Abstract
The genus Burkholderia comprises metabolically diverse and adaptable Gram-negative bacteria, which thrive in often adversarial environments. A few members of the genus are prominent opportunistic pathogens. These include B. mallei and B. pseudomallei of the B. pseudomallei complex, which cause glanders and melioidosis, respectively. B. cenocepacia, B. multivorans, and B. vietnamiensis belong to the B. cepacia complex and affect mostly cystic fibrosis patients. Infections caused by these bacteria are difficult to treat because of significant antibiotic resistance. The first line of defense against antimicrobials in Burkholderia species is the outer membrane penetration barrier. Most Burkholderia contain a modified lipopolysaccharide that causes intrinsic polymyxin resistance. Contributing to reduced drug penetration are restrictive porin proteins. Efflux pumps of the resistance nodulation cell division family are major players in Burkholderia multidrug resistance. Third and fourth generation β-lactam antibiotics are seminal for treatment of Burkholderia infections, but therapeutic efficacy is compromised by expression of several β-lactamases and ceftazidime target mutations. Altered DNA gyrase and dihydrofolate reductase targets cause fluoroquinolone and trimethoprim resistance, respectively. Although antibiotic resistance hampers therapy of Burkholderia infections, the characterization of resistance mechanisms lags behind other non-enteric Gram-negative pathogens, especially ESKAPE bacteria such as Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa.
Keywords: Burkholderia, Burkholderia cepacia complex, glanders, melioidosis, antibiotics resistance
1. Introduction
Antimicrobial resistance is rapidly becoming an unavoidable public health crisis with the potential to radically alter the standard of medical care across the globe (Brown and Wright, 2016; Lushniak, 2014; Michael et al., 2014). Without an increased understanding of the mechanisms that drive resistance to therapy for bacterial infection, especially those considered drugs of last resort, the treatment obstacles posed by these factors could ultimately prove insurmountable. If current resistance trends continue and the pipeline for new drugs with activity against Gram-negative remains stagnant, infections caused by these organisms possess the most potential for complete emergence into a post-antibiotic state. Species within this group have been established as organisms of concern in both environmental and nosocomial settings, with members of the Pseudomonas, Acinetobacter, Klebsiella and Burkholderia genera already known to be intransigent to standard first-line therapy as a result of both acquired and intrinsic resistance factors (Boucher et al., 2009; Michael et al., 2014). Many of these factors are found in all four clades and some members, including P. aeruginosa, A. baumannii and K. pneumonia, are ESKAPE bacteria (Boucher et al., 2009).
The Burkholderia genus is large clade within the β-proteobacteriaceae class, containing over 70 species (Sawana et al., 2014). Of these, only B. mallei is considered an obligate parasite of eukaryotic hosts, while the rest are found as environmental saprophytes (Galyov et al., 2010). Despite this, many species other than B. mallei are opportunistic pathogens and capable of causing disease. B. pseudomallei is the etiologic agent of a serious and often fatal syndrome known as melioidosis (Cheng and Currie, 2005; Limmathurotsakul and Peacock, 2011; Peacock, 2006; Wiersinga et al., 2012; Wiersinga et al., 2006). The B. cepacia complex (BCC) consists of a group of at least 17 species (Mahenthiralingam et al., 2005; Mahenthiralingam and Vandamme, 2005; Vandamme and Dawyndt, 2011; Vanlaere et al., 2009). Several members of this complex including B. cenocepacia, B. multivorans, and B. vietnamiensis, are opportunistic pathogens. These organisms are particularly problematic in patients suffering from cystic fibrosis, and are responsible for high mortality rates within this patient cohort (Drevinek and Mahenthiralingam, 2010; Jassem et al., 2011). An underlying theme within the genus is the ability to evade the action of multiple classes of antimicrobials, a capacity that is partially responsible for the seriousness of infections caused by its member species (Dance, 2014; Golini et al., 2004; Jassem et al., 2011; Peeters et al., 2009; Rajendran et al., 2010; Schweizer, 2012a; Wuthiekanun and Peacock, 2006).
2. Bacterial antibiotic resistance factors
Before we review antibiotic resistance in Burkholderia species, it is prudent to present a brief overview of the factors that govern bacterial resistance. This will provide a perspective of the commonalities and differences of the types of resistance determinants that Burkholderia species employ when compared to other bacteria, especially other Gram-negatives.
Three types of mechanisms contribute to antimicrobial resistance in bacteria. Intrinsic antibiotic resistance is caused by physicochemical properties of a bacterium that are not subject to genetic change in response to antibiotic exposure. Possibly the best recognized example of intrinsic resistance is exclusion of drug molecules from Gram-negative bacteria by constraints of the cell envelope, mostly the outer membrane and its lipopolysaccharide and porin constituents. Acquired resistance is caused by acquisition of a previously absent resistance trait, for instance mutation of a chromosomally encoded target, transfer of foreign resistance genes via mobile plasmids, phage-mediated transduction, and transformation using free DNA (Fig. 1). Mutations of a regulatory element for a normally not expressed resistance trait such as an enzyme, an efflux pump, etc., also contribute to acquired resistance.
Figure 1. Genetic mechanisms contributing to acquired resistance in bacteria.
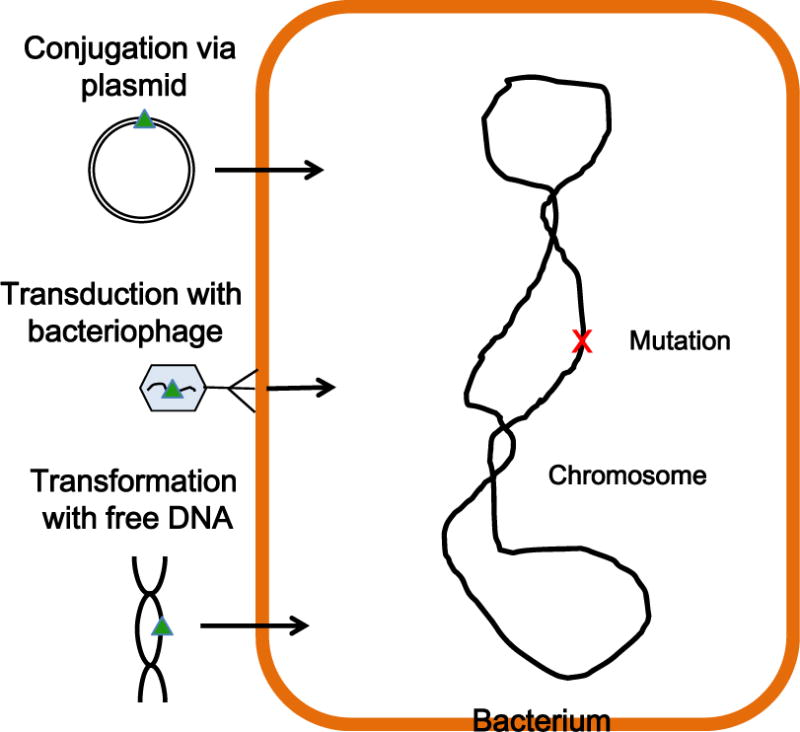
Genetic mechanisms that lead to acquisition of a resistance trait (indicated by the green triangle) include mutations on the chromosome (indicated by the red x) that can either affect drug targets or regulatory factors for normally not expressed resistance proteins, conjugal transfer of a plasmid-encoded resistance marker, bacteriophage-mediated transduction of a resistance marker, or acquisition of a resistance trait via natural DNA transformation, e.g. with chromosomal DNA. This figure is adapted from (Walsh and Wencewicsz, 2016).
Bacteria can exhibit tolerance to antimicrobials that is either dependent or independent of genetic change. This includes bacterial lifestyle adaptations, for instance planktonic versus biofilm growth, intracellular anaerobiosis or persister cell formation (Balaban et al., 2013; Mah, 2012; Monroe, 2007).
An overview of mechanisms that bacteria use to resist antibiotics is presented in Fig. 2. (Blair et al., 2015; Walsh and Wencewicsz, 2016). They include the following: 1) Exclusion from or reduced penetration into the cell by constraints mediated by the cell envelope; 2) Active efflux from the cell; 3) Enzymatic inactivation, either by substrate cleavage or chemical modification, for instance acetylation, adenylation, glucosylation and phosphorylation; 4) Target alteration by point mutations or, rarely, deletion. Targets can also be enzymatically modified, for instance by methylation of ribosomal RNA; 5) Metabolic bypass by substitution of a susceptible target with a resistant target; 6) Target overproduction by either increased transcription or gene multiplication; and 7) Drug sequestration by specific binding proteins (Schweizer, 2012a; Walsh and Wencewicsz, 2016).
Figure 2. Bacterial mechanisms of resistance.
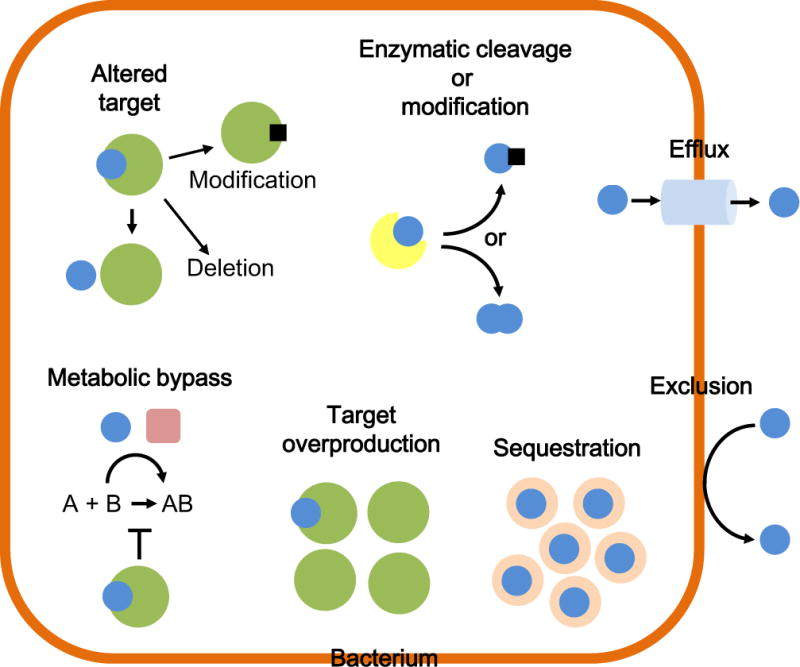
Bacterial antimicrobial resistance mechanisms include – in counterclockwise order – drug (blue sphere) exclusion from or reduced penetration into the cell; active extrusion via efflux pumps; inactivation either by drug cleavage or chemical modification via group (black square) transfer catalyzed by a specific enzyme (indicated in yellow); target (indicated in green) alteration via mutation or enzymatic modification via group transfer (e.g. methylation, black square); metabolic bypass via substitution of a susceptible target with a resistant target; target overproduction via increased transcription or gene multiplication; and sequestration by a binding protein (indicated in rose color). Frequently, different resistance mechanisms act in concert or synergistically, e.g. exclusion and efflux. Resistance mechanisms are either intrinsic or can be acquired, e.g. by chromosomal regulatory mutations that can cause expression of a normally not expressed resistance trait such as an enzyme, an efflux pump, etc. This figure is adapted from (Schweizer, 2012a).
Bacteria frequently employ disparate mechanisms that act synergistically to achieve elevated resistance. The often high-level acquired or intrinsic resistance of non-enteric bacteria such as P. aeruginosa and Burkholderia species is in no small part attributable to synergy between reduced penetration into and efflux from the cell (Schweizer, 2012b).
In this review we will provide an overview of major resistance mechanisms described in select pathogenic members of the genera, specifically the Burkholderia pseudomallei complex (Bpc) and the Burkholderia cepacia complex (Bcc).
3. Burkholderia pseudomallei complex organisms
Members of the Bpc consist of B. pseudomallei, B. mallei, B. humptydoensis and B. thailandensis and are characterized by their similarities to B. pseudomallei at a genetic and phenotypic level (Brett et al., 1998; Ginther et al., 2015; Holden et al., 2004; Nierman et al., 2004; Yu et al., 2006). These organisms are thought to share a common progenitor, most likely very similar to B. pseudomallei itself. B. mallei is considered a clone of B. pseudomallei having diverged from the latter in an animal host approximately 3.5 million years ago and developed into an obligate pathogen by in-host evolution (Losada et al., 2010; Song et al., 2010). Both B. pseudomallei and B. mallei are capable of causing serious disease (Galyov et al., 2010). B. thailandensis, although generally considered non-pathogenic, is known to cause sporadic human disease (Glass et al., 2006).
Mortality rates of B. pseudomallei infections still can exceed 50% without initiation of rapid and appropriate antibiotic therapy, especially in patients with underlying risk factors such as diabetes (Cheng and Currie, 2005; Limmathurotsakul and Peacock, 2011; Peacock, 2006; Wiersinga et al., 2012; Wiersinga et al., 2006). The majority of the resistance factors encoded by B. pseudomallei are also found in B. mallei, and B. thailandensis. However, B. mallei is generally more susceptible to antibiotics than B. pseudomallei, presumably because ongoing in-host evolution leads to genome reduction, including loss of antibiotic resistance determinants (Nierman et al., 2004).
Glanders, caused by B. mallei, is a rare but frequently fatal infection mostly affecting solipeds but occasionally also humans (Whitlock et al., 2007). The organism is known to have been weaponized during both modern and ancient conflicts, most likely because of its potentially devastating effect on cavalry horses and pack animals (Cheng et al., 2005; Dance, 2005; Van Zandt et al., 2013). Few modern human cases have been documented, but symptoms in both animals and humans range from suppurating abscess of the mucosa and solid organs, to sepsis, and pneumonia, depending on the route of infection (Van Zandt et al., 2013).
Melioidosis, caused by B. pseudomallei, is characterized by a variety of symptoms ranging from self-limiting abscess, to sepsis, necrotizing pneumonia, osteomyelitis, and dissemination to the solid organs and brain (Bartley et al., 1999; Caldera et al., 2013; Currie et al., 2010; Jane et al., 2012; Maguire et al., 1998; McLeod et al., 2015; Morse et al., 2013; St John et al., 2014; Wiersinga et al., 2012). Current recommended therapy for melioidosis consists of two weeks of intravenous ceftazidime or a carbapenem, followed by up to six months of oral trimethoprim+sulfamethoxazole (co-trimoxazole)(Chetchotisakd et al., 2014; Dance, 2014; Lipsitz et al., 2012; Pitman et al., 2015). In either phase of therapy, the administration of amoxicillin+clavulanate (co-amoxiclav) is used in instances where other drugs are contraindicated (Dance, 2014; Lipsitz et al., 2012; McLeod et al., 2015). Based largely on the similarity of the in vitro antibiotic susceptibility patterns of B. mallei and B. pseudomallei treatment of glanders in humans follows a similar scheme (Kenny et al., 1999; Lipsitz et al., 2012; Peacock et al., 2008).
3.1 β-lactam resistance in Bpc bacteria
Treatment failure during the administration of ceftazidime occurs in approximately 11–17 percent of clinical cases, although it was found that only a small minority of these cases were due to primary resistance when isolates are assessed using in vitro methods (Chierakul et al., 2005; Wuthiekanun and Peacock, 2006). A survey of over 4,000 B. pseudomallei patient isolates obtained over a period spanning 20 years showed that primary resistance to β-lactam antibiotics is rare, but does exist. For instance, 24 of 4,021 (or 0.6%) of isolates were resistant to ceftazidime (n=8), amoxicillin+clavulanic acid (n=8) or both drugs (n=13). None of the isolates was resistant to carbapenems.
A Class A β-lactamase encoded by penA located on chromosome 2 is responsible for primary resistance to β-lactam antibiotics in the majority of clinical B. pseudomallei isolates (Godfrey et al., 1991)(Fig. 3). Two types of mutations have been implicated in PenA-mediated ceftazidime resistance in clinical B. pseudomallei isolates. The majority of these cause changes to or near conserved β-lactamase domains, the so-called Ambler motifs (Ambler et al., 1991). Notable amino acid substitutions implicated in β-lactam resistance in clinical isolates include Cys69Tyr, Ser72Phe and Pro167Ser, which cause ceftazidime (Cys69Tyr and Pro167Ser) and clavulanic acid (Ser72Phe) resistance, respectively (Rholl et al., 2011; Sam et al., 2009; Sarovich et al., 2012a; Sarovich et al., 2012b; Tribuddharat et al., 2003). Several strains have been identified that contain amino acid substitutions leading to simultaneous ceftazidime and clavulanate resistance, thereby rendering β-lactamase inhibitors of the clavulanate family ineffective in their presence. Some ceftazidime resistant clinical B. pseudomallei isolates also contain a point mutation in the penA upstream region, which probably causes an increased expression of PenA via increased penA transcription (Sarovich et al., 2012b). This notion is supported by results of a recent study with in vitro selected ceftazidime resistant B. thailandensis mutants (Yi et al., 2012a). Treatment of ceftazidime resistant B. pseudomallei is still possible as these isolates remain susceptible to carbapenems. A recent study characterizing B. pseudomallei PenA found that it is a membrane bound lipoprotein, secreted by the twin-arginine transport (TAT) system (Randall et al., 2015; Rholl et al., 2011). The unique membrane association of the enzyme could be a productive route of inquiry as investigative drugs capable of targeting lipoprotein synthesis, as well as TAT-mediated protein export may exhibit efficacy against ceftazidime resistant penA strains. Indeed, tat mutants are susceptible to β-lactam antibiotics and TAT inhibitors could potentially be used to sensitize B. thailandensis to β-lactams (Rholl et al., 2011; Vasil et al., 2012).
Figure 3. Genetic organization of Class A β-lactamase encoding genes implicated in β-lactam resistance in B. pseudomallei and B. cenocepacia.
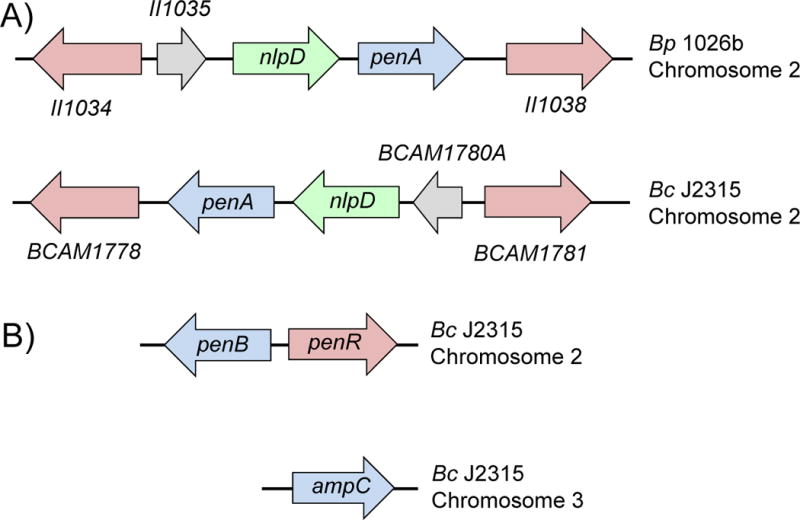
Chromosome location, gene organization and annotation are shown for representative B. pseudomallei (Bp) strain 1026b and B. cenocepacia (Bc) strain J2315. A) PenA confers β-lactam resistance in B. pseudomallei and B. mallei. The role of PenA in B. cenocepacia β-lactam resistance is unknown. B) PenB (originally known and annotated as PenA) and AmpC confer β-lactam resistance in B. cenocepacia. Genes encoding LysR regulatory proteins are indicated with magenta arrows. PenR (sometimes referred to as AmpR) regulates the expression of PenB and AmpC in B. cenocepacia. The nlpD gene encodes an outer membrane protein with peptidase and peptidoglycan-binding domains. Grey arrows indicate genes for hypothetical proteins. Annotations and coordinates are taken from (Winsor et al., 2008) and (Hwang and Kim, 2015). II1034, II1035 and II1038 are short for BP1026B_II1034, BP1026B_II1035, and BP1026B_II1038 used in annotation of the B. pseudomallei strain 1026b genome (Winsor et al., 2008).
Both B. mallei and B. thailandensis express a conserved PenA Class A β-lactamase, and therefore can be anticipated to exhibit similar molecular properties and phenotypes (Winsor et al., 2008). Most observations made with B. pseudomallei β-lactamase have been corroborated by in vitro studies with B. thailandensis (Yi et al., 2012a; Yi et al., 2012b). Studies on PenA β-lactamase expression and its contribution to β-lactam resistance are hampered by the diversity of potential functionally important PenA structural features and factors governing penA transcription in sequenced B. pseudomallei strains. While we are beginning to understand the roles that structural features of PenA play in its function and substrate profile, virtually nothing is known about factors governing the enzyme’s expression. Reliable information about the role of PenA in clinically significant β-lactam resistance continues to be obtained by analysis of isogenetic pre- and post-ceftazidime therapy B. pseudomallei patient isolates (Tribuddharat et al., 2003; Sam et al., 2009; Sarovich et al., 2012a; Sarovich et al., 2012b).
β-lactam resistance in this organism can also occur as the result of large scale rearrangements at the chromosomal level. In a 2011 study examining ceftazidime treatment failures in a Thai hospital, several B. pseudomallei isolates were found to have deleted large segments of chromosome 2. All isolates encompassed deletion of a common 71 kb segment (Chantratita et al., 2011). This segment contained three genes encoding putative penicillin-binding proteins (PBPs), which are known targets for β-lactam antibiotics. Of the two genes encoding PBP3s and one gene encoding a putative PBP6, one PBP3 homolog was shown to be responsible for the severe growth defect manifested as a filamentous appearance by microscopy, as well as high-level ceftazidime resistance (Chantratita et al., 2011). Because of the propagation of distinct populations arising in vivo, the highly-resistant subtype had initially been overlooked during diagnostic testing and was only discernable on specialized Ashdown’s selective medium, which contained glycerol that supported growth of the otherwise unstable mutants (Ashdown, 1979). This may represent a common persistence mechanism in cases where clinical ceftazidime resistance occurs, but cannot be corroborated by standard susceptibility analysis ex vivo.
3.2. Efflux pump mediated multidrug resistance in Bpc members
Efflux is a major resistance mechanism in Bpc organisms, more so in B. pseudomallei and B. thailandensis than in B. mallei. A recent review focused on efflux-mediated drug resistance in Burkholderia, which will be covered herein in an abbreviated manner (Podnecky et al., 2015).
Bacterial genomes encode at least six efflux pump families (Fernandez and Hancock, 2012ß; Hassan et al., 2015; Nikaido and Pages, 2012; Piddock, 2006). These include: 1) The major facilitator (MFS) superfamily; 2) The resistance nodulation cell division (RND) family; 3) The small multidrug resistance (SMR) family; 4) The multi-drug and toxic compound extrusion (MATE) family; 5) The ATP-binding cassette (ABC) family; and 6) The proteobacterial antimicrobial compound efflux (PACE) family. Most bacteria encode several members of each of these efflux pump families, and Burkholderia species are no exception. In Gram-negative bacteria, efflux pumps of the RND family are of major significance because of their unique ability to span the entire cell envelope, which can lead to high-level resistance by synergy between the outer membrane permeability barrier and efflux into the extracellular milieu (Fernandez and Hancock, 2012; Schweizer, 2012b).
All B. pseudomallei strains encode at least 10 RND systems, seven of which are encoded by chromosome 1 and three by chromosome 2 (Holden et al., 2004; Kumar et al., 2008; Podnecky et al., 2015). Three of these RND pumps have been characterized in B. pseudomallei. AmrAB-OprA is expressed in most B. pseudomallei strains and responsible for intrinsic resistance to aminoglycosides and macrolides, and also confers some resistance to tetracyclines (Moore et al., 1999; Trunck et al., 2009). AmrAB-OprA overexpression confers high-level resistance to cethromycin (Mima et al., 2011). Rare aminoglycoside-susceptible B. pseudomallei environmental or clinical isolates either do not express AmrAB-OprB due to regulatory mutations, or lack the amrAB-oprA operon as a result of a chromosomal deletion, or express a non-functional efflux pump due to mutations that affect the AmrB RND transporter (Podin et al., 2014; Trunck et al., 2009). Several strains of B. mallei lack AmrAB-OprA and are therefore aminoglycoside and macrolide susceptible (Nierman et al., 2004).
In regulatory mutants, BpeAB-OprB confers low-level resistance to chloramphenicol, fluoroquinolones, macrolides, and tetracyclines (Chan et al., 2004; Mima and Schweizer, 2010). In some isolates this pump has been implicated in additional functions in host adaptation and quorum sensing (Chan and Chua, 2005). The clinical significance of BpeAB-OprB remains unclear. BpeEF-OprC is expressed in regulatory mutants, and extrudes chloramphenicol, fluoroquinolones, sulfamethoxazole, tetracyclines, and trimethoprim (Hayden et al., 2012; Kumar et al., 2006; Podnecky et al., 2015; Podnecky et al., 2013). It was shown that the multidrug resistant phenotype in a clinical relapse isolate is likely due to constitutive BpeEF-OprC expression as a result of a regulatory mutation caused by a 800 kb chromosomal inversion (Hayden et al., 2012; Podnecky et al., 2015).
The pump repertoire, expression pattern and ensuing drug resistance profile of B. thailandensis parallels that of B. pseudomallei: AmrAB-OprA confers resistance to aminoglycosides, macrolides, and tetracyclines; BpeAB-OprB extrudes tetracyclines; and and BpeEF-OprC effluxes chloramphenicol, fluoroquinolones, sulfamethoxazole, tetracyclines, and trimethoprim (Biot et al., 2013; Biot et al., 2011).
Together, AmrAB-OprA, BperEF-OprB, BpeEF-OprC effectively render six entire classes of compounds at least partially inactive depending on expression level, and evidence obtained mainly with B. thailandensis suggests that they may act in concert to tightly control the intrusion of these compounds into the bacterial cell (Biot et al., 2013).
3.3. Bpc species outer membrane permeability barrier
Although it has been known for some time that the relative outer membrane permeation of non-enteric bacteria like Burkholderia is significantly lower that that of E. coli, the underlying mechanisms remain poorly understood (Hancock, 1998). However, alterations to membrane permeability and structure have been implicated as resistance determinants in these species. For instance, atypical lipopolysaccharide (LPS) structure plays a crucial role in intrinsic resistance of Burkholderia species to cationic peptides, notably polymyxin B (Loutet and Valvano, 2011). A common LPS modification leading to resistance to polymyxin B in Gram-negative bacteria is the modification of lipid A with a positively charged 4-amino-4-deoxy-arabinose (Ara4N) moiety that masks the negative charges of the two phosphate moieties attached to the lipid A (Loutet and Valvano, 2011; Olaitan et al., 2014). The major lipid A species of B. pseudomallei and B. thailandensis was reported to contain a biphosphorylated disaccharide backbone modified with Ara4N at both phosphate groups (Novem et al., 2009). The presence of Ara4N effectively reduces the net negative charge of the cell envelope, reducing the permeation of cationic antimicrobials like polymyxins. The LPS core structure also plays a role in B. pseudomallei polymyxin B resistance. One gene (waaF) encodes a protein required for LPS core oligosaccharide biosynthesis that when mutated increases polymyxin B susceptibility. However, resistance to polymyxins is not restricted to a single pathway but is highly complex. Other mutants exhibiting a polymyxin susceptible phenotype had mutations in a predicted UDP-glucose dehydrogenase and an enzyme called IspH (formerly LytB) necessary for the synthesis of isoprenoids (Rohdich et al., 2002).
Outer membrane porin proteins also play a role in antimicrobial resistance, especially in concert with resistance enzymes or efflux pump expression (Fernandez and Hancock, 2012; Pages et al., 2008). Little is known about porins and their possible roles in B. pseudomallei drug resistance. In vitro studies with B. pseudomallei Omp38 in reconstituted liposomes indicated that this protein functions as a diffusion porin for neutral sugars and charged antibiotics (Siritapetawee et al., 2004). Subsequent black lipid membrane reconstitution studies, liposome swelling assays and expression in a porin-deficient E. coli strain confirmed translocation of antibiotics through Omp38 and suggested a possible contribution of this porin in B. pseudomallei ceftazidime and carbapenem resistance (Aunkham et al., 2014; Suginta et al., 2011). However, assessment of the true contribution of this porin to B. pseudomallei antibiotic resistance awaits genetic and functional analyses in the source organism.
3.4. Alterations in drug targets
Point mutations that alter drug targets are one of the most common means that bacteria utilize to resist antibiotics (Blair et al., 2015; Walsh and Wencewicsz, 2016). Aside from the aforementioned PenA mutations that increase affinity to β-lactams that were previously poor substrates and the PBP3 deletion causing ceftazidime resistance in B. pseudomallei, reports on target mutations in B. pseudomallei leading to resistance to other antibiotics are scarce. To date there is one report on target mutation-based fluoroquinolone resistance in B. pseudomallei. Fluoroquinolones selectively target bacterial topoisomerase type II enzymes, DNA gyrase and topoisomerase IV (Walsh and Wencewicsz, 2016). Each enzyme is an A2B2-type heterotetramer, consisting of two GyrA and GyrB monomers for DNA gyrase and two ParC and ParD monomers for topoisomerase IV. Fluoroquinolones inhibit the respective “A” subunit of the topoisomerase heterotetramer, e.g. GyrA and ParC. Resistance to fluoroquinolones is frequently due to mutations affecting the quinolone resistance-determining regions (QRDR) of GyrA or ParC. In E. coli the GyrA QRDR encompasses a relatively small region (amino acids 67 to 106) of the protein (Yoshida et al., 1990). Quinolone resistance in E. coli and other bacteria is caused most often by mutations affecting amino acids 81 through 87. In E. coli roughly 50% of mutations affect Ser83 (Yoshida et al., 1990). In B. pseudomallei, all in vitro selected fluoroquinolone resistant mutants contained a Thr83Ile mutation (Viktorov et al., 2008). This is consistent with the finding that most Burkholderia species contain a GyrA Thr83 instead of the Ser83 found in other bacteria (Winsor et al., 2008). Notably, P. aeruginosa also contains Thr83 in GyrA and a frequent amino acid change seen in fluoroquinolone resistant clinical isolates is a Thr83Ile mutation (Kureishi et al., 1994).
4. Burkholderia cepacia complex organisms
The Burkholderia cepacia complex (Bcc) organisms are opportunistic nosocomial pathogens capable of causing severe disease in immunocompromised individuals, especially those with cystic fibrosis (CF) (Mahenthiralingam et al., 2005). In these patients, infection with Bcc organisms causes “cepacia syndrome”, typically characterized by necrotizing pneumonia, sepsis and an overall negative prognosis (Mahenthiralingam and Vandamme, 2005). Although several species from this group have been isolated from the lungs of CF patients, B. cenocepacia and B. multivorans appear to cause the most serious forms of disease in these individuals and account for 85% of all Bcc infections (Drevinek and Mahenthiralingam, 2010; Mahenthiralingam and Vandamme, 2005). Treatment of Bcc infections relies on ceftazidime and other extended-spectrum cephalosporins, as intrinsic resistance prevents the action of many other classes of antimicrobials. One study of several thousand clinical isolates isolated from CF patients found that resistance to many standard therapies was overwhelming; greater than 50 percent of isolates were resistant to chloramphenicol, co-trimoxazole, ciprofloxacin, tetracycline, rifampin, avibactam, and co-amoxiclav (Zhou et al., 2007). However, these data may overestimate the occurrence of resistance in Bcc organisms as the study was carried out on patient isolates solicited because they were in fact multidrug resistant. Despite this caveat, resistance patterns, both intrinsic and acquired, must not be discounted in these organisms.
4.1 β-lactam resistance in Bcc bacteria
Resistance to β-lactam antibiotics such as ceftazidime is caused by class A β-lactamases encoded by Bcc organisms, first described in the PenA-PenR system of B. cepacia 249 (Trepanier et al., 1997), which are now named PenB and PenR (AmpR) (Hwang and Kim, 2015). A recent study in B. cenocepacia identified ceftazidime-driven mutations to the peptidoglycan recycling enzyme AmpD as a putative cause for up-regulation of the PenB and AmpC β-lactamases (Hwang and Kim, 2015). PenB is a Class A penicillinase with broad spectrum carbapenemase character that highly conserved across the Bcc and shares significant protein homology with PenA of Bpc organisms (Poirel et al., 2009; Hwang and Kim, 2015). Both penB and ampC promoters were associated with binding sites for LysR-type repressor PenR (AmpR). PenR is a bifunctional protein. It is a repressor when it binds the D-alanine-D-alanine pentapeptide stem terminus of the peptidoglycan precursor UDP-MurNAc-pentapeptide. This protein was previously found to bind 1,6-anhydro-MurNac peptides, which are produced in the presence of β-lactam antibiotics or after disruption of AmpD, an enzyme that normally degrades them. This binding causes PenR to become an activator and leads to transcription of its penB and ampC targets (Hwang and Kim, 2015; Vadlamani et al., 2015). As a result, susceptibilities to ceftazidime, cefotaxime, and meropenem were greatly reduced.
B. cenocepacia also contains a Class A PenA β-lactamase, which is 55% identical and 67% similar to B. pseudomallei strain 1026b PenA (Winsor et al., 2008). As in the Bpc bacteria, the penA gene is located on chromosome 2 and its genetic surroundings are very similar to that of B. pseudomallei penA (Fig. 3). However, unlike B. pseudomallei or B. thailandensis PenA (the B. thailandensis PenA is now also referred to as PenL in some publications (Hwang and Kim, 2015)) the B. cenocepacia enzyme has not yet been shown to be involved in β-lactam resistance. B. thailandensis mutants with in vitro selected ceftazidime resistance solely contain PenA (PenL) structural and/or regulatory mutations, whereas in B. cenocepacia the same selection leads to regulatory mutants that either overexpress PenB or AmpC (Hwang and Kim, 2015; Yi et al., 2012a). PenB and AmpC exhibit similar substrate spectra and both are under control of the LysR-type PenR regulatory protein (Fig. 3). PenB and AmpC are absent from Bpc bacteria. Of note is that the sequenced B. cenocepacia strain J2315 PenB β-lactamase contains a Ser72Tyr substitution, which may explain this strain’s intrinsic clavulanate resistance (Hwang and Kim, 2015).
B. multivorans contains a PenA enzyme (Bmul_3689 in B. multivorans ATCC 17616), that is closely related to PenB in BCC bacteria (Poirel et al., 2009; Hwang and Kim, 2015). It is also similar to KPC-2, which is the most clinically significant serine carbapenemase (Papp-Wallace et al., 2013). B. multivorans PenA is closely related to B. pseudomallei PenA (also called PenI by Dr. Bonomo’s group (Papp-Wallace et al., 2013)). However, the B. multivorans enzyme is an inhibitor-resistant carbapenemase, whereas the B. pseudomallei enzyme is an extended spectrum β-lactamase. The role of PenA in clinically significant B. multivorans β-lactam resistance compromising therapy is not well established.
4.2. Efflux pump mediated multidrug resistance in Bcc members
The roles of efflux pumps in antibiotic resistance of members of the Bcc have recently been reviewed (Podnecky et al., 2015). In B. cenocepacia, at least six efflux pumps of the RND family have been implicated in drug resistance – RND-1, RND-3, RND-4, RND-8, RND-9, and RND-10 (Bazzini et al., 2011; Buroni et al., 2014; Buroni et al., 2009; Coenye et al., 2011; Nair et al., 2004). Of these RND-3, RND-4, and RND-10 (also known as CeoAB-OpcM) correspond to B. pseudomallei AmrAB-OprA, BpeAB-OprB and BpeEF-OprC, respectively, although the resistance patterns bestowed by the respective pumps are somewhat different, with the exception of RND-10 and BpeEF-OprC whose resistance profiles are very similar (Podnecky et al., 2015). An inventory of B. cenocepacia resistance mechanisms showed that efflux pump activity is prevalent in this bacterium and that mutations in the RND-3 pump regulator are the major cause of efflux pump activity and RND-3 mediated antibiotic resistance (Tseng et al., 2014). In B. vietnamiensis, aminoglycoside resistance emerges during chronic infection or after in vitro exposure to aminoglycosides and is the result of AmrAB-OprM efflux pump expression, which is most likely a homolog of B. pseudomallei and B. thailandensis AmrAB-OprA (Jassem et al., 2014; Jassem et al., 2011).
The B. vietnamiensis NorM pump, a member of the multidrug and toxic compound extrusion family of efflux systems, was shown to contribute to polymyxin B resistance, but curiously only in the presence of exogenously added tetracycline (Fehlner-Gardiner and Valvano, 2002).
4.3. Bcc species outer membrane permeability barrier
Similar to Bpc organisms, Bcc bacteria are typically resistant to polymyxins (Loutet and Valvano, 2011; Olaitan et al., 2014). As described above, this is thought to be partially attributable to a unique LPS structure that inhibits polymyxin binding to the outer membrane (Loutet and Valvano, 2011). In B. cenocepacia, an amino arabinose biosynthesis operon is responsible for the synthesis of 4-amino-4-deoxy-L-arabinose (Ara4N) used for modifications of lipid A that alter the total charge of the LPS molecule, thereby decreasing susceptibility to cationic antimicrobial peptides and polymyxins (Isshiki et al., 1998; Loutet and Valvano, 2011; Olaitan et al., 2014; Ortega et al., 2007).
The alternative sigma factor RpoE, which controls the expression of a regulon of genes required for bacteria to respond to extracytoplasmic stress, plays a significant role in polymyxin B resistance in B. cenocepacia at 37°C but not at 30°C (Loutet and Valvano, 2011).
As in Bpc bacteria, polymyxin resistance in Bcc species is complex and the pathways found in the former are also found in Bcc bacteria, as reviewed in Loutet et al. (Loutet and Valvano, 2011).
In B. multivorans, genes needed for the synthesis of hopanoid compounds are also implicated in polymyxin resistance (Malott et al., 2012). Interestingly, the use of fosmidomycin may potentiate colistin activity against B. multivorans by interrupting this process. Through disruption of the isoprenoid synthesis pathway, fosmidomycin prevents hopanoid synthesis and alters membrane composition, ultimately resulting in a 64 fold decrease in the colistin MIC (Malott et al., 2014).
The role of porins in decreased antibiotic susceptibility of Bcc species has long been established. Bcc complex bacteria such as B. cenocepacia contain general porins that exhibit a permeability that is similar of P. aeruginosa, and approximately 10 times less than E. coli (Parr et al., 1987). β-lactam resistant CF B. cenocepacia isolates and a resistant mutant were shown to have decreased porin content (Aronoff, 1988).
4.4. Alterations in Bcc bacteria drug targets
Drug target modification in these species has been mostly associated with resistance to fluoroquinolones. Depending on the selection scheme and drug concentration used, in vitro selection of ciprofloxacin resistant B. cenocepacia mutants yielded mostly Thr83Ile or Asp87Asn mutations in the GyrA QRDR (Pope et al., 2008). These resulted in a 12–64 fold increase in the ciprofloxacin minimal inhibitory concentration (MIC). High-level ciprofloxacin resistance (MIC>256 μg/ml) required an additional Ser80Leu mutation in the ParC QRDR (Pope et al., 2008). Similar gyrA mutations were identified in a majority of levofloxacin resistant isolates studied in a survey of resistance mechanisms in Bcc (Tseng et al., 2014). The resistant isolates contained Gly81Asp, Thr83Ile, and Asp87His mutations. None of the isolates contained mutations in the parC QRDR.
Dihydrofolate reductase is the target of trimethoprim. Purification of the enzyme from trimethoprim susceptible and trimethoprim resistant B. cepacia strains indicated that the protein from the resistant strain was indeed refractory to trimethoprim inhibition (Burns et al., 1989). While this finding is consistent with target alteration, the molecular and genetic basis for this reduced inhibition was not established.
5. Conclusions
Although the resilience of Burkholderia species to antimicrobials has been recognized for quite some time, a literature review quickly reveals that our overall understanding of resistance in these bacteria is still rather rudimentary. Fig. 4 summarizes the current state of knowledge of the various resistance determinants that have been observed in antibiotic resistant Burkholderia species. First, the outer membrane permeability barrier is an important contributor to Burkholderia drug resistance. The two major players involved are lipopolysaccharide (LPS) and outer membrane porins. LPS typically plays a general role in drug resistance, but in most Burkholderia species it plays a major role in resistance to cationic peptides. Lipid A modification by aminoarabinose and changes to the LPS core are major determinants for the widespread intrinsic polymyxin resistance in Burkholderia. Restrictive porin proteins are contributing factors to drug resistance, especially in combination with other determinants such as efflux. RND pump-mediated efflux makes major contributions to intrinsic and acquired multidrug resistance. Depending on species, either periplasmic or membrane-bound β-lactamases play important roles in intrinsic β-lactam resistance and acquired resistance to clinically significant β-lactam antibiotics. Resistance due to antibiotic target mutations has mostly been associated with fluoroquinolones, but has also been implicated with resistance to other antibiotics, e.g. trimethoprim. Of note is that most, if not all, of the Burkholderia resistance determinants identified to date are encoded by the genome of the respective organisms. The genomes of Burkholderia species consist of at least two chromosomes, e.g. Bpc bacteria, but other species contain additional genetic elements, e.g. B. cenocepacia, whose sequenced prototype strain J2315 contains a third chromosome and a plasmid (Holden et al., 2009; Holden et al., 2004; Nierman et al., 2004). Plasmids have not yet been demonstrated in Bpc bacteria. The constellation of both intrinsic and acquired resistance mechanisms in this genus combines to create a unique and often difficult challenge for researchers and clinicians. Further study is necessary to understand the interplay of these factors and their effect on antimicrobial therapy.
Figure 4. Summary of resistance determinants identified in Burkholderia species.
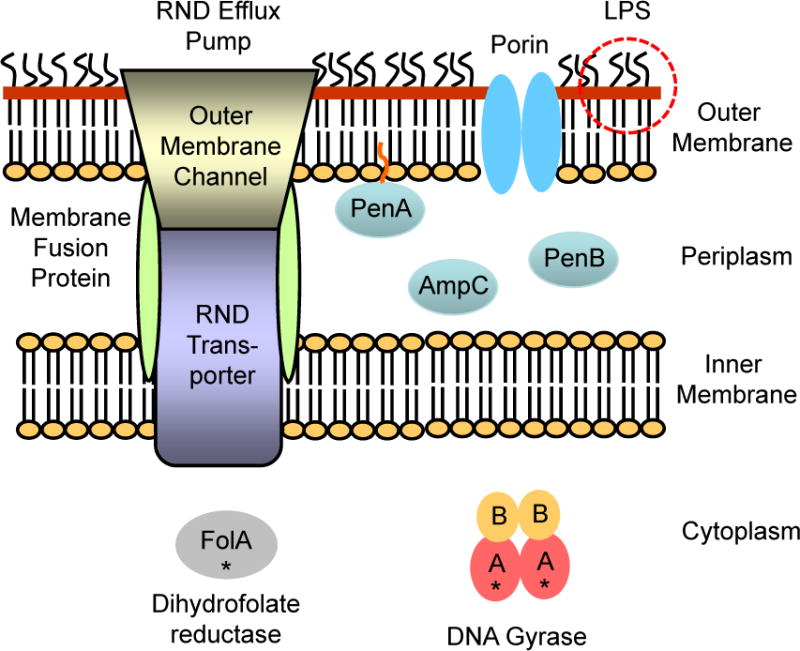
The outer membrane provides a major penetration barrier, mainly because of the physicochemical properties of lipopolysaccharide (LPS) and restrictive porins. Efflux via tripartite RND pumps consisting of an RND transporter, a membrane fusion protein and an outer membrane channel plays a major role in multidrug resistance. Depending on species, acquired and intrinsic resistance to β-lactam antibiotics is provided by periplasmic β-lactamases, e.g. AmpC and PenB, or a membrane-bound β-lactamase, e.g. PenA. Target mutations (symbolized by black asterisks) in in the DNA gyrase A subunit and dihydrofolate reductase cause resistance to fluoroquinolones and trimethoprim, respectively.
Acknowledgments
The authors acknowledge the contributions of several talented graduate students (Kyoung-Hee Choi, Carolina Lopez, Nicole Podnecky, Katie Propst, Linnell Randall, Drew Rholl, Lily Trunck) and postdocs (Sunisa Chirakul, Ayush Kumar, Takehiko Mima, Nawarat Somprasong) to antibiotic resistance research performed in the Schweizer laboratory. Work in the HPS laboratory was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and contracts from the United States Defense Threat Reduction Agency and the United States Broad Agency Research Development Authority.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronoff SC. Outer membrane permeability in Pseudomonas cepacia: diminished porin content in a beta-lactam-resistant mutant and in resistant cystic fibrosis isolates. Antimicrob Agents Chemother. 1988;32:1636–1639. doi: 10.1128/aac.32.11.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathol. 1979;11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- Aunkham A, Schulte A, Winterhalter M, Suginta W. Porin involvement in cephalosporin and carbapenem resistance of Burkholderia pseudomallei. PLoS One. 2014;9:e95918. doi: 10.1371/journal.pone.0095918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Microbiol. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- Bartley PP, Pender MP, Woods ML, 2nd, Walker D, Douglas JA, Allworth AM, Eisen DP, Currie BJ. Spinal cord disease due to melioidosis. Trans Roy Soc, Trop Med Hyg. 1999;93:175–176. doi: 10.1016/s0035-9203(99)90299-7. [DOI] [PubMed] [Google Scholar]
- Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One. 2011;6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biot FV, Lopez MM, Poyot T, Neulat-Ripoll F, Lignon S, Caclard A, Thibault FM, Peinnequin A, Pages JM, Valade E. Interplay between three RND efflux pumps in doxycycline-selected strains of Burkholderia thailandensis. PLoS One. 2013;8:e84068. doi: 10.1371/journal.pone.0084068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biot FV, Valade E, Garnotel E, Chevalier J, Villard C, Thibault FM, Vidal DR, Pages JM. Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One. 2011;6:e16892. doi: 10.1371/journal.pone.0016892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Brett PJ, DeShazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Burns JL, Lien DM, Hedin LA. Isolation and characterization of dihydrofolate reductase from trimethoprim-susceptible and trimethoprim-resistant Pseudomonas cepacia. Antimicrob Agents Chemother. 1989;33:1247–1251. doi: 10.1128/aac.33.8.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother. 2014;58:7424–7429. doi: 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. Assessment of three Resistance-Nodulation-Cell Division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol. 2009;9:200. doi: 10.1186/1471-2180-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldera AS, Kumanan T, Corea E. A rare cause of septic arthritis: melioidosis. Trop Doctor. 2013;43:164–166. doi: 10.1177/0049475513505091. [DOI] [PubMed] [Google Scholar]
- Chan YY, Chua KL. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J Bacteriol. 2005;187:4707–4719. doi: 10.1128/JB.187.14.4707-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YY, Tan TMC, Ong YM, Chua KL. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother. 2004;48:1128–1135. doi: 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Thanwisai A, Chua HH, Ooi WF, Holden MTG, Day NP, Tan P, Schweizer HP, Peacock SJ. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2011;108:17165–17170. doi: 10.1073/pnas.1111020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Dance DA, Currie BJ. Bioterrorism, Glanders and melioidosis. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2005;10:E1–2. author reply E1–2. [PubMed] [Google Scholar]
- Chetchotisakd P, Chierakul W, Chaowagul W, Anunnatsiri S, Phimda K, Mootsikapun P, Chaisuksant S, Pilaikul J, Thinkhamrop B, Phiphitaporn S, Susaengrat W, Toondee C, Wongrattanacheewin S, Wuthiekanun V, Chantratita N, Thaipadungpanit J, Day NP, Limmathurotsakul D, Peacock SJ. Trimethoprim-sulfamethoxazole versus trimethoprim-sulfamethoxazole plus doxycycline as oral eradicative treatment for melioidosis (MERTH): a multicentre, double-blind, non-inferiority, randomised controlled trial. Lancet. 2014;383:807–814. doi: 10.1016/S0140-6736(13)61951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chierakul W, Anunnatsiri S, Short JM, Maharjan B, Mootsikapun P, Simpson AJ, Limmathurotsakul D, Cheng AC, Stepniewska K, Newton PN, Chaowagul W, White NJ, Peacock SJ, Day NP, Chetchotisakd P. Two randomized controlled trials of ceftazidime alone versus ceftazidime in combination with trimethoprim-sulfamethoxazole for the treatment of severe melioidosis. Clin Infect Dis. 2005;41:1105–1113. doi: 10.1086/444456. [DOI] [PubMed] [Google Scholar]
- Coenye T, Van Acker H, Peeters E, Sass A, Buroni S, Riccardi G, Mahenthiralingam E. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother. 2011;55:1912–1919. doi: 10.1128/AAC.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance D. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents. 2014;43:310–318. doi: 10.1016/j.ijantimicag.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance DAB. Melioidosis and glanders as possible biological weapons. In: Fong W, Alibek K, editors. Bioterrorism and infectious agents A new dilemma for the 21st century. Springer Science and Business Media; New York: 2005. pp. 99–145. [Google Scholar]
- Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- Fehlner-Gardiner CC, Valvano MA. Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux system. FEMS Microbiol Lett. 2002;215:279–283. doi: 10.1111/j.1574-6968.2002.tb11403.x. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Ginther JL, Mayo M, Warrington SD, Kaestli M, Mullins T, Wagner DM, Currie BJ, Tuanyok A, Keim P. Identification of Burkholderia pseudomallei near-neighbor species in the Northern Territory of Australia. PLoS Negl Trop Dis. 2015;9:e0003892. doi: 10.1371/journal.pntd.0003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass MB, Gee JE, Steigerwalt AG, Cavuoti D, Barton T, Hardy RD, Godoy D, Spratt BG, Clark TA, Wilkins PP. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J Clin Microbiol. 2006;44:4601–4604. doi: 10.1128/JCM.01585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AJ, Wong S, Dance DA, Chaowagul W, Bryan LE. Pseudomonas pseudomallei resistance to beta-lactam antibiotics due to alterations in the chromosomally encoded beta-lactamase. Antimicrob Agents Chemother. 1991;35:1635–1640. doi: 10.1128/aac.35.8.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golini G, Favari F, Marchetti F, Fontana R. Bacteriostatic and bactericidal activity of levofloxacin against clinical isolates from cystic fibrosis patients. Eur J Clin Microbiol. 2004;23:798–800. doi: 10.1007/s10096-004-1216-3. [DOI] [PubMed] [Google Scholar]
- Hancock REW. Resistance mechanisms in Pseudomonas aeruginosa and other non-fermentative bacteria. Clin Infect Dis. 1998;27(Suppl 1):S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Liu Q, Henderson PJ, Paulsen IT. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio. 2015;6 doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One. 2012;7:e36507. doi: 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins TP, Crossman LC, Pitt TL, Churcher C, Mungall KL, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kim HS. Cell Wall Recycling-Linked Coregulation of AmpC and PenB beta-Lactamases through ampD Mutations in Burkholderia cenocepacia. Antimicrob Agents Chemother. 2015;59:7602–7610. doi: 10.1128/AAC.01068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki Y, Kawahara K, Zahringer U. Isolation and characterisation of disodium (4-amino-4-deoxy-beta-L- arabinopyranosyl)-(1–>8)-(D-glycero-alpha-D-talo-oct-2-ulopyranosylona te)- (2–>4)-(methyl 3-deoxy-D-manno-oct-2-ulopyranosid)onate from the lipopolysaccharide of Burkholderia cepacia. Carbohydrate Res. 1998;313:21–27. doi: 10.1016/s0008-6215(98)00179-7. [DOI] [PubMed] [Google Scholar]
- Jane L, Crowe A, Daffy J, Gock H. Burkholderia pseudomallei osteomyelitis: An unusual cause of fever in a returned traveller. Austral Med J. 2012;5:141–143. doi: 10.4066/AMJ.2012.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassem AN, Forbes CM, Speert DP. Investigation of aminoglycoside resistance inducing conditions and a putative AmrAB-OprM efflux system in Burkholderia vietnamiensis. Ann Clin Microbiol Antimicrob. 2014;13:2. doi: 10.1186/1476-0711-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassem AN, Zlosnik JE, Henry DA, Hancock RE, Ernst RK, Speert DP. In vitro susceptibility of Burkholderia vietnamiensis to aminoglycosides. Antimicrob Agents Chemother. 2011;55:2256–2264. doi: 10.1128/AAC.01434-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny DJ, Russell P, Rogers D, Eley SM, Titball RW. In vitro susceptibilities of Burkholderia mallei in comparison to those of other pathogenic Burkholderia spp. Antimicrob Agents Chemother. 1999;43:2773–2775. doi: 10.1128/aac.43.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chua KL, Schweizer HP. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother. 2006;50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. Expression of resistance-nodulation-cell division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from Northern Australia. Trans Roy Soc Trop Med Hyg. 2008;102(S1):S145–S151. doi: 10.1016/S0035-9203(08)70032-4. [DOI] [PubMed] [Google Scholar]
- Kureishi A, Diver JM, Beckthold B, Schollaardt T, Bryan LE. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 1994;38:1944–1952. doi: 10.1128/aac.38.9.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D, Peacock SJ. Melioidosis: a clinical overview. Brit Med Bull. 2011;99:125–139. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, Cheng AC, Currie BJ, Dance D, Gee JE, Larsen J, Limmathurotsakul D, Morrow MG, Norton R, O’Mara E, Peacock SJ, Pesik N, Rogers LP, Schweizer HP, Steinmetz I, Tan G, Tan P, Wiersinga WJ, Wuthiekanun V, Smith TL. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei Infection, 2010. Em Infect Dis. 2012;18:e2. doi: 10.3201/eid1812.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, Kim HS, Shabalina SA, Pearson TR, Brinkac L, Tan P, Nandi T, Crabtree J, Badger J, Beckstrom-Sternberg S, Saqib M, Schutzer SE, Keim P, Nierman WC. Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol Evol. 2010;2:102–116. doi: 10.1093/gbe/evq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Valvano MA. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol. 2011;2:159. doi: 10.3389/fmicb.2011.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushniak BD. Antibiotic resistance: a public health crisis. Pub Health Rep. 2014;129:314–316. doi: 10.1177/003335491412900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire GP, Flavell HD, Burrow JN, Currie BJ. Relapsing neurological melioidosis from the top end of the Northern Territory. Austral New Zealand J Med. 1998;28:219–220. doi: 10.1111/j.1445-5994.1998.tb02978.x. [DOI] [PubMed] [Google Scholar]
- Mah TF. Biofilm-specific antibiotic resistance. Fut Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Antimicrob. Agents Chemother. complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Vandamme P. Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chr Resp Dis. 2005;2:209–217. doi: 10.1191/1479972305cd053ra. [DOI] [PubMed] [Google Scholar]
- Malott RJ, Steen-Kinnaird BR, Lee TD, Speert DP. Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans. Antimicrob Agents Chemother. 2012;56:464–471. doi: 10.1128/AAC.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott RJ, Wu CH, Lee TD, Hird TJ, Dalleska NF, Zlosnik JE, Newman DK, Speert DP. Fosmidomycin decreases membrane hopanoids and potentiates the effects of colistin on Burkholderia multivorans clinical isolates. Antimicrob Agents Chemother. 2014;58:5211–5219. doi: 10.1128/AAC.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod C, Morris PS, Bauert PA, Kilburn CJ, Ward LM, Baird RW, Currie BJ. Clinical presentation and medical management of melioidosis in children: a 24-year prospective study in the Northern Territory of Australia and review of the literature. Clin Infect Dis. 2015;60:21–26. doi: 10.1093/cid/ciu733. [DOI] [PubMed] [Google Scholar]
- Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front Pub Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima T, Schweizer HP. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother. 2010;54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima T, Schweizer HP, Xu ZQ. In vitro activity of cethromycin against Burkholderia pseudomallei and investigation of mechanism of resistance. J Antimicrob Chemother. 2011;66:73–78. doi: 10.1093/jac/dkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007;5:e307. doi: 10.1371/journal.pbio.0050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse LP, Smith J, Mehta J, Ward L, Cheng AC, Currie BJ. Osteomyelitis and septic arthritis from infection with Burkholderia pseudomallei: A 20-year prospective melioidosis study from northern Australia. J Orthop. 2013;10:86–91. doi: 10.1016/j.jor.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair BM, Cheung KJ, Jr, Griffith A, Burns JL. Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia) J Clin Invest. 2004;113:464–473. doi: 10.1172/JCI19710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci USA. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Pages JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novem V, Shui G, Wang D, Bendt AK, Sim SH, Liu Y, Thong TW, Sivalingam SP, Ooi EE, Wenk MR, Tan G. Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin Vaccine Immunol. 2009;16:1420–1428. doi: 10.1128/CVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, Govan JR, Valvano MA. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J Bacteriol. 2007;189:3639–3644. doi: 10.1128/JB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M. Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem. 2013;288:19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr TR, Jr, Moore RA, Moore LV, Hancock RE. Role of porins in intrinsic antibiotic resistance of Pseudomonas cepacia. Antimicrob Agents Chemother. 1987;31:121–123. doi: 10.1128/aac.31.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SJ. Melioidosis. Curr Opinion Infect Dis. 2006;19:421–428. doi: 10.1097/01.qco.0000244046.31135.b3. [DOI] [PubMed] [Google Scholar]
- Peacock SJ, Schweizer HP, Dance DAB, Smith TL, Gee JE, Wuthiekanun V, DeShazer D, Steinmetz I, Tan P, Currie BJ. Consensus guidelines on the management of accidental laboratory exposure to Burkholderia pseudomallei and Burkholderia mallei. Em Infect Dis. 2008;14 doi: 10.3201/eid1407.071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E, Nelis HJ, Coenye T. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J Antimicrob Chemother. 2009;64:801–809. doi: 10.1093/jac/dkp253. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman MC, Luck T, Marshall CS, Anstey NM, Ward L, Currie BJ. Intravenous therapy duration and outcomes in melioidosis: a new treatment paradigm. PLoS Negl Trop Dis. 2015;9:e0003586. doi: 10.1371/journal.pntd.0003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Wong J, Tuanyok A, Keim P, Giffard PM, Currie BJ. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother. 2014;58:162–166. doi: 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky NL, Rhodes KA, Schweizer HP. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol. 2015;6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky NL, Wuthiekanun V, Peacock SJ, Schweizer HP. The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob Agents Chemother. 2013;57:4381–4386. doi: 10.1128/AAC.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Rodriguez-Martinez JM, Plesiat P, Nordmann P. Naturally occurring Class A ss-lactamases from the Burkholderia cepacia complex. Antimicrob Agents Chemother. 2009;53:876–882. doi: 10.1128/AAC.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CF, Gillespie SH, Pratten JR, McHugh TD. Fluoroquinolone-resistant mutants of Burkholderia cepacia. Antimicrob Agents Chemother. 2008;52:1201–1203. doi: 10.1128/AAC.00799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Quinn RF, Murray C, McCulloch E, Williams C, Ramage G. Efflux pumps may play a role in tigecycline resistance in Burkholderia species. Int J Antimicrob Agents. 2010;36:151–154. doi: 10.1016/j.ijantimicag.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Randall LB, Dobos K, Papp-Wallace KM, Bonomo RA, Schweizer HP. Membrane-Bound PenA beta-Lactamase of Burkholderia pseudomallei. Antimicrob Agents Chemother. 2015;60:1509–1514. doi: 10.1128/AAC.02444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, Schweizer HP. Molecular Investigations of PenA-mediated beta-lactam Resistance in Burkholderia pseudomallei. Front Microbiol. 2011;2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F, Hecht S, Gartner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam IC, See KH, Puthucheary SD. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J Clin Microbiol. 2009;47:1556–1558. doi: 10.1128/JCM.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarovich DS, Price EP, Limmathurotsakul D, Cook JM, Von Schulze AT, Wolken SR, Keim P, Peacock SJ, Pearson T. Development of ceftazidime resistance in an acute Burkholderia pseudomallei infection. Infecti Drug Resist. 2012a;5:129–132. doi: 10.2147/IDR.S35529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarovich DS, Price EP, Von Schulze AT, Cook JM, Mayo M, Watson LM, Richardson L, Seymour ML, Tuanyok A, Engelthaler DM, Pearson T, Peacock SJ, Currie BJ, Keim P, Wagner DM. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One. 2012b;7:e30789. doi: 10.1371/journal.pone.0030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana A, Adeolu M, Gupta RS. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 2014;5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Fut Microbiol. 2012a;7:1389–1399. doi: 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. When it comes to drug discovery not all Gram-negative bacterial biodefence pathogens are created equal: Burkholderia pseudomallei is different. Microb Biotechnol. 2012b;5:581–583. doi: 10.1111/j.1751-7915.2012.00334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siritapetawee J, Prinz H, Krittanai C, Suginta W. Expression and refolding of Omp38 from Burkholderia pseudomallei and Burkholderia thailandensis, and its function as a diffusion porin. Biochem J. 2004;384:609–617. doi: 10.1042/BJ20041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathogens. 2010;6:e1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JA, Ekberg JA, Dando SJ, Meedeniya AC, Horton RE, Batzloff M, Owen SJ, Holt S, Peak IR, Ulett GC, Mackay-Sim A, Beacham IR. Burkholderia pseudomallei penetrates the brain via destruction of the olfactory and trigeminal nerves: implications for the pathogenesis of neurological melioidosis. mBio. 2014;5:e00025. doi: 10.1128/mBio.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suginta W, Mahendran KR, Chumjan W, Hajjar E, Schulte A, Winterhalter M, Weingart H. Molecular analysis of antimicrobial agent translocation through the membrane porin BpsOmp38 from an ultraresistant Burkholderia pseudomallei strain. Biochim Biophys Acta. 2011;1808:1552–1559. doi: 10.1016/j.bbamem.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Trepanier S, Prince A, Huletzky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997;41:2399–2405. doi: 10.1128/aac.41.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribuddharat C, Moore RA, Baker P, Woods DE. Burkholderia pseudomallei class a beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob Agents Chemother. 2003;47:2082–2087. doi: 10.1128/AAC.47.7.2082-2087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunck LA, Propst KL, Wuthiekanun V, Tuanyok A, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Peacock SJ, Keim P, Dow SW, Schweizer HP. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis. 2009;3:e0000519. doi: 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SP, Tsai WC, Liang CY, Lin YS, Huang JW, Chang CY, Tyan YC, Lu PL. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: an emphasis on efflux pump activity. PLoS One. 2014;9:e104986. doi: 10.1371/journal.pone.0104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. The beta-lactamase gene regulator AmpR is a tetramer that recognizes and binds the D-Ala-D-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem. 2015;290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zandt KE, Greer MT, Gelhaus HC. Glanders: an overview of infection in humans. Orphanet J Rare Dis. 2013;8:131. doi: 10.1186/1750-1172-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Dawyndt P. Classification and identification of the Burkholderia cepacia complex: Past, present and future. Syst Appl Microbiol. 2011;34:87–95. doi: 10.1016/j.syapm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- Vasil ML, Tomaras AP, Pritchard AE. Identification and evaluation of twin-arginine translocase inhibitors. Antimicrob Agents Chemother. 2012;56:6223–6234. doi: 10.1128/AAC.01575-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorov DV, Zakharova IB, Podshivalova MV, Kalinkina EV, Merinova OA, Ageeva NP, Antonov VA, Merinova LK, Alekseev VV. High-level resistance to fluoroquinolones and cephalosporins in Burkholderia pseudomallei and closely related species. Trans Roy Soc Trop Med Hyg. 2008;102(Suppl 1):S103–110. doi: 10.1016/S0035-9203(08)70025-7. [DOI] [PubMed] [Google Scholar]
- Walsh C, Wencewicsz T. Challenges, Mechanisms, Opportunities. ASM Press; Washington, DC: 2016. Antibiotics. [Google Scholar]
- Whitlock GC, Estes DM, Torres AG. Glanders: off to the races with Burkholderia mallei. FEMS Microbiol Lett. 2007;277:115–122. doi: 10.1111/j.1574-6968.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. New Engl J Med. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthiekanun V, Peacock SJ. Management of melioidosis. Exp Rev Anti Infect Ther. 2006;4:445–455. doi: 10.1586/14787210.4.3.445. [DOI] [PubMed] [Google Scholar]
- Yi H, Cho KH, Cho YS, Kim K, Nierman WC, Kim HS. Twelve positions in a beta-lactamase that can expand its substrate spectrum with a single amino acid substitution. PLoS One. 2012a;7:e37585. doi: 10.1371/journal.pone.0037585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Kim K, Cho KH, Jung O, Kim HS. Substrate spectrum extension of PenA in Burkholderia thailandensis with a single amino acid deletion, Glu168del. Antimicrob Agents Chemother. 2012b;56:4005–4008. doi: 10.1128/AAC.00598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chen Y, Tabibi S, Alba L, Garber E, Saiman L. Antimicrobial susceptibility and synergy studies of Burkholderia cepacia complex isolated from patients with cystic fibrosis. Antimicrob Agents Chemother. 2007;51:1085–1088. doi: 10.1128/AAC.00954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


