Abstract
In August 2012, a wildlife biologist became ill immediately following a 6-wk field trip to collect bats and rodents in South Sudan and Uganda. After returning to the US, the biologist was admitted to the hospital with multiple symptoms including fever, malaise, headache, generalized myalgia and arthralgia, stiffness in the neck, and sore throat. Soon after admission, the patient developed a maculopapular rash and oropharynx ulcerations. The patient remained hospitalized for 14 d. Several suspect pathogens, including viral hemorrhagic fever viruses such as Ebola viruses and Marburg viruses, were ruled out through standard diagnostic testing. However, deep sequencing and metagenomic analyses identified a novel paramyxovirus, later named Sosuga virus, in the patient’s blood. To determine the potential source, bat tissues collected during the 3-wk period just prior to the onset of symptoms were tested for Sosuga virus, and several Egyptian rousette bats (Rousettus aegyptiacus) were found to be positive. Further analysis of archived Egyptian rousette tissues collected at other localities in Uganda found additional Sosuga virus–positive bats, suggesting this species could be a potential natural reservoir for this novel paramyxovirus.
Keywords: Bats, paramyxovirus, Rousettus aegyptiacus, Sosuga virus, spillover, wildlife biologist
In late August 2012, a wildlife biologist returned to the US from Africa infected with a novel paramyxovirus, provisionally named Sosuga virus (Albariño et al. 2014). Initially, the biologist worked for 3 wk in remote areas of South Sudan collecting bats and rodents, but later, the individual traveled to Kibaale, Uganda, for a second 3-wk period collecting only bats (Fig. 1). Altogether, the patient handled >20 bat and rodent species while working in Africa. Two days after return to the US, the patient developed a severe but nonfatal disease that included high fever, malaise, generalized myalgia and arthralgia, neck stiffness, sore throat, and a maculopapular rash that became confluent over time. Initial diagnostic tests for known African viral hemorrhagic fevers were negative, including those caused by Ebola viruses, Marburg viruses, Crimean Congo hemorrhagic fever virus, and Lassa virus. Using deep sequencing and metagenomic analysis, the etiologic agent was found to be a novel paramyxovirus most closely related to rubula-like viruses found in several species of Asian and African fruit bats (Leschenault’s rousette, Rousettus leschesnaulti; variable flying fox, Pteropus hypomelanus; and the straw-colored fruit bat, Eidolon helvum; Chua et al. 2002; Lau et al. 2010; Drexler et al. 2012; Baker et al. 2013; Albariño et al. 2014).
Figure 1.
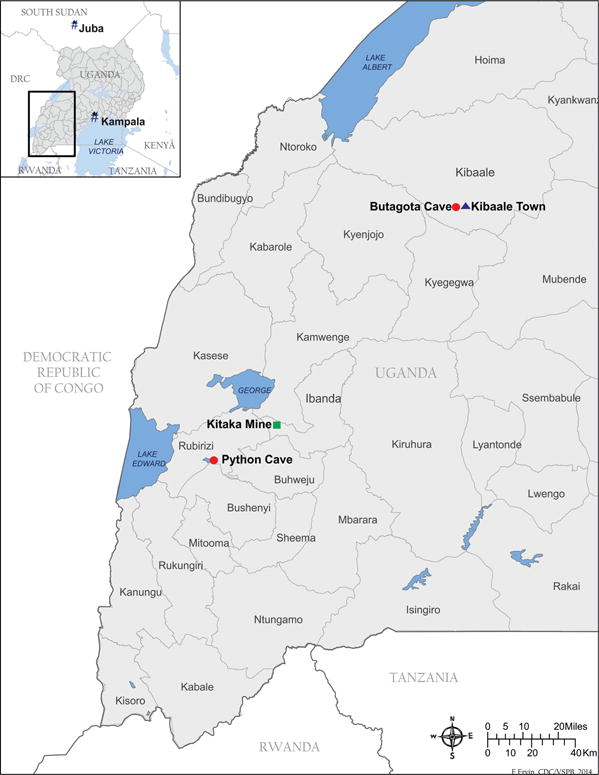
Map of Uganda showing capture locations of bats tested for Sosuga virus.
It is unclear how the biologist became infected with Sosuga virus. Interviews with the patient revealed that appropriate levels of personal protective equipment (PPE) were used during animal capture and processing in Kibaale, Uganda, including the use of disposable Tyvek suits coupled with powered air-purifying respirators (PAPRs; 3M, St. Paul, Minnesota, USA). However, inconsistent adherence to PPE practices did occur during the earlier South Sudan work. Incubation periods with other human paramyxovirus infections vary greatly and are generally in the range of 1–3 wk (Sartwell 1950; Goh et al. 2000; Playford et al. 2010). This fact made the field work in Kibaale, the 3-wk period just prior to symptom onset, the most plausible time for exposure to the virus. Taking these variables into account, combined with the close genetic relationship between Sosuga virus and the other fruit bat–borne rubula-like viruses, efforts to identify the virus source were focused on bats caught and necropsied at the Kibaale field site in Uganda (Table 1). There, bats were captured using a harp trap (Bat Conservation and Management, Inc., Carlisle, Pennsylvania, USA) placed at the entrance of a cave roost (Butogota Cave; 0°47′51.30″N, 31°2′27.42″E). The specific use of PPE is detailed by Towner et al. (2011). Briefly, PPE, consisting of a caving helmet, full face respirator with P100 filters, Tyvek coveralls, rubber gum boots, and bite-resistant leather gloves over double-layered latex gloves, was worn at all times during bat captures. Necropsies were performed at a central processing station away from public access. Liver, spleen, heart, lung, and kidney tissue aliquots were taken and placed directly in liquid nitrogen or in chaotropic lysis buffer known to have viracidal properties. During necropsies, PPE included double latex gloves, disposable gowns, and PAPRs.
Table 1.
Bats captured in southwestern and central Uganda and tested for Sosuga virus by quantitative reverse transcriptase PCR (qRT-PCR) on pooled liver/spleen samples. Shown are total number and percent of positive bats by species. Results for Egyptian rousettes (Rousettus aegyptiacus) are further subdivided by sex and age. QENP represents bats collected at Python Cave, Queen Elizabeth National Park. Egyptian rousettes caught in Kibaale were captured at Butogota Cave. All animal work was performed in accordance with a Centers for Disease Control and Prevention Animal Care and Use Committee approved protocol.
| Locality and species | n | No. qRT-PCR positive (%) |
|---|---|---|
| Kibaale August 2012 | ||
| Epomophorus labiatus | 262 | 0 (0) |
| Lissonycteris angolensis | 18 | 0 (0) |
| Hipposideros spp. | 1 | 0 (0) |
| Rousettus aegyptiacus | 122 | 3 (2.5) |
| Female | 68 | 3 (4.4) |
| Male | 54 | 0 (0.0) |
| Adult | 77 | 2 (2.6) |
| Juvenile | 45 | 1 (2.2) |
| QENP August 2009 | ||
| Rousettus aegyptiacus | 401 | 3 (0.7) |
| Female | 196 | 2 (1.0) |
| Male | 205 | 1 (0.5) |
| Adult | 237 | 0 (0.0) |
| Juvenile | 161 | 3 (1.9) |
| QENP November 2009 | ||
| Rousettus aegyptiacus | 408 | 15 (3.6) |
| Female | 187 | 4 (2.1) |
| Male | 221 | 11 (5.0) |
| Adult | 165 | 9 (5.5) |
| Juvenile | 243 | 6 (2.5) |
| Kitaka November 2012 | ||
| Rousettus aegyptiacus | 400 | 41 (10.2) |
| Female | 203 | 19 (9.4) |
| Male | 197 | 22 (11.2) |
| Adult | 233 | 22 (9.4) |
| Juvenile | 167 | 19 (11.4) |
Testing for Sosuga virus in bat specimens was carried out using a highly sensitive quantitative reverse transcriptase PCR (qRT-PCR) assay targeting the NP gene, which had been initially developed for detection and quantitation of Sosuga virus in patient blood (Albariño et al. 2014). Briefly, total nucleic acid from pooled bat liver/spleen tissue was extracted as described by Amman et al. (2012). All tissues were flash-frozen in liquid nitrogen in the field during necropsy and stored continuously frozen until processing. Of all the Egyptian rousettes, also known as Egyptian fruit bats (Rousettus aegyptiacus), caught at Butogota Cave (Fig. 1), 2.5% (3/122) were PCR positive for Sosuga virus, whereas the 262 Ethiopian epauletted fruit bats (Epomophorus labiatus), 18 Angolan rousettes (Lissonycteris angolensis), and one round-leaf bat (Hipposideros spp.) caught in the same general vicinity were negative (Table 1). To determine if testing of other bat tissues was more sensitive, liver, spleen, heart, kidney, lung, and blood from each of the three virus-positive bats (bats 841, 867, and 926) were tested separately by qRT-PCR, and only spleen was positive for Sosuga virus RNA.
To determine if Sosuga virus infection of Egyptian rousettes was common across Uganda, pooled liver/spleen tissue samples from approximately 1,200 archived Egyptian rousette bats from other locations were tested by qRT-PCR. Egyptian rousettes are a reservoir for Marburg viruses, and extensive bat samples were still available from previous studies at Python Cave in Queen Elizabeth National Park (0°16′37.92″S, 30°3′7.20″E in August 2009 and November 2009; Amman et al. 2012), and more recently from Kitaka Mine (0°7′50.34″S, 30°18′32.18″E) in October 2012 (Amman et al. 2014). Evidence of Sosuga virus was found in bats from all three Egyptian rousette collections tested, dating back to August 2009. The highest number of positive bats, 41 total (10% overall), was found in the Kitaka Mine in October 2012 (Table 1). Both Kitaka Mine and Python Cave are approximately 130 km from Butogota Cave in Kibaale and well within reported Egyptian rousette dispersal ranges of up to 500 km (Jacobsen and Du Plessis 1976; Amman et al. 2012), thus making intermixing between the populations likely.
All samples with Sosuga virus qRT-PCR cycle threshold (Ct) values <35 were additionally subjected to reverse transcriptase PCR (RT-PCR) using primers specific for a 331-nucleotide region in the HN gene as well as heminested RT-PCR using primers specific for a 127-nucleotide region in the NP gene. Because of the low levels of RNA found in tissues, the sequence was determined from only 11 bats: one bat caught at the Kibaale field site and 10 from the Kitaka Mine in 2012. These sequences were subjected to a BLAST (NCBI 2014) search to confirm identity and analyzed with other known rubula-like paramyxoviruses, including true rubula viruses (mumps), to generate phylogenies showing the inclusion of Sosuga virus within the rubula-like virus clade in the family Paramyxoviridae (Fig. 2). A more detailed phylogenetic placement of Sosuga virus within the Paramyxoviridae was described by Albariño et al. (2012). The bat from the Kibaale field site (bat 926) was positive in the NP assay only, and the sequence was identical to the patient isolate. For the 10 bats from the Kitaka Mine that were positive in the NP assay (Fig. 2A), seven had sequences identical to the patient isolate, while three bats differed by one nucleotide. Four Kitaka bats were additionally positive in the HN assay and differed by 6/331 (2%) nucleotides or less from each other and the sequence of the virus found in the infected biologist (Fig. 2B). Parallel attempts at virus isolation in Vero E6 cells and suckling mice were performed on those specimens with Ct values less than 35 using methods described in Albariño et al. (2012). Unfortunately, isolation attempts were negative (data not shown), presumably due to the low viral loads in the tissues.
Figure 2.
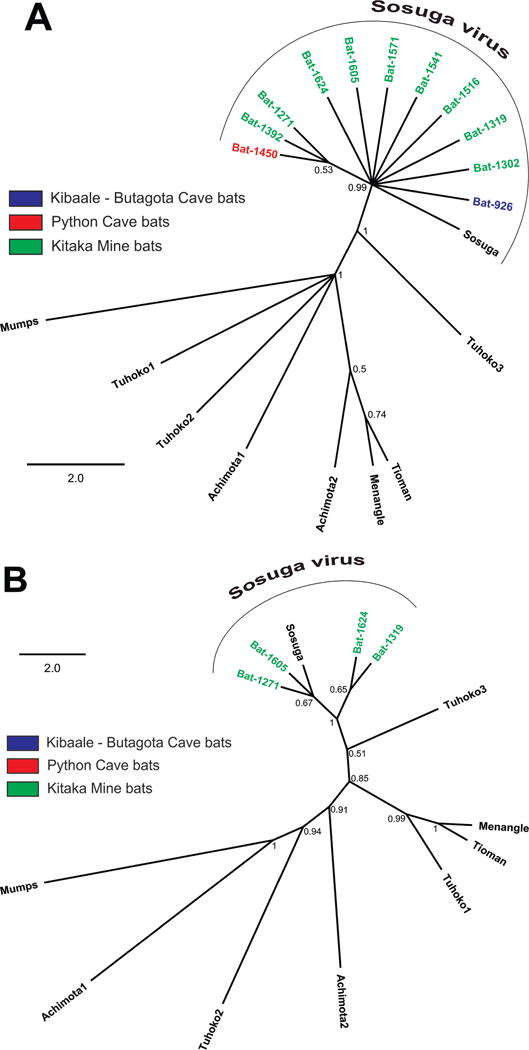
Phylogenetic analysis of Sosuga virus sequences determined from reverse transcriptase PCR amplification of NP and HN genes from bats. Nucleotide sequences corresponding to (A) 127-nucleotide and (B) 331-nucleotide fragments of the NP and HN genes, respectively, and those from eight representative rubula-like viruses, including comparable sequence fragments from the patient (Sosuga), were aligned using the MUSCLE algorithm (CLC Genomics Workbench version 6.0.1; CLC Bio, Cambridge, Massachusetts, USA). Mumps virus sequence was used as an out-group. Phylogenetic analysis was conducted with a Bayesian algorithm (Mr. Bayes, Geneious version 6.1.5, www.geneious.com). Bat sample localities are represented by color: Blue, Kiballe (Butagota Cave); Red, Python Cave; Green, Kitaka Mine. HN sequences were extracted from the complete genomic sequences in GenBank: KF774436 (Sosuga virus [SosV]), GU128082 (Tuhoko virus 3), U128081 (Tuhoko virus 2), GU128080 (Tuhoko virus 1), AF298895 (Tioman virus), NC_007620 (Menangle virus), JX051319 (Achimota virus 1), JX051320 (Achimota virus 2), NC_002200 (mumps virus). Posterior probability values are shown at each node. Scale is in substitutions/site. A more detailed phylogenetic placement of Sosuga virus within the virus family Paramyxoviridae was described in Albariño et al. (2012). GenBank accession numbers are as follows: Sosuga virus KF774436, Mumps NC_002200, Achimota virus 1 JX051319, Achimota virus 2 JX051320, Menangle virus NC_007620, Tioman virus AF298895, Tuhoko virus 1 GU128080, Tuhoko virus 2 GU128081, Tuhoko virus 3 GU128082, Sosuga virus from bats HN gene partial sequence, Bat-1605 KP150637, Bat-1319 KP150638, Bat-1271 KP150639, Bat-1624 KP150640, Sosuga virus from bats NP gene partial sequence, Bat-926 KP150641, Bat-1302 KP150642, Bat-1319 KP150643, Bat-1516 KP150644, Bat-1541 KP150645, Bat-1571 KP150646, Bat-1605 KP150647.
The sequence data presented herein are limited in information regarding the exact placement of Sosuga virus within the Paramyxoviridae. They exhibit very little variation (≤2% for the HN and <1% for the NP sequences) but clearly identify Sosuga as a rubula-like virus. For comparison, Hendra virus exhibited ≤1% variation during multiple separate introductions over a 2-yr period (Marsh et al. 2010). Moreover, we show that Egyptian rousette bats caught at multiple locations across Uganda over a 3-yr period were actively infected with Sosuga virus. This finding is consistent with Tuhoko3 virus, the nearest known relative of Sosuga virus, being found in Leschenault’s rousette in Asia (Lau et al. 2010).
Given the wildlife biologist’s exposure to bats in Uganda during the 3 wk prior to onset of illness, these Egyptian rousettes were the probable source of the infection. The wide distribution and detection of the virus at multiple time points suggest the Egyptian rousette could be a reservoir species, although that was not formally demonstrated here. If so, the extensive range of these bats across Sub-Saharan Africa would predict a wide distribution of the Sosuga virus. It is difficult to predict if a paramyxovirus closely related to Sosuga virus, such as Tuhoko virus, is capable of productively infecting humans. However, Drexler et al. (2012) report that bats appear to be the ancestral source of paramyxoviruses and that viruses in this family are known for their promiscuity, having spilled over into multiple orders of mammalian fauna.
We thank the Uganda Virus Research Institute, the Uganda Ministry of Health, and the Uganda Wildlife Authority for their assistance during past collection efforts. We also thank E. Ervine for assistance with creating the map of Uganda. Funding for this study was provided by the US Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or Health and Human Services.
LITERATURE CITED
- Albariño CG, Foltzer M, Towner JS, Rowe LA, Campbell S, Jaramillo CM, Bird BH, Reeder DM, Vodzak ME, Rota P. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerg Infect Dis. 2014;20:211–216. doi: 10.3201/eid2002.131620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, Kemp A, Erickson BR, Comer JA, Campbell S, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Nyakarahuka L, McElroy AK, Dodd KA, Sealy TK, Schuh AJ, Shoemaker TR, Balinandi S, Atimnedi P, Kaboyo W, et al. Marburgvirus resurgence in Kitaka mine bat population after extermination attempts, Uganda. Emerg Infect Dis. 2014;20:1761–1762. doi: 10.3201/eid2010.140696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, Todd S, Marsh GA, Crameri G, Barr J, Kamins AO, Peel AJ, Yu M, Hayman DT, Nadjm B. Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum. J Virol. 2013;87:1348–1358. doi: 10.1128/JVI.01202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, Wang LF, Lam SK, Eaton BT. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Arch Virol. 2002;147:1323–1348. doi: 10.1007/s00705-002-0815-5. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:1–12. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KJ, Tan CT, Chew NK, Tan PSK, Kamarulzaman A, Sarji SA, Wong KT, Abdullah BJJ, Chua KB, Lam SK. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342:1229–1235. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- Jacobsen NHG, Du Plessis E. Observations on the ecology and biology of the Cape fruit bat Rousettus aegyptiacus leachi in the eastern Transvaal. S Afr J Sci. 1976;72:270–273. [Google Scholar]
- Lau S, Woo P, Wong B, Wong A, Tsoi H, Wang M, Lee P, Xu H, Poon R, Guo R. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology. 2010;404:106–116. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, Todd S, Foord A, Hansson E, Davies K, Wright L, Morrissy C, Halpin K, Middleton D, Field HE, et al. Genome sequence conservation of Hendra virus isolates during spillover to horses, Australia. Emerg Infect Dis. 2010;16:1767–1769. doi: 10.3201/eid1611.100501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) BLAST. 2014 http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome. Accessed December 2014.
- Playford EG, McCall B, Smith G, Slinko V, Allen G, Smith I, Moore F, Taylor C, Kung YH, Field H. Human Hendra virus encephalitis associated with equine outbreak, Australia, 2008. Emerg Infect Dis. 2010;16:219–223. doi: 10.3201/eid1602.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartwell PE. The distribution of incubation periods of infectious disease. Am J Epidemiol. 1950;51:310–318. doi: 10.1093/oxfordjournals.aje.a119397. [DOI] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Nichol ST. Significant zoonotic diseases identified in bats: Filoviruses. In: Newman SH, Field H, Epstein J, de Jong C, editors. Investigating the role of bats in emerging zoonoses. Food and Agriculture Organisation of the United Nations; Rome, Italy: 2011. pp. 123–135. [Google Scholar]


