280 were significantly upregulated in roots (Fig. 3 and Fig. 5A-B). It would be interesting to investigate if the peptides encoded by sORFs specifically up-regulated in root tissue could contribute to signaling between roots and shoots during the course of a drought stress. RT-qPCR analysis also confirmed that ATRIKEN27811, ATRIKEN28145 and ATRIKEN29195 were upregulated at day 7 and 9 of the drought stress treatment in both roots and shoots (Fig. 4 and Fig. 5C-E). Interestingly, ATRIKEN29193, ATRIKEN29195, and ATRIKEN29358 are tandemly located on chromosome 3 of Arabidopsis (Fig. S3A) and there appears to be 2 copies of ATRIKEN29193 and ATRIKEN29195. The two copies of ATRIKEN29195 are also tandemly located, separated by a 1788 bp sequence. It is also interesting to note that the sequence of ATRIKEN29193, ATRIKEN29195, and ATRIKEN29358 is strikingly similar (Fig. S3B). Thus, primers used to monitor the expression of ATRIKEN29195, also monitor the expression of ATRIKEN29193 and ATRIKEN29358. A sequence similarity among ATRIKEN29357, ATRIKEN29359, ATRIKEN29360, and ATRIKEN29361, as well as other sORFs, was also observed (Fig. S3C). Gene duplication is a prominent feature of plant genome which might have played an important role in the evolution of phenotypic novelty within plants.23 Moreover, gene duplication may be serving as a mechanism of genomic adaptation to a changing environment.24 Thus sORFs may have been duplicated during evolution, and sORFs having more than one copy may have a greater likelihood of playing a role in the physiology of a plant as well as in response to abiotic stresses.
Figure 3.
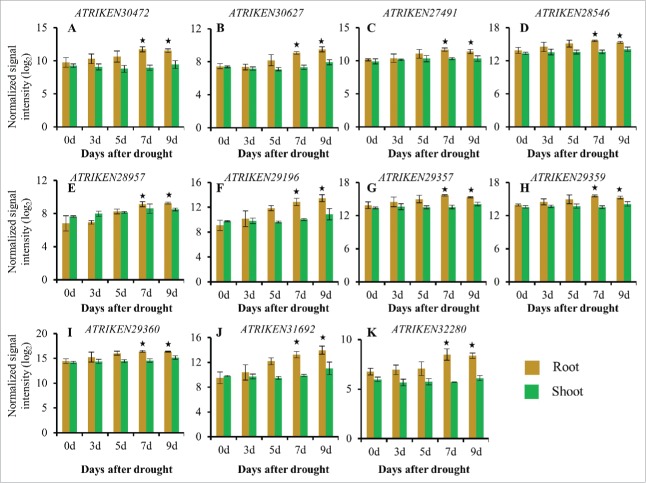
sORFs upregulated specifically in roots in response to a progressive drought stress treatment. Normalized log2 values were used to plot the expression of sORFs up-regulated specifically in roots in response to a progressive drought stress treatment. (A): ATRIKEN30472, (B): ATRIKEN30627, (C): ATRIKEN27491, (D): ATRIKEN28546, (E): ATRIKEN28957, (F): ATRIKEN29196, (G): ATRIKEN29357, (H): ATRIKEN29359, (I): ATRIKEN29360, (J): ATRIKEN31692, (K): ATRIKEN32280. Data presented are the mean ± sd, n = 3. Bars with an asterisk above are significantly different from Day 0 based on a q value <0.01, fold change ≥2.
Figure 4.
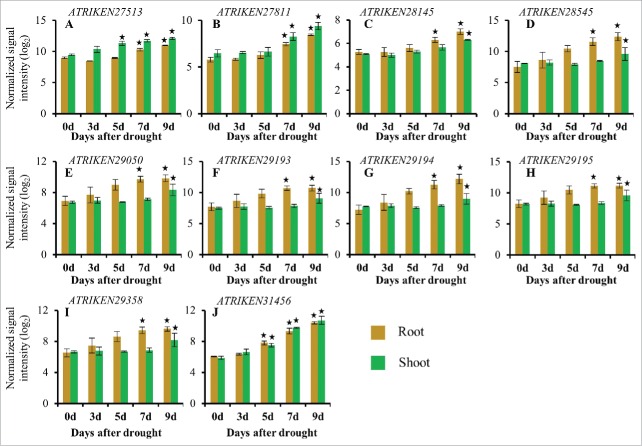
Changes in the expression of sORFs in response to a progressive drought stress treatment. Normalized log2 values were used to plot the expression of sORFs responsive to a progressive drought stress treatment. (A): ATRIKEN27513, (B): ATRIKEN27811, (C): ATRIKEN28145, (D): ATRIKEN28545, (E): ATRIKEN29050, (F): ATRIKEN29193, (G): ATRIKEN29194, (H): ATRIKEN29195, (I): ATRIKEN29358, (J): ATRIKEN31456. Data presented are the mean ± sd, n = 3. Bars with an asterisk above are significantly different from Day 0 based on a q value <0.01, fold change ≥2.
Figure 5.
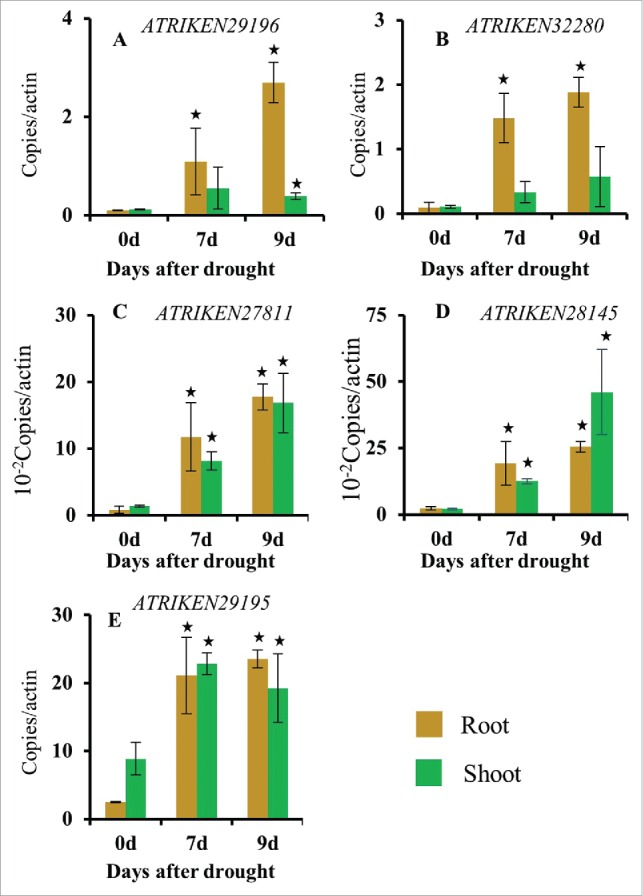
RT-qPCR analysis of sORFs expression in response to a progressive drought stress treatment. Changes in the expression of (A): ATRIKEN29196, (B): ATRIKEN32280, (C): ATRIKEN27811, (D): ATRIKEN28145 and (E): ATRIKEN29195 in response to a progressive drought stress treatment. Bars with an asterisk above are significantly different from Day 0 based on a t-test (p < 0.05). n = 4.
sORFs have been identified in several plant species, including rice and Arabidopsis. The expression level of the majority of sORFs in Arabidopsis tends to be significantly lower as compared to other known coding genes. As a result, it is generally assumed that sORFs do not play a significant role in regulating plant processes.19 It was recently demonstrated, however, that sORFs could play a significant role in various morphological processes.11,19,25 More than 48,000 putative sORFs have been identified in rice, which represents a much greater number than the sORFs that have been identified in Arabidopsis.26 sORFs have also been identified in legumes.27,28 The regulation of sORFs in rice in response to abiotic stresses has also been discussed.26,29 In the current study, several sORFs were identified in roots and shoots that are differentially regulated in response to a progressive drought stress treatment. Since these sORFs does not possess any conserved sequences, it is extremely difficult at this stage of our knowledge to predict the function of these sORFs. Although the exact function of the peptides putatively encoded by these sORFs remains unclear, it is plausible that some are involved in root to shoot signaling in response to drought stress. Cell-to-cell and root-to-shoot signaling is extremely important in plants and both function to regulate developmental changes10 and the response to abiotic stresses. Secreted peptides are reported to play an important role in cell-to-cell communication and are believed to coordinate and specify cellular functions in plants during development and the response to environmental stresses.30 For example, peptides belonging to the GLV/RGF/CLEL secreted peptide family have been demonstrated to be involved in plant developmental-related phenomenon, such as the development of root hairs, root meristem maintenance; as well as gravitropism.20,31 Further experimentation is required to confirm whether or not drought-responsive sORFs encode functional peptides and to elucidate the mechanisms through which these peptides work. In summary, the current study identified sORFs that may represent new players that are involved in the response of soil grown Arabidopsis roots and shoots to drought stress.
Supplementary Material
Disclosure of potential conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest.
Funding
This work was supported by the Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST), and grants from RIKEN, Japan [to MS]. K.B is supported through the RIKEN FPR program and S.R. is supported by the RIKEN JRA program. We are grateful to Maho Tanaka, Satoshi Takahashi (RIKEN CSRS) and Kei Iida (Graduate School of Medicine, Kyoto University) for supporting the microarray analysis.
References
- 1.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot 2007; 58:221-7; PMID:17075077; http://dx.doi.org/ 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- 2.Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002; 14:S165-S83; PMID:12045276; http://dx.doi.org/ 10.1105/tpc.010278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 2006; 17:113-22; PMID:16495045; http://dx.doi.org/ 10.1016/j.copbio.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 4.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 2010; 61:1041-52; PMID:20409277; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annl Rev Plant Biol 2006; 57:781-803; PMID:16669782; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- 6.Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 2007; 10:296-302; PMID:17468040; http://dx.doi.org/ 10.1016/j.pbi.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 7.Tardieu F, Parent B, Caldeira CF, Welcker C. Genetic and physiological controls of growth under water deficit. Plant Physiol 2014; 164:1628-35; PMID:24569846; http://dx.doi.org/ 10.1104/pp.113.233353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front Plant Sci 2016; 7:180; PMID:26941754; http://dx.doi.org/ 10.3389/fpls.2016.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang S, Li X-F, Liu z, Wang C-J. ZmGT1 transports glutathione conjugates and its expression is induced by herbicide atrazine. Prog Biochem Biophys 2010; 37:1120-7; http://dx.doi.org/ 10.3724/SP.J.1206.2010.00188 [DOI] [Google Scholar]
- 10.Katsir L, Davies KA, Bergmann DC, Laux T. Peptide signaling in plant development. Curr Biol 2011; 21:R356-R64; PMID:21549958; http://dx.doi.org/ 10.1016/j.cub.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada K, Zhang X, Borevitz JO, Li W-H, Shiu S-H. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 2007; 17:632-40; PMID:17395691; http://dx.doi.org/ 10.1101/gr.5836207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci 2013; 110:2395-400; PMID:23341627; http://dx.doi.org/ 10.1073/pnas.1213958110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olexiouk V, Crappé J, Verbruggen S, Verhegen K, Martens L, Menschaert G. sORFs. org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 2016; 44:D324-D9; PMID:26527729; http://dx.doi.org/ 10.1093/nar/gkv1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, Couso J-P. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 2014; 3:e03528; PMID:25144939; http://dx.doi.org/12164808 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 2002; 31:279-92; PMID:12164808; http://dx.doi.org/ 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- 16.Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 2008; 49:1135-49; PMID:18625610; http://dx.doi.org/ 10.1093/pcp/pcn101 [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological) 1995; 57:289-300 [Google Scholar]
- 18.Bechtold U, Penfold CA, Jenkins DJ, Legaie R, Moore JD, Lawson T, Matthews JS, Vialet-Chabrand SR, Baxter L, Subramaniam S. Time-series transcriptomics reveals that AGAMOUS-LIKE22 links primary metabolism to developmental processes in drought-stressed Arabidopsis. Plant Cell 2016; 28:345-66; PMID:26842464; http://dx.doi.org/ 10.1105/tpc.15.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci 2013; 110:2395-400; PMID:23341627; http://dx.doi.org/ 10.1073/pnas.1213958110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng L, Buchanan BB, Feldman LJ, Luan S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci 2012; 109:1760-5; PMID:22307643; http://dx.doi.org/ 10.1073/pnas.1119864109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006; 313:845-8; PMID:16902141; http://dx.doi.org/ 10.1126/science.1128439 [DOI] [PubMed] [Google Scholar]
- 22.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010; 329:1065-7; PMID:20798316; http://dx.doi.org/ 10.1126/science.1191132 [DOI] [PubMed] [Google Scholar]
- 23.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytologist 2009; 183:557-64; PMID:19555435; http://dx.doi.org/ 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- 24.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Royal Soc London B: Biol Sci 2012; 279:5048-57; PMID:22977152; http://dx.doi.org/ 10.1098/rspb.2012.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu S-H. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics 2010; 26:399-400; PMID:20008477; http://dx.doi.org/ 10.1093/bioinformatics/btp688 [DOI] [PubMed] [Google Scholar]
- 26.Okamoto M, Higuchi-Takeuchi M, Shimizu M, Shinozaki K, Hanada K. Substantial expression of novel small open reading frames in Oryza sativa. Plant Signaling Behav 2014; 9:e27848; PMID:24526015; http://dx.doi.org/ 10.4161/psb.27848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan B, Sheng J, Sun W, Zhao Y, Hao P, Li X. OrysPSSP: a comparative platform for small secreted proteins from rice and other plants. Nucleic Acids Res 2013; 41:D1192-D8; PMID:23203890; http://dx.doi.org/ 10.1093/nar/gks1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillén G, Diaz-Camino C, Loyola-Torres CA, Aparicio-Fabre R, Hernández-López A, Díaz-Sánchez M, Sanchez F. Detailed analysis of putative genes encoding small proteins in legume genomes. Front Plant Sci 2013; 4:208; PMID:23802007; http://dx.doi.org/ 10.3389/fpls.2013.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bashir K, Hanada K, Shimizu M, Seki M, Nakanishi H, Nishizawa NK. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 2014; 7:18; PMID:26224551; http://dx.doi.org/ 10.1186/s12284-014-0018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsubayashi Y. Posttranslationally modified small-peptide signals in plants. Annl Rev Plant Biol 2014; 65:385-413; PMID:24779997; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120122 [DOI] [PubMed] [Google Scholar]
- 31.Fernandez A, Hilson P, Beeckman T. GOLVEN peptides as important regulatory signalling molecules of plant development. J Exp Bot 2013; 64:5263-8; PMID:23975768; http://dx.doi.org/ 10.1093/jxb/ert248 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.