Abstract
Breaking the Abbe diffraction limit has opened the door to imaging the nanometer-scale structure of living cells with fluorescence. New advances have pushed the resolution of structured illumination microscopy even further to gently and rapidly image the complex molecular structure of the plasma membrane in living cells.
The closer we are to observing, measuring, and studying the activities of life directly in living cells the closer we will get to correctly understanding them. Currently, imaging living cells is the domain of optical microscopy. But because of diffraction, the resolution of the standard optical microscope is limited to around half the wavelength of visible light. Given that many cellular and molecular processes occur at the nanometer scale, resolving the structure and dynamics of these processes requires more refined methods. Over the last twenty years three primary approaches have emerged that break the classic Abbe diffraction limit of optical microscopy [1]. These methods include those that rely on successively localizing single molecules (Photo-activation localization microscopy, PALM, stochastic optical reconstruction microcopy, STORM), scanning point spread function shaping methods (Stimulated emission depletion microscopy, STED), and wide-field imaging methods that use patterned illumination (Structured Illumination Microscopy, SIM) [1–3]. The most highly-resolved of these methods can approach a precision near the molecular-scale (iPALM and STED) but require long acquisition times (minutes to hours for iPALM) or very high laser powers (STED), which can be extremely destructive to cells and tissues. Thus, they are not optimal for live cell imaging over time.
The most gentle super-resolution approach that can operate at speeds that match many biological processes is SIM [2, 4]. SIM works by taking advantage of a physical process called the morié effect [4, 5]. A morié pattern is an interference pattern that occurs when two finely-structured patterns are overlaid (Figure 1A). The combination of the two patterns contains a beat pattern with high frequency information about the structure of the individual samples [5]. Thus, if a known, finely structured pattern is used to illuminate an unknown sample on a microscope, the details of the sample can be extracted below the diffraction limit by observing the morié patterns in the combined image. The illumination of samples with these spatial patterns also dramatically enhances contrast particularly in the axial dimension. There are, however, limits to the resolution that can be obtained in SIM. The patterns used to image the sample are generated by diffraction optics, which have their own resolution limits described by the Abbe equation. In practice, this has limited SIM to a resolution about half that of a standard optical microscope or around ~100 nm [5].
Figure 1.
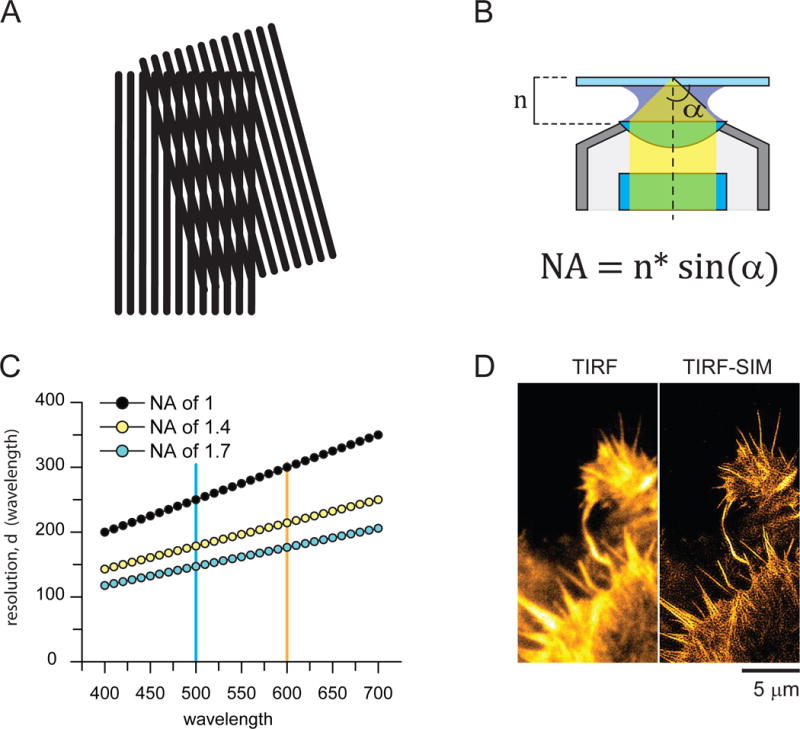
Imaging with structured illumination. A) Example cartoon of a morié effect when two finely spaced patterns are overlaid. B) Cartoon of an objective showing the components that contribute to the numerical aperture value (NA) including n (refractive index of the objective, oil, and cover-glass material) and α (half angle aperture of the cone of light the objective collects and produces). C) Relationship between NA and resolution at different wavelengths. D) Example of a living Hela cell labeled for actin imaged with TIRF (left) and TIRF-SIM. The images are courtesy of Kem Sochacki, Leanna Ferrand, and Patrina Pellett.
In a recent study in Science, Betzig and colleagues succeeded in improving on the resolution of SIM in two interesting ways. Their goal was to create a super-resolution imaging method that would be most useful for biologists working in the dynamic and delicate environment of living cells [6]. First, they used the highest Numerical Aperture (NA) objective lens commercially available to generate the SIM patterns with total internal reflection florescence microscopy (TIRF) and then collect the fluorescence data (NA of 1.7). The NA of an objective is a number that relates to the size of the cone of light that an objective can collect or produce and the refractive index of the lens’ optical materials. Generally, higher NA objectives have higher refractive index materials and wider possible cones of light (Figure 1B). Extremely high NA objectives with NA values above 1.4 have been around for some time [7]. They were first popularized in the development of through-the-objective TIRF microscopy. These objectives allow for the glancing angles necessary to generate very shallow evanescent fields [7]. Because of their high NA values they also can produce finely spaced 2D illumination patterns useful for SIM. This can be seen in the equation for resolution (d = λ/2NA) where d is resolution and NA is numerical aperture. Because resolution is inversely proportion to twice the NA at a given wavelength, the larger the NA the finer the resolution of the light (Figure 1C). The use of TIRF illumination further allowed the authors to dramatically improve axial contrast and reduce the amount of potentially damaging light the sample was exposed to during the experiment (Figure 1D). The authors report an impressive 84 nm resolution in TIRF-SIM with a 1.7 NA objective [6]. In a second set of experiments, the authors took advantage of the non-linear response of photo-activatable proteins to further compress the “effective” pattern of illumination in SIM. These non-linear SIM methods, advanced by Gustafsson and Heintzmann, have theoretically unlimited resolution [8, 9]. Betzig and colleagues applied this additional twist to SIM to further improve high-NA SIM into the 40 nm-resolution range [6]. These non-linear methods, however, come at a cost of more intense illumination and a larger number of frames needed to generate the SIM images.
The future lies in the application of these powerful methods to important biological questions. For example, how the cytoskeleton drives cell movement, how exocytic and endocytic vesicles traffic at the surface of the cell, how calcium-triggered vesicles fuse with the membrane, and how immune cells are activated by antigens can now be imaged at the nanometer scale [6]. To fully achieve these goals, several advances would help including the development of brighter and more photo-stable fluorescent probes that would allow for faster imaging, which is particularly important for rapidly moving objects such as vesicles, filaments, or proteins. If an object moves more than the resolution limit of the microscope within the amount of time it takes to collect the multiple images needed for SIM, a distinctive SIM structured blurring artifact can arise [6, 10]. These artifacts can make it difficult to correctly quantitate these images. Also, these extremely high resolution SIM methods work best with TIRF, where the refractive index of the cell does not disturb the highly structured illumination patterns. Advancing these methods into 3D could be challenging. One might imagine merging other methods with SIM to push these techniques in new directions. Combinations of SIM and FRET, lifetime measurements, polarization, 3D TIRF, interferometry, adaptive optics, or spectral detection could further bridge the gap between super-resolution fluorescence microscopy and the atomic structure of molecules in living cells. As we move closer to imaging the dynamic behavior of individual molecules in living cells we can hope for a more complete understanding of how these complex machines are spatially organized and function.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schermelleh L, et al. A guide to super-resolution fluorescence microscopy. The Journal of cell biology. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, et al. Imaging live-cell dynamics and structure at the single-molecule level. Molecular cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 3.Heintzmann R, Cremer C. Laterally modulated excitation microscopy: Improvement of resolution by using a diffraction grating. P Soc Photo-Opt Ins. 1999;3568:185–196. [Google Scholar]
- 4.Gustafsson MG. Extended resolution fluorescence microscopy. Current opinion in structural biology. 1999;9:627–634. doi: 10.1016/s0959-440x(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 5.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Journal of microscopy. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 6.Li D, et al. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axelrod D. Selective imaging of surface fluorescence with very high aperture microscope objectives. Journal of biomedical optics. 2001;6:6–13. doi: 10.1117/1.1335689. [DOI] [PubMed] [Google Scholar]
- 8.Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzmann R, et al. Saturated patterned excitation microscopy - a concept for optical resolution improvement. J Opt Soc Am A. 2002;19:1599–1609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- 10.Kner P, et al. Super-resolution video microscopy of live cells by structured illumination. Nature methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]


