See Mormann and Andrzejak (doi:10.1093/brain/aww091) for a scientific commentary on this article.
Seizures are thought to arise from an identifiable pre-ictal state. Brinkmann et al. report the results of an online, open-access seizure forecasting competition using intracranial EEG recordings from canines with naturally occurring epilepsy and human patients undergoing presurgical monitoring. The winning algorithms forecast seizures at rates significantly greater than chance.
Keywords: epilepsy, intracranial EEG, refractory epilepsy, experimental models
See Mormann and Andrzejak (doi:10.1093/brain/aww091) for a scientific commentary on this article.
Seizures are thought to arise from an identifiable pre-ictal state. Brinkmann et al. report the results of an online, open-access seizure forecasting competition using intracranial EEG recordings from canines with naturally occurring epilepsy and human patients undergoing presurgical monitoring. The winning algorithms forecast seizures at rates significantly greater than chance.
Abstract
See Mormann and Andrzejak (doi:10.1093/brain/aww091) for a scientific commentary on this article.
Accurate forecasting of epileptic seizures has the potential to transform clinical epilepsy care. However, progress toward reliable seizure forecasting has been hampered by lack of open access to long duration recordings with an adequate number of seizures for investigators to rigorously compare algorithms and results. A seizure forecasting competition was conducted on kaggle.com using open access chronic ambulatory intracranial electroencephalography from five canines with naturally occurring epilepsy and two humans undergoing prolonged wide bandwidth intracranial electroencephalographic monitoring. Data were provided to participants as 10-min interictal and preictal clips, with approximately half of the 60 GB data bundle labelled (interictal/preictal) for algorithm training and half unlabelled for evaluation. The contestants developed custom algorithms and uploaded their classifications (interictal/preictal) for the unknown testing data, and a randomly selected 40% of data segments were scored and results broadcasted on a public leader board. The contest ran from August to November 2014, and 654 participants submitted 17 856 classifications of the unlabelled test data. The top performing entry scored 0.84 area under the classification curve. Following the contest, additional held-out unlabelled data clips were provided to the top 10 participants and they submitted classifications for the new unseen data. The resulting area under the classification curves were well above chance forecasting, but did show a mean 6.54 ± 2.45% (min, max: 0.30, 20.2) decline in performance. The kaggle.com model using open access data and algorithms generated reproducible research that advanced seizure forecasting. The overall performance from multiple contestants on unseen data was better than a random predictor, and demonstrates the feasibility of seizure forecasting in canine and human epilepsy.
Introduction
The apparently random nature of seizures is a significant factor affecting the quality of life for patients with epilepsy (Fisher, 2000; Schulze-Bonhage and Kuhn, 2008). Despite taking daily medications many patients with epilepsy continue to have seizures (Kwan et al., 2010; Kwan and Brodie, 2010). Accurate seizure forecasting could transform epilepsy care, allowing patients to modify activities to avoid risk and take antiepileptic drugs only when needed to stop seizures before they develop. However, to achieve clinically relevant seizure forecasting, better methods are needed for identifying periods when seizures are likely to occur (Cook et al., 2013). Significant evidence has emerged supporting the idea that seizures arise from an identifiable preictal brain state (Stacey et al., 2011; Cook et al., 2013). Clinical studies describe patients self-reporting seizure-prone states prior to seizure at a rate greater than chance (Haut et al., 2007), and changes in cerebral blood flow, oxygenation, and cortical excitability have been reported prior to seizures (Baumgartner et al., 1998; Adelson et al., 1999; Aarabi et al., 2008; Badawy et al., 2009).
While many early seizure forecasting studies using EEG features suffered from inadequate statistical analysis, particularly with regards to adequate sampling of the interictal period (Mormann et al., 2007; Andrzejak et al., 2009), recent studies have demonstrated in a rigorous statistical framework (Snyder et al., 2008) that human and canine seizure forecasting is possible (Cook et al., 2013; Howbert et al., 2014; Teixeira et al., 2014; Brinkmann et al., 2015). A major challenge for seizure forecasting research has been the lack of long duration recordings with adequate interictal data and number of seizures for rigorous statistical testing (Mormann et al., 2007; Andrzejak et al., 2009). The majority of early studies were limited to relatively short human intracranial EEG (iEEG) recordings obtained as part of epilepsy surgery evaluations. These clinical iEEG studies from the epilepsy monitoring units rarely extend beyond 10 days and are enriched with seizures because the antiepileptic drugs are tapered to expedite the evaluation (Duncan et al., 1989). These clinical records rarely yield an adequate number of seizures separated by clear interictal periods for rigorous statistical testing, and thus are limited in their usefulness to develop predictors of patients’ habitual seizures (Marciani et al., 1985; Duncan et al., 1989). Longer-duration iEEG recordings have been analysed from epileptic animal models where an artificial epileptic focus is created (Bower and Buckmaster, 2008; Fujita et al., 2014), but the usefulness of these models to develop algorithms for forecasting naturally occurring focal epilepsy remains unclear (Loscher, 2011).
Recent studies have applied machine learning techniques to seizure forecasting with promising results (Mirowski et al., 2009; Park et al., 2011; Howbert et al., 2014). While many apply rigorous statistics to their results (Snyder et al., 2008), the scarcity of long duration recordings with adequate seizures remains an obstacle, as does the inability to directly compare algorithm performance from different research groups using common data. Recently an implantable seizure advisory system developed by NeuroVista Inc. made possible wireless telemetry of 16 channels of iEEG (sampling at 400 Hz) to a patient advisory device capable of running a real-time seizure forecasting algorithm (Davis et al., 2011; Cook et al., 2013). Initially the device was validated in canines with naturally-occurring epilepsy (Davis et al., 2011; Coles et al., 2013; Howbert et al., 2014). Naturally-occurring canine epilepsy is an excellent platform for human epilepsy device development (Leppik et al., 2011; Patterson, 2014) as dogs can be large enough to accommodate human devices, and their epilepsy is similar clinically (Potschka et al., 2013; Packer et al., 2014) and neurophysiologically (Berendt et al., 1999; Berendt and Dam, 2003; Pellegrino and Sica, 2004) to human epilepsy. Canine epilepsy is treated with many of the same medications at dosages comparable to human epilepsy (Farnbach, 1984; Dowling, 1994), and canine epilepsy is refractory to these medications at a comparable rate to human epilepsy (Govendir et al., 2005; Munana et al., 2012; Kiviranta et al., 2013). In a recent landmark clinical pilot study, NeuroVista and a team of Australian researchers implanted this device in 15 patients with drug-resistant epilepsy (http://ClinicalTrials.gov, study NCT01043406), and achieved seizure forecasting sensitivity of 65–100% in 11 patients during algorithm training, and eight patients prospectively after 4 months. In addition, the seizure advisory system was able to forecast low seizure likelihood periods with >98% negative predictive value in five patients tested (Cook et al., 2013).
Despite these advances, improvements are needed in sensitivity and specificity of seizure forecasting algorithms to attain clinically useful performance, and publicly available chronic iEEG datasets are needed to directly compare algorithms in a model relevant to human epilepsy. To stimulate reproducible research and improve the state of the art in seizure forecasting algorithms, the American Epilepsy Society, Epilepsy Foundation of America, and National Institutes of Health sponsored an open invitation competition on kaggle.com in 2014 using iEEG data from canines and humans with epilepsy. Contestants were provided with labelled interictal and preictal iEEG training data, and unlabelled testing iEEG data from ambulatory recordings taken with the NeuroVista seizure advisory system device in five canines with naturally occurring epilepsy, and wide bandwidth (5 kHz) presurgical iEEG recordings from two patients with epilepsy. The contestants used a wide range of supervised machine learning algorithms of their choice that were trained on available labelled training data and attempted to accurately label the unknown ‘testing data’ clips as preictal or interictal. Following the competition, the top performing algorithms were further tested on held-out, unseen data clips to assess the generalizability and robustness of algorithms developed via the kaggle.com forum.
Materials and methods
Subjects and data
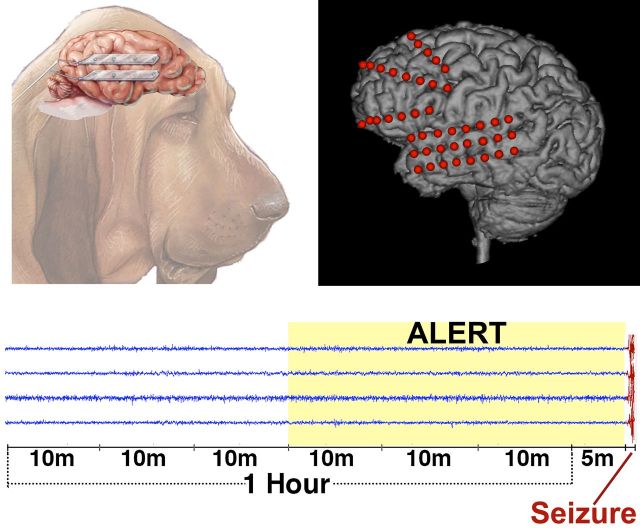
Intracranial EEG data were recorded chronically from eight canines with naturally occurring epilepsy using the NeuroVista seizure advisory system implanted device described previously (Davis et al., 2011; Coles et al., 2013). The dogs were housed at the veterinary hospitals at the University of Minnesota and University of Pennsylvania. Sixteen subdural electrodes were implanted intracranially in each canine in a bilaterally symmetrical arrangement (Fig. 1), with paired four-contact strips oriented from anterior to posterior on each hemisphere. The electrode wires were tunnelled caudally through openings in the cranium, anchored, looped and passed under the skin to the implanted telemetry unit medial to the dog’s shoulder. Wires were connected to a recording device, which was implanted under the latissimus dorsi muscle and iEEG data were wirelessly telemetered to a receiver and storage unit in a vest worn by the dog. Recorded data were stored on removable flash media, which were periodically removed and copied via the internet to a cloud storage platform for subsequent analysis. The implanted recording device was powered by a rechargeable battery unit, which was charged daily by monitoring personnel. Recorded iEEG from the 16 electrode contacts was referenced to the group average. Of the eight implanted canines, five produced high quality iEEG data and had an adequate number of seizures recorded for analysis. Two of the eight dogs had no seizures, and one dog had two seizures following implantation surgery.
Figure 1.
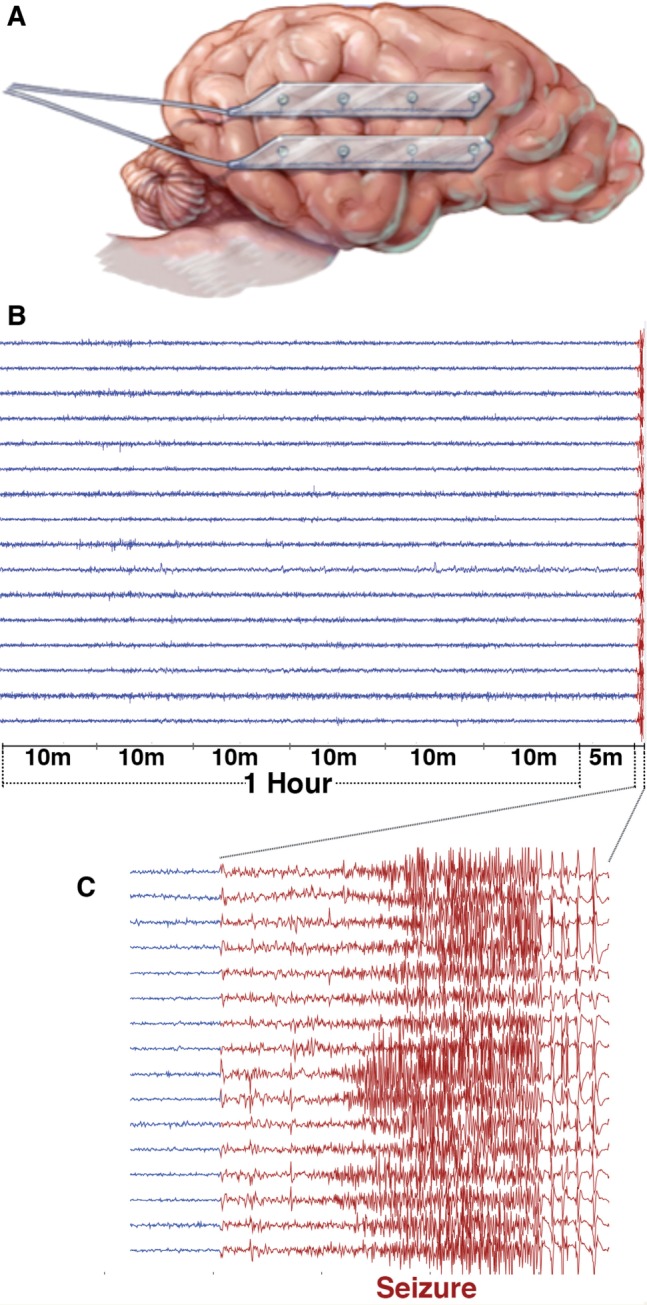
Canine electrode locations and data segments. (A) For the canine subjects, bilateral pairs of 4-contact strips were implanted oriented along the anterior-posterior direction. Electrode wires were tunnelled through the neck and connected to an implanted telemetry device secured beneath the latissimus dorsi muscle. (B) An hour of data with a 5-min offset before each lead seizure was extracted and split into 10-min segments for analysis. (C) The expanded view illustrates a ∼35-s long seizure.
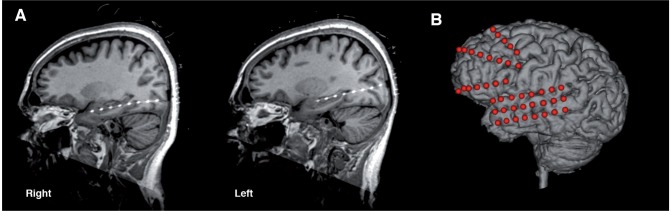
Epilepsy patients who underwent wide bandwidth (5 kHz sampling) iEEG monitoring for drug-resistant epilepsy at Mayo Clinic Rochester were reviewed. Subjects with poor data quality or other technical issues were excluded from further analysis, as were patients with fewer than four recorded lead seizures, defined as seizures occurring without a preceding seizure for a minimum of 4 h. Two patients were chosen with long recordings of high quality iEEG data and maximum possible separation between lead seizures. The patients’ electrode configurations and placement had been determined by clinical considerations, and are illustrated in Fig. 2. Patient 1 was a 70-year-old female with intractable epilepsy who underwent intracranial monitoring with 8-contact depth electrodes placed from a posterior approach in each temporal lobe and into the hippocampus. There were 71.3 h of iEEG data with five annotated seizures, four of which were lead seizures. Patient 2 was a 48-year-old female with intractable epilepsy who had a 3 × 8-contact subdural electrode grid placed over her left temporal lobe in addition to two 4-contact depth electrodes in each of the right and left temporal lobes, two left subtemporal 4-contact strip electrodes, and three left frontal 8-contact strips. This patient was monitored for 158.5 h recording 41 seizures, six of which met criteria for lead seizures. To limit data size, only data from the 3 × 8 subdural grid were used in the competition, as this grid covered both seizure onset zone and non-pathological tissue. These research iEEG data were acquired in parallel with the patient’s clinical recording as described previously (Brinkmann et al., 2009).
Figure 2.
Human implanted electrode locations. Implanted electrodes are visible in X-ray CT images coregistered to the space of the patient’s MRI for the two epilepsy patients whose data was used in this competition. (A) Patient 1 had bitemporal 8-contact penetrating depth electrodes implanted along the axes of the left and right hippocampus. (B) Patient 2 had a 3 × 8 subdural electrode grid placed along the axis of the left temporal lobe and frontal lobe strip electrodes. Spheres represent approximate electrode positions due to post-craniotomy brain surface shift in the CT. Electrodes not used in these experiments have been omitted from this illustration.
All iEEG data records were reviewed and seizures annotated by a board certified epileptologist (G.A.W.). Preictal data clips were extracted from the 66 min prior to lead seizures in six 10-min data clips. The preictal data clips were spaced 10 s apart in time, and offset by 5 min prior to the marked seizure onset to prevent subtle early ictal activity from contaminating the final preictal data clip. Interictal clips were selected similarly in groups of six 10-min clips with 10-s spacing beginning from randomly selected times a minimum of 1 week from any seizure. Each extracted data segment was individually mean centred. Data segments were stored as ordered structures including sample data, data segment length, iEEG sampling frequency, and channel names in uncompressed MATLAB format data files. Training data files also included a sequence number indicating the clip’s sequential position in the series of six 10-min data clips. The temporal sequence of the training and testing data was not made available to the contestants. The full data record was divided approximately in half, with labelled training interictal and preictal data clips taken from the first portion and unlabelled testing data clips from the last portion of the record. The division of testing and training data was selected to make an adequate number of lead seizures available for both training and testing (Table 1). Data clips for each subject were stored in separate folders and bundled into separate zip-compressed file archives which ranged between 2.6 GB and 14.83 GB. The total size of the data for the seven subjects was 59.64 GB. Compressed file archives were linked on the contest page at kaggle.com (https://www.kaggle.com/c/seizure-prediction/data) and made available for download by contestants. All data remain available for download at ieeg.org and msel.mayo.edu/data.html.
Table 1.
Data characteristics for the Kaggle.com seizure forecasting contest and held-out data experiment
| Subject | Sampling rate (Hz) | Recorded data (h) | Seizures | Lead seizures | Training clips (% interictal) | Testing clips (% interictal) | Held-out clips (% interictal) |
|---|---|---|---|---|---|---|---|
| Dog 1 | 400 | 1920 | 22 | 8 | 504 (95.2) | 502 (95.2) | 2000 (99.7) |
| Dog 2 | 400 | 8208 | 47 | 40 | 542 (92.3) | 1000 (91.0) | 1000 (100) |
| Dog 3 | 400 | 5112 | 104 | 18 | 1512 (95.2) | 907 (95.4) | 1000 (100) |
| Dog 4 | 400 | 7152 | 29 | 27 | 901 (89.2) | 990 (94.2) | 1000 (95.8) |
| Dog 5 | 400 | 5616 | 19 | 8 | 480 (93.8) | 191 (93.7) | 0 |
| Patient 1 | 5000 | 71.3 | 5 | 4 | 68 (73.5) | 195 (93.9) | 0 |
| Patient 2 | 5000 | 158.5 | 41 | 6 | 60 (70.0) | 150 (90.7) | 0 |
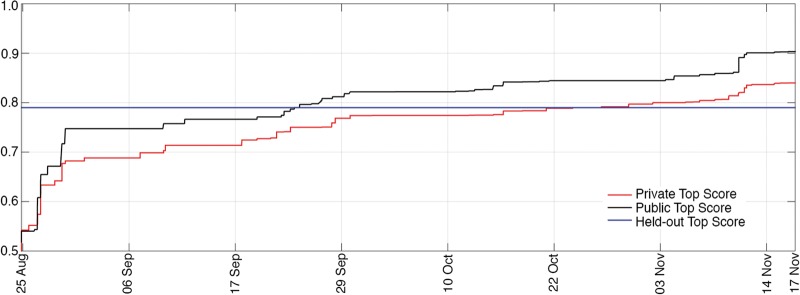
The contest ran from 25 August to 17 November 2014. Contestants were permitted to develop algorithms in any computer language and using any features, classification and data processing methods they chose, but classifications were required to come directly from an algorithm—classification by visual review was prohibited. Algorithms were also required to use a uniform data processing method for all subjects, but were permitted to modify data processing methods based on data parameters, such as sampling frequency. Contestants uploaded preictal probability scores (a floating point number between 0 and 1 indicating the probability of each clip being preictal) for the 3935 testing data clips in a comma separated values file, and a real-time public leader board on kaggle.com provided immediate feedback on classification accuracy. Public leader board scores were computed on a randomly sampled 40% subset of the test data clips, but official winners were determined based on the remaining 60% of the testing data (Fig. 3). Classification scores were computed by Kaggle as the area under the receiver operating characteristic (ROC) curve created by applying varying threshold values to the probability scores. Contestants were permitted five submissions per day at the beginning of the contest, and 10 submissions per day for the final 2 weeks. Prizes were awarded for first ($15 000), second ($7000), and third ($3000) place finishers as determined by the private leader board scores. Winning teams were required to submit their algorithms under an open source license to be made publicly available on via the IEEG portal (ieeg.org) and the Mayo Systems Electrophysiology Lab (MSEL.mayo.edu/data.html).
Figure 3.
Leading scores during the competition. Plots of the leading score on the kaggle.com public (black line) and private (red line) leader boards for the duration of the competition. The top score from the held-out data experiment is represented by the horizontal blue line.
Following the competition, the top 10 finishing teams were invited to run their algorithms on a held-out set of unseen data clips to assess the robustness of the algorithms developed on new data. An additional 5000 unlabelled data clips from four of the five original dogs (Table 1) were provided to these contestants. These clips were from the same data records but represented new, unseen, iEEG data from the original dataset. For this dataset a higher proportion of interictal to interictal data (100:1) was selected in an attempt to more closely approximate the preictal:interictal ratio in patients having a few seizures per month. Participants again submitted probability scores for the holdout data in a comma separated values format, and results were scored as the area under the ROC curve. Participants who used aggregations of multiple machine learning techniques also submitted separate classifications for each technique. Six of the top 10 teams (Table 2), including the three winners, agreed to participate in the holdout data experiment and provide detailed descriptions of their algorithms. The team with the top overall score, area under the curve (AUC) = 0.84, chose to forfeit the prize to avoid disclosing source code and pursue an algorithm patent. This team did not participate in the subsequent analysis of held-out data.
Table 2.
AUC scores for top ten Kaggle.com finalists in the public and private leaderboards
| Place | Team name | Public leader board | Private leader board | Entries |
|---|---|---|---|---|
| 1 | QMSDP | 0.86 | 0.82 | 501 |
| 2 | Birchwood | 0.84 | 0.80 | 160 |
| 3 | ESAI CEU-UCH | 0.82 | 0.79 | 182 |
| 4 | Michael Hills | 0.86 | 0.79 | 427 |
| 5 | KPZZ | 0.82 | 0.79 | 196 |
| 6 | Carlos Fernandez | 0.84 | 0.79 | 299 |
| 7 | Isaac | 0.84 | 0.79 | 253 |
| 8 | Wei Wu | 0.82 | 0.79 | 140 |
| 9 | Golondrina | 0.82 | 0.78 | 171 |
| 10 | Sky Kerzner | 0.84 | 0.78 | 97 |
The public leader board score was computed on a randomly-chosen 40% subset of the data, while the private leader board was computed on the remaining 60%.
Data used in this competition as well as the source code for the top performing algorithms are freely available on the International IEEG Portal (http://ieeg.org), and the Mayo Systems Electrophysiology Lab ftp site (http://msel.mayo.edu/data.html).
Algorithms
Algorithms are described below and summarized in Table 3 in order of performance on the private leader board. More detailed information regarding the top finishers’ algorithms can be found in the Supplementary material.
Table 3.
AUC scores for the held-out data experiment compared to scores on the public and private leader boards
| Team name | Window (overlap) | Features | Machine learning algorithm | Ensemble method | Public leader board | Private leader board | Held-out data | Per cent change | Sensitivity at 75% specificity |
|---|---|---|---|---|---|---|---|---|---|
| QMSDP | 60s (0%), 8 s (97%) | Spectral power, spectral entropy, correlation, fractal dimensions, Hjorth parameters, distribution statistics, signal variance | LassoGLM, Bagged SVM, Random Forest | Weighted average | 0.86 | 0.82 | 0.75 | −7.97 | 0.71 |
| QMSDP | 60 s (0%) | Spectral entropy, correlation, fractal dimensions, Hjorth parameters, distribution statistics | LassoGLM | 0.84 | 0.81 | 0.73 | −9.26 | 0.69 | |
| QMSDP | 8 s (97%) | Spectral power, correlation, signal variance | Bagged SVM | 0.79 | 0.76 | 0.76 | 0.91 | 0.73 | |
| QMSDP | 8 s (97%) | Spectral power, correlation, signal variance | Random Forest | 0.79 | 0.72 | 0.59 | −17.88 | 0.33 | |
| Birchwood | 50 s (0%) | Log spectral power, covariance | SVM | Platt scaling | 0.84 | 0.80 | 0.74 | −8.01 | 0.60 |
| ESAI CEU-UCH | 60 s (50%) | Spectral power, correlation, signal derivativePCA and ICA preprocessing | Neural Network and K Nearest Neighbour clustering | Bayesian combination | 0.82 | 0.79 | 0.72 | −9.77 | 0.54 |
| Michael Hills | Spectral power, correlation, spectral entropy, fractal dimensions, Hurst exponent Genetic algorithm feature selection | SVM | 0.86 | 0.79 | 0.79 | −0.29 | 0.73 | ||
| Wei Wu | 60 s (0%) | Spectral power, statistical measures, covariance matrices | SVM and GLMNet | Weighted average of rank scores | 0.82 | 0.79 | 0.77 | −1.86 | 0.69 |
| Golondrina | 60 s (0%) | Spectral power, signal standard deviation | Convolutional neural networks (test data calibration) | 0.82 | 0.78 | 0.76 | −2.77 | 0.73 | |
| Golondrina | 60 s (0%) | Spectral power, signal standard deviation | Convolutional neural networks (not calibrated on test data) | 0.81 | 0.78 | 0.77 | −1.39 | 0.75 |
Rows in bold represent algorithm variations submitted after the competition as part of the held-out data experiment. Additional information on algorithms is available in the Supplementary material.
First place team
The first place team’s approach consisted of an ensemble of three distinct algorithms:
Algorithm 1
Intracranial EEG data were sampled in sequential 1-min windows, in which were calculated spectral entropy and Shannon’s entropy (MacKay, 2003) at six frequency bands: delta (0.1–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz), low-gamma (30–70 Hz) and high gamma (70–180 Hz), and Shannon’s entropy in dyadic (between 0.00167 and 109 Hz spaced by factors of 2n) frequency bands. The feature set also included the spectral edge at 50% power below 40 Hz, spectral correlation between channels in dyadic frequency bands, the time series correlation matrix and its eigenvalues, fractal dimensions, Hjorth activity, mobility and complexity parameters (Hjorth, 1970), and the statistical skewness and kurtosis of the distribution of time series values. These features were used to train a LassoGLM classifier implemented in MATLAB (MathWorks Inc, Natick MA).
Algorithm 2
The iEEG data were analysed in 8-s windows with 7.75 s of overlap. Sums of fast Fourier Transform (FFT) power over bands spanning the fundamental frequency of the FFT, 1 Hz, 4 Hz, 8 Hz, 16 Hz, 32 Hz, 64 Hz, 128 Hz and Nyquist, yielding nine bands per channel, time series correlation matrix, and time series variance were computed for the feature set. A support vector machine (SVM) model (Vapnik and Vapnik, 1998) with a linear kernel was trained with bootstrap aggregation (Breiman, 1996) training on 10% of the data, and a kernel principal component analysis (PCA) (Hotelling, 1933) decomposition of the features was performed with basis truncation. The algorithm was implemented in python using the scikit-learn toolkit (http://scikit-learn.org/stable/modules/svm.html.)
Algorithm 3
This algorithm used the same 8-s overlapping iEEG windows and features as Algorithm 2 above. Classification was accomplished using a random forest algorithm with 80 trees implemented in MATLAB. For this model adjacent window scores were interpolated by a factor of 8 using a cubic spline algorithm before ensembling.
The three numerical models were median centred and an ensemble of the three models was created using an empirically determined weighted average: (1/4 × Random Forest + 1/4 × Bagged SVM + 1/2 × LassoGLM). In the held-out data experiment this team submitted classifications produced separately by each of these algorithms to assess their relative contributions, as well as the final ensembled result.
Second place team
The second place algorithm downsampled the iEEG data to 100 Hz and analysed the data in 50-s non-overlapping windows. The set of iEEG-derived features consisted of the logarithm of the FFT magnitude in 18 equal frequency bands between 1 and 50 Hz, the inter-channel covariance and eigenvalues of these frequency bands, and the interchannel covariance and eigenvalues in the time domain. A SVM machine learning algorithm with a radial basis function (RBF) kernel (C = 10−6, gamma = 0.01) was trained and used to classify the power-in-band features in each analysis window. A combination of the arithmetic and harmonic means of individual analysis windows with Platt scaling (Platt, 1999) was used to aggregate analysis windows into a single probability score for each segment. Algorithms were coded in python using the scikit-learn toolkit.
Third place team
The third place team analysed the iEEG data in 60-s windows with 30 s of overlap. A Hamming window was applied to the data segments, and the FFT was divided into six frequency bands: delta (0.1–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz), low gamma (30–70 Hz) and high gamma (70–180 Hz). PCA (Hotelling, 1933) and independent component analysis (ICA) (Kruskal, 1969) were applied to the six frequency bands across the sequence of 60-s windows. Eigenvalues of the frequency domain interchannel correlation matrix were computed from the original iEEG signal and the derivative of the iEEG signal over the full 10-min segment length. A Bayesian model combination of artificial neural networks with different depths and a k-nearest neighbour (k = 40) classification algorithm was used to provide the final classification of each segment. Algorithms were coded in R (http://www.R-project.org) and used the APRIL-ANN machine learning toolkit (.https://github.com/pakozm/april-ann).
Fourth place team
The fourth place team used non-overlapping 75-s windows, and the feature set included the upper right triangle (non-redundant coefficients) of the time domain correlation matrix with sorted eigenvalues, the upper right triangle of the frequency domain correlation matrix with sorted eigenvalues, the FFT magnitude with logarithmic scaling for frequency bands up to 48 Hz (0.5, 2.25, 4, 5.5, 7, 9.5, 12, 21, 30, 39, and 48 Hz), spectral entropies up to 24 Hz, as well as the Higuchi fractal dimension (Higuchi, 1988), Petrosian fractal dimension (Petrosian, 1995), and Hurst exponent (Feder, 1988). A genetic algorithm (population 30, 10 generations) was used to select features within the Petrosian fractal dimension features, the Hurst exponent features, and the Higuchi fractal dimension and spectral entropy features, using a 3-fold cross validation in the training data. A SVM with RBF kernel (gamma = 0.0079, C = 2.7) was used to classify the data segments.
Fifth, sixth and seventh place teams
The fifth, sixth, and seventh place teams did not participate in the held-out data experiment and did not provide additional detail about their algorithms.
Eighth place team
The eighth place team downsampled the data to 200 Hz and analysed each 10-min data clip in non-overlapping 1-min windows. In each window the mean, maximum, and standard deviation in both the time and frequency domains were calculated for each channel, and for the average of all channels. The frequency with maximum amplitude in the FFT was identified for each individual channel as well. The interchannel covariance matrices were calculated in the time and frequency domains, and the mean, three highest covariances, and standard deviation were added to the set of features. The lower 20% (up to 40 Hz for the dogs and 500 Hz for the humans) of the frequency spectrum below the Nyquist limit of each channel was divided into 24 equally spaced frequency bands, and the average spectral power in each bands was included as well. The GLMNet (Friedman et al., 2010) classifier (http://cran.r-project.org/web/packages/glmnet/index.html) and a SVM (RBF kernel, C = 100, gamma = 0.001) were trained globally across all training data, as well as separately on individual subjects using all features with a 2-fold cross validation with 10 shuffles. The classifiers were ensembled by ranking data clip probabilities from each model and computing a weighted average of all the ranks. The mean of the 1-min data windows was taken as the probability for each 10 min data clip. This algorithm was implemented in R.
Ninth place team
The ninth place algorithm partitioned the raw iEEG data clips into non-overlapping 1-min windows. The standard deviation and average spectral power in delta (0.1–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz), low gamma (30–70 Hz) and high gamma (70–180 Hz) frequency bands (Howbert et al., 2014) were computed for each channel. A convolutional neural network (CNN) (LeCun et al., 1998) was used for classification, with convolutions done in the time domain. The neural network consisted of two convolutional layers followed by a temporal global pooling layer, a fully-connected layer, and a logistic regression layer. During algorithm training, additional data windows were generated by resampling data to span consecutive data clips. The final clip probability was determined by the average of the scores generated by 11 CNNs with variations in analysis window sizes, frequency bands, and CNN architecture.
Tenth place team
The 10th place team did not participate in the held-out data experiment.
Results
Public and private leader board results from the competition are plotted in Fig. 2, for the duration of the contest. In total 505 teams comprising 654 individuals entered the competition and submitted classifications. A total of 17 856 classifications of the test data were submitted. Statistics for the top scoring teams are listed in Table 2. For teams participating in the held-out data experiment, the mean (max–min) public leader board score was 0.84 (0.86–0.82), private leader board score was 0.79 (0.82–0.78), and contestants made a mean (max–min) of 242.6 (501–140) entries. The mean (max–min) AUC score on the held-out data was 0.74 (0.79–0.59), representing a mean 6.85% (standard deviation 2.45%) decline relative to the mean private leader board score. AUC scores and algorithm sensitivity at 75% specificity are reported in Table 3. Full ROC curves for the contest algorithms on the held-out data are included in the Supplementary material.
Discussion
Formulating the seizure forecasting problem as a contest on kaggle.com proved a unique way to engage a large pool of data scientists worldwide on an important problem. The opportunity for a group of independent data scientists to analyse a large, freely available dataset from humans and canines with epilepsy yielded reproducible and directly comparable results from a range of seizure forecasting approaches. There is now widespread recognition that many published claims in biomedical research are not reproducible. (Ioannidis, 2005; Landis et al., 2012; Button et al., 2013) The consequences of the lack of reproducibility are profound, and inefficient use of limited resources may slow the development of therapies for patients. In the computational science and engineering communities in particular, reproducible research requires open source data and algorithms (Buckheit and Donoho, 1995; Donoho, 2010) in addition to published methods and results. Early studies in seizure forecasting were limited by both inadequate datasets and flawed statistical testing (Mormann et al., 2007; Andrzejak et al., 2009), and lack of openly available data and algorithms hindered investigators from challenging these results. Making the data and algorithm source code from the present study freely available (http://ieeg.org and http://msel.mayo.edu/data.html), facilitates reproducibility and provides a benchmark for future algorithm development.
This study demonstrates that seizures are not random events and supports the feasibility of real-time seizure forecasting. All six algorithms in the held-out data experiment achieved performance greater than a random chance predictor (P < 0.0001, z-score computed relative to AUC of 0.5), as was the top scoring algorithm on the private leader board (P < 0.0001). On the private leader board 359 teams scored above the upper 95% confidence limit AUC relative to a random classifier (0.531, Hanley-McNeil method). While no published study yet has used this full data set as a benchmark, the results compare favourably to a recent study (0.72 AUC) computing on the full continuous data from the five canines (Brinkmann et al., 2015) .
At a time when skills in analytics and machine learning command a high premium in the marketplace and research labs face reduced funding, an online competition can represent a cost-effective method of achieving progress on difficult problems. Access to contestants with different backgrounds and approaches can quickly and efficiently evaluate a broad range of features and algorithms. There are, however, some limitations to the online kaggle.com competition format that should be noted. First, the ability to submit multiple trials may contribute to overtraining on the contest dataset. While determining winners by the private leader board score computed on the majority of data reduces this risk somewhat, it is critical in this type of forum to provide as broad a sampling of data as possible to ensure extensibility of solutions to the real-world problem. Here this issue was further mitigated by running a post-contest analysis using withheld data not seen during algorithm development. The fact that there was a modest decline in forecasting performance suggests overtraining was not a significant factor.
Second, the necessity of providing contestants with the full set of testing data in an unlabelled form provides both an advantage and a disadvantage to contestants. Having the testing set available gave contestants the opportunity to directly measure the full statistical range of future data, aiding normalization of models in a way not possible in prospective real-time seizure forecasting. In contrast, timing information about the testing clips could not be provided in this format, which prevented contestants from deploying background normalization strategies commonly used in time series analysis. A third limitation of the competition format is that algorithms and source code are not required to be fast, modular, or well documented, and significant development effort may be required to make even the best competition algorithm suitable for application on a broader range of data.
Algorithms developed for the competition used a wide range of time domain and frequency domain features, in addition to more complex features. Most participants developed their approaches empirically, and with machine learning approaches it is difficult to identify which features contribute predictive value to the model and which features are primarily ignored. All six algorithms used some form of spectral power in discreet frequency bands, and five of the six algorithms used time domain and/or frequency domain interchannel correlations. Both power in band and bivariate interchannel correlation have previously been shown to be independently capable of forecasting (Park et al., 2011; Howbert et al., 2014; Brinkmann et al., 2015). While six different machine learning algorithms were used individually or as part of an ensemble in the held out data experiment, it is interesting to note that SVM was the most commonly used algorithm, appearing in four of the six participating entries. Further investigation is needed however, to assess the relative predictive value of different feature classes, and the relative capabilities of different machine learning algorithms in this context.
A large-scale online competition aimed at developing novel algorithms for seizure forecasting was successfully conducted using open access datasets from canines and humans. The kaggle.com competition format enabled direct comparison between different seizure forecasting algorithms on a common dataset, and provides a benchmark for future forecasting studies. Multiple groups using different approaches succeeded in independently developing successful algorithms for seizure forecasting, supporting the hypothesis that seizures are not random but arise from an observable preictal state. Open access to data, methods, and algorithms creates a platform for reproducible seizure forecasting research. Future studies are required to clarify what percentage of patients with epilepsy have seizures that can be forecast using iEEG, and the level of forecasting performance needed for improving outcome and quality of life.
Supplementary Material
Acknowledgements
The authors acknowledge Cindy Nelson, Mark Bower PhD, Karla Crockett, Daniel Crepeau, and Matt Stead MD, PhD for data collection and assistance with data processing. The canine data was recorded using devices developed by NeuroVista Inc., and we acknowledge the contributions of NeuroVista’s former management and employees.
Glossary
Abbreviations
- AUC =
area under the curve
- FFT =
fast Fourier transform
- iEEG =
intracranial electroencephalography
- SVM =
support vector machine
Funding
The authors acknowledge the generous support of the American Epilepsy Society, The Epilepsy Foundation, Kaggle.com (which waived a portion of its normal fee for this competition), and the National Institutes of Health. Data collection, processing, analysis, and manuscript preparation were supported by NeuroVista Inc. and grants NIH-NINDS UH2/UH3 95495 (G.W.), U01-NS 73557 (G.W.), U24-NS063930 (B.L., G.W.), K01 ES025436-01 (J.W.), and R01-NS92882 (G.W.), the Mirowski family foundation, and Mayo Clinic.
Supplementary material
Supplementary material is available at Brain online.
References
- Aarabi A, Wallois F, Grebe R. Does spatiotemporal synchronization of EEG change prior to absence seizures? Brain Res 2008; 1188: 207–21. [DOI] [PubMed] [Google Scholar]
- Adelson PD, Nemoto E, Scheuer M, Painter M, Morgan J, Yonas H. Noninvasive continuous monitoring of cerebral oxygenation periictally using near-infrared spectroscopy: a preliminary report. Epilepsia 1999; 40: 1484–9. [DOI] [PubMed] [Google Scholar]
- Andrzejak RG, Chicharro D, Elger CE, Mormann F. Seizure prediction: any better than chance? Clin Neurophysiol 2009; 120: 1465–78. [DOI] [PubMed] [Google Scholar]
- Badawy R, Macdonell R, Jackson G, Berkovic S. The peri-ictal state: cortical excitability changes within 24 h of a seizure. Brain 2009; 132: 1013–21. [DOI] [PubMed] [Google Scholar]
- Baumgartner C, Serles W, Leutmezer F, Pataraia E, Aull S, Czech T, et al. Preictal SPECT in temporal lobe epilepsy: regional cerebral blood flow is increased prior to electroencephalography-seizure onset. J Nucl Med 1998; 39: 978–82. [PubMed] [Google Scholar]
- Berendt M, Dam M. Re: clinical presentations of naturally occurring canine seizures: similarities to human seizures. Epilepsy Behav 2003; 4: 198–9; author repyl 9–201. [DOI] [PubMed] [Google Scholar]
- Berendt M, Hogenhaven H, Flagstad A, Dam M. Electroencephalography in dogs with epilepsy: similarities between human and canine findings. Acta Neurol Scand 1999; 99: 276–83. [DOI] [PubMed] [Google Scholar]
- Bower MR, Buckmaster PS. Changes in granule cell firing rates precede locally recorded spontaneous seizures by minutes in an animal model of temporal lobe epilepsy. J Neurophysiol 2008; 99: 2431–42. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Mach Learn 1996; 24: 123–40. [Google Scholar]
- Brinkmann BH, Bower MR, Stengel KA, Worrell GA, Stead M. Large-scale electrophysiology: acquisition, compression, encryption, and storage of big data. J Neurosci Methods 2009; 180: 185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BH, Patterson EE, Vite C, Vasoli VM, Crepeau D, Stead M, et al. Forecasting seizures using intracranial EEG measures and SVM in naturally occurring canine epilepsy. PLoS One 2015; 10: e0133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckheit J, Donoho D. WaveLab and Reproducible Research In: Antoniadis A, Oppenheim G, editors. Wavelets and Statistics. New York: Springer; 1995. p. 55–81. [Google Scholar]
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci 2013; 14: 365–76. [DOI] [PubMed] [Google Scholar]
- Coles LD, Patterson EE, Sheffield WD, Mavoori J, Higgins J, Michael B, et al. Feasibility study of a caregiver seizure alert system in canine epilepsy. Epilepsy Res 2013; 106: 456–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MJ, O'Brien TJ, Berkovic SF, Murphy M, Morokoff A, Fabinyi G, et al. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: a first-in-man study. Lancet Neurol 2013; 12: 563–71. [DOI] [PubMed] [Google Scholar]
- Davis KA, Sturges BK, Vite CH, Ruedebusch V, Worrell G, Gardner AB, et al. A novel implanted device to wirelessly record and analyze continuous intracranial canine EEG. Epilepsy Res 2011; 96: 116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho DL. An invitation to reproducible computational research. Biostatistics 2010; 11: 385–8. [DOI] [PubMed] [Google Scholar]
- Dowling PM. Management of canine epilepsy with phenobarbital and potassium bromide. Can Vet J 1994; 35: 724–5. [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Smith SJ, Forster A, Shorvon SD, Trimble MR. Effects of the removal of phenytoin, carbamazepine, and valproate on the electroencephalogram. Epilepsia 1989; 30: 590–6. [DOI] [PubMed] [Google Scholar]
- Farnbach GC. Serum concentrations and efficacy of phenytoin, phenobarbital, and primidone in canine epilepsy. J Am Vet Med Assoc 1984; 184: 1117–20. [PubMed] [Google Scholar]
- Feder J. Fractals. New York: Plenum Press; 1988. [Google Scholar]
- Fisher RS. Epilepsy from the patient's perspective: review of results of a community-based survey. Epilepsy Behav 2000; 1: S9–S14. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1. [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Toyoda I, Thamattoor AK, Buckmaster PS. Preictal activity of subicular, CA1, and dentate gyrus principal neurons in the dorsal hippocampus before spontaneous seizures in a rat model of temporal lobe epilepsy. J Neurosci 2014; 34: 16671–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govendir M, Perkins M, Malik R. Improving seizure control in dogs with refractory epilepsy using gabapentin as an adjunctive agent. Aust Vet J 2005; 83: 602–8. [DOI] [PubMed] [Google Scholar]
- Haut SR, Hall CB, LeValley AJ, Lipton RB. Can patients with epilepsy predict their seizures? Neurology 2007; 68: 262–6. [DOI] [PubMed] [Google Scholar]
- Higuchi T. Approach to an irregular time series on the basis of the fractal theory. Physica D 1988; 31: 277–83. [Google Scholar]
- Hjorth B. EEG analysis based on time domain properties. Electroencephalogr Clin Neurophysiol 1970; 29: 306–10. [DOI] [PubMed] [Google Scholar]
- Hotelling H. Analysis of a complex of statistical variables into principal components. J Educ Psychol 1933; 24: 417 [Google Scholar]
- Howbert JJ, Patterson EE, Stead SM, Brinkmann B, Vasoli V, Crepeau D, et al. Forecasting seizures in dogs with naturally occurring epilepsy. PLoS One 2014; 9: e81920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Why most published research findings are false. PLoS Med 2005; 2: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviranta AM, Laitinen-Vapaavuori O, Hielm-Bjorkman A, Jokinen T. Topiramate as an add-on antiepileptic drug in treating refractory canine idiopathic epilepsy. J Small Anim Pract 2013; 54: 512–20. [DOI] [PubMed] [Google Scholar]
- Kruskal JB. Toward a practical method which helps uncover the structure of a set of multivariate observations by finding the linear transformation which optimizes a new ‘index of condensation’ In: Statistical computation. New York: Academic Press; 1969. p. 427–40. [Google Scholar]
- Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia 2010; 51: 1069–77. [DOI] [PubMed] [Google Scholar]
- Kwan P, Brodie MJ. Definition of refractory epilepsy: defining the indefinable? Lancet Neurol 2010; 9: 27–9. [DOI] [PubMed] [Google Scholar]
- Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 2012; 490: 187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun YBL, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE November 1998; 86: 2278–324. [Google Scholar]
- Leppik IE, Patterson EN, Coles LD, Craft EM, Cloyd JC. Canine status epilepticus: a translational platform for human therapeutic trials. Epilepsia 2011; 52 (Suppl 8): 31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W. Critical review of current animal models of seizures and epilepsy used in the discovery and development of new antiepileptic drugs. Seizure 2011; 20: 359–68. [DOI] [PubMed] [Google Scholar]
- MacKay DJ. Information theory, inference and learning algorithms. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- Marciani MG, Gotman J, Andermann F, Olivier A. Patterns of seizure activation after withdrawal of antiepileptic medication. Neurology 1985; 35: 1537–43. [DOI] [PubMed] [Google Scholar]
- Mirowski P, Madhavan D, Lecun Y, Kuzniecky R. Classification of patterns of EEG synchronization for seizure prediction. Clin Neurophysiol 2009; 120: 1927–40. [DOI] [PubMed] [Google Scholar]
- Mormann F, Andrzejak RG, Elger CE, Lehnertz K. Seizure prediction: the long and winding road. Brain 2007; 130: 314–33. [DOI] [PubMed] [Google Scholar]
- Munana KR, Thomas WB, Inzana KD, Nettifee-Osborne JA, McLucas KJ, Olby NJ, et al. Evaluation of levetiracetam as adjunctive treatment for refractory canine epilepsy: a randomized, placebo-controlled, crossover trial. J Vet Intern Med 2012; 26: 341–8. [DOI] [PubMed] [Google Scholar]
- Packer RM, Shihab NK, Torres BB, Volk HA. Clinical risk factors associated with anti-epileptic drug responsiveness in canine epilepsy. PLoS One 2014; 9: e106026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Luo L, Parhi KK, Netoff T. Seizure prediction with spectral power of EEG using cost-sensitive support vector machines. Epilepsia 2011; 52: 1761–70. [DOI] [PubMed] [Google Scholar]
- Patterson EE. Canine epilepsy: an underutilized model. Ilar J 2014; 55: 182–6. [DOI] [PubMed] [Google Scholar]
- Pellegrino FC, Sica RE. Canine electroencephalographic recording technique: findings in normal and epileptic dogs. Clin Neurophysiol 2004; 115: 477–87. [DOI] [PubMed] [Google Scholar]
- Petrosian A. Kolmogorov complexity of finite sequences and recognition of different preictal EEG patterns. 1995 Proceedings of the Eighth IEEE Symposium on Computer-Based Medical Systems. Lubbock, TX: IEEE; 1995. p. 212–7. [Google Scholar]
- Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv Large Margin Classifiers 1999; 10: 61–74. [Google Scholar]
- Potschka H, Fischer A, von Ruden EL, Hulsmeyer V, Baumgartner W. Canine epilepsy as a translational model? Epilepsia 2013; 54: 571–9. [DOI] [PubMed] [Google Scholar]
- Schulze-Bonhage A, Kuhn A. Unpredictability of Seizures and the Burden of Epilepsy. in Seizure Prediction in Epilepsy: From Basic Mechanisms to linical Applications Wiley VCH: Verlag GmbH & Co. KGaA, Weinheim; 2008. pp. 1–10. [Google Scholar]
- Snyder DE, Echauz J, Grimes DB, Litt B. The statistics of a practical seizure warning system. J Neural Eng 2008; 5: 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey W, Le Van Quyen M, Mormann F, Schulze-Bonhage A. What is the present-day EEG evidence for a preictal state? Epilepsy Res 2011; 97: 243–51. [DOI] [PubMed] [Google Scholar]
- Teixeira CA, Direito B, Bandarabadi M, Le Van Quyen M, Valderrama M, Schelter B, et al. Epileptic seizure predictors based on computational intelligence techniques: a comparative study with 278 patients. Comput Methods Programs Biomed 2014; 114: 324–36. [DOI] [PubMed] [Google Scholar]
- Vapnik VN, Vapnik V. Statistical learning theory: New York: Wiley; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.