Abstract
Bile acids are potential carcinogens in gastrointestinal cancer, and interact with nuclear and membrane receptors to initiate downstream signaling. The effect of TGR5 [also known as G protein-coupled bile acid receptor 1 (GPBAR1)] on cancer progression is dependent on the tissue where it is activated. In this report, the function of TGR5 expression in cancer was studied using a bioinformatic approach. TGR5 expression in ampullary adenocarcinoma and normal duodenum was compared by western blotting, reverse transcription polymerase chain reaction, and immunohistochemistry (IHC). High GPBAR1 gene expression was found to be an indicator of worse prognosis in gastric and breast cancer patients, and an indication of better prognosis in ovarian cancer patients. The level of GPBAR1 gene expression was higher in bile-acid exposed cancer than in other types of cancer, and was increased in well-differentiated ampullary adenocarcinoma. Negative, weak or mild expression of TGR5 was correlated with younger age, higher plasma level of total/direct bilirubin, higher plasma concentration of CA-125, advanced tumor stage and advanced AJCC TNM stage. The disease-specific survival rate was highest in ampullary adenocarcinoma patients with high TGR5 expression and high total bilirubin level. In summary, TGR5 functions as a tumor-suppressor in patients with ampullary adenocarcinoma and preoperative hyperbilirubinemia. Further study of the suppressive mechanism may provide a new therapeutic option for patients with ampullary adenocarcinoma.
Keywords: ampullary adenocarcinoma, bile acids, TGR5, GPBAR1, Kaplan-Meier plotter, PrognoScan, cBioPortal
Introduction
Carcinoma of the ampulla of Vater is the most common cancer of the small intestine and is commonly adenocarcinoma (1). The ampulla of Vater is located in the second portion of the duodenum, at the confluence of the common bile and pancreatic duct. Long-term exposure to bile acids is a possible reason for malignant transformation. Bile acids are potential carcinogens in gastrointestinal cancer, including gastric, esophageal and colon cancer, and cholangiocarcinoma (2). Incidences of intestinal metaplasia and gastric cardia cancer are increased in patients with gastroesophageal reflux disease (3). Treatment of squamous cell carcinoma cell lines of the esophagus with bile acids induces cell cycle progression and production of G1-regulating molecules (4). The secondary bile acids, deoxycholate and lithocholate, are the most well-known carcinogens in human colon cancer and are associated with the generation of reactive oxygen/nitrogen species (ROS/RNS) (5). The accumulation of ROS/RNS was found to cause oxidative DNA damage and further mutation in colon cancer (6). The conjugated bile acid glycochenodeoxycholate (GCDA) induced expression of cyclooxygenase 2 (COX-2) and genes that are related to cell proliferation in cholangiocytes (7). Another type of conjugated bile acid, taurochenodeoxycholate (TCDA), induced phosphorylation of epidermal growth factor receptor (EGFR) and its downstream signaling in a human cholangiocarcinoma cell line (8).
Bile acids interact with several types of nuclear receptors, including farnesoid X receptor (FXR), vitamin D receptor, pregnane X receptor, and constitutive androstane receptor (9). Nuclear receptors are ligand-modulated transcription factors and regulate uptake, detoxification and secretion of bile acids. FXR is the main form of nuclear receptor of bile acids expressed in the gastrointestinal tract where it mediates homeostasis of bile acids, lipids and glucose. Expression of FXR is reduced in human colon cancer or Barrett's esophageal cancer. FXR functions as a tumor suppressor in colon, liver and esophageal cancer (10–12). The oncosuppressive roles of FXR in these cancers include suppression of proliferation and induction of apoptosis after exposure to bile acids. Bile acids also act as systemic hormones when interacting with membrane receptors, such as G protein-coupled bile acid receptor 1 (gene GPBAR1; also known as TGR5, M-BAR and BG37). Bile acid-dependent TGR5 activation is involved in the immunomodulatory properties of bile acids, synthesis of endothelial nitric oxide, and mitochondrial energy homeostasis (2,9). In the normal physiologic condition, the functions of TGR5 include modulation of gallbladder filling, improvement of insulin sensitivity, maintenance of glucose homeostasis, and increased energy expenditure to attenuate diet-induced obesity (13,14). The function of TGR5 is variable in different types of cancer (15). Expression of TGR5 is increased in the intestinal subtype of gastric adenocarcinoma and intestinal metaplasia, but not in normal gastric epithelium. Mild to strong TGR5 staining is associated with poor patient survival, and TCDA increased proliferation of a gastric adenocarcinoma cell line through the TGR5-dependent pathway (16). TDCA-induced ROS production and cell proliferation are mediated through TGR5 in Barrett's esophageal and esophageal adenocarcinoma cell lines (17). Stimulation with bile acids prompted the proliferation of an endometrial cell line by activating TGR5 and inducing cyclin D1 expression (18). In contrast to patients with gastric, esophageal and endometrial cancer, the binding of bile acids to TGR5 induced c-Jun-N terminal kinase (JNK) activation and enhanced apoptosis in hepatocytes (19). TGR5-deficient mice are much more susceptible to chemically induced acute liver injury with increased incidence of liver cancer (20). TGR5 may promote or suppress carcinogenesis after stimulation by bile acids (2). Since the role of bile acids in ampullary cancer is largely unknown, in the present study, we investigated the role of TGR5 in ampullary adenocarcinoma.
Materials and methods
Bioinformatic analysis
First, we conducted a search of the Kaplan-Meier plotter database (http://kmplot.com/analysis/) to systematically assess the expression level of the GPBAR1 gene in gastric, breast, lung and ovarian cancer patients (21–23). Kaplan-Meier Plot survival curves were drawn. Second, a PrognoScan database (http://www.abren.net/PrognoScan/) analysis was conducted. The expression level of the GPBAR1 gene was correlated with the survival of cancer patients. Third, data of GPBAR1 gene expression from genomics studies of 30 types of human cancer in the cBioPortal database (http://www.cbioportal.org/index.do) were examined (24,25). Mutation of the GPBAR1 gene was documented by oncogenomic analysis.
Patients
A total of 99 patients who were diagnosed as having ampullary adenocarcinoma and who underwent radical resection at National Cheng Kung University Hospital from January 1990 to January 2010 were enrolled. Patients who received conservative treatment or exhibited other cell types of ampullary cancer were excluded. Demographics, histopathological findings and clinical outcomes were collected by conducting a retrospective chart review. A formal written informed consent was obtained from each patient. Their medical charts were reviewed until January 2016. The disease-specific survival rate was defined as the period from surgery until cancer-related death. The present study was approved by the Institutional Review Board of the National Cheng Kung University Hospital (NCKUH IRB no. A-ER-101-390 and B-ER-103-408).
Western blotting
Total protein lysates from tumor specimens and corresponding specimens of normal duodenum were obtained from the same patient and the protein concentration of the supernatants was measured using the amido black method. Equivalent amounts of protein (30 µg) were separated on 10–15% polyacrylamide gels by SDS-gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with the antibody against TGR5 (Abcam Biotechnology, Cambridge, UK), FXR (R&D, Abingdon, UK), and GAPDH (Cell Signaling Technology, Danvers, MA, USA) proteins. Protein expression was visualized by ECL chemiluminescence (Promega, Madison, WI, USA) and quantitated by comparison with GAPDH.
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR)
The fresh cancer tissues and normal duodenum from the same patient, were obtained for RT-PCR. The total RNA was extracted from fresh tissues, and single-stranded cDNA was synthesized using oligo(dT) as the random primer. The cDNA was amplified using the primers for β-actin, GPBAR1 and FXR genes, which were: β-actin sense, 5′-AGCGGGAAATCGTGCGTG-3′; and β-actin antisense, 5′-CAGGGTACATGGTGGTGGTGCC-3′; GPBAR1 sense, 5′-CCCAGGCTATCTTCCCAGC-3′ and GPBAR1 antisense, 5′-GCCAGGACTGAGAGGAGCA-3′; FXR sense, 5′-GACTTTGGACCATGAAGACCAG-3′ and FXR antisense, 5′-GCCCAGACGGAAGTTTCTTATT-3′. The RT-PCR products were analyzed using agarose gel electrophoresis, and the GPBAR1 or FXR bands were semi-quantified using densitometric analysis and subsequently normalized relative to the β-actin bands.
Immunohistochemical (IHC) staining
Samples of ampullary adenocarcinoma and the surrounding duodenum were fixed in 4% formalin and embedded in paraffin. IHC staining was performed using a monoclonal mouse anti-human TGR5 antibody (Abcam Biotechnology). The sections were incubated using an avidin-biotin complex reagent (Dako, Carpinteria, CA, USA), incubated with 3-amino-9-ethyl carbazole (Zymed Laboratories, South San Francisco, CA, USA) to develop the final color, and counterstained with hematoxylin. The immunoreactivity of the TGR5 protein was assessed using a semi-quantitative method and according to the Remmele and Stegner immunoreactive scoring (IRS) system (26). The IRS scores ranged from 0 to 12 and immunoreactivity was characterized as negative, weak, mild and strong. One researcher assessed the lesions (H.P. Hsu).
Statistical analysis
All statistical analyses were conducted using SPSS version 12.0 (SPSS, Inc., New York, NY, USA). A univariate analysis of the categorical variables was performed using the Chi-square test. The continuous variables were compared using the non-parametric Kruskal-Wallis H test. Any association between specific markers and the recurrence-free survival of patients was assessed using the Kaplan-Meier method, and the level of significance was tested using the log-rank test. A P-value of <0.05 was considered to indicate a statistically significant result.
Results
Analysis of microarray GAPBR1 gene expression data
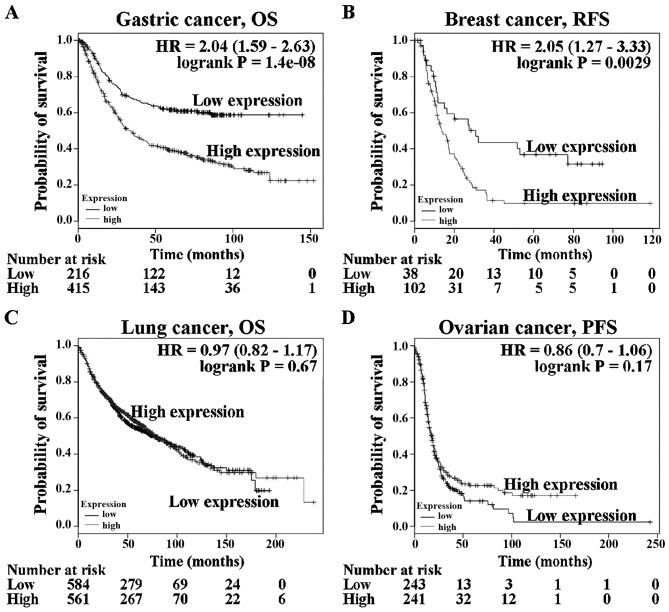
Several gene expression databases of human cancer genetics are available at websites, including the Kaplan-Meier plotter, PrognoScan and cBioPortal system. There is no public database of ampullary adenocarcinoma genetics. Other human cancer types were used to study the function of TGR5 (gene name: GPBAR1). These three databases were used to assess GPBAR1 gene expression and correlate it with clinical outcome. Analysis of the relationship of the GPBAR1 gene expression level (based on Kaplan-Meier plotter data) to survival in patients with gastric, breast, lung and ovarian cancer (Fig. 1) revealed that prognosis was poorer in gastric cancer and breast cancer patients with high GPBAR1 gene expression than in those with low expression (Fig. 1A and B). GPBAR1 gene expression was not correlated with overall survival of patients with lung cancer (Fig. 1C). Ovarian cancer patients with high GPBAR1 gene expression tended to have a better prognosis than those with low expression (Fig. 1D). The function of GPBAR1 gene in tumor development differed between these four types of cancer.
Figure 1.
Kaplan-Meier survival analysis of GPBAR1 gene expression in cancer patients. The data were obtained from the Kaplan-Meier plotter database (http://kmplot.com/analysis/). High-GPBAR1 gene expression is indicated by the red line and low expression by the black line. (A) Overall survival of patients with gastric cancer. (B) Recurrence-free survival of patients with breast cancer. (C) Overall survival of patients with lung cancer. (D) Progression-free survival of patients with ovarian cancer. OS, overall survival; RFS, relapse-free survival; PFS, progression-free survival.
Prognoscan is a collection of human cancer microarray data-sets
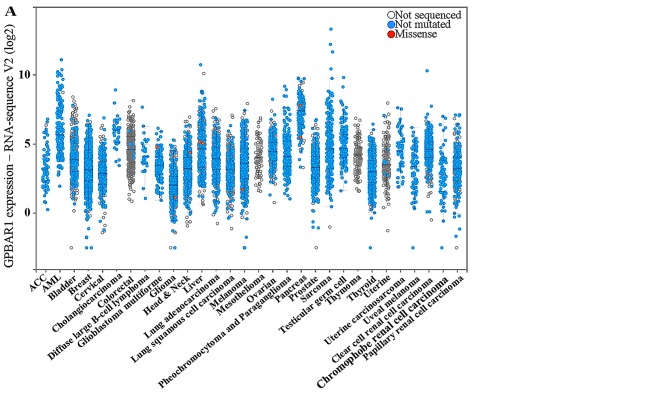
High GPBAR1 gene expression predicted a trend toward poor prognosis in 12 datasets (Table I) and good prognosis in 15 datasets (Table II). In most datasets, the GPBAR1 gene expression was not significantly correlated with survival and only one dataset in each group displayed predictive power (GSE13507 and GSE8894). The level of GPBAR1 expression in human cancer was investigated using the cBioPortal system and analyzed in 30 types of cancer (Fig. 2A). Three specific types of cancer (cholangiocarcinoma, pancreatic and colorectal cancer) were compared (Fig. 2B). Cholangiocarcinoma and pancreatic cancer are continuously exposed to bile and toxic bile acids are well-established carcinogens for cholangiocarcinoma and colorectal cancer. Cholangiocarcinoma and pancreatic cancer, compared to other cancers, had a higher level of GPBAR1 expression (Fig. 2B). Thus the function of GPBAR1 is distinct in different cancers, and its expression may be correlated with bile exposure.
Table I.
PrognoScana microarray analysis of the prognostic value of GPBAR1 in human cancer. (High GPBAR1 gene expression predicted poor prognosis in these datasets).
| Dataset | Cancer type | End point | Cohort | Contributor | Array type | Probe ID | No. | Cut point | Minimum P-value | Corrected P-value | ln (HR-high/HR-low) | COX P-value | ln (HR) | HR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE13507 | Bladder | OS | CNUH | Kim | Human-6 v2 | ILMN_1727709 | 165 | 0.61 | 0.000 | 0.014b | 0.83 | 0.80 | −0.06 | 0.94 (0.57–1.54) |
| GSE2658 | Blood | DSS | Arkansas Z | han | HG-U133_Plus_2 | 1552501_a_at | 559 | 0.53 | 0.030 | 0.358 | 0.44 | 0.62 | 0.08 | 1.08 (0.79–1.48) |
| GSE7696 | Brain | OS | Lausanne | Murat | HG-U133_Plus_2 | 1552501_a_at | 70 | 0.89 | 0.127 | – | 0.61 | 1.00 | 0.00 | 1.00 (0.18–5.57) |
| GSE19615 | Breast | DMFS | DF/HCC | Li | HG-U133_Plus_2 | 1552501_a_at | 115 | 0.30 | 0.194 | – | 0.95 | 0.77 | 0.18 | 1.20 (0.36–3.99) |
| GSE12276 | Breast | RFS | EMC | Bos | HG-U133_Plus_2 | 1552501_a_at | 204 | 0.43 | 0.051 | – | 0.28 | 0.58 | 0.05 | 1.05 (0.89–1.24) |
| GSE6532-GPL570 | Breast | RFS | GUYT | Loi | HG-U133_Plus_2 | 1552501_a_at | 87 | 0.83 | 0.232 | – | 0.55 | 0.67 | −0.33 | 0.72 (0.16–3.31) |
| DMFS | Loi | 87 | 0.83 | 0.232 | – | 0.55 | 0.67 | −0.33 | 0.72 (0.16–3.31) | |||||
| GSE9195 | Breast | RFS | GUYT2 | Loi | HG-U133_Plus_2 | 1552501_a_at | 77 | 0.16 | 0.102 | – | 15.30 | 0.29 | 1.50 | 4.47 (0.29–69.89) |
| GSE17537 | Colorectal | DFS | VMC | Smith | HG-U133_Plus_2 | 1552501_a_at | 55 | 0.49 | 0.047 | 0.472 | 1.03 | 0.48 | 1.21 | 3.36 (0.12–96.71) |
| OS | 55 | 0.67 | 0.288 | – | 0.48 | 0.90 | −0.18 | 0.83 (0.05–14.36) | ||||||
| GSE3141 | Lung | OS | Duke | Bild | HG-U133_Plus_2 | 1552501_a_at | 111 | 0.22 | 0.024 | 0.304 | 0.88 | 0.08 | 0.28 | 1.33 (0.96–1.83) |
| GSE17710 | Lung | RFS | UNC | Wilkerson | Agilent-UNC-custom-4X44K | 25074 | 56 | 0.68 | 0.159 | – | 0.50 | 0.56 | 0.32 | 1.38 (0.46–4.11) |
P<0.05 is defined as statistically significant. OS, overall survival; DSS, disease-specific survival; DMFS, disease metastasis-free survival; RFS, relapse-free survival; HR, hazard ratio; CI, confidence interval.
Table II.
PrognoScana microarray analysis of the prognostic value of GPBAR1 in human cancer. (High GPBAR1 gene expression predicted good prognosis in these datasets.)
| Dataset | Cancer type | End point | Cohort | Contributor | Array type | Probe ID | No. | Cut point | Minimum P-value | Corrected P-value | ln (HR-high/HR-low) | COX P-value | ln (HR) | HR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE12417-GPL570 | Blood | OS | AMLCG (2004) | Metzeler | HG-U133_Plus_2 | 1552501_a_at | 79 | 0.27 | 0.114 | – | −0.50 | 0.41 | −0.28 | 0.76 (0.39–1.47) |
| GSE16581 | Brain | OS | UCLA | Lee | HG-U133_Plus_2 | 1552501_a_at | 67 | 0.25 | 0.021 | 0.275 | −1.44 | 0.63 | −0.81 | 0.44 (0.02–12.73) |
| GSE9195 | Breast | DMFS | GUYT2 | Loi | HG-U133_Plus_2 | 1552501_a_at | 77 | 0.74 | 0.058 | – | −15.50 | 1.00 | −0.01 | 0.99 (0.06–16.54) |
| GSE17536 | Colorectal | DFS | MCC | Smith | HG-U133_Plus_2 | 1552501_a_at | 145 | 0.70 | 0.005 | 0.087 | −1.39 | 0.12 | −1.87 | 0.15 (0.01–1.62) |
| OS | 177 | 0.85 | 0.035 | 0.389 | −0.94 | 0.25 | −0.95 | 0.39 (0.08–1.96) | ||||||
| DSS | 177 | 0.84 | 0.023 | 0.295 | −1.27 | 0.58 | −0.52 | 0.60 (0.10–3.68) | ||||||
| GSE14333 | Colorectal | DFS | Melbourne | Jorissen | HG-U133_Plus_2 | 1552501_a_at | 226 | 0.85 | 0.076 | – | −1.01 | 0.25 | −0.17 | 0.85 (0.64–1.13) |
| GSE22138 | Eye | DMFS | BRCIC | Laurent | HG-U133_Plus_2 | 1552501_a_at | 63 | 0.81 | 0.015 | 0.216 | −1.60 | 0.35 | −6.72 | 0.00 (0–1599.31) |
| GSE2837 | Head and neck | RFS | VUMC, VAMC, UTMDACC (1992–2005) | Chung | U133_X3P | Hs2.160954.1. S1_3p_s_at | 28 | 0.25 | 0.117 | – | −0.90 | 0.23 | −3.90 | 0.02 (0.00–12.57) |
| GSE13213 | Lung | OS | Nagoya (1995–1999, 2002–2004) | Tomida | G4112F | A_23_P400378 | 117 | 0.31 | 0.107 | – | −0.47 | 0.97 | −0.01 | 0.99 (0.62–1.59) |
| GSE31210 | Lung | OS | NCCRI | Okayama | HG-U133_Plus_2 | 1552501_a_at | 204 | 0.87 | 0.121 | – | −1.45 | 0.81 | 0.05 | 1.05 (0.70–1.58) |
| RFS | 204 | 0.88 | 0.052 | – | −1.30 | 0.48 | −0.11 | 0.90 (0.67–1.21) | ||||||
| GSE17537 | Colorectal | DSS | VMC | Smith | HG-U133_Plus_2 | 1552501_a_at | 49 | 0.90 | 0.151 | – | −15.26 | 0.87 | 0.30 | 1.35 (0.04–44.67) |
| GSE8894 | Lung | RFS | Seoul (1995–2005) | Lee | HG-U133_Plus_2 | 1552501_a_at | 138 | 0.26 | 0.002 | 0.048b | −0.75 | 0.08 | −10.60 | 0.00 (0.00–3.68) |
| GSE17710 | Lung | OS U | NC | Wilkerson | Agilent-UNC-custom-4X44K | 25074 | 56 | 0.21 | 0.267 | – | −0.44 | 0.83 | −0.11 | 0.89 (0.32–2.53) |
| GSE9891 | Ovarian | OS | AOCS, RBH, WH, NKI-AVL (1992–2006) | Tothill | HG-U133_Plus_2 | 1552501_a_at | 278 | 0.74 | 0.024 | 0.304 | −0.50 | 0.37 | −0.35 | 0.71 (0.33–1.52) |
| GSE17260 | Ovarian | PFS | Niigata (1997–2008) | Yoshihara | G4112A | A_23_P400378 | 110 | 0.70 | 0.077 | – | −0.46 | 0.99 | 0.00 | 1.00 (0.58–1.73) |
| OS | 110 | 0.69 | 0.106 | – | −0.57 | 0.56 | −0.20 | 0.82 (0.41–1.63) | ||||||
| GSE19234 | Skin | OS | NYU | Bogunovic | HG-U133_Plus_2 | 1552501_a_at | 38 | 0.79 | 0.093 | – | −1.20 | 0.31 | −0.66 | 0.52 (0.14–1.86) |
P<0.05 is defined as statistically significant. OS, overall survival; DSS, disease-specific survival; DMFS, disease metastasis-free survival; RFS, relapse-free survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval.
Figure 2.
Analysis of GPBAR1 expression level data from the cBioPortal database (http://www.cbioportal.org/index.do). Every spot represents a single study. White spots represent those analyzed without gene sequencing, blue spots represent normal results of gene sequencing and red spots represent missense mutations. (A) Level of GPBAR1 expression in 30 types of human cancer. (B) Level of GPBAR1 expression in three specific-types of cancer (bile acid-exposed cholangiocarcinoma and pancreatic cancer vs. colorectal cancer). The median and interquartile range are presented. The median level of GPBAR1 gene expression was higher in cholangiocarcinoma and pancreatic cancer than in colorectal cancer.
Expression of TGR5 protein in ampullary cancer
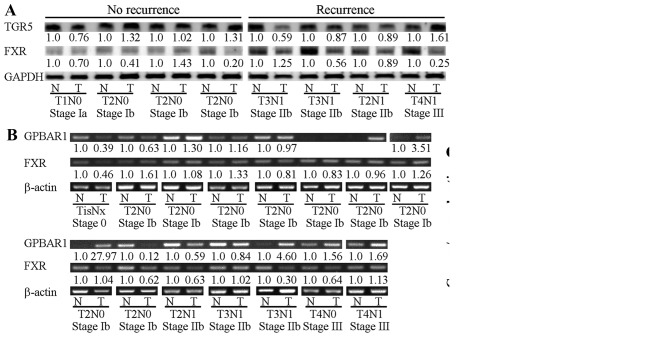
The ampulla of Vater is located in the second part of the duodenum and is exposed to bile acids under normal physiological conditions. TGR5 protein (product of the GPBAR1 gene) and GPBAR1 mRNA were detected in clinical samples of ampullary cancer and the surrounding normal duodenum (Fig. 3). In the patients with cancer recurrence, the TGR5 protein level was lower in the tumor than that noted in the normal duodenum. In the patients without cancer recurrence, the tumor TGR5 level was similar to the normal tissue level. In contrast, no such pattern was found for FXR protein (Fig. 3A). Increased GPBAR1 mRNA was detected in 7 of 15 specimens of ampullary cancer, particularly in well-differentiated ampullary adenocarcinoma (Fig. 3B and C). Expression of FXR mRNA was not correlated with histological differentiation (Fig. 3C).
Figure 3.
Expression of TGR5 and FXR in ampullary adenocarcinoma. Sample pairs consisting of ampullary adenocarcinoma (T) and its corresponding normal duodenum (N) were collected. (A) Eight sample pairs were assessed by western blotting of TGR5 and FXR proteins with GAPDH serving as a loading control. The fold-change of TGR5/GAPDH or FXR/GAPDH is indicated below the band. (B) A total of 15 sample pairs were assessed by semi-quantitative RT-PCR, with β-actin serving as a loading control. The fold-change of TGR5/β-actin or FXR/β-actin is indicated below the band. (C) The proportion of TGR5/β-actin or FXR/β-actin expressed in tumors relative to normal control tissues was correlated with histological differentiation.
Immunohistochemical staining of TGR5 in ampullary cancer
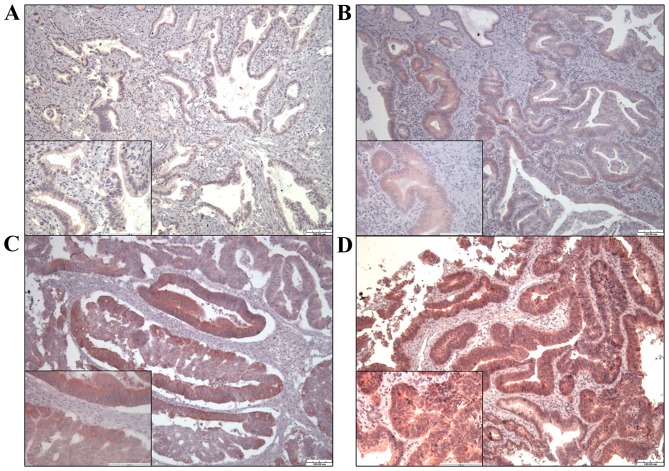
To study the relationship of TGR5 protein expression with clinical outcome, 99 specimens of ampullary adenocarcinoma were immunostained for TGR5. TGR5 was detected in the cytoplasm and nucleus of each cancer cell (Fig. 4). We divided the result as negative, weak, mild and strong expression of TGR5. Expression of TGR5 was negative in 14 patients, weak in 33, mild in 29, and strong in 23 (Table I). Negative, weak or mild expression of TGR5 was correlated with younger age (P=0.043) and higher level of direct bilirubin (P=0.023, separately) and tended to be correlated with higher level of total bilirubin (P=0.059), higher plasma level of cancer antigen-125 (CA-125) (P=0.099), advanced tumor stage and AJCC TNM stage (P=0.063 and 0.062, separately) (Table III).
Figure 4.
Expression of TGR5 in ampullary adenocarcinoma was assessed by immunohistochemical (IHC) staining and rated using the Remmele and Stegne immunoreactive scoring system. Expression was either (A) 0–1 (negative); (B) 2–3 (weak); (C) 4–8 (mild) or (D) 9–12 (strong).
Table III.
Correlation of TGR5 expression with demographics and histopathological findings in patients with ampullary adenocarcinoma who underwent radical resection.
| Expression of TGR5
|
P-value | ||
|---|---|---|---|
| Negative, weak, mild | Strong | ||
| Patients, n (%) | 76 (77) | 23 (23) | |
| Gender, n (%) | 0.481 | ||
| Female | 30 (73) | 11 (27) | |
| Male | 46 (79) | 12 (21) | |
| Age at surgery (years)a | 65 (32–90) | 68 (35–83) | 0.043 |
| Total bilirubin (mg/dl)a | 3.4 (0.2–19.6) | 1.3 (0.4–16.3) | 0.059 |
| Direct bilirubin (mg/dl)a | 2.6 (0–18.0) | 0.6 (0–7.3) | 0.023 |
| Preoperative bile decompression, n (%) | 41 (77) | 12 (23%) | 1.000 |
| CEA (ng/ml)a | 1.9 (0.1–296.3) | 2.5 (0–13.0) | 0.306 |
| CA-125 (U/ml)a | 15.4 (5.2–164.1) | 11.5 (0.5–66.7) | 0.099 |
| CA-199 (U/ml)a | 55.4 (0.3–7512.9) | 44.2 (1.4–1860) | 0.892 |
| Subtype, n (%)b | 0.206 | ||
| Intestinal type | 37 (74) | 13 (26) | |
| Pancreaticoduodenal type | 17 (90) | 2 (10) | |
| Tumor type, n (%) | 0.315 | ||
| Polypoid | 40 (74) | 14 (26) | |
| Ulcerative | 21 (87) | 3 (13) | |
| Mixed | 15 (71) | 6 (29) | |
| Resection margin, n (%) | 1.000 | ||
| Free | 66 (76) | 21 (24) | |
| Microscopically positive | 8 (80) | 2 (20) | |
| Lymph node metastasis, n (%)b | 0.176 | ||
| Negative | 42 (75) | 14 (25) | |
| Positive | 30 (88) | 4 (12) | |
| Lymphovascular invasion, n (%)b | 0.192 | ||
| Negative | 27 (69) | 12 (31) | |
| Positive | 34 (83) | 7 (17) | |
| Perineural invasion, n (%)b | 0.373 | ||
| Negative | 28 (68) | 13 (32) | |
| Positive | 18 (82) | 4 (18) | |
| Histological differentiation, n (%)b | 0.847 | ||
| Well | 31 (74) | 11 (26) | |
| Moderate | 37 (77) | 11 (23) | |
| Poor | 5 (83) | 1 (17) | |
| Pancreatic invasion, n (%)b | 0.149 | ||
| Negative | 34 (71) | 14 (29) | |
| Positive | 42 (84) | 8 (16) | |
| Tumor size (cm)a | 2.4 (0.7–8.0) | 2.5 (1.0–6.0) | 0.783 |
| Tumor stage, n (%) | 0.063 | ||
| T1 | 5 (45) | 6 (55) | |
| T2 | 27 (75) | 9 (25) | |
| T3 | 29 (83) | 6 (17) | |
| T4 | 15 (88) | 2 (12) | |
| AJCC TNM stage, n (%) | 0.062 | ||
| I | 26 (63) | 15 (37) | |
| II | 34 (85) | 6 (15) | |
| III | 15 (88) | 2 (12) | |
| IV | 1 (100) | 0 (0) | |
Values are expressed as median (range).
Excluding patients without detailed records. AJCC TNM stage, American Joint Committee on Cancer tumor-node-metastases (TNM) staging system.
Correlation of TGR5 expression with clinical outcomes of ampullary cancer patients
In 95 patients with regular follow-up (range, 3–249 months), 55 patients developed recurrence. The recurrences in patients with negative, weak or mild TGR5 expression tended to be earlier (within postoperative 12 months) (P=0.089), although the level of TGR5 expression was not associated with recurrence patterns (Table IV). The disease-specific survival rate tended to be better in patients with strong TGR5 expression (P=0.1118; Fig. 5A).
Table IV.
Correlation between disease recurrence and TGR5 expression in patients with ampullary adenocarcinoma who underwent radical resection.
| Expression of TGR5
|
P-value | ||
|---|---|---|---|
| Negative, weak, mild | Strong | ||
| Patients, n (%) | 76 (77) | 23 (23) | |
| No recurrence, n (%)a | 28 (70) | 12 (30) | |
| Recurrence, n (%)a | 44 (80) | 11 (20) | 0.089 |
| Delayed recurrence, n (%)(after postoperative 12 months) | 16 (70) | 7 (30) | |
| Early recurrence, n (%)(within postoperative 12 months) | 28 (90) | 3 (10) | |
| Patterns of recurrence, n (%)a,b | |||
| Liver metastasis, n (%) | 19 (86) | 3 (14) | 0.260 |
| Local recurrence, n (%) | 28 (85) | 5 (15) | 0.205 |
| Peritoneal carcinomatosis, n (%) | 12 (86) | 2 (14) | 0.506 |
| Bone metastasis, n (%) | 6 (67) | 3 (33) | 0.437 |
| Other metastasis, n (%)c | 14 (82) | 3 (18) | 0.754 |
Excludes two patients who died due to surgical complications and two patients who were lost in the follow-up in our hospital.
Some patients developed more than one type of metastases.
Including brain, lung and ovary metastases.
Figure 5.
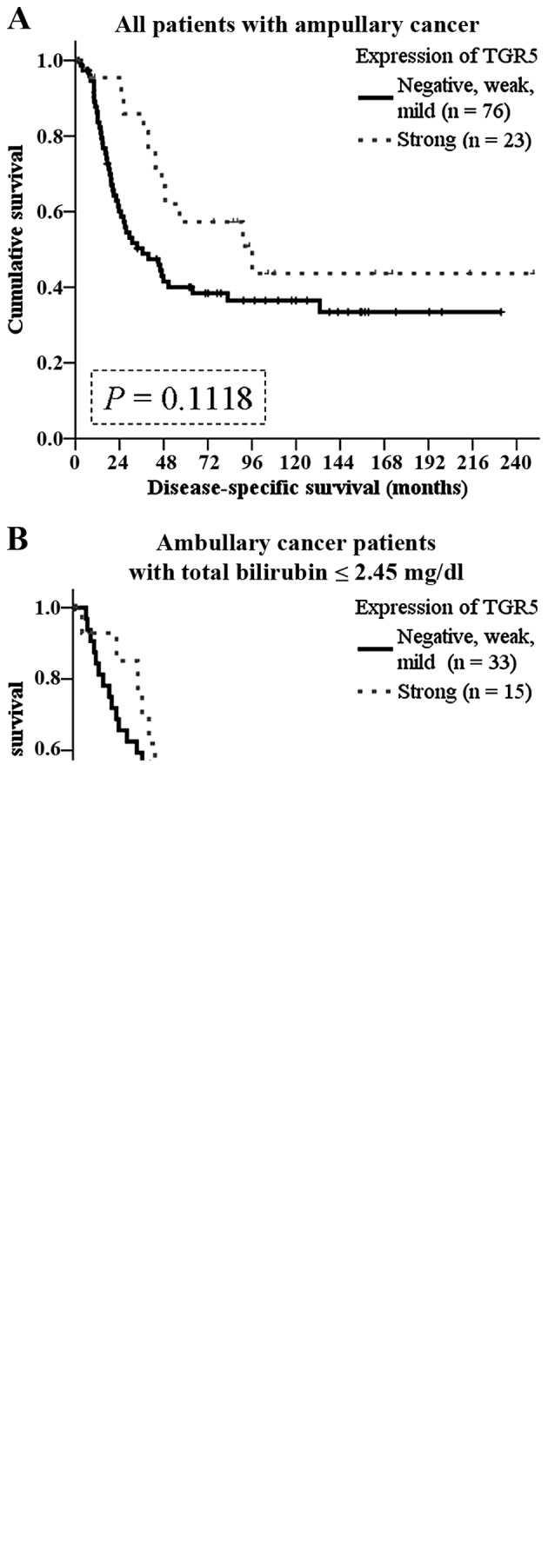
Kaplan-Meier analysis of the impact of TGR5 expression on disease-specific survival in patients with ampullary adeno carcinoma. (A) Disease-specific survival of all patients with ampullary adenocarcinoma who underwent surgery according to TGR5 expression level (P=0.1118). (B) Disease-specific survival of ampullary cancer patients with plasma total bilirubin concentration ≤2.45 mg/dl. TGR5 expression level was not correlated with survival (P=0.8428). (C) Disease-specific survival of ampullary cancer patients with plasma total bilirubin concentration >2.45 mg/dl. Strong TGR5 expression predicted a better survival (P=0.0464).
In the literature, the function of TGR5 depends on dysregulation of bile acid homeostasis (2). We hypothesized that high levels of preoperative bilirubin interacts with TGR5 in ampullary cancer. We grouped the patients according to the median level of total bilirubin (2.45 mg/dl). Although not correlated with survival in the patients with total bilirubin ≤2.45 mg/dl (Fig. 5B), TGR5 expression predicted a better prognosis in patients with higher than 2.45 mg/dl (P=0.0464; Fig. 5C).
Discussion
The ampullary of Vater is normally exposed to bile. This is the first study to investigate expression of the membrane bile acid receptor, TGR5, in ampullary adenocarcinoma. The results of analysis of GPBAR1 gene expression data (gene of TGR5) in microarray databases was correlated with clinical outcomes and varied between types of cancers. The pathological function of TGR5 in cancer is regulated by a complex mechanism. In ampullary adenocarcinoma, the present study detected TGR5 protein and GPBAR1 mRNA in the tumor and surrounding normal duodenum. Negative, weak or mild TGR5 expression was correlated with elevation of plasma bilirubin. In the patients with plasma total bilirubin higher than the median, strong TGR5 expression predicted a better prognosis.
There are two types of ampullary adenocarcinoma: intestinal and pancreaticobiliary types. These differ in clinical behavior (27) and the differences may be intrinsic. Nuclear accumulation of β-catenin promotes WNT activation and cancer progression; however, loss of the β-catenin protein in ampullary cancer is correlated with poor prognosis (28). Nestin, a stemness protein, performs a dual role in ampullary adenocarcinoma, as a predictor of good prognosis in early cancer and as a promoter of metastasis in advanced cancer (29). The epithelial cell marker, EpCAM, is one of the signatures of cancer stem cells with oncogenic potential mediated via upregulation of c-myc and cyclins. However, loss of EpCAM is linked to a more aggressive phenotype of ampullary cancer, suggesting that EpCAM may play a different role in ampullary cancer than in other cancers (30). Ampullary adenocarcinoma is a unique cancer with a 5-year survival rate <50% after curative resection (27,31,32). Further study of ampullary adenocarcinoma is required to develop new treatment modalities and improve clinical outcomes.
The ampullary of Vater is located at the confluence of the common bile and pancreatic ducts, and second portion of the duodenum. Long-term exposure to bile acids increases oxidative stress, generates ROS/RNS, and induces cell damage and mutation rates in gastrointestinal cancer (2,5,6). Alteration of the bile contents of gastroesophageal reflux is correlated with the increased incidence of cancer in cell culture, animal models and epidemiology studies (3,4,33). Toxic bile acids induce expression of COX-2 or activation of EGFR and promote carcinogenesis in cholangiocarcinoma (7,8). In our previous study, preoperative plasma concentration of total bilirubin was elevated in non-survivors of ampullary cancer (32). However, no relationship was found between hyperbilirubinemia and ampullary cancer recurrence.
Bile acids interact not only with nuclear receptors, but also with membrane receptors. TGR5 is a G protein-coupled bile acid receptor that mediates bile acid-regulated energy and glucose homeostasis (2,9,34). Bile acids induce cell proliferation and cell cycle progression through the TGR5-dependent pathway and TGR5 acts such as an oncoprotein (16–18). In hepatocytes, suppression of TGR5 enhances chemical-induced carcinogenesis and activation of TGR5 promotes cell apoptosis (19,20). Whether TGR5 promotes or suppresses carcinogenesis depends on the composition of the bile acids (2). We analyzed multiple microarray datasets and found that high GPBAR1 gene expression predicted poor prognosis in some datasets, but good prognosis in others (Tables I and II; Fig. 1). Bile acid-exposed cancers (such as cholangiocarcinoma and pancreatic cancer) had a higher ratio of GPBAR1 expression (Fig. 2B). Since the ampulla of Vater is also exposed to bile acids under normal physiological conditions, the study of bile acid receptors, such as TGR5 and FXR, is indicated in ampullary cancer. In the present study, the mRNAs or proteins of TGR5 or FXR were detected in specimens of ampullary adenocarcinoma (Fig. 3). Increased expression of GPBAR1 mRNA but not FXR mRNA was correlated with histological differentiation and well-differentiated adenocarcinoma (Fig. 3C). Taken together, our results indicate that the membrane receptor of bile acids, TGR5, may be activated in ampullary cancer.
Activation of TGR5 plays a role in cyclic adenosine monophosphate (cAMP), EGFR, mitogen-activated protein kinase (MAPK, such as JNK, ERK-1/2), cyclooxygenase-2 (COX-2) or signal transducer and activator of transcription 3 (STAT3) signaling (35). TGR5 functions in a cell type-dependent and context-dependent manner in cancer. In gastric and esophageal cancer, the TGR5-dependent pathway mediates bile acid-induced ROS production and cell proliferation as well as deoxycholate-induced EGFR phosphorylation and ERK1/2 activation. Moreover, TGR5 expression is associated with the poor prognosis of patients (36–38), suppresses STAT3 signaling and inhibits cell cycle progression, angiogenesis, metastasis and evasion of the immune system in gastric cancer (36). Interaction of TGR5 and EGFR depends on lipid rafts. Deoxycholate induces EGFR phosphorylation and ERK1/2 activation through the TGR5-dependent pathway (38). However, TGR5 performs as a tumor-suppressor in liver cancer. TGR5-deficient mice have an increased incidence of liver cancer (19). Bile acids conjugate TGR5 to induce JNK activation and enhance apoptosis in hepatocytes (20). In the present study, strong TGR5 expression was correlated with lower plasma concentration of total and direct bilirubin. The patients with strong TGR5 expression tended to have a lower plasma level of CA-125, earlier tumor stage, and earlier AJCC TNM stage and also a better disease-specific survival rate, particularly those patients with total bilirubin concentration higher than 2.45 mg/dl. We conclude that TGR5 performs as a tumor suppressor in hyperbilirubinemic patients with ampullary adenocarcinoma.
In summary, high TGR5 expression was correlated with lower plasma concentration of total/direct bilirubin, lower plasma level of CA-125, early tumor stage and AJCC TNM stage. High TGR5 expression also predicted a good survival in patients with total bilirubin levelss higher than 2.45 mg/dl. TGR5 performs as a tumor suppressor in hyperbilirubinemia condition of ampullary adenocarcinoma patients.
Acknowledgments
The present study was supported by grants from the National Science Council (grant NSC-102-B-2314-B-006-043), the Ministry of Science and Technology (grant MOST-104-B-2314-B-006-049), the National Cheng Kung University Hospital (to H.-P.H.), and the Chi Mei Medical Center (to M.-C.C.). The authors are grateful for the support from the Human Biobank, Research Center of Clinical Medicine, National Cheng Kung University Hospital. We were blessed with support from the late superintendent, Professor Pin-Wen Lin. Furthermore, we thank Mr. Chih-Yang Wang and Ms. Yu-Hsuan Hung for their support.
Abbreviations
- AJCC TNM stage
American Joint Committee on Cancer tumor-node-metastases staging system
- CA-125
cancer antigen-125
- CA-199
cancer antigen-199
- cAMP
cyclic adenosine monophosphate
- CEA
carcinoembryonic antigen
- COX-2
cyclooxygenase-2
- EGFR
epidermal growth factor receptor
- FXR
farnesoid X receptor
- GCDC
glycochenodeoxycholate
- GPBAR1
G protein-coupled bile acid receptor 1
- IHC
immunohistochemistry
- IRS
immunoreactive score of Remmele and Stegner
- JNK
c-Jun-N terminal kinase
- MAPK
mitogen-activated protein kinase
- PDGFR
platelet-derived growth factor receptor
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RT-PCR
reverse transcription polymerase chain reaction
- STAT3
signal transducer and activator of transcription 3
- TCDA
taurochenodeoxycholate
References
- 1.Albores-Saavedra J, Schwartz AM, Batich K, Henson DE. Cancers of the ampulla of vater: Demographics, morphology, and survival based on 5,625 cases from the SEER program. J Surg Oncol. 2009;100:598–605. doi: 10.1002/jso.21374. [DOI] [PubMed] [Google Scholar]
- 2.Tsuei J, Chau T, Mills D, Wan YJ. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med. 2014;239:1489–1504. doi: 10.1177/1535370214538743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye W, Chow WH, Lagergren J, Yin L, Nyrén O. Risk of adenocarcinomas of the esophagus and gastric cardia in patients with gastroesophageal reflux diseases and after antireflux surgery. Gastroenterology. 2001;121:1286–1293. doi: 10.1053/gast.2001.29569. [DOI] [PubMed] [Google Scholar]
- 4.Nishioka K, Doki Y, Miyata H, Tamura S, Yasuda T, Kimura Y, Kishi K, Yoshida K, Fujiwara Y, Yano M, et al. Bile acid promotes the proliferation of squamous cell carcinoma of the esophagus, independent of its inducing COX-2 expression. J Surg Res. 2006;132:130–135. doi: 10.1016/j.jss.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein C, Payne CM, Bernstein H. Bile acids: Promoters or carcinogens in colon cancer? J Carcinogene Mutagene. 2011;2:101e. doi: 10.4172/2157-2518.1000101e. [DOI] [Google Scholar]
- 6.Payne CM, Bernstein C, Dvorak K, Bernstein H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin Exp Gastroenterol. 2008;1:19–47. doi: 10.2147/CEG.S4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komichi D, Tazuma S, Nishioka T, Hyogo H, Chayama K. Glycochenodeoxycholate plays a carcinogenic role in immortalized mouse cholangiocytes via oxidative DNA damage. Free Radic Biol Med. 2005;39:1418–1427. doi: 10.1016/j.freeradbiomed.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Yoon JH, Higuchi H, Werneburg NW, Kaufmann SH, Gores GJ. Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology. 2002;122:985–993. doi: 10.1053/gast.2002.32410. [DOI] [PubMed] [Google Scholar]
- 9.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 10.Lax S, Schauer G, Prein K, Kapitan M, Silbert D, Berghold A, Berger A, Trauner M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130:2232–2239. doi: 10.1002/ijc.26293. [DOI] [PubMed] [Google Scholar]
- 11.van de Winkel A, van Zoest KP, van Dekken H, Moons LM, Kuipers EJ, van der Laan LJ. Differential expression of the nuclear receptors farnesoid X receptor (FXR) and pregnane X receptor (PXR) for grading dysplasia in patients with Barrett's oesophagus. Histopathology. 2011;58:246–253. doi: 10.1111/j.1365-2559.2011.03743.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol. 2011;25:1066–1071. doi: 10.1210/me.2010-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao W, Tian W, Hong J, Li D, Tavares R, Noble L, Moss SF, Resnick MB. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2013;304:G322–G327. doi: 10.1152/ajpgi.00263.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong J, Behar J, Wands J, Resnick M, Wang LJ, DeLellis RA, Lambeth D, Souza RF, Spechler SJ, Cao W. Role of a novel bile acid receptor TGR5 in the development of oesophageal adenocarcinoma. Gut. 2010;59:170–180. doi: 10.1136/gut.2009.188375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casaburi I, Avena P, Lanzino M, Sisci D, Giordano F, Maris P, Catalano S, Morelli C, Andò S. Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle. 2012;11:2699–2710. doi: 10.4161/cc.21029. [DOI] [PubMed] [Google Scholar]
- 19.Yang JI, Yoon JH, Myung SJ, Gwak GY, Kim W, Chung GE, Lee SH, Lee SM, Kim CY, Lee HS. Bile acid-induced TGR5-dependent c-Jun-N terminal kinase activation leads to enhanced caspase 8 activation in hepatocytes. Biochem Biophys Res Commun. 2007;361:156–161. doi: 10.1016/j.bbrc.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Chen WD, Yu D, Forman BM, Huang W, Wang YD. Deficiency of G-protein-coupled bile acid receptor Gpbar1 (TGR5) enhances chemically induced liver carcinogenesis. Hepatology. 2013;57:656–666. doi: 10.1002/hep.26019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Györffy B, Lánczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Győrffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241.1–e82241.8. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS) Pathol Res Pract. 1993;189:862–866. doi: 10.1016/S0344-0338(11)81095-2. [DOI] [PubMed] [Google Scholar]
- 27.Schiergens TS, Reu S, Neumann J, Renz BW, Niess H, Boeck S, Heinemann V, Bruns CJ, Jauch KW, Kleespies A. Histomorphologic and molecular phenotypes predict gemcitabine response and overall survival in adenocarcinoma of the ampulla of Vater. Surgery. 2015;158:151–161. doi: 10.1016/j.surg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Hsu HP, Shan YS, Jin YT, Lai MD, Lin PW. Loss of E-cadherin and beta-catenin is correlated with poor prognosis of ampullary neoplasms. J Surg Oncol. 2010;101:356–362. doi: 10.1002/jso.21493. [DOI] [PubMed] [Google Scholar]
- 29.Shan YS, Chen YL, Lai MD, Hsu HP. Nestin predicts a favorable prognosis in early ampullary adenocarcinoma and functions as a promoter of metastasis in advanced cancer. Oncol Rep. 2015;33:40–48. doi: 10.3892/or.2014.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piscuoglio S, Lehmann FS, Zlobec I, Tornillo L, Dietmaier W, Hartmann A, Wünsch PH, Sessa F, Rümmele P, Baumhoer D, et al. Effect of EpCAM, CD44, CD133 and CD166 expression on patient survival in tumours of the ampulla of Vater. J Clin Pathol. 2012;65:140–145. doi: 10.1136/jclinpath-2011-200043. [DOI] [PubMed] [Google Scholar]
- 31.Rostain F, Hamza S, Drouillard A, Faivre J, Bouvier AM, Lepage C. Trends in incidence and management of cancer of the ampulla of Vater. World J Gastroenterol. 2014;20:10144–10150. doi: 10.3748/wjg.v20.i29.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu HP, Yang TM, Hsieh YH, Shan YS, Lin PW. Predictors for patterns of failure after pancreaticoduodenectomy in ampullary cancer. Ann Surg Oncol. 2007;14:50–60. doi: 10.1245/s10434-006-9136-3. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein SR, Yang GY, Curtis SK, Reuhl KR, Liu BC, Mirvish SS, Newmark HL, Yang CS. Development of esophageal metaplasia and adenocarcinoma in a rat surgical model without the use of a carcinogen. Carcinogenesis. 1997;18:2265–2270. doi: 10.1093/carcin/18.11.2265. [DOI] [PubMed] [Google Scholar]
- 34.Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5: A valuable metabolic target. Dig Dis. 2011;29:37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stepanov V, Stankov K, Mikov M. The bile acid membrane receptor TGR5: A novel pharmacological target in metabolic, inflammatory and neoplastic disorders. J Recept Signal Transduct Res. 2013;33:213–223. doi: 10.3109/10799893.2013.802805. [DOI] [PubMed] [Google Scholar]
- 36.Guo C, Su J, Li Z, Xiao R, Wen J, Li Y, Zhang M, Zhang X, Yu D, Huang W, et al. The G-protein-coupled bile acid receptor Gpbar1 (TGR5) suppresses gastric cancer cell proliferation and migration through antagonizing STAT3 signaling pathway. Oncotarget. 2015;6:34402–34413. doi: 10.18632/oncotarget.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda H, Hirata S, Inoue K, Mashima H, Ohnishi H, Yoshiba M. Involvement of membrane-type bile acid receptor M-BAR/TGR5 in bile acid-induced activation of epidermal growth factor receptor and mitogen-activated protein kinases in gastric carcinoma cells. Biochem Biophys Res Commun. 2007;354:154–159. doi: 10.1016/j.bbrc.2006.12.168. [DOI] [PubMed] [Google Scholar]
- 38.Jensen DD, Godfrey CB, Niklas C, Canals M, Kocan M, Poole DP, Murphy JE, Alemi F, Cottrell GS, Korbmacher C, et al. The bile acid receptor TGR5 does not interact with β-arrestins or traffic to endosomes but transmits sustained signals from plasma membrane rafts. J Biol Chem. 2013;288:22942–22960. doi: 10.1074/jbc.M113.455774. [DOI] [PMC free article] [PubMed] [Google Scholar]