SUMMARY
Mutations in the isocitrate dehydrogenase-1 gene (IDH1) are common drivers of acute myeloid leukemia (AML) but their mechanism is not fully understood. It is thought that IDH1 mutants act by inhibiting TET2 to alter DNA methylation, but there are significant unexplained clinical differences between IDH1- and TET2-mutant diseases. We have discovered that mice expressing endogenous mutant IDH1 have reduced numbers of hematopoietic stem cells (HSC), in contrast to Tet2 knockout (TET2-KO) mice. Mutant IDH1 downregulates the DNA damage (DD) sensor ATM by altering histone methylation, leading to impaired DNA repair, increased sensitivity to DD, and reduced HSC self-renewal, independent of TET2. ATM expression is also decreased in human IDH1-mutated AML. These findings may have implications for treatment of IDH-mutant leukemia.
INTRODUCTION
IDH1 is commonly mutated at the codon R132 in human myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). Mutation in isocitrate dehydrogenase-1 (IDH1-R132) confers an enzymatic activity that converts α-ketoglutarate (αKG) to the oncometabolite D-2-hydroxyglutarate (D2HG) (Cairns and Mak, 2013). D2HG is a competitive inhibitor of aKG–dependent dioxygenase reactions, which are catalyzed by more than 80 human enzymes. Many of these aKG–dependent enzymes have been implicated in aspects of HSC biology and cancer: the TET family proteins affect DNA methylation; the JumonjiC domain-containing (JmjC) histone demethylases alter histone methylation; and the prolyl hydroxylases and lysyl hydroxylases are required for collagen folding and regulation of hypoxia-inducible factor (HIF) protein stability (Cairns and Mak, 2013). Thus, it is thought that mutant IDH alters the epigenetic state of cells, perturbs collagen maturation, and affects oxygen homeostasis, although these effects may be more prominent in some tissues and cell types than in others (Sasaki et al., 2012a; Sasaki et al., 2012b). In particular, the epigenetic effects of IDH on DNA methylation and on both repressive and activating methylation marks on Histone H3 have been shown to affect cellular differentiation (Figueroa et al., 2010; Lu et al., 2012). Although IDH1 has been established as an oncogene in the myeloid lineage, the specific mechanisms underlying its tumorigenic effects are still not well understood.
Mutations in TET2 are also common in MDS and AML and cause a characteristic hypermethylated DNA phenotype (Cancer Genome Atlas Research, 2013; Xie et al., 2014). IDH1 and TET2 mutations are almost always mutually exclusive in these diseases, which has led to the widely accepted proposition that IDH mutation may act primarily via inhibition of TET2 that results in altered DNA methylation and blocked differentiation (Cairns and Mak, 2013; Figueroa et al., 2010). However, there are differences in the clinical features of IDH1- and TET2-mutant hematopoietic disease (Busque et al., 2012; Patel et al., 2012; Shih et al., 2012; Xie et al., 2014), suggesting that the oncogenic mechanisms of mutant IDH and mutant TET2 are not equivalent, although no experimental evidence supporting this hypothesis has been reported.
The hematopoietic system is organized hierarchically, beginning with HSC that give rise to a series of progenitors and eventually, mature terminally differentiated cells. HSC is the only cell type within the hematopoietic system that self-renew throughout life, whereas progenitor cells are more short-lived and differentiate in order to produce mature cells. It is assumed that HSC possess cyto- and geno-protective mechanisms to secure genomic integrity to maintain functional potential. This concept has been demonstrated by multiple studies showing that genetically engineered mice lacking components of DNA damage response (DDR) and repair pathways have impaired HSC function. Unrepaired DNA damage in the form of γH2AX foci also accumulates in the HSC of aged normal mice (Rossi et al., 2007), and this accumulation is more severe in DDR-deficient animals (Nijnik et al., 2007; Nitta et al., 2011). In humans, defective DNA repair is associated with premalignant and malignant hematological conditions such as MDS and AML (Jan et al., 2012; Welch et al., 2012), in keeping with the concept that genomic instability is an important hallmark of cancer (Hanahan and Weinberg, 2011).
The DDR involves a network of proteins that sense and respond appropriately to DD, and perturbation of this network can promote tumorigenesis. A potentially lethal form of DD is the double strand break (DSB). Upon DSB formation, the MRN complex (Mre11/Rad50/Nbs1) recognizes the lesion and activates the sensor kinase encoded by the ataxia telangiectasia mutated gene (ATM). ATM in turn recruits the DNA repair machinery and activates downstream signaling pathways (Stracker et al., 2013). DSB are repaired either by the homology-directed repair (HR) pathway or the more error-prone non-homologous end-joining (NHEJ) pathway. ATM activates the cell cycle checkpoint or induces apoptosis by phosphorylating itself and Chek2, an effector kinase that phosphorylates and activates the tumor suppressor p53. ATM is known to play an important role in HSC because Atm-deficient murine HSC show self-renewal defects, decreased survival, enhanced accumulation of γH2AX foci with age, and increased sensitivity to irradiation (IR)-induced cell death (Ito et al., 2004; Ito et al., 2007; Mohrin et al., 2010; Nitta et al., 2011). In humans, ATM is mutated in several cancer types, including hematological malignancies. Collectively, these studies show that a lack of functional ATM in hematopoietic tissues impairs HSC homeostasis and function, and increases the risk of lymphoma and leukemia (Chen et al., 2014; Takagi et al., 2013).
We previously showed, using a myeloid lineage-specific conditional IDH1-R132-KI (LysM-IDH1-KI) mouse model, that this mutation increases the level of D2HG; affects epigenetics by altering both global DNA methylation and histone methylation; partially blocks differentiation at the HSC/progenitor stage; and produces a hematopoietic phenotype reminiscent of human MDS (Sasaki et al., 2012b). However, the critical molecular mechanisms responsible for these phenotypes have not been fully characterized, and the effects of IDH mutation on DDR signaling have not been investigated. Furthermore, it has recently been found that IDH mutations can occur early during progression to AML and can be present in a population of pre-leukemic stem cells in some patients (Corces-Zimmerman et al., 2014; Shlush et al., 2014), further highlighting the importance of understanding the effects of IDH1 mutations in HSC.
RESULTS
Impaired DDR signaling in HSC and progenitor cells expressing mutant IDH1
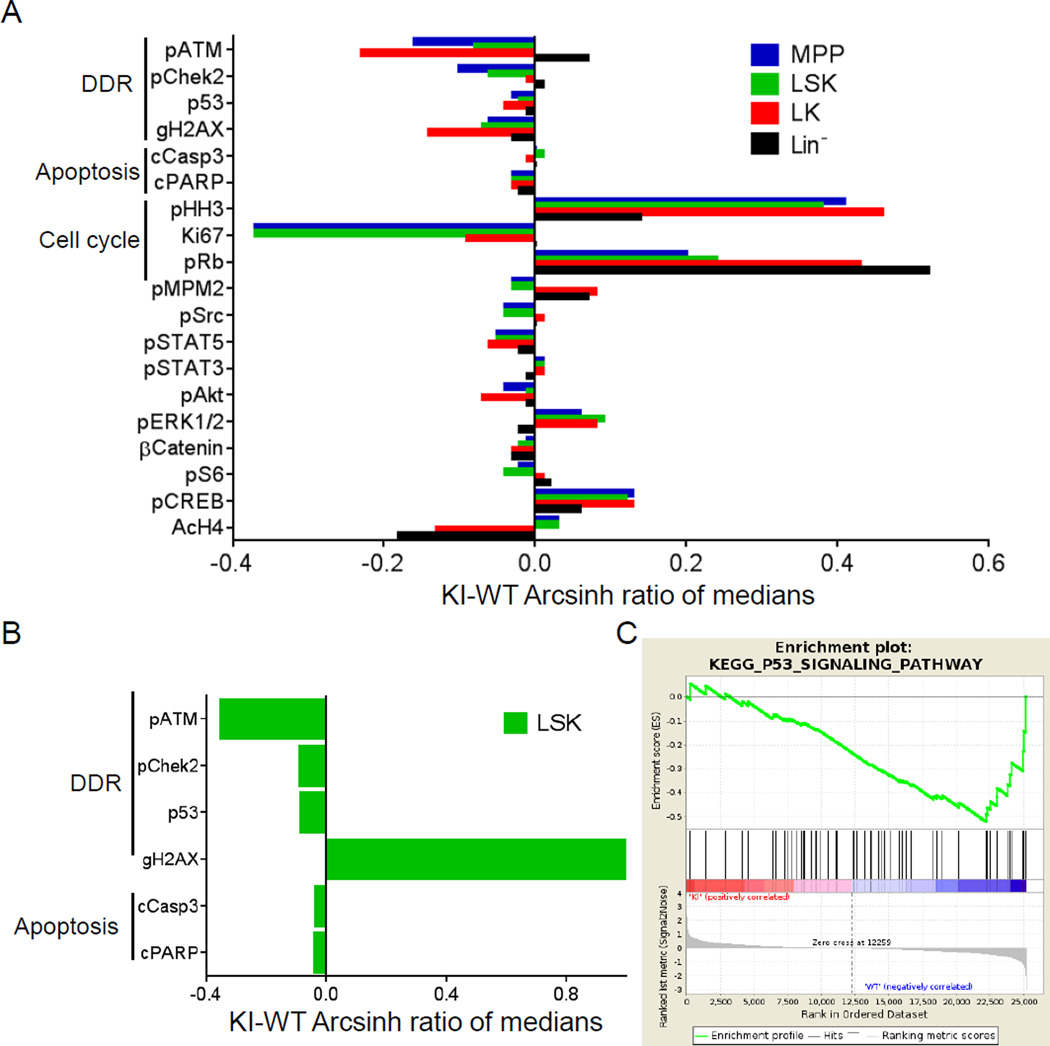
To characterize the effects of mutant IDH1 on the hematopoietic system, we used CyTOF mass cytometry of mouse bone marrow (BM), which enables simultaneous measurement of the abundance and phosphorylation state of multiple proteins in single cells. Undifferentiated hematopoietic cells from young (3–4 months) LysM-IDH1-KI mice exhibited lower levels of phospho-ATM, γH2AX and phospho-Chek2 in populations of i) LK cells, which contain granulocyte macrophage progenitors (GMP), common myeloid progenitors (CMP), and megakaryocyte erythrocyte progenitors (MEP), ii) LSK cells, which contain long-term hematopoietic stem cells (LT-HSC), short-term hematopoietic stem cells (ST-HSC), and multipotent progenitors (MPP), and iii) MPP cells, compared to wild-type cells (Figure 1A). LSK cells of aged (7–10 months) LysM-IDH1-KI mice displayed decreased phospho-ATM and phospho-Chek2 but increased γH2AX (Figure 1B), suggesting a defect in DDR signaling downstream of ATM and the accumulation of unrepaired DNA damage with age. Microarray analysis confirmed that p53 signaling components downstream of ATM-mediated DDR signaling (Stracker et al., 2013) were decreased in IDH1-mutant LSK cells (Figure 1C). Thus, DDR signaling downstream of ATM appears to be compromised in HSC and progenitors expressing mutant IDH1.
Figure 1. Decreased phosphorylation of ATM/Chek2/γH2AX in LysM-IDH1-KI hematopoietic cells.
(A, B) Representative CyTOF analyses of BM cells from young (A) and aged (B) LysM-IDH1-KI (KI) and WT mouse. The indicated cell populations were immunostained with metal-conjugated Abs to detect the indicated signaling molecules. Histograms represent expression levels in the KI cell relative to the WT. (C) Gene set enrichment analysis of the KEGG p53 pathway DDR gene set in LSK cells of LysM-IDH1-KI and WT mice (n=4/group). Top: x-axis shows genes ordered from high expression in LysM-IDH1-KI samples (left end) to high expression in WT samples (right end). The y-axis shows the running enrichment score (ES) along the ranked gene list. The negative enrichment score near the WT end indicates downregulation of the pathway in the LysM-IDH1-KI samples. Middle: black lines indicate expression of the P53 pathway genes in the LysM-IDH1-KI samples relative to the ranked gene list. Bottom: metric for ranking genes based on the LysM-IDH1-KI or WT phenotype. ES: −0.52. Normalized ES: −1.63. Nominal p value: 0.0032. False Discovery Rate (FDR) q value: 0.088.
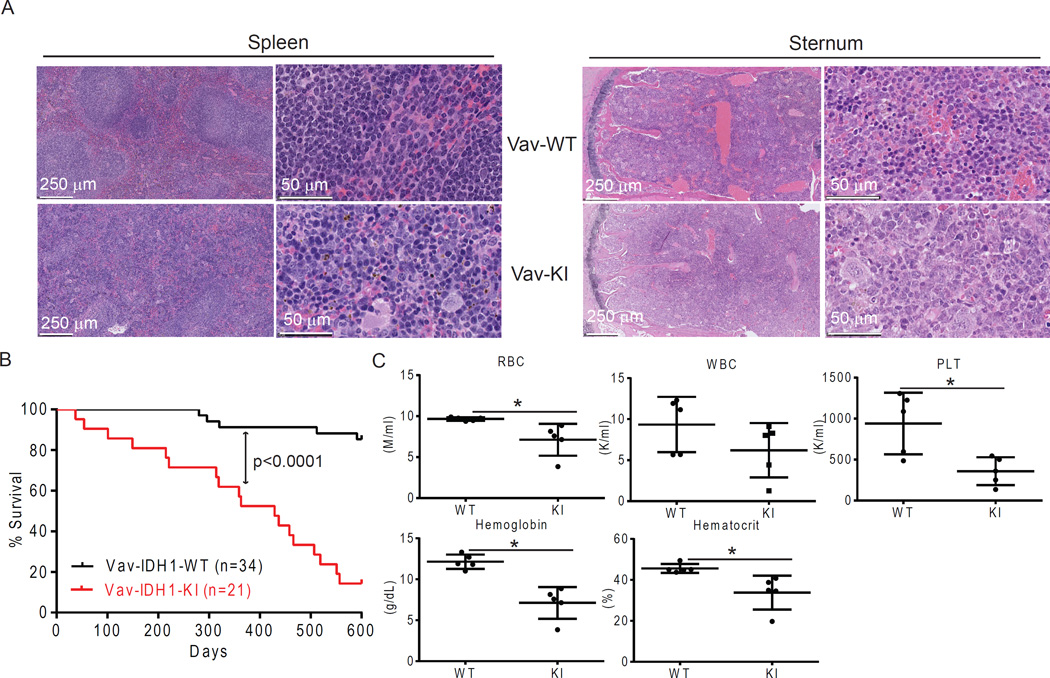
To explore this observation in all hematopoietic cell types, we first characterized our Vav-IDH1-KI mice (Figure S1A). Vav-IDH1-KI mice were born and developed normally despite producing high urinary levels of D2HG (Figure S1B). Aged Vav-IDH1-KI mice showed an increase in immature cKit+ hematopoietic cells in BM and spleen (Figure S1C), and Vav-IDH1-KI BM showed histologic evidence of myeloid hyperplasia and erythroid hypoplasia (Figure 2A). The architecture of Vav-IDH1-KI spleen was severely disrupted, with evidence of hematopoietic cell hyperplasia in the red pulp (Figure 2A). This phenotype is similar to that of LysM-IDH1-KI mice, which is reminiscent of human MDS (Sasaki et al., 2012b). Notably, in contrast to LysM-IDH1-KI mice (Sasaki et al., 2012b), Vav-IDH1-KI mice had a significantly shorter lifespan than littermate controls (Figure 2B). About 50% of Vav-IDH1-KI mice died by age 12 months, showing splenomegaly, anemia and thrombocytopenia (Figures 2B–C, S1D).
Figure 2. Vav-IDH1-KI mice die prematurely with splenomegaly and anemia.
(A) Representative H&E-stained sections of spleen and sternum from aged Vav-IDH1-KI (Vav-KI) and control littermate (Vav-WT) mice. (B) Kaplan-Meier plot of Vav-IDH1-KI and littermate Vav-IDH1-WT mice. p value, log-rank test. (C) Complete blood count (CBC) parameters in peripheral blood from aged Vav-IDH1-KI (KI) and WT mice (n=5). WBC, white blood cells; RBC, red blood cells; PLT, platelet count. Data for individual mice are shown as well as the group mean ± SD. *p<0.05, unpaired Student’s t-test. See also Figure S1.
Mutant IDH1 triggers an unexpected TET2-independent reduction in LT-HSC
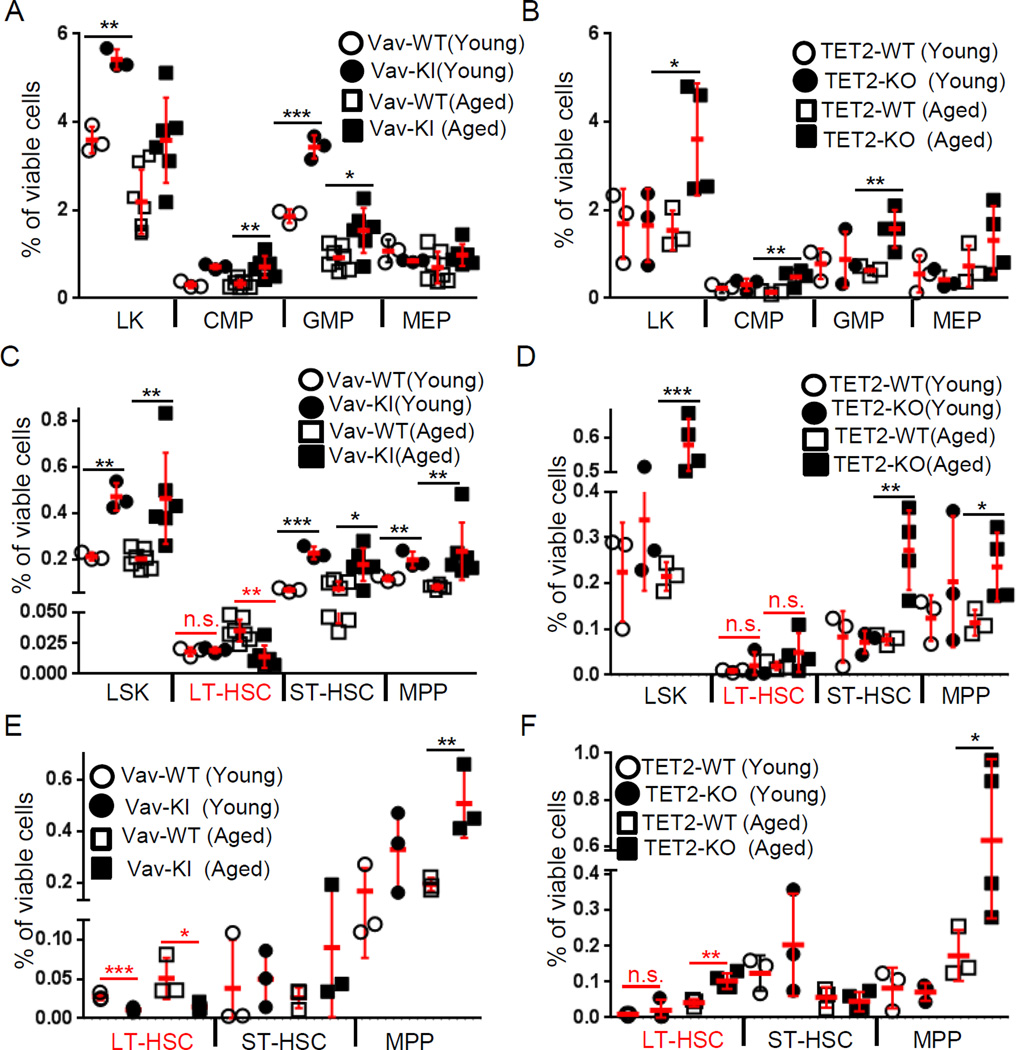
As IDH mutations are thought to affect hematopoietic differentiation via inhibition of TET2 by D2HG, we measured BM cell populations in Vav-IDH1-KI and Tet2 knockout (TET2-KO) mice. As expected, both the LSK and LK populations were increased in Vav-IDH1-KI and TET2-KO mice (Figures 3A–D and S2). Both Vav-IDH1-KI and TET2-KO mice showed significant increases in ST-HSC/MPP and GMP/CMP but not MEP (Figures 3 and S2). Surprisingly, Vav-IDH1-KI mice displayed a dramatic reduction in LT-HSC (Figures 3C, 3E, S2C, S2E) that progressed with age, similar to that reported for Atm knockout (ATM-KO) mice (Chen et al., 2014; Ito et al., 2004). This phenotype stands in contrast to TET2-KO mice, which showed normal or slightly increased LT-HSC numbers (Figures 3D, 3F, S2D, S2F). Thus, mutant IDH1 not only blocks differentiation at the ST-HSC/MPP stage (increased progenitor cells) and induces myeloid skewing (elevated GMP/CMP relative to MEP), like loss of TET2 (Moran-Crusio et al., 2011; Quivoron et al., 2011), but also triggers an unexpected and striking TET2-independent reduction in LT-HSC, reminiscent of ATM loss (Chen et al., 2014; Ito et al., 2004).
Figure 3. IDH1-R132 mutation reduces LT-HSC.
(A–F) Quantitation of flow cytometric analyses of (LK cells, CMP (FcγR−CD34+), GMP (FcγR+CD34+) and MEP (FcγR−CD34−) (A, B); LSK, LT-HSC (CD34−Flt3−), ST-HSC (CD34+Flt3−) and MPP (CD34+Flt3+) (C, D); and LT-HSC (CD150+CD48−), ST-HSC (CD150+CD48+) and MPP (CD150−CD48+) (E, F) among viable Lin− BM cells from young and aged Vav-IDH1-KI (Vav-KI) and control (Vav-WT) mice (n=3–7) (A, C, E) and young and aged TET2-KO and control (TET2-WT) mice (n=3–4) (B, D, F). Data points are percentage of viable cells in individual mice. Red lines, group means ± SD. * p<0.05; **p<0.01; ***p<0.001 by unpaired Student’s t-test. n.s., not significant. See also Figure S2.
ATM downregulation is induced by mutant IDH1-R132
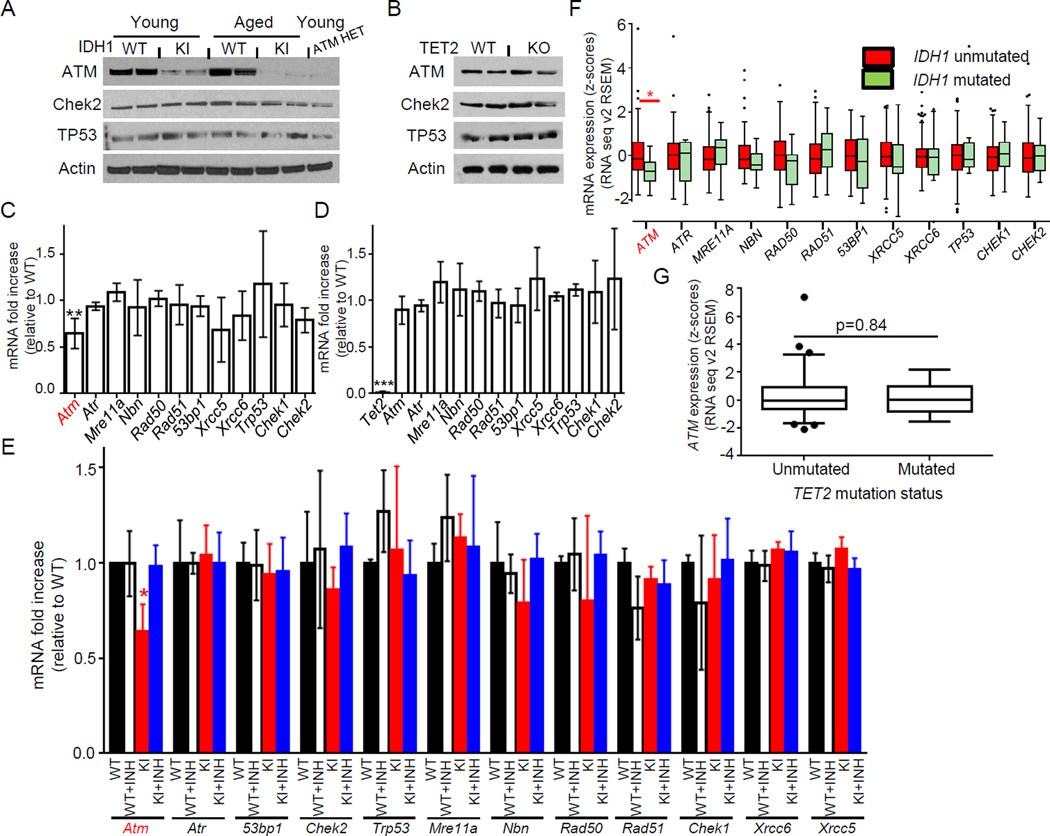
To investigate whether mutant IDH1 affects DDR gene expression in hematopoietic cells, we measured mRNA and protein levels of DDR signaling molecules. Consistent with our CyTOF analysis, a marked decrease in ATM protein was observed in lineage negative (Lin−) cells of Vav-IDH1-KI mice (Figures 4A–B). qPCR analysis of Vav-IDH1-KI LSK cells revealed significant downregulation of ATM mRNA (but not mRNAs of other DDR genes) in IDH1 mutant cells (Figure 4C). ATM protein level was decreased to a significantly greater degree than the ATM mRNA in Vav-IDH1-KI cells (Figures 4A, 4C), consistent with previous observation in Atm heterozygote (ATM-HET) mice (Spring et al., 2002; Zhan et al., 2010). We investigated ATM protein levels in lineage-depleted (Lin−) BM cells (HSC/progenitor-enriched population) from our ATM-HET mice and, indeed, observed a striking reduction in ATM protein in Lin− cells that was similar to that in IDH1-KI samples (Figure 4A, lane 9). This reduction occurred despite the expected levels of ATM mRNA in ATM-HET Lin− cells compared to Atm WT controls (51.8±4.4%; n=3, data not shown). Thus, although the mechanism underlying this effect is uncertain, it is clear that ATM mRNA and protein levels are not always directly correlated. No decrease in ATM mRNA or protein was observed in TET2-KO cells (Figures 4B, 4D), indicating that these effects are not mediated by TET2 inhibition. Treatment of IDH1-KI cells with AGI-5198, a competitive inhibitor of mutant IDH1-R132, restored Atm mRNA levels, confirming that Atm repression was due to mutant IDH1 activity (Figure 4E).
Figure 4. TET2-independent ATM downregulation by mutant IDH1.
(A, B) Immunoblot detecting the indicated proteins in Lin− cells from young and aged Vav-IDH1-KI (KI) and WT mice, plus young ATM-HET mice (A) and aged TET2-KO and TET2-WT mice (B). (C, D) qRT-PCR determination of the indicated mRNAs in LSK cells from young Vav-IDH1-KI (C) and TET2-KO (D) mice relative to controls. Data are the mean ± SD (n=3). (E) qRT-PCR determination of the indicated mRNAs from LSK cells isolated from Vav-IDH1-KI and WT mice and cultured with (+INH) or without mutant IDH1 inhibitor (n=3/group). Data are the mean ± SD. For (C–E), *p<0.05; **p<0.01; ***p<0.001 by unpaired Student’s t-test. (F) Z-score plot of mRNA levels of the indicated genes in human IDH1-unmutated (n=127) and IDH1-mutated (n=13) AML samples from TCGA. RNASeq version 2 values were quantified using the expectation-maximization method (RSEM) and converted to z-scores. IDH2- and TET2-mutated samples were excluded. *p<0.05 by Wilcoxon rank-sum test adjusted with the Benjamini and Hochberg method. (G) Z-score plot, generated and analyzed as in (F), of ATM mRNA levels in TET2-unmutated (n=128) and TET2-mutated (n=13) human AML samples from TCGA. IDH1- and IDH2-mutated samples were excluded. The boxplots are drawn using the method of Tukey (F, G). Briefly, central horizontal lines on the boxplots indicate the median, the box edges indicate the 25th and 75th percentiles, and the whiskers indicate the largest value above or below these percentiles within 1.5 times the interquartile range. Data beyond the whiskers are plotted as individual points.
To relate our findings to human AML, we queried the TCGA database (Cancer Genome Atlas Research, 2013) and found that expression levels of ATM, but not those of 11 other DDR genes, were significantly decreased in IDH1-mutated AML but not in TET2-mutated AML (Figures 4F–G). Thus, the ATM downregulation induced by mutant IDH1-R132 in mouse models is also observed in human IDH1-R132 mutated AML.
Histone methylation-dependent and TET2-independent ATM downregulation
To determine how IDH1-R132 mutant downregulates ATM, we first examined the levels of methylation and hydroxymethylation of DNA in mutant IDH1 cells. As expected, we observed a dramatic decrease in global 5-hydroxymethylcytosine (5hmC), but not 5-methylcytosine (5mC), in Lin− BM cells from both Vav-IDH1-KI and TET2-KO mice (Figure S3A). The magnitude of 5hmC reduction was similar in Vav-IDH1-KI and TET2-KO cells, suggesting that TET2 is the predominant regulator of 5-hydroxymethylation in this cell type. We next examined DNA methylation in Vav-IDH1-KI LT-HSC, ST-HSC and MPP using enhanced reduced representation bisulfite sequencing (eRRBS) (Figure S3B–E). Consistent with our previous analysis of LysM-IDH1-KI mice (Sasaki et al., 2012b), there was an increase in DNA methylation at the majority of CpG sites with altered methylation in Vav-IDH1-KI LT-HSC, ST-HSC and MPP compared to controls (Figure S3C), confirming that mutant IDH1 increases DNA methylation at these sites. However, no significant alterations to DNA methylation of the Atm promoter were detected in these Vav-IDH1-KI cell populations (Figure S3D–E), suggesting that a mechanism independent of both TET2 and DNA methylation is responsible for the decrease in Atm expression. As eRRBS analysis does not distinguish between 5mC and 5hmC, we used qPCR plus hMeDIP (hydroxymethylated DNA immunoprecipitation) (Taiwo et al., 2012) to specifically examine 5hmC levels in the promoters of a set of DDR genes, including Atm, in Vav-IDH1-KI, TET2-KO and control cells. No significant differences in 5hmC levels at the Atm promoter were observed (Figure S3F). Thus, the downregulation of Atm by mutant IDH1 does not depend on inhibition of TET2 activity or on changes in DNA methylation or hydroxymethylation.
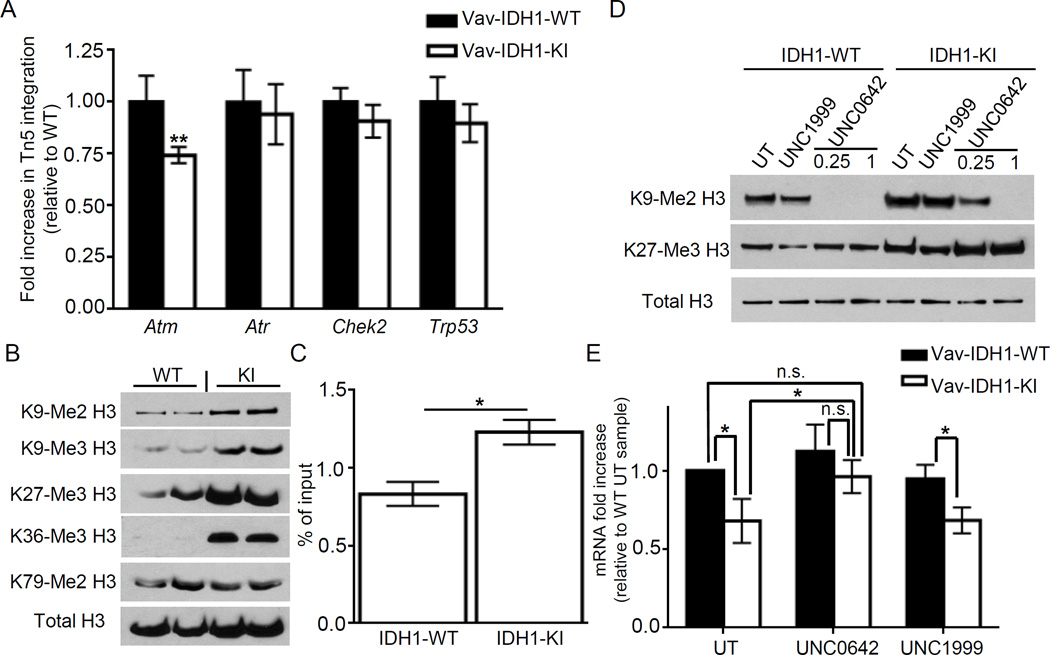
To examine the chromatin structure of the Atm promoter region, we subjected Vav-IDH1-KI and control cells to ATAC (Assay for Transposase-Accessible Chromatin)-qPCR (Buenrostro et al., 2013). We detected a ~30% reduction in open chromatin measured based on the Tn5 transposase integration in the Atm promoter region, but not at the promoters of other DDR genes, in Vav-IDH1-KI cells (Figure 5A), indicating a more closed chromatin structure in this region.
Figure 5. Partially closed chromatin structure and increased H3K methylation in the ATM promoter in Vav-IDH1-KI cells.
(A) ATAC-qPCR to analyze the indicated genes in LSK cells from Vav-IDH1-KI and WT mice (n=3). Data are the mean ± SD. (B) Immunoblot to detect the indicated methylated forms of histone H3 in Lin− cells from Vav-IDH1-KI and WT mice. (C) ChIP-qPCR analysis of the Atm promoter in LSK cells from Vav-IDH1-KI and WT mice (n=3) using anti-histone H3-K9me3 Ab. Data are the mean ± SD. (D) Immunoblot to detect the indicated proteins in LSK cells isolated from Vav-IDH1-KI and WT mice and cultured with/without UNC1999 (1 µM) or UNC0642(0.25 or 1 µM). UT, untreated. (E) qRT-PCR determination of Atm mRNA in the mice in (D) (1 µM UNC0642). Data are the mean ± SD (n=4). For A,C,E, *p<0.05, **p<0.01 by unpaired Student’s t-test. See also Figure S3.
We next investigated the importance of histone modifications known to alter chromatin structure. To ascertain whether histone methylation is increased by mutant IDH1, we performed immunoblotting in Lin− cells from Vav-IDH1-KI and controls. There was a significant global increase in some repressive histone marks in Vav-IDH1-KI Lin− cells, including lysine 9 and 27 of Histone H3 (H3K9 and H3K27) relative to controls (Figure 5B). In addition, chromatin immunoprecipitation (ChIP)-qPCR revealed increases in the repressive tri-methylated H3-K9 (H3K9Me3) mark at the Atm promoter in Vav-IDH1-KI LSK cells (Figure 5C), raising the possibility that this alteration causes a more closed chromatin structure, which downregulates Atm. To further investigate the function of H3-K9 methylation at the Atm promoter, freshly isolated LSK cells from Vav-IDH1-KI and controls were exposed to UNC0642 (an inhibitor of G9A, a methyltransferase of H3-K9) or UNC1999 (an inhibitor of EZH2, a methyltransferase of H3-K27). As expected, the treatment of cultured LSK cells with UNC0642 and UNC1999 cells caused a decrease in methylation of H3-K9 and H3-K27, respectively (Figure 5D). Treatment with UNC0642, but not UNC1999, largely restored ATM mRNA/protein levels (Figure 5E, S3G), suggesting that increased H3-K9 methylation contribute to downregulation of Atm by mutant IDH1. This is consistent with previous data demonstrating that D2HG can directly inhibit the class of histone lysine demethylases (KDM4) responsible for maintaining appropriate levels of methylated H3-K9 (Lu et al., 2012).
Mutant IDH1 LT-HSC show impaired DD repair, accumulation of DD with age, and impaired self-renewal
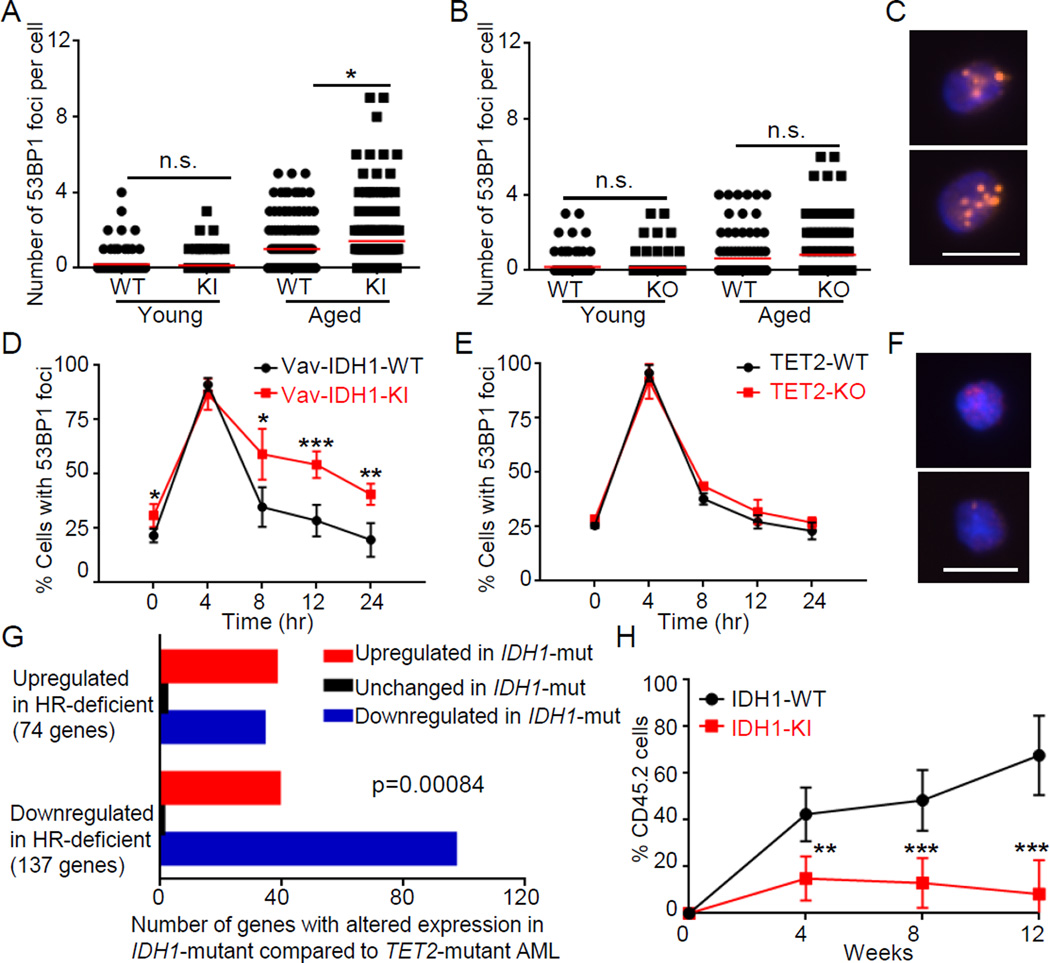
Whole exome-sequencing has revealed that human HSC accumulate DD with age and are the cell of origin of some hematopoietic malignancies, including AML (Jan et al., 2012). To determine whether the mutant IDH1 dependent reduction in ATM impairs DD repair, we measured 53BP1 and γH2AX foci in Vav-IDH1-KI LT-HSC and LSK cells. Like ATM-KO mice (Nitta et al., 2011) but in contrast to aged TET2-KO mice, cells from aged (but not young) Vav-IDH1-KI mice showed increased number of DD foci compared to controls (Figures 6A–C, S4A–C). It has been shown that in LT-HSC, DD is not repaired efficiently due to cell cycle-dependent downregulation of DDR genes (Beerman et al., 2014) and the predominance of the error-prone NHEJ mechanism of DNA repair (Mohrin et al., 2010). Downregulation of ATM could thus leave DD unrepaired, which may induce premature HSC differentiation (Santos et al., 2014) leading over time to the low number of LT-HSC in aged Vav-IDH1-KI mice and contributing to higher numbers of later progenitors (Figures 3C, 3E). To compare the DNA repair capacities of IDH1-KI and TET2-KO LT-HSC, we examined the kinetics of DD foci disappearance in LT-HSC exposed to 0.5 Gy IR. Disappearance of 53BP1 and γH2AX foci was delayed and decreased in IDH1-KI LT-HSC compared to IDH1-WT or TET2-KO LT-HSC (Figures 6D–F, S4D–E). This impaired DNA repair phenotype may also be present in human IDH1-mutated AML since gene downregulated in IDH1-mutated AML compared to TET2-mutated AML are associated with downregulation in HR-deficient cells compared to HR-normal cells (Figure 6G) (Peng et al., 2014).
Figure 6. Vav-IDH1-KI LT-HSC show elevated accumulation of age-related DD, impaired DNA repair and repopulation capacities.
(A, B) Quantitation of 53BP1 DD foci in LT-HSC (~100 cells from 3 mice/group) isolated from Vav-IDH1-KI and Vav-IDH1-WT (A), and TET2-KO and TET2-WT mice (B) (~100 cells from 3 mice/group). Data are values for individual LT-HSC. Red lines, group means. (C) Representative fluorescent images of 53BP1 DD foci in LT-HSC isolated from aged Vav-IDH1-KI mice. Scale bar, 5 µm. (D–E) Kinetics of disappearance of 53BP1 foci over the indicated period in LT-HSC (n=3/group) that were isolated from aged Vav-IDH1-KI and Vav-IDH1-WT (D) or TET2-KO and TET2-WT (E) mice (n=3/group) and exposed in vitro to IR (0.5 Gy). Data are the mean ± SD. (F) Representative images of 53BP1 foci in LT-HSC isolated from aged Vav-IDH1-KI at 24 hr post-IR. Scale bar, 5 µm. (G) Number of genes by direction of expression change in IDH1-mutated AML (n=5) relative to TET2-mutated AML (n=7) that were present in a gene expression signature derived from HR-deficient cells (Peng et al., 2014). The direction of change in IDH1-mutated samples correlated positively with the direction of change in HR-deficient cells. IDH2 mutants, DNMT3A mutants, and IDH1;TET2 double mutants were excluded. p=0.00084, Fisher’s exact test. (H) Quantitation of a competitive repopulation assay in which CD45.2+ cells from Vav-IDH1-KI or Vav-IDH1-WT mice were transferred to irradiated CD45.1+ recipients. Data are the mean ± SD (n=6/group. For A,B,D,H, *p<0.05, **p<0.01, ***p<0.005 by unpaired Student’s t test. n.s., not significant. See also Figure S4.
To examine the impact of decreased DNA repair capacity on HSC function, we performed LT-HSC competitive repopulation assays. In keeping with the phenotype of other DD repair-deficient HSC, including those lacking ATM (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007), Vav-IDH1-KI LT-HSC showed reduced in vivo reconstitution potential compared to IDH-WT LT-HSC (Figure 6H), a deficit not observed in comparable assays of TET2-KO (Moran-Crusio et al., 2011). To investigate if this deficit was due to H3-K9-mediated downregulation of ATM triggered by mutant IDH1, we quantified DD foci in LT-HSC freshly isolated from aged Vav-IDH1-KI mice and treated with UNC0642 or UNC1999. UNC0642, but not UNC1999, largely blocked the increase in DD foci in Vav-IDH1-KI LT-HSC cultures (Figure S4F, 0 hr). As expected, 0.5 Gy IR caused a rapid increase in DD foci in both mutant and control LT-HSC cultures, with or without these inhibitors (Figure S4F, 4 hr). By 24 hr post-IR, aged Vav-IDH1-KI LT-HSC showed impaired disappearance of DD foci compared to controls, a defect that was largely rescued by UNC0642 but not UNC1999 (Figure S4F, 24 hr), suggesting that ATM downregulation mediated by H3-K9 methylation is an important factor in the DDR deficit of IDH1-mutant hematopoietic cells.
Enhanced DD-sensitivity in mutant IDH1 expressing LT-HSC
We next explored the effect of mutant IDH1 on key molecules in the DDR signaling pathway induced by IR. In control mouse embryonic fibroblast (MEF) exposed to IR (5 Gy), we observed the expected time-dependent increase in the phosphorylation of ATM, Chek2 and p53, followed by p21 upregulation. In irradiated IDH1-KI cells, these events were abrogated (Figure S5A), suggesting that DDR signaling pathway is impaired by mutant IDH1.
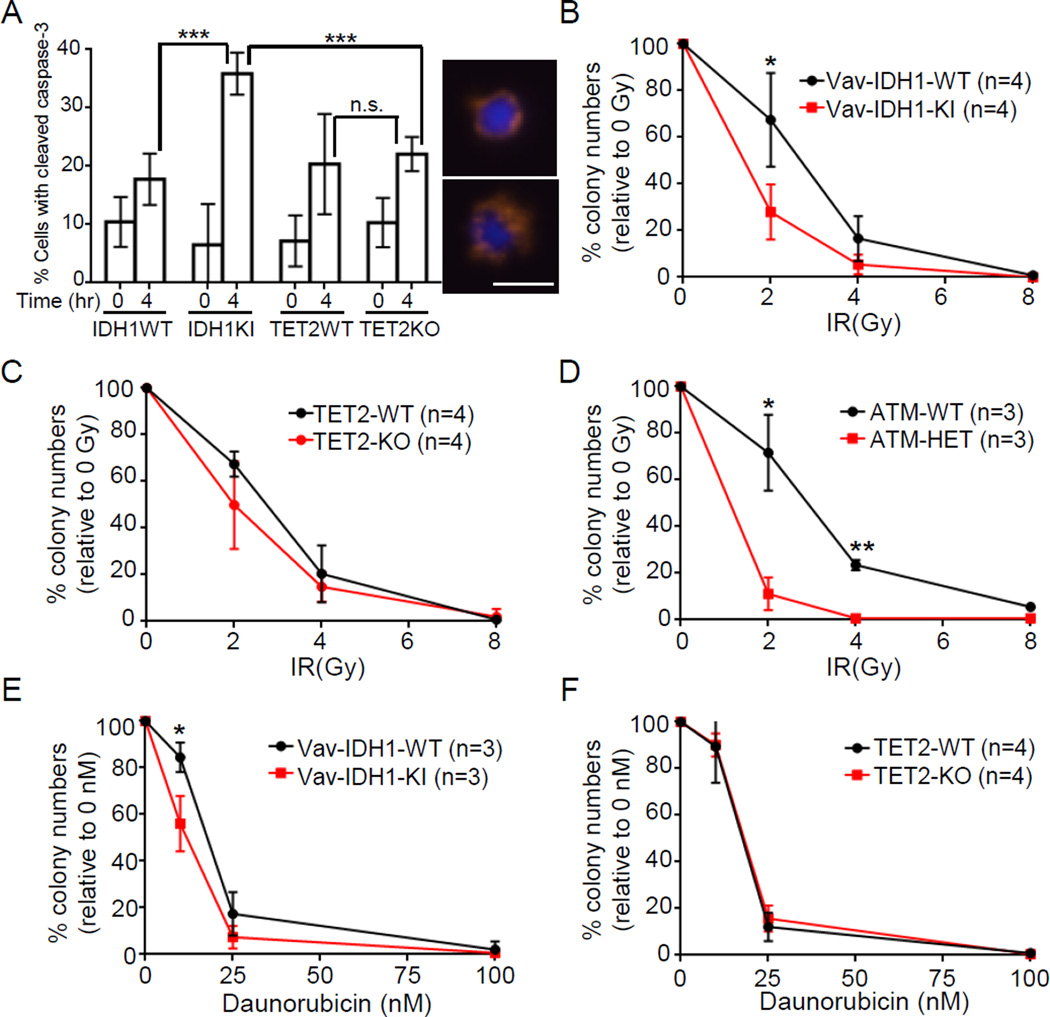
We also subjected Vav-IDH1-KI and TET2-KO LT-HSC to assays of caspase-3 cleavage and ex vivo clonogenic survival. Caspase-3 cleavage was increased in Vav-IDH1-KI, but not TET2-KO, LT-HSC, strongly suggesting that mutant IDH1 expression enhances IR-induced apoptosis (Figure 7A). To confirm enhanced apoptosis after IR results in increase DD-sensitivity, LT-HSC were exposed to either IR (Figures 7B–D) or daunorubicin (Figures 7E–F), a DNA-damaging compound commonly used to treat AML. Both IR and daunorubicin exposure significantly reduced colony numbers in Vav-IDH1-KI, but not TET2-KO, LT-HSC cultures (Figures 7B–C, 7E–F). This increased sensitivity of IDH1-mutant LT-HSC to IR parallels that of ATM-HET LT-HSC, which were included as a positive control (Figure 7D).
Figure 7. Enhanced IR and daunorubicin sensitivity in Vav-IDH1-KI LT-HSC.
(A) Left: Quantitation of percentages of LT-HSC that were isolated from aged Vav-IDH1-KI, Vav-IDH1-WT, TET2-KO or TET2-WT mice (n=3/group) and were positive for cleaved caspase-3 at 4 hr post-IR (1 Gy). Data are the mean ± SD. Right: Two representative images of cleaved caspase-3 in the irradiated Vav-IDH1-KI LT-HSC in the left panel. Scale bar, 5 µm. (B–F) Quantitation of colonies remaining 1 week after LT-HSC from aged Vav-IDH1-KI and Vav-IDH1-WT mice (n=3 mice/group) (B, E), TET2-KO and TET2-WT mice (n=4 mice/group) (C, F) and ATM WT and ATM-HET mice (n=3 mice/group) (D), after irradiation with 2, 4 or 8 Gy (B–D), or treated in vitro with the indicated concentrations of daunorubicin (E, F). Data are the mean ± SD and expressed as the percentage of colonies/plate relative to unirradiated or untreated controls. *p<0.05, **p<0.01, ***p<0.005 by unpaired Student’s t test. n.s., not significant. See also Figure S5.
Lack of ATM downregulation and age-associated accumulation of DD in TET1 KO
It was recently shown that loss of TET1 results in DD accumulation in murine pro-B cells (Cimmino et al., 2015). Although not commonly mutated in myeloid malignancies, it is possible that inhibition of TET1 could have contributed to the phenotype observed in the IDH1-KI cells. Therefore, we examined levels of ATM and DD in Tet1 knockout (TET1-KO) cells. Like TET2-KO but in contrast to aged IDH1-KI, there was neither a reduction of Atm mRNA in LSK cells, nor a reduction in ATM protein in Lin− cells, nor an accumulation of DD foci in aged TET1-KO (Figures S5B–E). These observations indicate that the reported mutant IDH1-mediated effects on ATM and subsequent DD do not depend on inhibition of TET2 or TET1.
DISCUSSION
In this study, we show that endogenous mutant IDH1 downregulates ATM, impairs DDR and alters LT-HSC homeostasis, all independent of its effects on TET2. Our findings clarify the effects of IDH mutations during the development of hematologic malignancies, which is important because IDH mutations can occur early during AML progression and persist in a pool of pre-leukemic stem cells (Corces-Zimmerman et al., 2014; Grove and Vassiliou, 2014; Shlush et al., 2014). We find that IDH1 mutations decrease ATM expression by increasing methylation of H3K9, a known repressive histone mark. Previous work has shown that KDM4A/C, which are histone lysine demethylases responsible for regulating H3K9 methylation, are particularly sensitive to D2HG inhibition (Chowdhury et al., 2011). Furthermore, increased H3K9 methylation is known to mediate effects of mutant IDH on adipocyte differentiation, independent of changes in DNA methylation (Lu et al., 2012). Taken together, these data provide evidence that mutant IDH can alter cellular differentiation via changes to the histone code rather than by changes to DNA methylation.
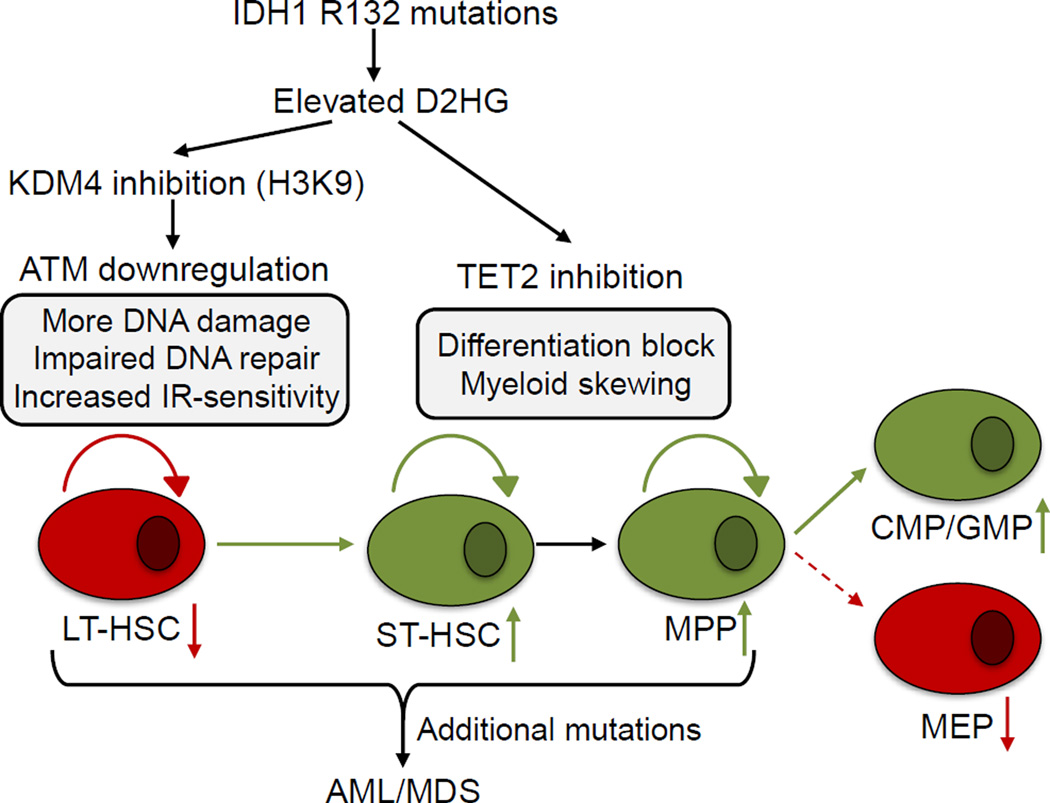
Our data, in combination with previous results, suggest a model in which mutated IDH1 has two distinct oncogenic effects on HSC and progenitors (Figure 8). First, mutant IDH1 induces impairments of DDR signaling and DNA repair that are most prominent in LT-HSC, are dependent on ATM, but are independent of TET2. Second, as suggested by previous clinical and experimental data, mutant IDH1 causes TET2-dependent alterations to DNA methylation that drive ST-HSC and MPP expansion accompanied by skewing towards CMP and GMP production (Ko et al., 2011; Kunimoto et al., 2012; Moran-Crusio et al., 2011; Quivoron et al., 2011). The TET2-independent, DDR-associated effects of mutant IDH1 may be responsible for some of the clinical differences between IDH1-mutated and TET2-mutated hematologic diseases. Several lines of evidence support this concept. First, TET2 (but not IDH1) mutations are observed in some healthy elderly individuals, suggesting that loss of TET2 can be tolerated without causing an overt hematological disorder (Busque et al., 2012; Xie et al., 2014). It is possible that the effects of mutant IDH1 on DDR signaling do not allow the long-term survival and maintenance of IDH1-mutant HSC in healthy individuals. Second, TET2 mutations are observed at higher frequencies in MDS (20–25%) and MDS/MPN (20–58%) than in AML (7–23%) (Shih et al., 2012), whereas IDH1 mutations are observed at higher rates in AML (6–16%) than in MDS (2–4%) and MDS/MPN (~1%) (Im et al., 2014). Therefore, it may be the TET2-independent effects of IDH mutations, particularly their effects on DDR, that increase the probability of progressing to more advanced disease. We believe that these effects of mutant IDH1 on DDR signaling in the crucial LT-HSC population may complement the known epigenetic changes affecting myeloid differentiation during IDH-associated tumorigenesis.
Figure 8. Proposed model of mutant IDH1 oncogenicity.
Mutant IDH1 increases the concentration of intracellular D2HG. In hematopoietic stem cells, D2HG inhibits histone lysine demethylases, leading to decreased expression of ATM and a DDR defect. In more differentiated cells, D2HG inhibition of TET2 leads to alterations in DNA methylation and altered myeloid differentiation. Upon acquisition of additional oncogenic mutations, these effects of mutant IDH1 may contribute to malignant transformation.
Although the ATM downregulation caused by mutant IDH1 is likely responsible for significant effects on LT-HSC homeostasis, our data do not exclude the possibility that other factors downstream of histone methylation changes may also affect the LT-HSC of IDH1-mutant mice (Lu et al., 2012). Tet2;Tet3 double knockout (DKO) hematopoietic cells (An et al., 2015) show decreased 5hmC, increased DD, and impaired DNA repair, raising the possibility that other mechanisms downstream of IDH and TET mutations affect DDR signaling. However, the DDR phenotypes in Tet2;Tet3 DKO mice were observed only in differentiated myeloid cells and not in the HSC-enriched LSK population (An et al., 2015), which is similar to what we observed in our TET2-KO LT-HSC and contrary to what we observed in our Vav-IDH-KI LT-HSC. Thus, a lack of TET2 and TET3 has little impact on DDR in HSC and these TET family members appear to be more important in differentiated cells. Similar context-dependent DDR phenotypes were observed in TET1-KO pro-B cells (Cimmino et al., 2015) that did not occur in TET1-KO LT-HSC. These results indicate that a decrease in 5hmC due to TET deficiency has an effect that is cell type-specific, is observed in differentiated cells, and does not affect DDR in HSC.
Although TET1 and TET3 mutations are extremely rare in AML (in TCGA: TET1, ~1%; TET2, ~9%; TET3, 0%) and co-mutation of TET2 with TET1 or TET3 does not occur in human AML (Cancer Genome Atlas Research, 2013), our results will be important for future investigation of the relationship between TET activity, 5hmC, and DDR in the hematopoietic system. Our dot blotting of HSC-enriched Lin− cells from Vav-IDH1-KI and TET2-KO mice revealed that the degree of 5hmC reduction is comparable in these two mutants, strongly suggesting that TET2 is the predominant TET enzyme influencing 5hmC levels in HSC/progenitors. Moreover, BM transplantation experiments for all published TET single or double deficient mice show that HSCs in these mutants exhibit enhanced repopulating potential in vivo (An et al., 2015; Cimmino et al., 2015; Moran-Crusio et al., 2011; Quivoron et al., 2011; Zhao et al., 2015), in contrast to the decreased potential of Vav-IDH1-KI LT-HSC. As numerous examinations of DDR-deficient mice have shown that an intact DDR system is crucial for maintenance of HSC repopulation/self-renewal (Garaycoechea et al., 2012; Ito et al., 2004; Niedernhofer, 2008; Nijnik et al., 2007; Rossi et al., 2007), impaired HSC repopulation capacity is widely considered a hallmark of DDR deficiency. These data reinforce our hypothesis that the DDR phenotypes observed in Vav-IDH1-KI LT-HSC are not due to loss of 5hmC and are independent of TET2.
The effects of the H3K9-mediated histone alterations regulated by mutant IDH1 are likely multifactorial. Studies of mice lacking G9A H3K9 methyltransferase as well as shRNA knockdown of Jmjd2 H3K9 demethylases in cell line systems, do not suggest a role for H3K9 alterations in HSC function (Cellot et al., 2013; Lehnertz et al., 2014). However, a lack of G9A does alter HoxA9-dependent gene expression, which is known to affect hematopoietic differentiation and leukemogenesis. Although we found that an H3K9 methyltransferase inhibitor rescued the DDR defect of IDH1-mutant LT-HSC using freshly explanted cells, the HSC undergo differentiation during these culture conditions. Therefore, additional in vivo studies will be required to fully understand this subset of phenotypic effects associated with IDH1 mutation.
Impaired DNA repair signaling in IDH1-mutated LT-HSC may increase the probability of both additional oncogenic mutation acquisition and clonal expansion of the IDH1-mutated LT-HSC pool. It is thought that a pathogenic AML clone arises from a heterogeneous pool of genetically diverse pre-leukemic subclones (Corces-Zimmerman et al., 2014; Jan et al., 2012; Shlush et al., 2014). Given that the effects of IDH1 mutation are most prominent in LT-HSCs, and appear to be cell type-specific, examination of genomic diversity in panels of single cells may be required in order to rigorously test whether IDH1-mutant HSCs accumulate increased numbers of additional mutations. Further studies of critical cell populations will be required to provide definitive answers to these questions.
Patients with IDH1-mutated AML appear to have a better prognosis following daunorubicin treatment than do patients with TET2-mutated AML (Patel et al., 2012). In keeping with this observation, our IDH1-mutated LT-HSC were more sensitive to IR and daunorubicin than were TET2-KO LT-HSC. In contrast, overexpression of mutant IDH2 impairs responses to cytarabine, another commonly used DNA-damaging agent (Chen et al., 2013). However, cytarabine and IR act via different mechanisms, as ATM knockdown results in increased sensitivity to IR but not to cytarabine (Karnitz et al., 2005). Thus, mutant IDH may affect responses to DNA-damaging agents via multiple mechanisms. These findings may be relevant for designing treatment strategies, including those where the use of IDH inhibitors might be combined with chemotherapy. Other targetable vulnerabilities in IDH1-mutant cells may also exist due to the complex relationship between DDR and hematopoietic differentiation.
In conclusion, our results reveal that mutant IDH1 has TET2-independent effects on DNA repair due to histone methylation-mediated downregulation of ATM. Our findings also point to DDR as a potential therapeutic target for patients with IDH1-mutant disorders.
Experimental Procedures
Mice
The generation of conditional Idh1-R132Q-LSL mice was as described. Idh1-R132Q-LSL mice (Sasaki et al., 2012b) were bred with Vav-Cre mice (Jackson Laboratories; Cat.No. 008610) to produce Vav-IDH1-KI mice (C57BL6/129Ola9; F10). Tet2 KO mice were from Jackson Laboratories (Cat.No: 023359) (Ko et al., 2011) or as described (Moran-Crusio et al., 2011). Atm KO and Tet1 KO mice were as described (Cimmino et al., 2015; Ito et al., 2004). CD45.1 mice were from Jackson Laboratories (Cat.No: 002014). The generation of conditional Idh1-R132H–LSL mice was as described (Hirata et al., 2015). Cre-ERT;Idh1-R132H-LSL mice used for primary MEF generation were produced by crossing Idh1-R132H-LSL mice with CreERT mice (Jackson Laboratories; Cat.No. 004847). For all experiments, young mice were analyzed at 3–4 months, and aged mice were analyzed at 7–10 months. All animal experiments were approved by the University Health Network Animal Care Committee (UHN-ACC) (ID: AUP985). Primers for genotyping PCR were: Idh1 (5’-ACC AGCACCTCCCAACTTGTAT-3’, 5’-AGGTTAGCTCTTGCCGATCCGT-3’, 5’-CAGCAGCCTCTGTTCCACATAC-3’); Vav-Cre (5’-AGATGCCAGGACATCAGGAACCTG-3’, 5’-ATCAGCCACACCAGACACAGAGATC-3’); and CreERT (5’-GCGGTCTGGCAGTAAAACTATC-3’, 5’-CTGAAACAGCATTGCTGTCACTT-3’).
Immunoblotting
Immunoblotting was performed as described (Inoue et al., 2013). Primary antibodies (Abs) used in this study recognized: ATM (Genetex, Cat.No: GTX70103 or Santa Cruz, Cat.No: sc-23921), phospho-ATM (Rockland, Cat.No: 200–301–400), Chek2 (Millipore, Cat.No: 05–649), phospho-p53 (Ser18) (Cell Signaling Technology, Cat. No: 9284), p53 (Vector Laboratories, Cat. No: VP-P956), p21 (BD Pharmingen, Cat.No: 556431), β-actin (Sigma; Cat.No: A2066), histone H3 (Abcam, Cat.No: ab10799), histone H3 (trimethyl K9) (Abcam, Cat.No: ab8898), histone H3 (dimethyl K9) (Abcam, Cat.No: ab1220), histone H3 (trimethyl K27) (Millipore, Cat.No: 07–449), histone H3 (dimethyl K79) (Cell Signalling Technology, Cat.No: 9757), and histone H3 (trimethyl K36) (Abcam, Cat.No: ab9050).
γH2AX and 53BP1 foci
LT-HSC (~200 cells/sample) or LSK cells (~300 cells/sample) were irradiated with or without 0.5 Gy and incubated at 37°C for 4, 8, 12 or 24 hr. Irradiated LT-HSC were plated onto poly-D-lysine-coated coverslips (BD Bioscience, Cat.No: CACB354087) for 30 min, fixed with 2% paraformaldehyde, 0.2% Triton X-100/PBS for 30 min, permeabilized with 0.5% NP-40/PBS for 20 min, and incubated overnight at 4°C with mouse anti-γH2AX Ab (Millipore, clone JBW301, Cat. No: 05–636, 1:100) or rabbit anti-53BP1 Ab (Bethyl Laboratories; 1:3,000). After 1 hr at room temperature, cells were washed with PBS and incubated for 35 min in goat Cy2-anti-mouse Ab (Jackson Immuno Research, Cat. No: 115–225–146, 1:750) and goat Cy3-anti-rabbit Ab (Jackson Immuno Research, Cat. No: 111–165–144, 1:750). Cells were again washed and counterstained with DAPI (0.2 µg/ml) for 10 min. Coverslips were dehydrated and mounted using Entellan (Millipore). Using an FV1000 scanning confocal microscope (Olympus) or Leica DM6000 microscope, γH2AX and 53BP1 foci were counted as described (Nitta et al., 2011).
Competitive repopulation assay
Repopulation capacity was assayed as described (Sasaki et al., 2012b). Briefly, FACS-sorted LT-HSC (250 cells) in PBS (200 µl) were injected into the tail veins of irradiated (10 Gy) CD45.1 mice along with competitor CD45.1+ BM cells (2×105). Peripheral blood cell samples were recovered 4, 8 and 12 weeks later and erythrocytes were lysed. Leukocytes were analyzed by FACs using APC-anti-CD45.1 Ab (eBioseience, Cat.No: 17–0453-82, 1:100) and FITC-anti-CD45.2 Ab (Biolegend, Cat.No: 109806, 1:100).
Colony formation
LT-HSC were left untreated, or irradiated with 2, 4 or 8 Gy, or treated with 0.01–0.1 µM daunorubicin. Cells were plated in 35 mm wells (100–200/well) containing 1 ml Methocult methylcellulose (Stem Cell Technologies, Cat.No. M3434). After 7 days, the number of colonies formed on each plate was counted using an inverted microscope.
Supplementary Material
SIGNIFICANCE.
IDH1 is often mutated at the codon R132 in myelodysplastic syndromes and in AML but how mutant IDH1 drives myeloid tumorigenesis is unclear, although alterations to DNA methylation via inhibition of TET2 are thought to play a major role. Using a mouse model, we have discovered TET2- and DNA methylation-independent effects of mutant IDH1 on the DNA damage response system in HSC. These effects are due to histone modifications that lead to downregulation of the DNA damage sensor ATM. We also find that ATM expression is lower in IDH1 mutant compared to wild-type human AML. These data provide information about the effects of IDH1 mutations in HSC that may be beneficial for tailoring therapy for IDH-mutant diseases.
Acknowledgments
We are grateful to Drs. John E. Dick (Princess Margaret Cancer Centre) and Atsushi Hirao (University of Kanazawa) and to all members of the Mak laboratory for their advice. We also appreciate the assistance of Ms. Irene Ng and the staff members of the flow cytometer facility, genotyping facility, and animal resource centre of the Princess Margaret Cancer Centre. Finally, we are grateful to Dr. Mary Saunders for scientific editing of the manuscript. This work was supported by grants to Drs. T.W.M. and R.A.C. from the Canadian Institutes of Health Research and the Leukemia and Lymphoma Society. K.E.Y. is an employee of and has an ownership interest in Agios Pharmaceuticals.
Footnotes
Accession numbers
The accession number of the eRRBS data at GEO is GSE66536.
Author contributions
W.Y.L., E.F.L., R.A.C. and T.W.M. initiated the project. S.I. and W.Y.L. performed most of the experiments, S.C.B., E.F.L., G.P.N. and R.A.C. designed and/or conducted the CyTOF experiments. A.T. conducted the bioinformatics analysis. I.B. and D.J.R. designed and conducted some flow cytometric experiments. S.I., K.J.K., N.T. and M.L. designed and conducted ATAC-qPCR experiments. A.J.E., F.L., Z.H., D.M.L., Q.L., B.E.S., A.W., J.H., KM. and ARE assisted with many experiments. A.C.P., S.Y.S. and D.D.D.C. designed and conducted the eRRBS and hMeDIP experiments. A.H.S. and R.L.L., and L.C. and I.A., provided bones of aged TET2-KO or TET1-KO mice, respectively. K.S. and K.E.Y. designed and conducted D2HG analyses. S.I. and R.A.C. wrote the manuscript with the help of W.Y.L., A.T., I.B., S.C.B., K.J.K., F.L., D.W.C., C.G., C.B., K.L.T., T.U., G.M., D.D.D.C., M.L., D.J.R. and T.W.M. All authors agree with the conclusions presented in the manuscript.
Other authors declare that no conflicts of interest exist.
References
- An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, Rao A. Acute loss of TET function results in aggressive myeloid cancer in mice. Nature communications. 2015;6:10071. doi: 10.1038/ncomms10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell stem cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer discovery. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot S, Hope KJ, Chagraoui J, Sauvageau M, Deneault E, MacRae T, Mayotte N, Wilhelm BT, Landry JR, Ting SB, et al. RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity. Blood. 2013;122:1545–1555. doi: 10.1182/blood-2013-04-496281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Lu C, Cross JR, Morris JPt, Shroff AS, Ward PS, Bradner JE, Thompson C, Lowe SW. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes & development. 2013;27:1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang R, Guo P, Ju Z. Gadd45a deletion aggravates hematopoietic stem cell dysfunction in ATM-deficient mice. Protein & cell. 2014;5:80–89. doi: 10.1007/s13238-013-0017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H, Lobry C, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nature immunology. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Disease models & mechanisms. 2014;7:941–951. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hirata M, Sasaki M, Cairns RA, Inoue S, Puviindran V, Li WY, Snow BE, Jones LD, Wei Q, Sato S, et al. Mutant IDH is sufficient to initiate enchondromatosis in mice. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2829–2834. doi: 10.1073/pnas.1424400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, Boyiadzis M. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28:1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Hao Z, Elia AJ, Cescon D, Zhou L, Silvester J, Snow B, Harris IS, Sasaki M, Li WY, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes & development. 2013;27:1101–1114. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Ito K, Takubo K, Arai F, Satoh H, Matsuoka S, Ohmura M, Naka K, Azuma M, Miyamoto K, Hosokawa K, et al. Regulation of reactive oxygen species by Atm is essential for proper response to DNA double-strand breaks in lymphocytes. Journal of immunology. 2007;178:103–110. doi: 10.4049/jimmunol.178.1.103. [DOI] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Science translational medicine. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnitz LM, Flatten KS, Wagner JM, Loegering D, Hackbarth JS, Arlander SJ, Vroman BT, Thomas MB, Baek YU, Hopkins KM, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Molecular pharmacology. 2005;68:1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto H, Fukuchi Y, Sakurai M, Sadahira K, Ikeda Y, Okamoto S, Nakajima H. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Scientific reports. 2012;2:273. doi: 10.1038/srep00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Pabst C, Su L, Miller M, Liu F, Yi L, Zhang R, Krosl J, Yung E, Kirschner J, et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes & development. 2014;28:317–327. doi: 10.1101/gad.236794.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, Passegue E. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell stem cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA repair. 2008;7:523–529. doi: 10.1016/j.dnarep.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Nitta E, Yamashita M, Hosokawa K, Xian M, Takubo K, Arai F, Nakada S, Suda T. Telomerase reverse transcriptase protects ATM-deficient hematopoietic stem cells from ROS-induced apoptosis through a telomere-independent mechanism. Blood. 2011;117:4169–4180. doi: 10.1182/blood-2010-08-297390. [DOI] [PubMed] [Google Scholar]
- Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. The New England journal of medicine. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM, Peng Y, Mo Q, Siwko S, Hu R, et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nature communications. 2014;5:3361. doi: 10.1038/ncomms4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, Onozawa M, Lee JE, Callen E, Gutierrez-Martinez P, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, Cairns RA, McCracken S, Wakeham A, Haight J, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes & development. 2012a;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012b;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nature reviews Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring K, Ahangari F, Scott SP, Waring P, Purdie DM, Chen PC, Hourigan K, Ramsay J, McKinnon PJ, Swift M, Lavin MF. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nature genetics. 2002;32:185–190. doi: 10.1038/ng958. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Roig I, Knobel PA, Marjanovic M. The ATM signaling network in development and disease. Frontiers in genetics. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM. Methylome analysis using MeDIP-seq with low DNA concentrations. Nature protocols. 2012;7:617–636. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
- Takagi M, Sato M, Piao J, Miyamoto S, Isoda T, Kitagawa M, Honda H, Mizutani S. ATM-dependent DNA damage-response pathway as a determinant in chronic myelogenous leukemia. DNA repair. 2013;12:500–507. doi: 10.1016/j.dnarep.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H, Suzuki T, Aizawa K, Miyagawa K, Nagai R. Ataxia telangiectasia mutated (ATM)-mediated DNA damage response in oxidative stress-induced vascular endothelial cell senescence. The Journal of biological chemistry. 2010;285:29662–29670. doi: 10.1074/jbc.M110.125138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Chen L, Dawlaty MM, Pan F, Weeks O, Zhou Y, Cao Z, Shi H, Wang J, Lin L, et al. Combined Loss of Tet1 and Tet2 Promotes B Cell, but Not Myeloid Malignancies, in Mice. Cell reports. 2015;13:1692–1704. doi: 10.1016/j.celrep.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.