Abstract
Cardiomyopathies are an important and heterogeneous group of common cardiac diseases. An increasing number of cardiomyopathies are now recognized to have familial forms, which result from single-gene mutations that render a Mendelian inheritance pattern, including hypertrophic cardiomyopathy, dilated cardiomyopathy, restrictive cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and left ventricular noncompaction cardiomyopathy. Recently, clinical genetic tests for familial cardiomyopathies have become available for clinicians evaluating and treating patients with these diseases, making it necessary to understand the current progress and challenges in cardiomyopathy genetics and diagnostics. In this review, we summarize the genetic basis of selected cardiomyopathies, describe the clinical utility of genetic testing for cardiomyopathies and outline the current challenges and emerging developments.
Keywords: arrhythmogenic right ventricular, cardiomyopathy, dilated, genetic testing, hypertrophic, left ventricular noncompaction, molecular diagnostics, mutations, restrictive
Cardiomyopathies are a heterogeneous group of heart muscle diseases associated with mechanical and/or electrical dysfunction that predispose patients to sudden cardiac death [1,2]. Familial cardiomyopathies are typically diagnosed in the third or fourth decades of life, but may present at any age. Over the last 20 years, and at an ever quickening pace in recent years, the association of specific genes involved with cardiomyopathies has illuminated their pathophysiology and identified potential therapeutic targets that may one day allow clinicians to stall, regress or even prevent certain cardiomyopathies. Hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), left ventricular noncompaction cardiomyopathy (LVNC), and arrhythmogenic right ventricular cardiomyopathy (ARVC) are all now recognized to have a genetic component (TABLE 1). The identification of cardiomyopathy-susceptibility genes has enabled the development of diagnostic tests that can identify genetic mutations underlying disease in a substantial portion of patients. The ability to identify disease-causing genetic mutations is still quite limited, largely due to the lack of complete knowledge of all the mutations and affected genes that lead to cardiomyopathies. Whenever disease-causing mutations are identified, family members at-risk for developing a cardiomyopathy can be determined, including those in which little or no clinical suspicion exists.
Table 1.
Genetic cardiomyopathies.
| Disease | Inheritance pattern | Estimated prevalence | Patients with mutations based on testing known causative genes (%) |
|---|---|---|---|
| Arrhythmogenic right ventricular cardiomyopathy | Autosomal dominant | 1:5000 | 50–55 |
| Dilated cardiomyopathy | Autosomal dominant, autosomal recessive, X-linked | 1:2500 | ~25 |
| Hypertrophic cardiomyopathy | Autosomal dominant, autosomal recessive, X-linked | 1:500 | 35–65 |
| Left ventricular noncompaction cardiomyopathy | Autosomal dominant, X-linked | Unknown | 20–25 |
| Restrictive cardiomyopathy | Autosomal dominant | Unknown | Unknown |
The recent transition of cardiomyopathy genetic testing from research laboratories into clinical practice highlights the need for the education of clinicians in the state of the art and the implications of cardiomyopathy genetic testing. This review summarizes the genetic basis of selected cardiomyopathies, describes the clinical utility of genetic testing for cardiomyopathies and outlines the current challenges and emerging developments in clinical genetic testing for cardiomyopathies.
Molecular basis of familial cardiomyopathies
Cardiomyopathies are largely monogenic disorders in which pathogenic mutations and disease susceptibility follow predictable Mendelian modes of transmission. In monogenic, or ‘single-gene’, disorders, disease-causing mutations in a particular individual are restricted to a single gene. The mutations typically consist of an alteration of a single nucleotide that causes one amino acid within the encoded protein to be substituted for another. Alternatively, the mutation may be the deletion or insertion of a short sequence of nucleotides that results in a truncated protein. Autosomal dominant inheritance, in which a single mutation affecting one copy of an autosomal gene causes disease that may affect either gender, is the most commonly observed inheritance pattern in cardiomyopathies. Although far less frequent and differing among the specific diseases and genes involved, autosomal recessive and X-linked inheritance patterns are also observed in cardiomyopathies. The phenomenon of age-dependent penetrance, where only a portion of carriers of a disease-causing mutation clinically manifest disease, can confound recognition of a cardiomyopa-thy as a familial disease. Familial cardiomyopathies affect all ethnicities. There is no broad ethnic predisposition outside of small, isolated populations affected by specific founder mutations. The precise molecular mechanisms and pathological processes leading from single-gene mutations to the development of clinically recognizable cardiomyopathies is still largely unknown; however, the identification and characterization of many mutations across many genes has begun to uncover the diverse molecular mechanisms leading to seemingly similar disease manifestations. Cardiomyopathies were initially defined clinically and their diagnosis and management continue to be based on clinical presentation, despite the genetic heterogeneity underlying these diseases. While little is still known regarding the causes of the heterogeneity of disease, it is known that having more than one type of mutation can increase the severity of disease [3]. A number of gene polymorphisms have also been shown to modify the severity of cardiomyopathy (for a recent review see [4]). Complicating the genetic diagnosis of cardiomyopathies is the realization that different mutations in a single gene can lead to two different cardiomyopathic phenotypes. For example, different mutations in cardiac troponin, cardiac troponin T, and α-cardiac actin can cause RCM or HCM (TABLES 2 & 3). Similarly, different mutations in lamin A/C can lead to LVNC or DCM phenotypes (TABLES 2 & 4).
Table 2.
Genetic causes of arrhythmogenic right ventricular cardiomyopathy, left ventricular noncompaction cardiomyopathy and restrictive cardiomyopathy.
| Protein | Frequency in patients (%) | Ref. |
|---|---|---|
| Restrictive cardiomyopathy | ||
| β-myosin heavy chain | Unknown | [104] |
| Cardiac troponin I | Unknown | [99,104] |
| Cardiac troponin T | Unknown | [100,102,167] |
| α-cardiac actin | Unknown | [100] |
| Left ventricular noncompaction cardiomyopathy | ||
| β-myosin heavy chain | 13 | [92,93] |
| α-cardiac actin | 3 | [92] |
| Cardiac troponin T | <2 | [92] |
| LIM domain-binding protein 3 (Cypher/ZASP) | Unknown | [87] |
| α-dystrobrevin | Unknown | [86] |
| Tafazzin | Unknown | [86] |
| Lamin A/C | Unknown | [168] |
| Arrhythmogenic right ventricular cardiomyopathy | ||
| Plakophilin 2 | 25–35 | [5,17] |
| Desmoplakin | 5 | [5,17] |
| Desmoglein 2 | 5 | [5,17] |
| Desmocollin 2 | Rare | [5,17] |
| Plakoglobin | Rare | [5,17] |
| TGFβ3 | Rare | [5,23] |
| Transmembrane protein 43 | Unknown | [24] |
Table 3.
Genetic causes of hypertrophic cardiomyopathy and hypertrophic cardiomyopathy phenocopy diseases.
| Gene | Description | Mutation frequency in familial forms of HCM (%) | Ref. |
|---|---|---|---|
| Hypertropic cardiomyopathy | |||
| MYH7 | β-myosin heavy chain | 15–25 | [130,169–171] |
| MYBPC3 | Myosin-binding protein C | 15–25 | [130,169–171] |
| TNNT2 | Cardiac troponin T | <5 | [130,169–171] |
| TPM1 | α-tropomyosin | <5 | [130,169–171] |
| TNNI3 | Cardiac troponin I | <5 | [130,169–171] |
| MYL2 | Myosin regulatory light chain | <2 | [130,169–171] |
| MYL3 | Myosin essential light chain | Rare | [130,169–171] |
| ACTC | α-cardiac actin | Rare | [130,169–171] |
| TNNC1 | Cardiac troponin C | Rare | [171,172] |
| TTN | Titin | Rare | [169,173] |
| MYH6 | α-myosin heavy chain | Rare | [169,174] |
| LDB3 | LIM binding domain 3 (Cypher/ZASP) | Rare | [175] |
| CSRP3 | Muscle LIM protein | Rare | [175,176] |
| TCAP | Telethonin | Rare | [175,177] |
| VCL | Vinculin/metavinculin | Rare | [175,178,179] |
| ACTN2 | α-actinin 2 | Rare | [175] |
| MYOZ2 | Myozenin 2 | Rare | [180] |
| ANKRD1 | Ankyrin repeat domain 1 | Rare | [181] |
| JPH2 | Junctophilin-2 | Rare | [43] |
| PLN | Phospholamban | Rare | [44,45] |
| Hypertropic cardiomyopathy phenocopy diseases (metabolic/ infiltrative diseases) | |||
| GLA | α-galactosidase A (Anderson-Fabry disease) | <5 | [182,183] |
| PRKAG2 | AMP-activated protein kinase subunit (WPW with LVH) | Rare | [133,169,184] |
| LAMP2 | Lysosome-associated membrane protein 2 (Danon syndrome) | Rare | [133,185] |
HCM: Hypertrophic cardiomyopathy; LVH: Left ventricular hypertrophy; WPW: Wolff–Parkinson–White syndrome.
Table 4.
Genetic causes of dilated cardiomyopathy.
| Gene | Description | Frequency in DCM patients (%) | Ref. |
|---|---|---|---|
| ACTC | α-cardiac actin | Rare | [57,186–188] |
| ANKRD1 | Ankyrin repeat domain 1 | <5 | [189,190] |
| LDB3 | LIM domain-binding protein 3 (Cypher/ZASP) | <5 | [87,141] |
| LMNA | Lamin A/C | 6 | [135,136,191–196] |
| MYBPC3 | Myosin-binding protein C | <5 | [197–200] |
| MYH7 | β-myosin heavy chain | <5 | [58,141,197,201] |
| PLN | Phospholamban | Rare | [201–203] |
| SCN5A | Sodium channel | <3 | [204–206] |
| TNNC1 | Cardiac troponin C | Rare | [61] |
| TNNI3 | Cardiac troponin I | Rare | [71,207] |
| TNNT2 | Cardiac troponin T | <2 | [58,60,61,141,201] |
| TPM1 | α-tropomyosin | Rare | [59] |
| TTN | Titin | Rare | [208] |
| MYH6 | α-myosin heavy chain | Rare | [174] |
| CSRP3 | Muscle LIM protein | Rare | [141,209] |
| TCAP | Telethonin | Rare | [141,177] |
| VCL | Vinculin/metavinculin | Rare | [201,210] |
| ACTN2 | α-actinin 2 | Rare | [211] |
| DES | Desmin | Rare | [188,212,213] |
| SGCD | δ-sarcoglycan | Rare | [214–216] |
| ABCC9 | ATP-binding cassette C member 9 | Rare | [217] |
| EYA4 | Eyes-absent 4 | Unknown | [218] |
| TMPO | Thymopoietin | Rare | [219] |
| PSNE1 | Presenilin 1 | Rare | [220] |
| PSNE2 | Presenilin 2 | Rare | [220] |
| DMD | Dystrophin | Rare | [142,199,221,222] |
| TAZ | Tafazzin | Rare | [86,223,224] |
DCM: Dilated cardiomyopathy.
Arrhythmogenic right ventricular cardiomyopathy
Arrhythmogenic right ventricular cardiomyopathy is estimated to affect one in 5000 individuals and is characterized by life-threatening arrythmias due to fatty infiltration and scarring of the right or both ventricles [5]. ARVC is an autosomal dominant disease with variable penetrance [6]. The replacement of right ventricular myocardium by fibrofatty tissue progresses over time, leading to ventricular wall thinning and aneurysms [7–9]. The fibrofatty infiltrates also interfere with the conduction of electrical impulses, causing characteristic changes in the electrocardiogram (ECG), including epsilon waves (FIGURE 1), late potentials, right bundle branch block and ventricular arrhythmias. Cell death can be seen by histology, frequently associated with inflammatory infiltrates and life-threatening arrhythmias [7–11]. The left ventricle is involved in nearly 50% of all cases, generally in the postero–lateral subepicardium [7,9]. Cell death can be seen by histology, frequently associated with inflammatory infiltrates and life-threatening arrhythmias [7–11]. The inflammatory cells may be a reaction to cell death and cardiotropic viruses have been reported in the myocardium of some patients with ARVC, suggesting a role for infectious etiologies in the pathogenesis of disease [12,13].
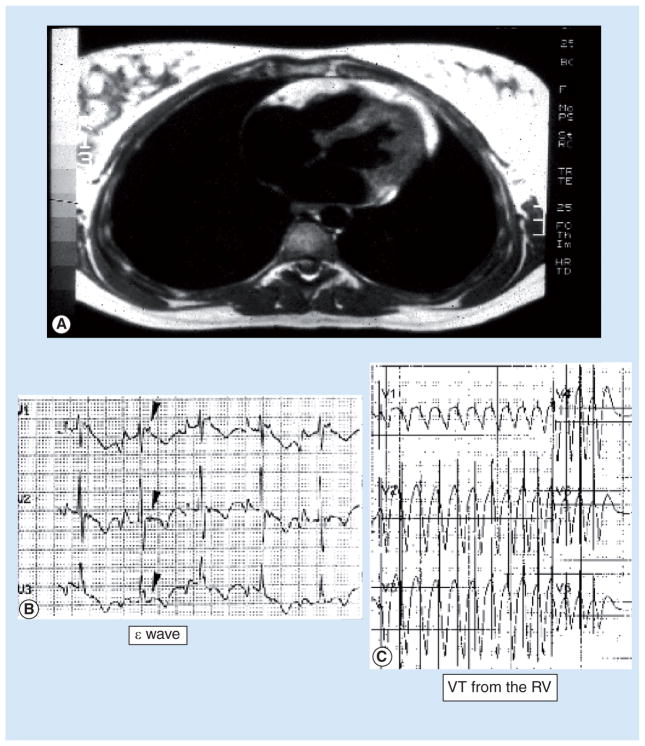
Figure 1. Arrhythmogenic right ventricular cardiomyopathy.
(A) Cardiac MRI reveals fibrofatty infiltration of the right ventricular free wall, with some involvement of the left ventricle. (B) Electrocardiogram in normal sinus rhythm exhibiting a repolarization abnormality characteristic of arrhythmogenic right ventricular cardiomyopathy, known as the ‘epsilon wave’ (indicated by arrows). (C) Electrocardiogram demonstrating ventricular tachycardia originating from the RV. RV: Right ventricle; VT: Ventricular tachycardia.
Recently, it has been discovered that ARVC is largely a disease of the desmosome. The desmosome is a group of cellular structures that mechanically couple cardiomyocytes to transmit contractile force (FIGURE 2). The prevailing view of ARVC pathogenesis is that disruption of the desmosome leads to progressive myocyte separation, myocyte death and subsequent replacement of dead cells with fat and scar tissue. However, mechanisms involving aberrant regulation of adipogenesis signaling pathways have also been proposed [14–16]. Mutations in five genes making up the desmosome (FIGURE 2) are found in approximately 50% of clinically diagnosed ARVC patients, including the genes desmoplakin (DSP), plakophilin-2 (PKP2), desmoglein-2 (DSG2), desmocollin-2 (DSC2), and plakoglobin (PKG) (TABLE 2) [17]. The PKP2 gene is most frequently mutated in 43% of cases (70% of proven familial cases) [18–22]. Mutations in extradesmosomal genes, such as TGFβ3 have also been associated with ARVC [23]. Recently, a founder mutation in the gene transmembrane protein 43 (TMEM43) was shown to be the cause of ARVC in 15 unrelated families, all with complete penetrance [24]. Whether the protein encoded by TMEM43 is a component of the desmosome is not yet known and its association with ARVC may lead to the discovery of a novel disease mechanism. There are a number of notable diseases that lead to clinical entities that mimic the findings of ARVC; that is, diseases that phenocopy ARVC. The closest phenocopy of ARVC is myotonic dystrophy [25], but sarcoidosis also can closely mimic ARVC [26]. This suggests an important role for genetic testing (for unstable CTG repeats in the DMPK gene) and biopsy (for sarcoid) also in distinguishing ARVC cases. The importance of the desmosome in the pathogenesis of ARVC may be highlighted in recent studies, which have identified a role for immunohistochemical analysis of conventional endomyocardial biopsies for desmosomal plakoglobin [27]. In ten out of 11 ARVC cases, a decrease in immunoreactive plakoglobin was identified, a finding not seen in control samples in blinded studies [27]. This diffuse reduction in plakoglobin demonstrated a sensitivity of 91% and a specificity of 82% (positive predictive value: [PPV] = 83%; negative predictive value [NPV] = 90%). While a number of proteins may be involved in the pathogenesis of ARVC, the ability of these proteins to stabilize the desmosome, regardless of their origin, may be key to the pathogenesis of ARVC.
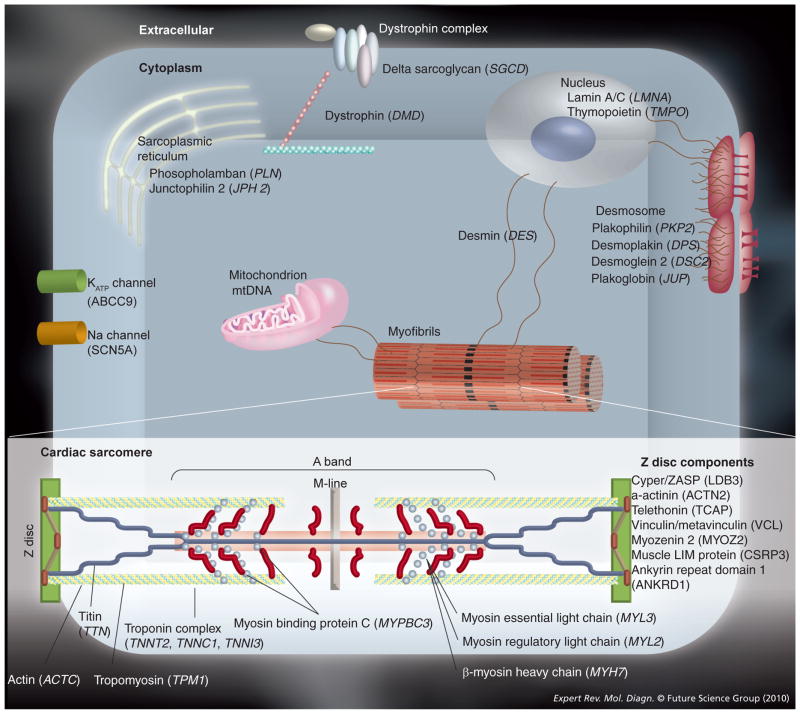
Figure 2. Selected proteins involved in the pathogenesis of cardiomyopathies shown in the context of their respective cellular structures.
Cardiomyopathies are a genetically heterogeneous group of diseases that result from dysfunction in a multitude of diverse biological processes, including contractile force generation and transmission, mechanical stretch sensing, nuclear structure and function, and ion channel function.
Hypertrophic cardiomyopathy
Hypertrophic cardiomyopathy is the most common genetic heart disease in the USA, affecting one in 500 people [28,29]. The diagnosis of HCM is made primarily on patient history, physical examination, echocardiography and ECG, identifying hypertrophy in the absence of underlying primary disease (FIGURE 3). There is a spectrum of symptoms in HCM patients: some patients may be asymptomatic their entire life, while others may present with dyspnea, syncope, chest pain or sudden cardiac death due to mechanical or electrical abnormalities [30–33]. Despite its namesake, sudden cardiac death can occur in HCM patients with little or no cardiac hypertrophy due to the histological hallmark of myocardial disarray, which can act as an arrhythmogenic substrate resulting in life-threatening arrhythmias [34–36]. There is a high degree of disease heterogeneity in HCM and in contrast to other genetic diseases, HCM has a preponderance of private, familial mutations and a lack of mutational hot spots within causative genes.
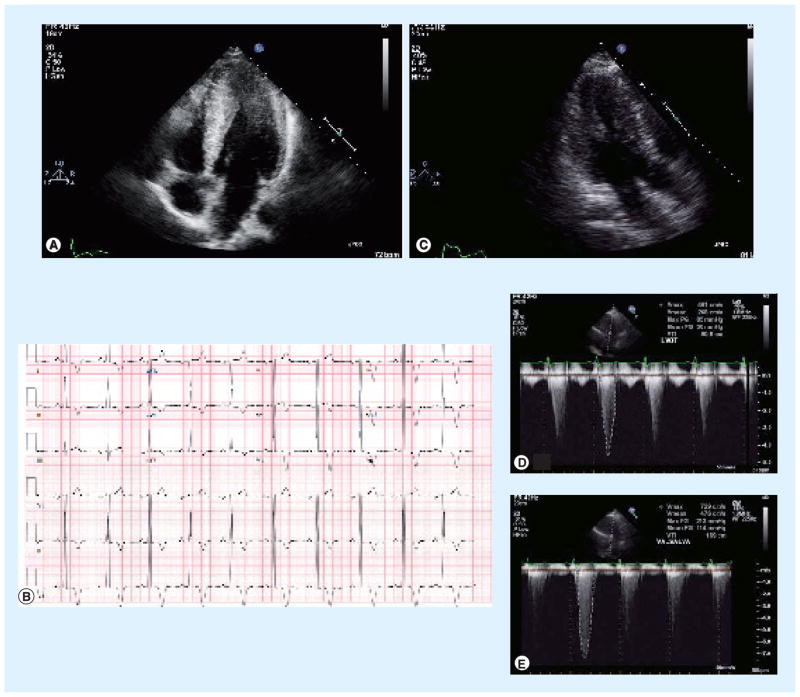
Figure 3. Hypertrophic cardiomyopathy (HCM).
(A) Transthoracic echocardiogram in the apical four-chamber view demonstrates pronounced thickening of the distal interventricular septum and lateral wall of the left ventricle, consistent with the apical variant of HCM. (B) 12-lead electrocardiogram with evidence of left ventricular hypertrophy and deep T-wave inversions throughout the precordium, characteristic of apical variant HCM. (C) Transthoracic echocardiogram in the apical long-axis view demonstrates left ventricular hypertrophy, most pronounced in the interventricular septum, consistent with HCM. (D) Transthoracic echocardiogram using Doppler signal in the left ventricular outflow tract reveals a resting pressure gradient of 85 mmHg between the cavity of the left ventricle and the aortic root. (E) This pressure gradient augments to 213 mmHg with Valsalva maneuver, characteristic of the dynamic left ventricular outflow tract gradient of obstructive HCM.
Nearly 50–60% of HCM cases have mutations in genes encoding proteins that constitute the sarcomere, the fundamental contractile unit of cardiac myocytes (FIGURE 1 & TABLE 3) [4]. More than 450 mutations in 20 genes that cause HCM have been described to date [37]. The genes most commonly affected are cardiac myosin binding protein C (MYBPC3), cardiac β-myosin heavy chain (MYH7), and cardiac troponin isoforms I (TNNI3) and T (TNNT2) (TABLE 3). Two mechanisms have been hypothesized to cause HCM. The affected protein may affect the heart by acting as a dominant negative ‘poison peptide’, disrupting function of the wild-type protein by its presence. Alternatively, mutations in sarcomere proteins may lead to haploinsufficiency [38]. In this scenario, it is hypothesized that mutations in one gene do not allow the sarcomere to be stoichiometrically balanced, resulting in poor performance of the remaining sarcomere [37,39]. Recent studies further suggest a disruption in protein quality control mechanisms may also play a causative role in disease [40]. Sarcomere mutations responsible for HCM result in impaired relaxation of the heart and significant changes in calcium signaling [41–45] and may also result in metabolic defects [46], which could play a role in the increased susceptibility of patients with HCM mutations to sudden cardiac death.
Dilated cardiomyopathy
Dilated cardiomyopathy is defined clinically by left ventricular dilation and reduced contractile performance (FIGURE 4). DCM is the most frequent diagnosis leading to heart transplantation in the USA [2,47]. There are a number of known causes of DCM, including coronary artery disease, thyroid disease, viral myocarditis and excessive alcohol intake. Idiopathic DCM, that is, DCM in which common acquired/nongenetic causes are excluded, is estimated to affect one in 2500 individuals with 20–50% of these estimated to have a genetic cause [48–56]. The genetic causes of DCM are particularly heterogeneous. To date, mutations in more than 30 genes account for disease in approximately 25% of patients (FIGURE 1 & TABLE 4). Candidate gene analyses have identified causative mutations in cardiac actin and other sarcomere genes [57], including MYH7 [58], MYBPC3, titin (TTN), α-tropomyosin (TPM1) [59] and TNNT2 and TNNC1 [58,60,61]. Several subgroups of DCM have been identified based on distinguishing clinical features associated with gene mutations [62–64]. Mutations in lamin A/C (LMNA) have been associated with autosomal dominant DCM with conduction disease [65,66], autosomal dominant and recessive Emery Dreifuss Muscular Dystrophy [67,68], and autosomal dominant limb-girdle muscular dystrophy type 1B [69,70]. While there appears to be a continuum of muscular dystrophy associated with DCM, the underlying mechanism for LMNA mutations in cardiac and extracardiac manifestations, such as the skeletal muscle, is not known.
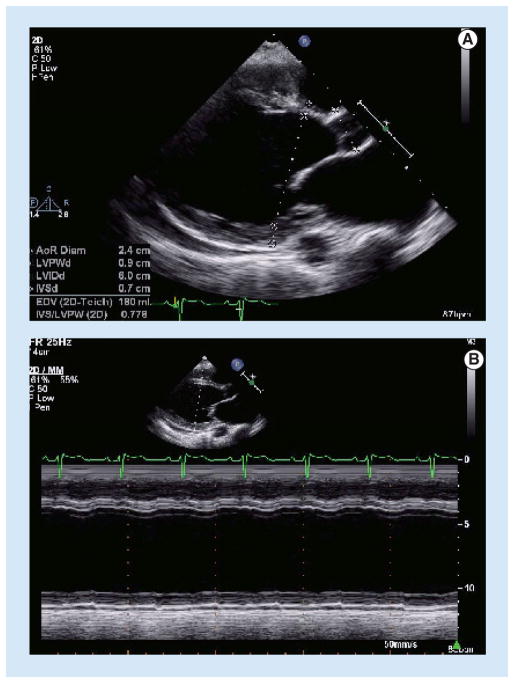
Figure 4. Dilated cardiomyopathy.
(A) Transthoracic echocardiogram in the parasternal long-axis view demonstrates dilation of the left ventricle. (B) M-mode echocardiography from the same view reveals markedly diminished systolic thickening of the myocardium.
Interestingly, HCM and DCM share partially overlapping molecular etiologies, as both may result from sarcomeric mutations, although the specific mutations associated with each and resulting molecular consequences can be different. Functional studies of different HCM- and DCM-associated mutations occurring within the same sarcomeric genes suggest that, in general, mutations that increase sensitivity of the cardiac sarcomere to calcium result in HCM, whereas desensitizing mutations result in DCM [71–74].
Left ventricular noncompaction cardiomyopathy & restrictive cardiomyopathy
Although LVNC was first described in 1900, it has only recently been recognized as a distinct clinical entity [1,2]. It is characterized by a pattern of prominent trabecular meshwork and deep intratrabecular recesses communicating with the cavity of the left ventricle (FIGURE 2). This is thought to be due to an arrest of myocardial morphogenesis during cardiac development [75,76]. LVNC is a rare disease, affecting less than 0.3% of the population [77,78], or an annual incidence in children of 0.1 per 100,000 [79,80]. The clinical manifestations of LVNC can range from asymptomatic to a progressive deterioration of cardiac function, arrhythmias, thromboembolic events and sudden cardiac death [75,77,81–83]. Approximately 40% of LVNC patients have evidence of familial disease, with a wide genetic heterogeneity [82].
The mainstay of diagnosis for LVNC has historically been echocardiography, as shown in FIGURE 5 [84]. The diagnostic criteria for LVNC include the lack of coexisting cardiovascular abnormalities, segmental left ventricular wall thickening with a thin compacted epicardial layer and a thicker noncompacted endocardial layer, an end-systolic noncompacted-to-compacted myocardial ratio greater than 2, and the identification of flow on color Doppler within the deep intratrabecular recesses [85]. Additional echocardiographic findings may include a decreased fractional shortening, impaired diastolic function, abnormal papillary muscle architecture and the presence of thrombi [85]. LVNC has been associated with mutations in a number of genes, including LIM domain binding protein 3 (LDB3/ZASP), dystrobrevin-α (DTNA), tafazzin (TAZ/G4.5) and lamin A/C (LMNA) [86–92]. Mutations in the sarcomere protein genes MYH7, TNNT2 and α-cardiac actin (ACTC) have also been associated with LVNC (TABLE 2) [86–92]. The more common genes reported in LVNC have been genes that encode for proteins found in the sarcomere (MYH7, TNNT2 and ACTC) [92,93].
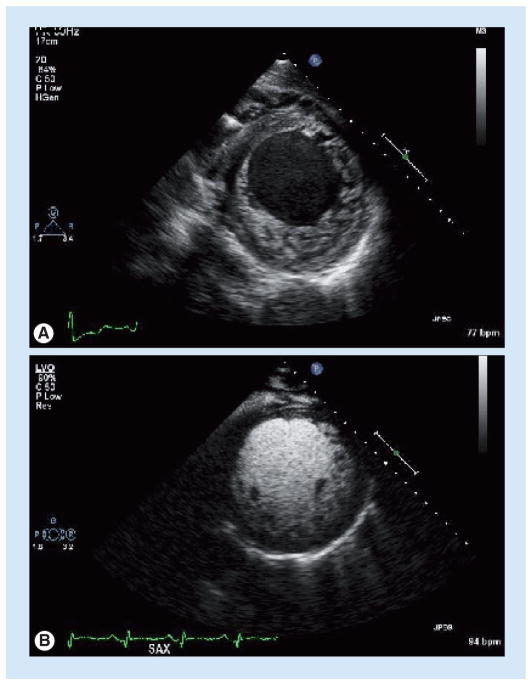
Figure 5. Left ventricular noncompaction cardiomyopathy.
(A)Transthoracic echocardiogram in the parasternal short-axis view reveals prominent myocardial noncompaction involving all segments of the left ventricle other than the interventricular septum. (B) Use of injectable echocardiographic contrast in the same view further defines the structural abnormalities.
Restrictive cardiomyopathy is characterized by an increase in cardiac wall stiffness, leading to decreased diastolic function with preserved systolic function. Patients generally develop symptoms of severe heart failure over a relatively short period of time, with the majority dying within a few years if they are unable to obtain a heart transplant [94]. Adult patients with RCM present with dyspnea, fatigue and a limited capacity to exercise [95], while children may present with failure to thrive, fatigue and sometimes syncope [96,97]. Chest radiography generally shows a normal sized heart with enlarged atria and varying degrees of pulmonary edema. On ECG, large P waves indicative of atrial enlargement may be present along with ST segment and T-wave abnormalities. By echocardiography, enlarged atria with impaired systolic function may be seen, and Doppler velocities may be indicative of a significant diastolic dysfunction. Many etiologies underlie RCM, including inflammatory (endomyocardial fibrosis), infiltrative (e.g., amyloidosis), storage (e.g., hemochromatosis) and idiopathic (reviewed in [94]). Since 1992, a number of reports have identified mutations in sarcomere genes underlying RCM, including TNNI3, TNNT2, MYH7 and ACTC [98–103].
Uncovering the genetic basis and molecular mechanisms of RCM and LVNC has been challenging since they are far less prevalent than HCM or DCM [104,105]. Several of the same genes described for HCM or DCM are also associated with RCM and LVNC (TABLE 2) [87,92,100,104,106,107]. This may indicate that RCM is a later spectrum of other clinically defined cardiomyopathies, or may indicate mutation-specific phenotypes. Much work is needed to delineate the molecular basis of familial RCM and LVNC in the context of other more common and well-defined cardiomyopathies.
General considerations for genetic testing of familial cardiomyopathies
Specific guidelines for the diagnosis, including the molecular diagnosis, and management of ARVC, HCM, DCM, LVNC and RCM have been written as expert opinions or consensus statements [1,108–110]. In general, physicians diagnosing and managing patients with cardiomyopathies should initially obtain a family history of at least three family generations [1]. Additional screening of at-risk family members with echocardiogram, ECG, history, physical examination and other tests may be warranted. It is important that genetic counseling takes place in parallel with these studies to ensure the patient understands the heritable basis of disease, the age at which the diseases might present and the presenting symptoms. The role of genetic counseling in genetic testing has never been more important because of our incomplete understanding of the genetic basis of cardiomyopathies, which continues to evolve. Patients with cardiomyopathies need to understand the utility, sensitivity, analytic validity and, most importantly, the implications of the test results [1]. A final component to the evaluation of cardiomyopathies is the consideration of genetic testing.
A recent practice guideline published by the Heart Failure Society of America provides graded recommendations for clinical genetic testing of cardiomyopathies [1]. For each cardiomyopathy evaluated by the guideline, the value of genetic testing of an affected individual is considered for its role in facilitating family screening and identification of at-risk relatives. The value was given a letter grade based on the current clinical and genetic knowledge, where the letter grade A corresponds to the highest score while C is the lowest. Genetic testing for HCM and ARVC were both given A level recommendations, genetic testing for DCM was given B level recommendation and genetic testing for RCM and LVNC were given C level recommendations [1].
If a disease-causing mutation is identified in a cardiomyopathy patient through genetic testing, then testing of family members can accurately predict the risk of those family members [1]. For an asymptomatic family member who is negative for their relative’s disease-causing mutation, that family member’s risk for developing the cardiomyopathy is considered to be the same level of risk as the general population and such individuals can forgo ongoing clinical screening for disease development [1]. Conversely, an asymptomatic family member who is positive for their relative’s disease-causing mutation is at substantially increased risk for developing the cardiomyopathy and should undergo continued clinical surveillance for disease development [1]. Genetic testing and targeted clinical surveillance of at-risk family members facilitates diagnosis during the early states of disease development and enables earlier clinical interventions. Nearly 40% of ARVC patients experience sudden cardiac death as their first clinical manifestation, however, with an early diagnosis and appropriate monitoring or treatment, often involving an implantable cardioverter defibrillator (ICD), most ARVC patients have an excellent prognosis [5,111–113]. Similarly, in DCM patients, where symptoms generally manifest after the disease has progressed to end-stage and the 5-year survival post-diagnosis is 50%, an early diagnosis allows for pharmacological treatments that may prevent disease progression and reduce complications or lead to transplantation [114]. In addition to confirming a diagnosis in a cardiomyopathy patient and identifying at-risk family members, genetic testing may also be useful for assessing the risk for conduction defects (ARVC, DCM), distinguishing the underlying cause of heart failure (DCM) and distinguishing other causes of adaptive hypertrophy with cardiomyopathies (HCM).
Genetic testing for ARVC
The Heart Failure Society of America assigned the grade of A for genetic testing of the one most clearly affected person in a family to facilitate screening and management of ARVC [1]. Although ARVC is a rare condition, analysis of a small number of genes can identify a significant proportion of cases. The testing modality most commonly applied by laboratories is DNA sequence analysis of the entire coding region or select exons of genes in which causative mutations have been identified (TABLE 5). Comprehensive sequence analysis of large regions of these genes is necessary due to allelic heterogeneity (multiple different mutations associated with disease) and the many ‘private’ mutations (mutations found in a single family) that can be present [5]. Sequencing of known ARVC-associated genes may identify mutations in as many as 50–55% of ARVC patients [5,17,115]. The 1994 Task Force diagnostic guidelines do not include genetic testing as a diagnostic criterion since they were developed in the pre-genetics era [116]. However, genetic testing may play an important role in ARVC diagnosis because the nonspecific features of ARVC render it difficult to diagnose, especially in the early disease stages [5]. Therefore, genetic testing for ARVC can fulfill an important role in the interpretation of borderline clinical investigations, as well as facilitate early diagnosis of family members potentially at-risk for developing ARVC. While risk for cardiac events or disease severity cannot be predicted based on specific mutations (i.e., genotype/ phenotype correlations), a recent genetic and clinical analysis of 82 clinically confirmed or suspected ARVC patients found that, in general, those patients with desmosomal mutations had an earlier onset of ARVC and were more likely to have ventricular tachycardia [17].
Table 5.
Current clinically available genetic testing modalities for the diagnosis of familial cardiomyopathies.
| Gene affected | Description | Analysis performed† | Laboratories performing test‡ (n) |
|---|---|---|---|
| Arrhythmogenic right ventricular cardiomyopathy | |||
| DSC2 | Desmocollin-2 | 1, 7, 10 | 6 |
| DSG2 | Desmoglein-2 | 1, 7, 10 | 5 |
| DSP | Desmoplakin | 1, 2, 7 | 6 |
| JUP | Junction plakoglobin | 1 | 2 |
| PKP2 | Plakophilin-2 | 1, 5, 7, 10 | 9 |
| RYR2 | Ryanodine receptor 2 | 1, 3, 4, 10 | 4 |
| TGFB3 | TGFβ3 | 1 | 1 |
| TMEM43 | Transmembrane protein 43 | 1, 2, 7 | 7 |
| Left ventricular noncompaction cardiomyopathy§ | |||
| DTNA | Dystrobrevin α | 1, 6, 7 | 3 |
| LDB3 | Lim domain-binding protein 3 | 1, 7 | 2 |
| LMNA | Lamin-A/C | 1, 3–5, 7 | 17 |
| TAZ | Tafazzin | 1, 6, 7 | 3 |
| Dilated cardiomyopathy | |||
| ABCC9 | ATP-binding cassette | 1, 7 | 1 |
| ACTC1 | Actin, α cardiac muscle 1 | 1, 7 | 4 |
| ACTN2 | α-actinin 2 | 1, 7 | 1 |
| CSRP3 | Cysteine and glycine-rich protein 3 | 1, 7 | 1 |
| DES | Desmin | 1, 7 | 2 |
| DMD | Dystrophin | 1–10 | 37 |
| LDB3 | Lim domain-binding protein 3 | 1, 7 | 2 |
| LMNA | Lamin-A/C | 1, 3–5, 7 | 17 |
| MYBPC3 | Myosin-binding protein C, cardiac type | 1, 7, 10 | 6 |
| MYH7 | Myosin 7 | 1, 7, 10 | 8 |
| PLN | Cardiac phospholamban | 1, 7 | 1 |
| SCN5A | Sodium channel protein type 5, subunit α | 1, 5, 7, 10 | 8 |
| SGCD | Sarcoglycan, δ | 1, 7, 10 | 3 |
| TAZ | Tafazzin | 1, 6, 7 | 6 |
| TCAP | Telethonin | 1, 7 | 2 |
| TNNI3 | Troponin I, cardiac muscle | 1, 2, 6, 7, 10 | 6 |
| TNNT2 | Troponin T, cardiac muscle | 1, 2, 7, 10¶ | 8 |
| TPM1 | Tropomyosin α-1 chain | 1, 7, 10 | 4 |
| TTN | Titin | 2 | 1 |
| VCL | Vinculin | 1, 7 | 1 |
| Hypertrophic cardiomyopathy | |||
| ACTC1† | Actin, α cardiac muscle 1 | 1, 5, 7 | 6 |
| CSRP3† | Cysteine and glycine-rich protein 3 | ||
| MYBPC3† | Myosin-binding protein C, cardiac type | 1, 2, 5, 7, 10 | 11 |
| MYH7† | Myosin heavy chain 7, cardiac muscle, β | 1, 2, 5, 7, 10 | 10 |
| MYL2 | Myosin light chain 2, regulatory, cardiac, slow | 1, 5, 7 | 7 |
| MYL3 | Myosin light chain 3, ventricular, slow | 1, 5, 6, 7 | 8 |
| TCAP† | Telethonin | 1, 7 | 2 |
| TNNC1 | Troponin C, cardiac muscle | 1, 5, 7 | 2 |
| TNNI3† | Troponin I, cardiac muscle | 1, 2, 5, 7, 10 | 9 |
| TNNT2† | Troponin T, cardiac muscle | 1, 2, 5, 7, 10§ | 10 |
| TPM1† | Tropomyosin α-1 chain | 1, 5, 7, 10 | 8 |
| TTN† | Titin | 2 | 1 |
| Restrictive cardiomyopathy | |||
| TNNI3† | Troponin I, cardiac muscle | 1, 2, 5, 7, 10 | 3 |
Types of analysis: 1: Analysis of entire coding region; 2: Sequence analysis of select exons; 3: Linkage analysis.; 4: Mutation scanning of select exons; 5: Deletion, duplication analysis; 6: Carrier testing; 7: Prenatal diagnosis; 8: FISH-metaphase; 9: FISH-anaphase; 10: Mutation scanning of entire coding region.
Covered in multiple disease phenotypes. The currently available testing methods were compiled from GeneTests. The designation of clinically available was made if the laboratory self-reported as being either a US CLIA-licensed laboratory or a non-US clinical laboratory. Verification must be made directly with the laboratory.
Testing for left ventricular noncompaction cardiomyopathy would include MYH7 as well, although not noted in GeneTests.
Testing procedures appear to be different for TNNT2 depending on what phenotype they are ordered for. Verification must be made directly with the laboratories performing the analysis.
Data from [140].
Several diseases mimic ARVC, which are important to recognize and rule out. These phenocopy diseases include myotonic dystrophy, which most closely mimics ARVC [25]. Myotonic dystrophy is caused by the unstable expansion of CTG trinucleotide repeats in the untranslated DMPK gene (encoding myotonic dystrophy protein kinase), localized to the intercalated disks of the cardiac muscle in proximity to gap junctions [117]. Similarly, sarcoidosis can mimic ARVC [26] and should be considered in the process of diagnosing disease.
Genetic testing for HCM
Molecular testing for HCM is the most established of the cardiomyopathies and has strong evidence to support clinical genetic testing [1]. The Heart Failure Society of America assigned the grade of A for genetic testing of the one most clearly affected person in a family to facilitate screening and management of HCM [1] (see also related recent reviews [118,119]). Clinical genetic testing is available for more than 20 HCM-susceptibility genes; however, analysis of two genes, MYH7 and MYBPC3, account for the majority of mutations that are identified in HCM patients (TABLES 3 & 5). Mutations in MYH7 and MYBPC3 account for approximately 80% of all genotype-positive HCM patients [120]. Genetic testing of the eight genes most commonly associated with HCM, which all encode components of the cardiac sarcomere, identify mutations in 35–65% of patients that meet the clinically accepted definition of HCM [118]. While strict gene- and genotype–phenotype correlations have been attempted, definitive relationships between the mutated gene and disease manifestation are not generally thought to exist across all populations. For example, based on studies of single mutations in large families or small cohorts, MYBPC3 mutations have gained the reputation of causing later onset disease, MYH7 mutations with earlier manifestation of disease, and TNNT2 mutations with mild LVH and increased risk of sudden death in some families [30,31,35,121–125]. However, in unrelated HCM cases where many rare, private mutations exist, the two most common forms of HCM caused by mutations of MYH7 and MYBPC3 are phenotypically indistinguishable in terms of age at diagnosis, extent of hypertrophy and family history [126,127]. HCM cases with MYH7 mutations have been reported with reputed TNNT2-like features of mild hypertrophy and early SCD, while other HCM cases with TNNT2 mutations have been shown to exhibit left ventricular hypertrophy with thickening of more than 3 cm [36,128]. Moreover, recurrent identification and analysis of identical mutations in unrelated HCM cases indicate that the prognostic implications of specific mutations, as well as of specific genes, must be assigned with great caution [126–129]. Interestingly, a recent prospective analysis of a large cohort of unrelated Italian HCM patients showed that HCM patients with a mutation in any one of eight myofilamentous sarcomeric genes (MYPBC3, MYH7, MYL2, MYL3, TNNT2, TNNI3, TPM1 and ACTC) were at increased risk for cardiovascular death, nonfatal ischemic stroke, or progression to severe heart failure symptoms compared with HCM patients with a negative genetic test result [130]. In a multivariate model that included established risk predictors in HCM, the presence of a myofilamentous sarcomeric gene mutation was associated with a more than fourfold independent increase in risk for unfavorable outcomes compared with patients with HCM and negative genetic test results [130], supporting a prognostic role for genetic testing in patients with clinically diagnosed HCM.
There are several rare diseases that mimic the phenotype of HCM that do not involve mutations in the sarcomere or sarcomere-associated genes. These diseases include familial Wolff–Parkinson–White syndrome, Anderson–Fabry disease, Pompe disease, Glycogen Storage disease Type III, Danon disease, LEOPARD syndrome/Noonan syndrome, and Frederich ataxia (for a review see [4]). It is important to recognize the differential phenotypes and/or perform the molecular test to identify these phenocopy diseases, as the treatments vary considerably. Several rare multisystem metabolic diseases, such as Danon disease and Anderson–Fabry disease, can present primarily with cardiac manifestations and mimic HCM caused by sarcomeric mutations; however, these metabolic diseases involve fundamentally different pathological processes and have different clinical courses and therapeutic strategies compared with typical HCM patients (TABLE 3) [131–133]. Anderson–Fabry disease is treated primarily with enzyme replacement [134], while Danon disease has a severe prognosis that may warrant earlier consideration of a heart transplant [131]. The recent Heart Failure Society of America recommends genetic testing when cardiomyopathy is associated with extra-cardiac manifestations (level of evidence grade A), as many of these HCM phenocopy diseases can be detected by genetic testing [1].
Genetic testing for DCM
The Heart Failure Society of America assigned the grade of B for genetic testing of the one most clearly affected person in a family to facilitate screening and management of DCM [1]. The recommendation for genetic testing for DCM was less strong because, although an ever-growing number of genes have been identified in association with DCM, none is associated with a significant proportion of DCM and together the currently known genes account for only a small percentage (~25%) of familial DCM. The analysis of 20 genes is currently available for patients suspected of having a genetic etiology underlying their idiopathic DCM, although most of these genes are infrequent causes of DCM (TABLES 4 & 5). Genetic testing of two genes that are the most common causes of DCM, LMNA and MYH7, is estimated to identify mutations in approximately 10% of idiopathic DCM patients, while testing of the more than 20 DCM-associated genes may account for an additional 10–15% of patients [1,56]. However, the frequency of LMNA mutations in DCM patients with conduction disease, particularly when skeletal muscle involvement is present, rises to 30–45% [64,135,136]. Thus, testing for mutations in LMNA is recommended for patients with idiopathic DCM and conduction system defects based on the overall higher mutation frequency in this subset of patients, even if a family history of DCM is not present [1]. Identification of a LMNA mutation portends a particularly poor prognosis, with nearly half of cases with LMNA-related cardiomyopathy suffering sudden cardiac death [137,138]. LMNA mutations may identify patients at risk for sudden cardiac death prior to presentation with heart failure: eight out of 19 (42%) patients who underwent permanent pacing and ICD therapy solely on the basis of the presence of LMNA mutations with cardiac conduction defects and normal ventricular function received appropriate ICD intervention [139]. Owing to the extensive genetic heterogeneity and the high frequency of private mutations in DCM, the mainstay of molecular diagnosis in DCM is sequence analysis of entire protein-coding regions of multiple DCM-associated genes either by direct Sanger DNA sequencing or with resequencing arrays [1,140,141].
As with ARVC and HCM, there are a number of diseases that mimic DCM and these should be considered in DCM diagnosis. Approximately 80% of patients with Duchenne’s muscular dystrophy and approximately 10% with Becker’s muscular dystrophy have DCM [142]. In addition to the mutations in dystrophin associated with these diseases, DCM has been associated with mutations in intermediate filament lamin A/C, which underlie Emery–Dreifuss muscular dystrophy with DCM [143].
Genetic testing for LVNC & RCM: is it worthwhile right now?
The Heart Failure Society of America assigned the lowest grade of C for their recommendation for genetic testing of the most clearly affected person in a family to facilitate screening and management for both LVNC and RCM [1]. These recommendations are largely due to the fact that there are few known genotype–phenotype correlations for either of these conditions [107], and only a fraction of the patients with disease have identifiable mutations. Collective testing of all known LVNC-associated genes may identify mutations in 20–25% of patients with LVNC. Identification of affected genes is primarily useful for identifying at-risk family members, including at-risk pregnancies. Genetic testing is available to identify LVNC-causing mutations in the DTNA, TAZ, LMNA and LDB3 genes by sequence analysis of the entire coding region (TABLE 5). It is not known what percentage of patients with LVNC have mutations in these four genes, but mutations in additional genes have been identified, including FK506-binding protein (FKBP-12), MYH7, ACTC and TNNT2 [92,107]. The most commonly affected proteins that can be tested for in LVNC include MYH7, TNNT2 and ACTC based on their reported prevalence. Similarly, no genotype–phenotype correlations have been described for RCM and estimates for the prevalence of mutations in MYH7 and TNNI3 are unavailable, as only a few studies have reported RCM families with mutations in these genes [99,104]. Since RCM is quite rare, it may be some time before large-scale studies can be performed to determine how common MYH7 and TNNI3 mutations are and to identify other causative genes. As more genes and mutations underlying LVNC and RCM are discovered, testing sensitivity will increase, thus, improving the clinical utility of testing. This will require identification of additional disease-associated genes and whether any genotype–phenotype correlations exist that may help in the prognosis and therapeutic strategies applied to patients with these clinical phenotypes.
Challenges in genetic testing for cardiomyopathies
There are a number of challenges that arise in genetic testing for cardiomyopathies. The major challenges are the difficulty and cost of testing due to the large number of genes (locus heterogeneity) and different mutations (allelic heterogeneity) associated with each of the cardiomyopathies, the poor sensitivity for diagnosis of familial cardiomyopathies and the interpretation of the significance of individual mutations identified, particularly novel variants. For these reasons, it is particularly important to have good clinical correlation and to rule out other causes of cardiomyopathy or demonstrate strong evidence of a family history prior to undergoing genetic testing. Critical to this process is genetic counseling to ensure that the patient understands the heritability of cardiomyopathies, family screening recommendations, genetic testing options, and the value and interpretation of testing based on recent guidelines for the genetic evaluation of cardiomyopathies [1,119].
Direct DNA sequence analysis of all protein-coding regions of selected genes is the most common methodology applied to cardiomyopathy genetic testing. Owing to the high cost and labor associated with DNA sequencing of multiple large genes, mutation scanning of the entire (or selected) coding regions is sometimes performed, which is a less costly method of identifying region(s) of the gene likely harboring mutation(s). These regions can then be targeted for subsequent sequence analysis to identify the mutation(s) (for a review see [4]). In addition, some laboratories have incorporated microarray chip-based ‘resequencing’, in which overlapping DNA oligonucleotides specific for every possible single nucleotide substitution are tiled onto a custom DNA microarray chip [144–146]. The major advantage of microarray resequencing approaches is that multiple genes can be analyzed on a single chip, which allows for a significant savings in labor and cost after the initial investment in capital equipment and assay development. A major disadvantage of resequencing arrays is that novel insertions and deletions (indels) are difficult to detect, thus decreasing the sensitivity for mutation detection, although previously identified indels can be detected using specific tiled probes [144,145,147]. In the future, next-generation sequencing (NGS) technologies may enable clinical large-scale testing of multiple genes; at the present time NGS approaches are expensive and have not been well validated for clinical accuracy, so they are primarily useful as a research tool to identify additional genes that may be associated with cardiomyopathy.
Despite extensive analysis of the entire coding regions of multiple genes, not all patients are found to have disease-causing mutations because the genetic basis of these diseases are not completely understood. For example, even in HCM, the best characterized of the cardiomyopathies, at least 40% of HCM patients have no identifiable mutation with current genetic tests [118,148]. The list of cardiomyopathy-associated genes is constantly evolving and sensitivity may be improved as additional genes are added; however, the inclusion of each additional gene may improve test sensitivity only slightly, as each gene individually may account for a small percentage of cases. Another possible explanation for decreased test sensitivity is the presence of mutations that are undetectable by commonly used approaches. Most genetic tests for cardiomyopathy-susceptibility genes are not designed to detect deep intronic mutations, large indels or gross genomic rearrangements, any of which may be deleterious and lead to cardiomyopathy. However, at least in the case of MYBPC3 and TNNT2, large deletions or rearrangements do not appear to play a role in the pathogenesis of HCM [149]. This contrasts with the RYR2 and LMNA gene deletions found to cause cardiomyopathy (ARVC and DCM, respectively), which were missed because standard sequencing techniques were unable to identify mutations [150,151].
Thus, because not all genetic causes of cardiomyopathy have been determined, genetic testing has limited sensitivity for diagnosis, despite high specificity. In practical terms, low test sensitivity and high specificity means that if a known disease-causing mutation is found, it should be considered as strong evidence for the diagnosis of familial cardiomyopathy. However, in cases where a disease-causing mutation is not found, the presence of familial cardiomyopathy cannot be ruled out, making clinical diagnosis still important. As the full spectrum of genetic mutations associated with the cardiomyopathies is identified, the sensitivity of genetic testing will continue to improve and may eventually evolve to the point that genetic testing is useful to ‘rule out’ a diagnosis of familial cardiomyopathy.
Another major challenge in clinical genetic testing is differentiating pathogenic mutations from benign ‘background’ genetic variation unrelated to disease. The human genome demonstrates significant genetic sequence variability, and differentiating deleterious mutations from benign sequence variation is a universal challenge for clinical genetic laboratories. The term ‘mutation’ is traditionally defined by the rarity of a genetic variant within a given population and not by an association with disease [152]. By this definition, any genetic variant with an allele frequency of less than 1% is considered a mutation, whereas variants present in the population with more than 1% frequency are considered common polymorphisms. Cardiomyopathies collectively result from mutations in more than 30 different genes in which hundreds of mutations are already known and new mutations are continuously identified in suspected cardiomyopathy patients. Most of these mutations are so rare that they are effectively ‘family-specific’ causes of disease (private mutations). Both novel sequence variants and previously identified variants whose association with disease is not definitively established are considered ‘variants of uncertain/unknown significance’ (VUS) [153]. A recent example of this was reported by Christensen et al. who screened 53 unrelated patients fulfilling Task Force criteria for ARVC for mutations in PKP2 by direct sequencing [154]. Seven patients carried missense mutations, which were also identified in healthy control populations, leaving their significance unknown at this time [154].
Multiple direct and predictive approaches have been used to distinguish disease-causing mutations from benign variants, including genetic testing of healthy individuals to determine allele frequency, cosegregation studies to determine if disease and mutation track together within a family, analysis of protein sequence conservation across different species to predict the importance of a specific mutated amino acid for a protein’s function, prediction of the effect of a mutation upon mRNA splicing or translation and functional characterization of the mutant gene product using in vitro and in vivo model systems.
Mutation analysis of control populations of different ethnic groups is critical for aiding interpretation of genetic test results, as the extent and type of genetic sequence variability varies between evolutionarily related populations. Cohort samples from control populations are available, both commercially and in individual genetics laboratories, and may consist of fewer than a hundred individuals to more than a thousand depending on the source and ethnic group. Sequencing the same genomic region in approximately 350 individuals in a population is estimated to identify all of the common polymorphisms (those variants with at least a 1% allele frequency) in the entire population. However, the real challenge lies in distinguishing pathogenic mutations from more rare benign ‘background’ variants with less than 1% frequency. While sequencing just 150 individuals in a control population is estimated to identify 80% of variants with a frequency of at least 0.1%, confidently identifying all variants with a frequency of at least 0.1% in a given population requires more than 3000 individuals [155]. Such large control datasets do not exist for cardiomyopathy genes today; however, large-scale genetic variation sequencing projects, such as the International HapMap Project and the 1000 Genomes Project, will make gene-specific variation data from large numbers of individuals universally available [156,157]. As increasing numbers of healthy individuals have specific disease-associated genes sequenced as part of these projects, knowledge of the extent of normal genetic variation within these disease-associated genes will increase and enable more informed interpretations of novel variants encountered in clinical genetic testing.
A promising population-based approach for distinguishing pathogenic mutations from benign variants was recently described for long QT syndrome (LQTS) [158]. Enabled by an extensive collection of mutations identified in LQTS patients and rare variants identified in more than 1000 control subjects, Kapa and colleagues identified regions of high and low specificity for predicting the pathogenicity of novel variants within the most common LQTS-susceptibility genes [158]. The practical outcome of such case–control variant analyses is to assign disease-causing probabilities to novel, uncharacterized variants. A prerequisite for such analyses is large genetic databases of both case and control populations. While large numbers of cases have been screened for HCM mutations, similar numbers of control subjects have not. Fortunately, the 1000 Genomes Project will soon enable such analyses in the cardiomyopathy genetic testing field by publicly providing variant data within HCM-susceptibility genes of healthy control subjects [156,157].
Another approach to determine whether a mutation is deleterious is to conduct family studies that track whether the cardiomyopathy and the mutation present in a family segregate together; however, such family studies are often hindered by the lack of relatives available for evaluation and the phenomena of reduced penetrance and/or age-related penetrance. Functional characterization of mutant gene products may help determine the pathogenic effect of sequence variants identified in cardiomyopathy patients. However, given the large number of novel putative pathogenic mutations still being discovered, it is impossible to expect timely functional characterization of all variants in the current labor-intensive in vitro and in vivo functional assays. Moreover, universally applicable ‘gold standard’ functional characterization assays do not exist for cardiomyopathy-associated proteins and a negative result in current assays may simply indicate the existence of novel and presently undetectable disease mechanisms.
Without ‘gold standard’ functional assays, reliable and accurate predictive algorithmic approaches that model the pathogenicity of novel mutations identified would be very useful. Sort Intolerant from Tolerant (SIFT) and Polymorphism Phenotype (PolyPhen) are two sequence homology-based software tools intended to predict the potential impact of a mutation on protein function [159,160]. While these tools are particularly useful for prioritizing research-based studies of specific mutations, they should be used cautiously for interpreting the significance of novel genetic variants identified in a clinical genetic test. An analysis of nearly 45,000 nonsynonymous polymorphisms by SIFT and PolyPhen demonstrated that each program independently predicted approximately a third of common polymorphisms to be deleterious or damaging with approximately 60% concordance [161]. This may indicate that these programs are overly sensitive and likely to label many benign variants as potentially disease-associated, resulting in false-positive interpretations of genetic test results. These tools should undergo rigorous evaluation in a disease- and gene-specific fashion by comparing the predictions produced for proven disease-associated mutations to the predictions for known benign variants in the same genes.
With whole-genome scale sequencing technologies promising affordable and overwhelming amounts of sequence data, resolving the significance of new genetic variants will continue to be a major challenge. Extending sequencing from protein coding regions to non-protein-coding regions of genes, including intronic regions, promoters, 5′ and 3′ untranslated regions, may identify synonymous deleterious variants that alter splicing or gene-expression levels, but as noncoding regions tend to harbor extensive genetic variability, such expanded analyses will greatly increase the number of VUSs identified. Until the extent of variability in relevant genes is cataloged and characterized, the interpretation of VUSs will continue to be a real challenge to interpretation of genetic testing for cardiomyopathies.
Conclusion
Over the past 20 years, there has been an increasing appreciation for the genetic basis of cardiomyopathies. The recognition of a number of clinically distinct cardiomyopathies (HCM, DCM, RCM, ARVC and LVNC) has led to intense investigation for underlying genetic defects. The genetic defects in HCM are the best characterized to date, with up to 65% of patients having identifiable disease-causing mutations. Similarly robust genetic testing for ARVC can identify an estimated 50–55% of ARVC patients. However, the collective testing of more than 20 genes identifies a genetic cause for only approximately 25% of idiopathic DCM patients. Since RCM and LVNC are relatively rare, studies determining the prevalence of mutations in these populations have not been performed. Thus, genetic testing for cardiomyopathies is currently limited by poor sensitivity for disease diagnosis, although the specificity of these tests is high. Despite these limitations, professional recommendations have been published for the use of genetic testing in the diagnosis of familial HCM, ARVC and DCM, with the caveat that negative test results do not rule out the presence of a heritable cardiomyopathy [1,108–110]. Furthermore, genotype–phenotype correlations are not well understood at this time, limiting the utility of genetic testing for prediction of disease course or severity in an affected individual and making genetic testing primarily useful for identifying the presence of known familial mutations in at-risk family members in order to target appropriate relatives for careful follow up. Although mutation- and disease-specific therapies have been suggested and may become available in the future, genetic testing for this use is currently premature [162].
Expert commentary
The diagnosis of cardiomyopathy often begins with the identification of characteristic structural and functional abnormalities of the heart, typically using clinical imaging techniques, such as echocardiography or MRI. Such findings should prompt a thorough discussion between the physician and the patient, directed at elucidating the clinical consequences and possible etiologies for the abnormalities. In the absence of other causative factors, the diagnosis of familial cardiomyopathy must be considered. The interplay between increasing diagnostic awareness and significant advances in DNA sequencing technology has revealed a rapidly expanding list of genes that cause heritable cardiomyopathies.
At present, the tools available to the clinician for diagnosing familial cardiomyopathies have outstripped the therapeutic options available for their treatment, although it is exciting to imagine a future in which the diagnosis of gene-specific abnormalities could prompt the initiation of gene-specific therapy. Nor is there currently a sufficient understanding of the relationship between the genotype and phenotype of these disorders to support the differential use of existing therapies for patients with familial cardiomyopathy. Thus, a patient with DCM due to a mutation in MYH7 is treated using the same clinical approach as a patient with DCM secondary to viral myocarditis. The true clinical benefit to making a diagnosis of familial cardiomyopathy arises from the possibility of identifying relatives of the proband who are presymptomatic and initiating life-extending evidence-based therapies.
Through little fault of their own, the vast majority of clinicians remain unaware of the ever-lengthening list of disease-causing mutations, let alone how to pursue their identification. The National Center for Biotechnology Information (NCBI) maintains a medical genetics website that compiles much of the present knowledge regarding the genetic causes of heritable disorders, including cardiomyopathies [301]. A search for ‘cardiomyopathy’ on the website reveals results organized by type of cardiomyopathy followed by lists of the known causative mutations (reviewed in TABLE 5). Many of the listed genes are accompanied by links to the contact information for clinical laboratories that will perform sequence analysis for the chosen mutation. While not completely contemporary, GeneTests is an extensive and important – if somewhat daunting – resource for clinicians interested in evaluating a patient for familial cardiomyopathy. In an effort to simplify the diagnostic interface, multiple laboratories have begun offering ‘cardiomyopathy panels’ based on the cardiac phenotype (e.g., DCM, HCM and ARVC) that use gene chips to screen a single sample for almost all of the mutations known to cause the cardiomyopathy of interest. Such an approach removes the onus from the provider for searching either their memory or the internet for a list of the genes to be sequenced. These sequencing services are available from both academically affiliated (Harvard/ Partners Healthcare) and commercial (Correlagen Diagnostics, Inc., GeneDx, PGxHealth/FAMILION) laboratories. Chip-based assays have also been developed and may provide cheaper and faster diagnosis of HCM [144,145] and DCM [146], although with some limitations. It seems likely that the availability of such efficient and user-friendly testing will increase the frequency with which clinicians will seek, and thus find, the diagnosis of familial cardiomyopathy.
Five-year view
It would be optimistic to predict that in 5 years we would recognize most of the genes and disease-causing mutations for the five clinically distinct cardiomyopathies. However, continued research efforts will probably result in a substantial increase in the number of genes identified that are associated with familial cardiomyopathies. It is expected that the identification of additional genes and the application of recent technical advances, such as next-generation sequencing and microarray-based ‘resequencing’ to genetic diagnosis (as recently reviewed in [4]) will lead to dramatic improvement in the clinical sensitivity and cost–effectiveness of genetic testing for cardiomyopathies. We also predict that the bioinformatic and functional approaches reviewed in this article will become valuable in distinguishing novel pathogenic mutations from novel benign variants. This will be assisted by the rapid increase in our understanding of the genetic variation in the human genome in both health and disease by large-scale resequencing efforts, which are currently underway or have yet to begin. While not discussed in detail here, genetic modifiers of HCM have been identified, which when present are associated with a greater severity of disease in some studies [4,163–165]. In the future, more conclusive genotype–phenotype relationships may be made by identifying mutations not only in specific structural genes but also in such comodifier genes, which may drive specific treatment modalities based on the spectrum of mutations identified. Lastly, we predict the identification and experimental application of mutation specific therapy to cardiomyopathies. For example, the use of drugs to read-through specific truncation mutations has been applied to Duchenne muscular dystrophy and cystic fibrosis [162,166]; its application to cardiomyopathies is certain to be tested in the near future. The prospect of specific therapies guided by mutation testing or targeting individual mutations is exciting given the lack of currently available therapies for most of the cardiomyopathies.
Key issues.
Cardiomyopathies are a clinically and genetically heterogeneous group of heart muscle diseases associated with mechanical and/or electrical dysfunction that may predispose patients to sudden cardiac death.
Over the last two decades, the association of specific genes involved with clinically distinct cardiomyopathies (hypertrophic cardiomyopathy, dilated cardiomyopathy, restrictive cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and left ventricular noncompaction cardiomyopathy) has shed light onto the pathophysiology and identification of these diseases. Not all disease-causing mutations have been identified, which currently limits the sensitivity of these tests. Mutations in the same genes may also underlie different cardiomyopathies.
With relatively cheaper and quicker whole-genome scale sequencing technologies promising overwhelming amounts of sequence data on the horizon, interpreting and resolving the significance of rare genetic variants will become a major challenge on which to focus.
Without ‘gold standard’ assays that can prove a pathogenic effect, it can be difficult to differentiate between novel disease-causing mutations and rare benign genetic variation that is seen in the general population.
Clinical genetic tests for several cardiomyopathies are commercially available. The major clinical utility shared by the different cardiomyopathies is the ability to accurately predict the risk for a family member for developing a familial cardiomyopathy who currently has little or no clinical evidence of disease.
The role of genetic counseling in genetic testing has never been more important because of our incomplete understanding of the genetic basis of cardiomyopathies that continues to evolve. Patients with cardiomyopathy need guidance to understand the complex and evolving issues of testing utility, sensitivity, analytic validity and implications of possible testing results.
Acknowledgments
The authors thank Melvin Scheinman from the University of California, San Francisco School of Medicine for assistance with the original clinical examples of familial ARVC included in the figures.
Footnotes
Financial & competing interests disclosure
The authors are supported by the American Heart Association (Scientist Development Grant to Monte Willis) and the NIH (1K08HL096836–01 to Brian Jensen). Thomas Callis is a Clinical Genetics Liaison in PGxHealth, a division of Clinical Data, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Thomas E Callis, PGxHealth Division, Clinical Data, Inc., 5 Science Park, New Haven, CT 06511, USA.
Brian C Jensen, McAllister Heart Institute, University of North Carolina, Chapel Hill, NC, 27599-7126, USA and Department of Internal Medicine, Section of Cardiology, University of North Carolina, Chapel Hill, NC 27599-7075, USA.
Karen E Weck, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599-7525, USA.
Monte S Willis, Email: monte_willis@med.unc.edu, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599-7525, USA and McAllister Heart Institute, University of North Carolina at Chapel Hill, 2340B Medical Biomolecular Research Building, 103 Mason Farm Road, Chapel Hill, NC 27599-7525, USA Tel.: +1 919 843 1938 Fax: +1 919 843 4585.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1•.Hershberger RE, Lindenfeld J, Mestroni L, et al. Genetic evaluation of cardiomyopathy – a Heart Failure Society of America practice guideline. J Card Fail. 2009;15(2):83–97. doi: 10.1016/j.cardfail.2009.01.006. Comprehensively evaluated the evidence for genetic evaluation, clinical screening, and molecular genetic testing of cardiomyopathies (hypertrophic cardiomyopathy [HCM], dilated cardiomyopathy [DCM], restrictive cardiomyopathy [RCM], arrhythmogenic right ventricular cardiomyopathy [ARVC] and left ventricular noncompaction cardiomyopathy [LVNC]) based in published studies. [DOI] [PubMed] [Google Scholar]
- 2•.Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113(14):1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. Presents the working framework for cardiomyopathies in the context of molecular genetics and their diverse phenotypes. Importantly, it recognizes the importance of molecular genetic testing and introduces several new entities, including LVNC, in the context of other cardiomyopathies. [DOI] [PubMed] [Google Scholar]
- 3.Ingles J, Doolan A, Chiu C, et al. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42(10):e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez JE, McCudden CR, Willis MS. Familial hypertrophic cardiomyopathy: basic concepts and future molecular diagnostics. Clin Biochem. 2009;42(9):755–765. doi: 10.1016/j.clinbiochem.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Sen-Chowdhry S, Syrris P, McKenna WJ. Role of genetic analysis in the management of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2007;50(19):1813–1821. doi: 10.1016/j.jacc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Nava A, Thiene G, Canciani B, et al. Familial occurrence of right ventricular dysplasia: a study involving nine families. J Am Coll Cardiol. 1988;12(5):1222–1228. doi: 10.1016/0735-1097(88)92603-4. [DOI] [PubMed] [Google Scholar]
- 7.Basso C, Thiene G, Corrado D, et al. Arrhythmogenic right ventricular cardiomyopathy. Dysplasia, dystrophy, or myocarditis? Circulation. 1996;94(5):983–991. doi: 10.1161/01.cir.94.5.983. [DOI] [PubMed] [Google Scholar]
- 8.Fontaine G, Frank R, Guiraudon G, et al. Significance of intraventricular conduction disorders observed in arrhythmogenic right ventricular dysplasia. Arch Mal Coeur Vaiss. 1984;77(8):872–879. [PubMed] [Google Scholar]
- 9.Thiene G, Basso C. Arrhythmogenic right ventricular cardiomyopathy: an update. Cardiovasc Pathol. 2001;10(3):109–117. doi: 10.1016/s1054-8807(01)00067-9. [DOI] [PubMed] [Google Scholar]
- 10.Corrado D, Basso C, Thiene G, et al. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J Am Coll Cardiol. 1997;30(6):1512–1520. doi: 10.1016/s0735-1097(97)00332-x. [DOI] [PubMed] [Google Scholar]
- 11.Thiene G, Corrado D, Nava A, et al. Right ventricular cardiomyopathy: is there evidence of an inflammatory aetiology? Eur Heart J. 1991;12(Suppl D):22–25. doi: 10.1093/eurheartj/12.suppl_d.22. [DOI] [PubMed] [Google Scholar]
- 12.Bowles NE, Ni J, Marcus F, Towbin JA. The detection of cardiotropic viruses in the myocardium of patients with arrhythmogenic right ventricular dysplasia/ cardiomyopathy. J Am Coll Cardiol. 2002;39(5):892–895. doi: 10.1016/s0735-1097(02)01688-1. [DOI] [PubMed] [Google Scholar]
- 13.Calabrese F, Basso C, Carturan E, Valente M, Thiene G. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: is there a role for viruses? Cardiovasc Pathol. 2006;15(1):11–17. doi: 10.1016/j.carpath.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Gras E, Lombardi R, Giocondo MJ, et al. Suppression of canonical Wnt/β-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116(7):2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Djouadi F, Lecarpentier Y, Hebert JL, et al. A potential link between peroxisome proliferator-activated receptor signalling and the pathogenesis of arrhythmogenic right ventricular cardiomyopathy. Cardiovasc Res. 2009;84(1):83–90. doi: 10.1093/cvr/cvp183. As ARVC is recognized as a defect in the desmin structure (desminopathy), novel underlying causes are still being identified as underlying causes of ARVC, such as mutations in PPAR signaling pathways described in this work. [DOI] [PubMed] [Google Scholar]
- 16.Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5(5):258–267. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Haan A, Tan B, Zikusoka M, Llado L. Comprehensive desmosome mutation analysis in North Americans with arrythmogenic right ventricular dysplasia/ cardiomyopathy. Circ Cardiovasc Genet. 2009;2(5):428–435. doi: 10.1161/CIRCGENETICS.109.858217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Tintelen JP, Entius MM, Bhuiyan ZA, et al. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation. 2006;113(13):1650–1658. doi: 10.1161/CIRCULATIONAHA.105.609719. [DOI] [PubMed] [Google Scholar]
- 19.Syrris P, Ward D, Asimaki A, et al. Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113(3):356–364. doi: 10.1161/CIRCULATIONAHA.105.561654. [DOI] [PubMed] [Google Scholar]
- 20.Antoniades L, Tsatsopoulou A, Anastasakis A, et al. Arrhythmogenic right ventricular cardiomyopathy caused by deletions in plakophilin-2 and plakoglobin (Naxos disease) in families from Greece and Cyprus: genotype–phenotype relations, diagnostic features and prognosis. Eur Heart J. 2006;27(18):2208–2216. doi: 10.1093/eurheartj/ehl184. [DOI] [PubMed] [Google Scholar]
- 21.Dalal D, James C, Devanagondi R, et al. Penetrance of mutations in plakophilin-2 among families with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2006;48(7):1416–1424. doi: 10.1016/j.jacc.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Corrado D, Thiene G. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: clinical impact of molecular genetic studies. Circulation. 2006;113(13):1634–1637. doi: 10.1161/CIRCULATIONAHA.105.616490. [DOI] [PubMed] [Google Scholar]
- 23.Beffagna G, Occhi G, Nava A, et al. Regulatory mutations in transforming growth factor-β3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65(2):366–373. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Merner ND, Hodgkinson KA, Haywood AF, et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82(4):809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen-Chowdhry S, Syrris P, McKenna WJ. Genetics of right ventricular cardiomyopathy. J Cardiovasc Electrophysiol. 2005;16(8):927–935. doi: 10.1111/j.1540-8167.2005.40842.x. [DOI] [PubMed] [Google Scholar]
- 26.Ott P, Marcus FI, Sobonya RE, et al. Cardiac sarcoidosis masquerading as right ventricular dysplasia. Pacing Clin Electrophysiol. 2003;26(7 Pt 1):1498–1503. doi: 10.1046/j.1460-9592.2003.t01-1-00217.x. [DOI] [PubMed] [Google Scholar]
- 27••.Asimaki A, Tandri H, Huang H, et al. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N Engl J Med. 2009;360(11):1075–1084. doi: 10.1056/NEJMoa0808138. Identifies for the first time that routine immunohistochemical analysis of conventional endomyocardial-biopsy samples appear to be a sensitive and specific test for ARVC. [DOI] [PubMed] [Google Scholar]
- 28.Maron BJ, Gardin JM, Flack JM, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 29.Maron BJ, Pelliccia A. The heart of trained athletes: cardiac remodeling and the risks of sports, including sudden death. Circulation. 2006;114(15):1633–1644. doi: 10.1161/CIRCULATIONAHA.106.613562. [DOI] [PubMed] [Google Scholar]
- 30.Niimura H, Bachinski LL, Sangwatanaroj S, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338(18):1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 31.Charron P, Dubourg O, Desnos M, et al. Genotype–phenotype correlations in familial hypertrophic cardiomyopathy. A comparison between mutations in the cardiac protein-C and the β-myosin heavy chain genes. Eur Heart J. 1998;19(1):139–145. doi: 10.1053/euhj.1997.0575. [DOI] [PubMed] [Google Scholar]
- 32.Fananapazir L, Epstein ND. Genotype–phenotype correlations in hypertrophic cardiomyopathy. Insights provided by comparisons of kindreds with distinct and identical β-myosin heavy chain gene mutations. Circulation. 1994;89(1):22–32. doi: 10.1161/01.cir.89.1.22. [DOI] [PubMed] [Google Scholar]
- 33.Havndrup O, Bundgaard H, Andersen PS, et al. The Val606Met mutation in the cardiac β-myosin heavy chain gene in patients with familial hypertrophic cardiomyopathy is associated with a high risk of sudden death at young age. Am J Cardiol. 2001;87(11):1315–1317. doi: 10.1016/s0002-9149(01)01532-6. [DOI] [PubMed] [Google Scholar]
- 34.Watkins H, McKenna WJ, Thierfelder L, et al. Mutations in the genes for cardiac troponin T and α-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332(16):1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 35.Moolman JC, Corfield VA, Posen B, et al. Sudden death due to troponin T mutations. J Am Coll Cardiol. 1997;29(3):549–555. doi: 10.1016/s0735-1097(96)00530-x. [DOI] [PubMed] [Google Scholar]
- 36.Christiaans I, Lekanne dit Deprez RH, van Langen IM, Wilde AA. Ventricular fibrillation in MYH7-related hypertrophic cardiomyopathy before onset of ventricular hypertrophy. Heart Rhythm. 2009;6(9):1366–1369. doi: 10.1016/j.hrthm.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 37.Keren A, Syrris P, McKenna WJ. Hypertrophic cardiomyopathy: the genetic determinants of clinical disease expression. Nat Clin Pract Cardiovasc Med. 2008;5(3):158–168. doi: 10.1038/ncpcardio1110. [DOI] [PubMed] [Google Scholar]
- 38.van Dijk SJ, Dooijes D, dos Remedios C, et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119(11):1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 39.Thierfelder L, Watkins H, MacRae C, et al. A-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77(5):701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 40••.Mearini G, Gedicke C, Schlossarek S, et al. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc Res. 2009;85(2):357–366. doi: 10.1093/cvr/cvp348. Reports specific mechanisms by which the ubiquitin proteasome system targets wild-type and mutant saromere MyBP-c differentially by cardiac specific ubiquitin ligases. This article may give vital insight to the underlying pathophysiology of a number of cardiomyopathies and may also help identify where therapies need to target to be specific and effective in treating sarcomere-based cardiomyopathies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirschner SE, Becker E, Antognozzi M, et al. Hypertrophic cardiomyopathy-related β-myosin mutations cause highly variable calcium sensitivity with functional imbalances among individual muscle cells. Am J Physiol Heart Circ Physiol. 2005;288(3):H1242–H1251. doi: 10.1152/ajpheart.00686.2004. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Xu Y, Kerrick WG, et al. Prolonged Ca2+ and force transients in myosin RLC transgenic mouse fibers expressing malignant and benign FHC mutations. J Mol Biol. 2006;361(2):286–299. doi: 10.1016/j.jmb.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Landstrom AP, Weisleder N, Batalden KB, et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42(6):1026–1035. doi: 10.1016/j.yjmcc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minamisawa S, Sato Y, Tatsuguchi Y, et al. Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2003;304(1):1–4. doi: 10.1016/s0006-291x(03)00526-6. [DOI] [PubMed] [Google Scholar]
- 45.Haghighi K, Kolokathis F, Gramolini AO, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103(5):1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crilley JG, Boehm EA, Blair E, et al. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol. 2003;41(10):1776–1782. doi: 10.1016/s0735-1097(02)03009-7. [DOI] [PubMed] [Google Scholar]
- 47.Mestroni L, Maisch B, McKenna WJ, et al. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20(2):93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 48.Franz WM, Muller OJ, Katus HA. Cardiomyopathies: from genetics to the prospect of treatment. Lancet. 2001;358(9293):1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- 49.Schonberger J, Seidman CE. Many roads lead to a broken heart: the genetics of dilated cardiomyopathy. Am J Hum Genet. 2001;69(2):249–260. doi: 10.1086/321978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104(4):557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 51.Codd MB, Sugrue DD, Gersh BJ, Melton LJ., 3rd Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation. 1989;80(3):564–572. doi: 10.1161/01.cir.80.3.564. [DOI] [PubMed] [Google Scholar]
- 52.Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326(2):77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 53.Honda Y, Yokota Y, Yokoyama M. Familial aggregation of dilated cardiomyopathy – evaluation of clinical characteristics and prognosis. Jpn Circ J. 1995;59(9):589–598. doi: 10.1253/jcj.59.589. [DOI] [PubMed] [Google Scholar]
- 54.Keeling PJ, Gang Y, Smith G, et al. Familial dilated cardiomyopathy in the United Kingdom. Br Heart J. 1995;73(5):417–421. doi: 10.1136/hrt.73.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunig E, Tasman JA, Kucherer H, et al. Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol. 1998;31(1):186–194. doi: 10.1016/s0735-1097(97)00434-8. [DOI] [PubMed] [Google Scholar]
- 56.Hershberger RE, Kushner JD, Parks SB. GeneReviews at GeneTests. Medical Genetics Information Resource; WA, USA: 2009. Dilated cardiomyopathy overview. [Google Scholar]
- 57.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280(5364):750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 58.Kamisago M, Sharma SD, DePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343(23):1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 59.Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of α-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol. 2001;33(4):723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 60.Li D, Czernuszewicz GZ, Gonzalez O, et al. Novel cardiac troponin T mutation as a cause of familial dilated cardiomyopathy. Circulation. 2001;104(18):2188–2193. doi: 10.1161/hc4301.098285. [DOI] [PubMed] [Google Scholar]
- 61.Mogensen J, Murphy RT, Shaw T, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44(10):2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 62.MacLeod HM, Culley MR, Huber JM, McNally EM. Lamin A/C truncation in dilated cardiomyopathy with conduction disease. BMC Med Genet. 2003;4:4. doi: 10.1186/1471-2350-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Antoniades L, Eftychiou C, Kyriakides T, Christodoulou K, Katritsis DG. Malignant mutation in the lamin A/C gene causing progressive conduction system disease and early sudden death in a family with mild form of limb-girdle muscular dystrophy. J Interv Card Electrophysiol. 2007;19(1):1–7. doi: 10.1007/s10840-007-9133-x. [DOI] [PubMed] [Google Scholar]
- 64.van Tintelen JP, Hofstra RM, Katerberg H, et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154(6):1130–1139. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 65.Graber HL, Unverferth DV, Baker PB, et al. Evolution of a hereditary cardiac conduction and muscle disorder: a study involving a family with six generations affected. Circulation. 1986;74(1):21–35. doi: 10.1161/01.cir.74.1.21. [DOI] [PubMed] [Google Scholar]
- 66.Nelson SD, Sparks EA, Graber HL, et al. Clinical characteristics of sudden death victims in heritable (chromosome 1p1–1q1) conduction and myocardial disease. J Am Coll Cardiol. 1998;32(6):1717–1723. doi: 10.1016/s0735-1097(98)00424-0. [DOI] [PubMed] [Google Scholar]
- 67.Bonne G, Di Barletta MR, Varnous S, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet. 1999;21(3):285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 68.Raffaele Di Barletta M, Ricci E, Galluzzi G, et al. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery–Dreifuss muscular dystrophy. Am J Hum Genet. 2000;66(4):1407–1412. doi: 10.1086/302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shackleton S, Lloyd DJ, Jackson SN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24(2):153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 70.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2000;9(1):109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 71.Carballo S, Robinson P, Otway R, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105(4):375–382. doi: 10.1161/CIRCRESAHA.109.196055. [DOI] [PubMed] [Google Scholar]
- 72.Chang AN, Harada K, Ackerman MJ, Potter JD. Functional consequences of hypertrophic and dilated cardiomyopathy-causing mutations in α-tropomyosin. J Biol Chem. 2005;280(40):34343–34349. doi: 10.1074/jbc.M505014200. [DOI] [PubMed] [Google Scholar]
- 73.Michele DE, Gomez CA, Hong KE, Westfall MV, Metzger JM. Cardiac dysfunction in hypertrophic cardiomyopathy mutant tropomyosin mice is transgene-dependent, hypertrophy-independent, and improved by β-blockade. Circ Res. 2002;91(3):255–262. doi: 10.1161/01.res.0000027530.58419.82. [DOI] [PubMed] [Google Scholar]
- 74.Mirza M, Marston S, Willott R, et al. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280(31):28498–506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 75.Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation. 1990;82(2):507–513. doi: 10.1161/01.cir.82.2.507. [DOI] [PubMed] [Google Scholar]
- 76.Dusek J, Ostadal B, Duskova M. Postnatal persistence of spongy myocardium with embryonic blood supply. Arch Pathol. 1975;99(6):312–317. [PubMed] [Google Scholar]
- 77.Ritter M, Oechslin E, Sutsch G, et al. Isolated noncompaction of the myocardium in adults. Mayo Clin Proc. 1997;72(1):26–31. doi: 10.4065/72.1.26. [DOI] [PubMed] [Google Scholar]
- 78.Oechslin EN, Attenhofer Jost CH, Rojas JR, Kaufmann PA, Jenni R. Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis. J Am Coll Cardiol. 2000;36(2):493–500. doi: 10.1016/s0735-1097(00)00755-5. [DOI] [PubMed] [Google Scholar]
- 79.Nugent AW, Daubeney PE, Chondros P, et al. The epidemiology of childhood cardiomyopathy in Australia. N Engl J Med. 2003;348(17):1639–1646. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 80.Lipshultz SE, Sleeper LA, Towbin JA, et al. The incidence of pediatric cardiomyopathy in two regions of the United States. N Engl J Med. 2003;348(17):1647–1655. doi: 10.1056/NEJMoa021715. [DOI] [PubMed] [Google Scholar]
- 81.Pignatelli RH, McMahon CJ, Dreyer WJ, et al. Clinical characterization of left ventricular noncompaction in children: a relatively common form of cardiomyopathy. Circulation. 2003;108(21):2672–2678. doi: 10.1161/01.CIR.0000100664.10777.B8. [DOI] [PubMed] [Google Scholar]
- 82.Ichida F, Hamamichi Y, Miyawaki T, et al. Clinical features of isolated noncompaction of the ventricular myocardium: long-term clinical course, hemodynamic properties, and genetic background. J Am Coll Cardiol. 1999;34(1):233–240. doi: 10.1016/s0735-1097(99)00170-9. [DOI] [PubMed] [Google Scholar]
- 83.Jenni R, Oechslin EN, van der Loo B. Isolated ventricular non-compaction of the myocardium in adults. Heart. 2007;93(1):11–15. doi: 10.1136/hrt.2005.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eidem BW. Noninvasive evaluation of left ventricular noncompaction: what’s new in 2009? Pediatr Cardiol. 2009;30(5):682–689. doi: 10.1007/s00246-008-9372-3. [DOI] [PubMed] [Google Scholar]
- 85.Jenni R, Oechslin E, Schneider J, Attenhofer Jost C, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart. 2001;86(6):666–671. doi: 10.1136/heart.86.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ichida F, Tsubata S, Bowles KR, et al. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103(9):1256–1263. doi: 10.1161/01.cir.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 87.Vatta M, Mohapatra B, Jimenez S, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42(11):2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 88.Kenton AB, Sanchez X, Coveler KJ, et al. Isolated left ventricular noncompaction is rarely caused by mutations in G4.5, α-dystrobrevin and FK binding protein-12. Mol Genet Metab. 2004;82(2):162–166. doi: 10.1016/j.ymgme.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 89.Chen R, Tsuji T, Ichida F, et al. Mutation analysis of the G4.5 gene in patients with isolated left ventricular noncompaction. Mol Genet Metab. 2002;77(4):319–325. doi: 10.1016/s1096-7192(02)00195-6. [DOI] [PubMed] [Google Scholar]
- 90.Xing Y, Ichida F, Matsuoka T, et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol Genet Metab. 2006;88(1):71–77. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 91.Hermida-Prieto M, Monserrat L, Castro-Beiras A, et al. Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am J Cardiol. 2004;94(1):50–54. doi: 10.1016/j.amjcard.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 92.Klaassen S, Probst S, Oechslin E, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117(22):2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 93.Hoedemaekers YM, Caliskan K, Majoor-Krakauer D, et al. Cardiac β-myosin heavy chain defects in two families with non-compaction cardiomyopathy: linking non-compaction to hypertrophic, restrictive, and dilated cardiomyopathies. Eur Heart J. 2007;28(22):2732–2737. doi: 10.1093/eurheartj/ehm429. [DOI] [PubMed] [Google Scholar]
- 94.Benotti JR, Grossman W, Cohn PF. Clinical profile of restrictive cardiomyopathy. Circulation. 1980;61(6):1206–1212. doi: 10.1161/01.cir.61.6.1206. [DOI] [PubMed] [Google Scholar]
- 95.Mogensen J, Arbustini E. Restrictive cardiomyopathy. Curr Opin Cardiol. 2009;24(3):214–220. doi: 10.1097/hco.0b013e32832a1d2e. [DOI] [PubMed] [Google Scholar]
- 96.Chen SC, Balfour IC, Jureidini S. Clinical spectrum of restrictive cardiomyopathy in children. J Heart Lung Transplant. 2001;20(1):90–92. doi: 10.1016/s1053-2498(00)00162-5. [DOI] [PubMed] [Google Scholar]
- 97.Russo LM, Webber SA. Idiopathic restrictive cardiomyopathy in children. Heart. 2005;91(9):1199–1202. doi: 10.1136/hrt.2004.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Angelini A, Calzolari V, Thiene G, et al. Morphologic spectrum of primary restrictive cardiomyopathy. Am J Cardiol. 1997;80(8):1046–1050. doi: 10.1016/s0002-9149(97)00601-2. [DOI] [PubMed] [Google Scholar]
- 99.Mogensen J, Kubo T, Duque M, et al. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111(2):209–216. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaski JP, Syrris P, Burch M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94(11):1478–1484. doi: 10.1136/hrt.2007.134684. [DOI] [PubMed] [Google Scholar]
- 101.Karam S, Raboisson MJ, Ducreux C, et al. A de novo mutation of the β cardiac myosin heavy chain gene in an infantile restrictive cardiomyopathy. Congenit Heart Dis. 2008;3(2):138–143. doi: 10.1111/j.1747-0803.2008.00165.x. [DOI] [PubMed] [Google Scholar]
- 102.Peddy SB, Vricella LA, Crosson JE, et al. Infantile restrictive cardiomyopathy resulting from a mutation in the cardiac troponin T gene. Pediatrics. 2006;117(5):1830–1833. doi: 10.1542/peds.2005-2301. [DOI] [PubMed] [Google Scholar]
- 103.Gambarin FI, Tagliani M, Arbustini E. Pure restrictive cardiomyopathy associated with cardiac troponin I gene mutation: mismatch between the lack of hypertrophy and the presence of disarray. Heart. 2008;94(10):1257. doi: 10.1136/hrt.2008.154203. [DOI] [PubMed] [Google Scholar]
- 104.Kubo T, Gimeno JR, Bahl A, et al. Prevalence, clinical significance, and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49(25):2419–2426. doi: 10.1016/j.jacc.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 105.Sen-Chowdhry S, McKenna WJ. Left ventricular noncompaction and cardiomyopathy: cause, contributor, or epiphenomenon? Curr Opin Cardiol. 2008;23(3):171–175. doi: 10.1097/HCO.0b013e3282fdc939. [DOI] [PubMed] [Google Scholar]
- 106.Kostareva A, Gudkova A, Sjoberg G, et al. Deletion in TNNI3 gene is associated with restrictive cardiomyopathy. Int J Cardiol. 2009;131(3):410–412. doi: 10.1016/j.ijcard.2007.07.108. [DOI] [PubMed] [Google Scholar]
- 107.Moric-Janiszewska E, Markiewicz-Loskot G. Genetic heterogeneity of left-ventricular noncompaction cardiomyopathy. Clin Cardiol. 2008;31(5):201–204. doi: 10.1002/clc.20202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maron BJ, McKenna WJ, Danielson GK, et al. American College of Cardiology/ European Society of Cardiology clinical expert consensus document on hypertrophic cardiomyopathy. A report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the European Society of Cardiology Committee for Practice Guidelines. J Am Coll Cardiol. 2003;42(9):1687–1713. doi: 10.1016/s0735-1097(03)00941-0. [DOI] [PubMed] [Google Scholar]
- 109.Semsarian C. Guidelines for the diagnosis and management of hypertrophic cardiomyopathy. Heart Lung Circ. 2007;16(1):16–18. doi: 10.1016/j.hlc.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 110.Fatkin D. Guidelines for the diagnosis and management of familial dilated cardiomyopathy. Heart Lung Circ. 2007;16(1):19–21. doi: 10.1016/j.hlc.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 111.Dalal D, Nasir K, Bomma C, et al. Arrhythmogenic right ventricular dysplasia: a United States experience. Circulation. 2005;112(25):3823–3832. doi: 10.1161/CIRCULATIONAHA.105.542266. [DOI] [PubMed] [Google Scholar]
- 112.Hamid MS, Norman M, Quraishi A, et al. Prospective evaluation of relatives for familial arrhythmogenic right ventricular cardiomyopathy/dysplasia reveals a need to broaden diagnostic criteria. J Am Coll Cardiol. 2002;40(8):1445–1450. doi: 10.1016/s0735-1097(02)02307-0. [DOI] [PubMed] [Google Scholar]
- 113.Nava A, Bauce B, Basso C, et al. Clinical profile and long-term follow-up of 37 families with arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2000;36(7):2226–2233. doi: 10.1016/s0735-1097(00)00997-9. [DOI] [PubMed] [Google Scholar]
- 114.Luk A, Ahn E, Soor GS, Butany J. Dilated cardiomyopathy: a review. J Clin Pathol. 2009;62(3):219–225. doi: 10.1136/jcp.2008.060731. [DOI] [PubMed] [Google Scholar]
- 115.Pilichou K, Nava A, Basso C, et al. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113(9):1171–1179. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- 116.McKenna WJ, Thiene G, Nava A, et al. Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Task Force of the Working Group Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology. Br Heart J. 1994;71(3):215–218. doi: 10.1136/hrt.71.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schiavon G, Furlan S, Marin O, Salvatori S. Myotonic dystrophy protein kinase of the cardiac muscle: evaluation using an immunochemical approach. Microsc Res Tech. 2002;58(5):404–411. doi: 10.1002/jemt.10223. [DOI] [PubMed] [Google Scholar]
- 118.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54(3):201–211. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 119.Hershberger RE, Cowan J, Morales A, Siegfried JD. Progress with genetic cardiomyopathies: screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/ cardiomyopathy. Circ Heart Fail. 2009;2(3):253–261. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cirino AL, Ho CY. Genetic testing in cardiac disease: from bench to bedside. Nat Clin Pract Cardiovasc Med. 2006;3(9):462–463. doi: 10.1038/ncpcardio0654. [DOI] [PubMed] [Google Scholar]
- 121.Niimura H, Patton KK, McKenna WJ, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105(4):446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 122.Watkins H, Rosenzweig A, Hwang DS, et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;326(17):1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 123.Anan R, Greve G, Thierfelder L, et al. Prognostic implications of novel β cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;93(1):280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Varnava AM, Elliott PM, Baboonian C, et al. Hypertrophic cardiomyopathy: histopathological features of sudden death in cardiac troponin T disease. Circulation. 2001;104(12):1380–1384. doi: 10.1161/hc3701.095952. [DOI] [PubMed] [Google Scholar]
- 125.Watkins H, Conner D, Thierfelder L, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11(4):434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 126.Van Driest SL, Ackerman MJ, Ommen SR, et al. Prevalence and severity of “benign” mutations in the β-myosin heavy chain, cardiac troponin T, and α-tropomyosin genes in hypertrophic cardiomyopathy. Circulation. 2002;106(24):3085–3090. doi: 10.1161/01.cir.0000042675.59901.14. [DOI] [PubMed] [Google Scholar]
- 127.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(9):1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 128.Ackerman MJ, VanDriest SL, Ommen SR, et al. Prevalence and age-dependence of malignant mutations in the β-myosin heavy chain and troponin T genes in hypertrophic cardiomyopathy: a comprehensive outpatient perspective. J Am Coll Cardiol. 2002;39(12):2042–2048. doi: 10.1016/s0735-1097(02)01900-9. [DOI] [PubMed] [Google Scholar]
- 129.Van Driest SL, Maron BJ, Ackerman MJ. From malignant mutations to malignant domains: the continuing search for prognostic significance in the mutant genes causing hypertrophic cardiomyopathy. Heart. 2004;90(1):7–8. doi: 10.1136/heart.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83(6):630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 131.Maron BJ, Roberts WC, Arad M, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Monserrat L, Gimeno-Blanes JR, Marin F, et al. Prevalence of Fabry disease in a cohort of 508 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2007;50(25):2399–2403. doi: 10.1016/j.jacc.2007.06.062. [DOI] [PubMed] [Google Scholar]
- 133.Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352(4):362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 134.Hoffmann B. Fabry disease: recent advances in pathology, diagnosis, treatment and monitoring. Orphanet J Rare Dis. 2009;4:21. doi: 10.1186/1750-1172-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arbustini E, Pilotto A, Repetto A, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39(6):981–990. doi: 10.1016/s0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 136.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341(23):1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 137.van Berlo JH, de Voogt WG, van der Kooi AJ, et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J Mol Med. 2005;83(1):79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- 138.Mestroni L, Taylor MR. Lamin A/C gene and the heart: how genetics may impact clinical care. J Am Coll Cardiol. 2008;52(15):1261–1262. doi: 10.1016/j.jacc.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meune C, Van Berlo JH, Anselme F, et al. Primary prevention of sudden death in patients with lamin A/C gene mutations. N Engl J Med. 2006;354(2):209–210. doi: 10.1056/NEJMc052632. [DOI] [PubMed] [Google Scholar]
- 140.University of Washington. GeneTests. Medical Genetics Information Resource; WA, USA: 1993–2009. database online. [Google Scholar]
- 141.Hershberger RE, Parks SB, Kushner JD, et al. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1(1):21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Muntoni F, Cau M, Ganau A, et al. Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med. 1993;329(13):921–925. doi: 10.1056/NEJM199309233291304. [DOI] [PubMed] [Google Scholar]
- 143.Decostre V, Ben Yaou R, Bonne G. Laminopathies affecting skeletal and cardiac muscles: clinical and pathophysiological aspects. Acta Myol. 2005;24(2):104–109. [PubMed] [Google Scholar]
- 144.Waldmuller S, Muller M, Rackebrandt K, et al. Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clin Chem. 2008;54(4):682–687. doi: 10.1373/clinchem.2007.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fokstuen S, Lyle R, Munoz A, et al. A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Hum Mutat. 2008;29(6):879–885. doi: 10.1002/humu.20749. [DOI] [PubMed] [Google Scholar]
- 146.Zimmerman RS, Cox S, Lakdawala N, et al. A novel custom resequencing array for dilated cardiomyopathy (DCM). Presented at: American College of Medical Genetics Annual Meeting; Tampa, FL, USA. 27 March 2009; Abstract 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kothiyal P, Cox S, Ebert J, et al. An overview of custom array sequencing. Curr Protoc Hum Genet. 2009;7(Unit 7):17. doi: 10.1002/0471142905.hg0717s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum Mol Genet. 2002;11(20):2499–2506. doi: 10.1093/hmg/11.20.2499. [DOI] [PubMed] [Google Scholar]
- 149•.Bagnall RD, Yeates L, Semsarian C. The role of large gene deletions and duplications in MYBPC3 and TNNT2 in patients with hypertrophic cardiomyopathy. Int J Cardiol. 2009 doi: 10.1016/j.ijcard.2009.07.009. (Epub ahead of print). Investigates how common large deletions and duplications are in hypertrophic cardiomyopathy in MYBP3 and TNNT2 genes. They failed to identify large deletions and duplications associated with disease, suggesting that standard methods are probably not missing many deletions and duplications in HCM. [DOI] [PubMed] [Google Scholar]
- 150.Bhuiyan ZA, van den Berg MP, van Tintelen JP, et al. Expanding spectrum of human RYR2-related disease: new electrocardiographic, structural, and genetic features. Circulation. 2007;116(14):1569–1576. doi: 10.1161/CIRCULATIONAHA.107.711606. [DOI] [PubMed] [Google Scholar]
- 151.Gupta P, Bilinska ZT, Sylvius N, et al. Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol. 2010;105(3):365–377. doi: 10.1007/s00395-010-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27(3):234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 153.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 154.Christensen AH, Benn M, Tybjaerg-Hansen A, Haunso S, Svendsen JH. Missense variants in plakophilin-2 in arrhythmogenic right ventricular cardiomyopathy patients – disease-causing or innocent bystanders? Cardiology. 2010;115(2):148–154. doi: 10.1159/000263456. [DOI] [PubMed] [Google Scholar]
- 155••.Ionita-Laza I, Lange C, Laird MN. Estimating the number of unseen variants in the human genome. Proc Natl Acad Sci USA. 2009;106(13):5008–5013. doi: 10.1073/pnas.0807815106. Based on the frequency of mutations, this study estimates the number of control patients that are needed to appreciate the genetic variation that exists in the absence of disease. The authors determine for the first time that an analysis of as little as 150 people are necessary to identify 80% of the variants for a disease frequency of at least 0.1%, whereas more than 3000 are needed to identify them all. This gives a rationale guideline for control sample numbers when analyzing novel variants that may be associated with disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kuehn BM. 1000 Genomes Project promises closer look at variation in human genome. JAMA. 2008;300(23):2715. doi: 10.1001/jama.2008.823. [DOI] [PubMed] [Google Scholar]
- 157.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kapa S, Tester DJ, Salisbury BA, et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120(18):1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jegga AG, Gowrisankar S, Chen J, Aronow BJ. PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res. 2007;35(Database issue):D700–D706. doi: 10.1093/nar/gkl826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carrier L, Schlossarek S, Willis MS, Eschenhagen T. Ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovasc Res. 2009;85(2):330–338. doi: 10.1093/cvr/cvp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lechin M, Quinones MA, Omran A, et al. Angiotensin-I converting enzyme genotypes and left ventricular hypertrophy in patients with hypertrophic cardiomyopathy. Circulation. 1995;92(7):1808–1812. doi: 10.1161/01.cir.92.7.1808. [DOI] [PubMed] [Google Scholar]
- 164.Wang SX, Fu CY, Zou YB, et al. Polymorphisms of angiotensin-converting enzyme 2 gene associated with magnitude of left ventricular hypertrophy in male patients with hypertrophic cardiomyopathy. Chin Med J (Engl) 2008;121(1):27–31. [PubMed] [Google Scholar]
- 165.Lieb W, Graf J, Gotz A, et al. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men. Results of the MONICA Augsburg echocardiographic substudy. J Mol Med. 2006;84(1):88–96. doi: 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- 166.Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24(11):552–563. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 167.Pinto JR, Parvatiyar MS, Jones MA, Liang J, Potter JD. A troponin T mutation that causes infantile restrictive cardiomyopathy increases Ca2+ sensitivity of force development and impairs the inhibitory properties of troponin. J Biol Chem. 2008;283(4):2156–2166. doi: 10.1074/jbc.M707066200. [DOI] [PubMed] [Google Scholar]
- 168.Rankin J, Auer-Grumbach M, Bagg W, et al. Extreme phenotypic diversity and nonpenetrance in families with the LMNA gene mutation R644C. Am J Med Genet A. 2008;146A(12):1530–1542. doi: 10.1002/ajmg.a.32331. [DOI] [PubMed] [Google Scholar]
- 169.Marian AJ, Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2001;33(4):655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80(6):739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 171.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 172.Landstrom AP, Parvatiyar MS, Pinto JR, et al. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol. 2008;45(2):281–288. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Satoh M, Takahashi M, Sakamoto T, et al. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem Biophys Res Commun. 1999;262(2):411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 174.Carniel E, Taylor MR, Sinagra G, et al. A-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112(1):54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- 175.Theis JL, Bos JM, Bartleson VB, et al. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2006;351(4):896–902. doi: 10.1016/j.bbrc.2006.10.119. [DOI] [PubMed] [Google Scholar]
- 176.Geier C, Perrot A, Ozcelik C, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107(10):1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 177.Hayashi T, Arimura T, Itoh-Satoh M, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44(11):2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 178.Vasile VC, Ommen SR, Edwards WD, Ackerman MJ. A missense mutation in a ubiquitously expressed protein, vinculin, confers susceptibility to hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2006;345(3):998–1003. doi: 10.1016/j.bbrc.2006.04.151. [DOI] [PubMed] [Google Scholar]
- 179.Vasile VC, Will ML, Ommen SR, et al. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87(2):169–174. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 180.Osio A, Tan L, Chen SN, et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100(6):766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Arimura T, Bos JM, Sato A, et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54(4):334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- 182.Chimenti C, Pieroni M, Morgante E, et al. Prevalence of Fabry disease in female patients with late-onset hypertrophic cardiomyopathy. Circulation. 2004;110(9):1047–1053. doi: 10.1161/01.CIR.0000139847.74101.03. [DOI] [PubMed] [Google Scholar]
- 183.Sachdev B, Takenaka T, Teraguchi H, et al. Prevalence of Anderson–Fabry disease in male patients with late onset hypertrophic cardiomyopathy. Circulation. 2002;105(12):1407–1411. doi: 10.1161/01.cir.0000012626.81324.38. [DOI] [PubMed] [Google Scholar]
- 184.Gollob MH, Green MS, Tang AS, et al. Identification of a gene responsible for familial Wolff–Parkinson–White syndrome. N Engl J Med. 2001;344(24):1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 185.Yang Z, McMahon CJ, Smith LR, et al. Danon disease as an underrecognized cause of hypertrophic cardiomyopathy in children. Circulation. 2005;112(11):1612–1617. doi: 10.1161/CIRCULATIONAHA.105.546481. [DOI] [PubMed] [Google Scholar]
- 186.Mayosi BM, Khogali S, Zhang B, Watkins H. Cardiac and skeletal actin gene mutations are not a common cause of dilated cardiomyopathy. J Med Genet. 1999;36(10):796–797. doi: 10.1136/jmg.36.10.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Takai E, Akita H, Shiga N, et al. Mutational analysis of the cardiac actin gene in familial and sporadic dilated cardiomyopathy. Am J Med Genet. 1999;86(4):325–327. doi: 10.1002/(sici)1096-8628(19991008)86:4<325::aid-ajmg5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 188.Tesson F, Sylvius N, Pilotto A, et al. Epidemiology of desmin and cardiac actin gene mutations in a European population of dilated cardiomyopathy. Eur Heart J. 2000;21(22):1872–1876. doi: 10.1053/euhj.2000.2245. [DOI] [PubMed] [Google Scholar]
- 189.Duboscq-Bidot L, Charron P, Ruppert V, et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009;30(17):2128–2136. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- 190.Moulik M, Vatta M, Witt SH, et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54(4):325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Becane HM, Bonne G, Varnous S, et al. High incidence of sudden death with conduction system and myocardial disease due to lamins A and C gene mutation. Pacing Clin Electrophysiol. 2000;23(11 Pt 1):1661–1666. doi: 10.1046/j.1460-9592.2000.01661.x. [DOI] [PubMed] [Google Scholar]
- 192.Hershberger RE, Hanson EL, Jakobs PM, et al. A novel lamin A/C mutation in a family with dilated cardiomyopathy, prominent conduction system disease, and need for permanent pacemaker implantation. Am Heart J. 2002;144(6):1081–1086. doi: 10.1067/mhj.2002.126737. [DOI] [PubMed] [Google Scholar]
- 193.Karkkainen S, Reissell E, Helio T, et al. Novel mutations in the lamin A/C gene in heart transplant recipients with end stage dilated cardiomyopathy. Heart. 2006;92(4):524–526. doi: 10.1136/hrt.2004.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Parks SB, Kushner JD, Nauman D, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156(1):161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sebillon P, Bouchier C, Bidot LD, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40(8):560–567. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41(5):771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 197.Daehmlow S, Erdmann J, Knueppel T, et al. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;298(1):116–120. doi: 10.1016/s0006-291x(02)02374-4. [DOI] [PubMed] [Google Scholar]
- 198.Ehlermann P, Weichenhan D, Zehelein J, et al. Adverse events in families with hypertrophic or dilated cardiomyopathy and mutations in the MYBPC3 gene. BMC Med Genet. 2008;9:95. doi: 10.1186/1471-2350-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shimizu M, Ino H, Yasuda T, et al. Gene mutations in adult Japanese patients with dilated cardiomyopathy. Circ J. 2005;69(2):150–153. doi: 10.1253/circj.69.150. [DOI] [PubMed] [Google Scholar]
- 200.Zeller R, Ivandic BT, Ehlermann P, et al. Large-scale mutation screening in patients with dilated or hypertrophic cardiomyopathy: a pilot study using DGGE. J Mol Med. 2006;84(8):682–691. doi: 10.1007/s00109-006-0056-2. [DOI] [PubMed] [Google Scholar]
- 201.Villard E, Duboscq-Bidot L, Charron P, et al. Mutation screening in dilated cardiomyopathy: prominent role of the β myosin heavy chain gene. Eur Heart J. 2005;26(8):794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 202.Haghighi K, Kolokathis F, Pater L, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111(6):869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schmitt JP, Kamisago M, Asahi M, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299(5611):1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 204.McNair WP, Ku L, Taylor MR, et al. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110(15):2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 205.Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293(4):447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shi R, Zhang Y, Yang C, et al. The cardiac sodium channel mutation delQKP 1507–1509 is associated with the expanding phenotypic spectrum of LQT3, conduction disorder, dilated cardiomyopathy, and high incidence of youth sudden death. Europace. 2008;10(11):1329–1335. doi: 10.1093/europace/eun202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Murphy RT, Mogensen J, Shaw A, et al. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363(9406):371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 208.Gerull B, Gramlich M, Atherton J, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30(2):201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 209.Knoll R, Hoshijima M, Hoffman HM, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111(7):943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 210.Olson TM, Illenberger S, Kishimoto NY, et al. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105(4):431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 211.Mohapatra B, Jimenez S, Lin JH, et al. Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80(1–2):207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 212.Li D, Tapscoft T, Gonzalez O, et al. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation. 1999;100(5):461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 213.Taylor MR, Slavov D, Ku L, et al. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115(10):1244–1251. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 214.Sylvius N, Duboscq-Bidot L, Bouchier C, et al. Mutational analysis of the β- and δ-sarcoglycan genes in a large number of patients with familial and sporadic dilated cardiomyopathy. Am J Med Genet A. 2003;120A(1):8–12. doi: 10.1002/ajmg.a.20003. [DOI] [PubMed] [Google Scholar]
- 215.Tsubata S, Bowles KR, Vatta M, et al. Mutations in the human δ-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest. 2000;106(5):655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Karkkainen S, Miettinen R, Tuomainen P, et al. A novel mutation, Arg71Thr, in the δ-sarcoglycan gene is associated with dilated cardiomyopathy. J Mol Med. 2003;81(12):795–800. doi: 10.1007/s00109-003-0480-5. [DOI] [PubMed] [Google Scholar]
- 217.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36(4):382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schonberger J, Wang L, Shin JT, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37(4):418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 219.Taylor MR, Slavov D, Gajewski A, et al. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat. 2005;26(6):566–574. doi: 10.1002/humu.20250. [DOI] [PubMed] [Google Scholar]
- 220.Li D, Parks SB, Kushner JD, et al. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet. 2006;79(6):1030–1039. doi: 10.1086/509900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Towbin JA, Hejtmancik JF, Brink P, et al. X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87(6):1854–1865. doi: 10.1161/01.cir.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 222.Milasin J, Muntoni F, Severini GM, et al. A point mutation in the 5′ splice site of the dystrophin gene first intron responsible for X-linked dilated cardiomyopathy. Hum Mol Genet. 1996;5(1):73–79. doi: 10.1093/hmg/5.1.73. [DOI] [PubMed] [Google Scholar]
- 223.Bione S, D’Adamo P, Maestrini E, et al. A novel X-linked gene, G4.5 is responsible for Barth syndrome. Nat Genet. 1996;12(4):385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 224.D’Adamo P, Fassone L, Gedeon A, et al. The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet. 1997;61(4):862–867. doi: 10.1086/514886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 301.Medical genetics information resource. www.genetests.org.