Abstract
Transforming growth factor-β (TGF-β) proteins are important cytokines in the occurrence and development of tumors. However, its neural functions in glioma are still not understood. In the present study, we evaluated the effects of TGF-β1 on glioma cell line U87. miR-205 and miR-195 were involved in TGF-β1 signaling pathway. Quantitative real-time PCR was used to detect miR-205 and miR-195 levels in human glioma tissue samples and U87 cells treated with different concentrations of TGF-β1. Enzyme-linked immunosorbent assay (ELISA) was performed to determine TGF-β1 in the glioma patients peripheral blood. In vitro, U87 cells were transfected with mimics or inhibitors of miR-205 and miR-195. SMAD proteins were assayed by western blotting. Luciferase assay and co-immunoprecipitation (Co-IP) were used to determine the relationships between miR-205 and SMAD2, miR-195 and SMAD7. Effects of miR-205 and miR-195 on glioma cell proliferation and invasion using colony forming and cell migration assays. It was shown that miR-205 was decreased in glioma tissue, but miR-195 and TGF-β1 was increased. In addition, TGF-β1 concentration was negatively correlated with miR-205 mRNA level, but positively correlated with miR-195 mRNA. In addition, miR-205 was downregulated and miR-195 was upregulated by TGF-β1 in a dose-dependent manner. miR-205 and miR-195 targeted and inhibited SMAD2 and SMAD7 expression, respectively, in U87. High expression of miR-205 but not miR-195 reduced SMAD2 and SMAD4 heteromer formation. In addition, it was also shown that miR-205 overexpression inhibited U87 proliferation and invasion efficiently. All the results suggested that miR-205 and miR-195 participated in the TGF-β1 signaling pathway and showed opposite effects in glioma. These findings contribute to the understanding of TGF-β1 function in glioma.
Keywords: transforming growth factor-β, miR-205, miR-195, SMAD, glioma
Introduction
Glioma is a malignant brain tumor lacking effective therapeutic strategies. Underlying biological pathogenic mechanisms need to be explored to supply new targets for glioma treatment. Gliomas are classified into four grades according to the World Health Organization (WHO) (1). Glioblastoma (GB) belonging to grade IV is the most malignant glioma with a poor prognosis. Therefore in order to develop novel therapeutic methods for GB, it is important to elucidate the fundamental molecular mechanisms leading to glioma formation.
Transforming growth factor-β (TGF-β) is a multi-functional cytokine regulating cell growth and differentiation (2). TGF-β promotes tumor growth in several ways, including regulating epithelial-to-mesenchymal transition, changing immune response, stimulating angiogenesis and promoting metastasis (3–6). TGF-β signaling pathway has been identified to participate in glioma progression by mediating cell proliferation and invasion (7,8). TGF-β is overexpressed in glioma tissues in human patients compared with normal tissues, which suggests TGF-β plays a critical role. In glioma, TGF-β1 produced by infiltrating microglia enhances glioma invasion (8). In addition, TGF-β1 prohibits tumor growth by upregulating miRNAs. It is reported that TGF-β regulates miRNAs including miR-200, miR-205, miR-182, miR-21 and miR-31 (9–11). In TGF-β signaling pathway, SMAD2, SMAD3, SMAD4 and SMAD7 are critical proteins (12). Alterations of SMAD proteins are contributed to induce tumor cell proliferation (13). These proteins can be regulated by many factors. Recently, microRNAs (miRNAs) were reported to be involved in regulating SMAD proteins (14–16).
miRNAs are usually short non-coding and single-stranded RNA containing 18–25 nucleotides, which regulate expressions of many proteins. miRNAs suppress gene expression by binding to target mRNAs at 3′ untranslated regions (3′UTRs). A confirmed miRNA target in multiple genes. By this mechanism, miRNAs are related to cell proliferation, invasion, differentiation and apoptosis in tumors. Thus, dysfunction of miRNAs is involved in various types of human cancers including glioma, and miRNAs can act as both oncogenes and tumor inhibitors during carcinogenesis.
miR-205 is often disordered in various solid tumors. miR-205 is overexpressed in non-small cell lung cancer and ovarian cancer cells, promoting tumor progression and chemoresistance (17–20). It can target the tumor suppressor gene phosphatase and tensin homolog (PTEN), CYR61 and CTGF, SH2-containing phosphoinositide 5-phosphatase 2 (SHIP2), then, promoting tumor growth (21,22). Markedly, miR-205 functions as a tumor inhibitor by targeting oncogenes. miR-205 decreases breast cancer cells proliferation by suppressing ErbB3 and VEGF-A, inhibits melanoma growth by targeting E2F1, hampers renal carcinomas by degrading Src protein (23–25). However, the effect of miR-205 on TGF-β1 signaling pathway in glioma is not understood.
Expression and function of miR-195 in tumor development is not confirmed. miR-195 level is reported to be decreased in solid tumors and increased in adenomas (26), which suggests that miR-195 reveals growth promotion or growth resistance in cancers. The roles of miR-195 in glioma need to be elucidated by TGF-β1 stimulation. Accordingly, it is pivotal to explore the biological functions and molecular mechanisms of miR-205 and miR-195 in the environment with high level of TGF-β1 to find a novel therapy for glioma.
Materials and methods
Ethics statement
All experiments were approved by the Ethics Committee of Wuhan University. Glioma cancer tissues were obtained from Inner Mongolia Autonomous Region People's Hospital, China. Tissues from patients after surgical procedures were sent for pathology diagnosis, and were then collected in TRIzol (Ambion, Austin, TX, USA). Each patient voluntarily participated in the present study. Non-cancer volunteers were patients without glioma, but with other brain disease and needed pathological diagnosis.
Cells and reagents
Human GBM cell line U87 was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) (no. HTB-14™) and was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% of fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) at 37°C in a humidified chamber. Primers for quantitative real-time PCR (qRT-PCR) were synthesized by Invitrogen (Waltham, MA, USA). miR-205 and miR-195 mimics were purchased from Ambion. Enzyme-linked immunosorbent assay (ELISA) kit of human TGF-β1 was obtained from BioLegend (San Diego, CA, USA). Anti-SMAD2 mAb, anti-SMAD3 mAb, anti-SMAD4 mAb and anti-SMAD7 mAb were purchased from Santa Cruz (Santa Cruz, CA, USA). SYBR-Green was purchased from Toyobo (Satte City, Saitama, Japan). CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay kit was obtained from Promega (Madison, WI, USA). TGF-β1 and its inhibitor LY364947 were obtained from Sigma (St. Louis, MO, USA).
Transfection
U87 cell line was used for transfection. Oligonucleotides miR-205 or miR-195 mimics (miR-205 or miR-195 mimics), miRNA mimic negative control (mimic NC), miR-205 inhibitor or miR-195 inhibitor, inhibitor negative control (inhibitor NC), were all purchased from Ambion. Cells (2×105) were cultured in 6-well plates for 24 h, and were then transfected with 100 nM miR-205 and miR-195 mimics, miR-205 and miR-195 inhibitors, or corresponding controls using the jetPEI transfection kit (Polyplus Transfection, New York, NY, USA) according to the manufacturer's instructions. Transfection efficiency was determined by qRT-PCR.
qRT-PCR
Total RNA was extracted as previously described (27). Briefly, RNA was isolated from tissues using TRIzol according to the manufacturer's instructions. For quantification of miRNA, RNA samples were first reverse-transcribed with miRNA-specific primers using TaqMan miRNA reverse transcription kit and qRT-PCR was performed to detect miR-205 and miR-195 using TaqMan microRNA assay kit and StepOne Plus Real-Time PCR system (both from (Applied Biosystems, Grand Island, NY, USA). U6 was used as internal control. The primers were as follows: miR-205 forward, 5′-CTTGTCCTTCATTCCACCGGA-3′ and reverse, 5′-TGCCGCCTGAACTTCACTCC-3′ (28); miR-195 forward, 5-GGGGAGCCAAAAGGGTCATCATCT-3′ and reverse, 5′-GAGGGGCCATCCACAGTCTTCT-3′ (29); U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGA ATTTGCGT (27). Data were analyzed using the 2−ΔΔCt method.
To detect the effects of TGF-β1 on miR-205 and miR-195, U87 cells were cultured in 6-well plates and treated with 0, 1, 2, 4 and 8 ng/ml of TGF-β1 for 12 h. mRNA levels of miR-205 and miR-195 were determined using qRT-PCR.
ELISA
TGF-β1 secreted into peripheral blood was detected using a TGF-β1 ELISA kit (Invitrogen). The serum of glioma patients and non-cancer volunteers were collected. TGF-β1 concentration was assayed with ELISA kit following the manufacturer's instructions. Absorbance detection was performed at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Western blot analysis
Total cellular lysates were prepared with RIPA buffer containing a 2.5% protease inhibitor cocktail, according to the manual. Concentration of protein was quantified by a Bradford assay. Then, 20 µg of the protein samples were separated by SDS-PAGE (10%) and transferred onto polyvinylidene fluoride (PVDF) membrane. SMAD proteins were detected by western blotting using anti-SMAD2 mAb (1:1,000), anti-SMAD3 mAb (1:1,000), anti-SMAD4 mAb (1:2,000) and anti-SMAD7 mAb (1:1,000), respectively. Horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce, Rockford, IL, USA) was used as a secondary antibody. ECL detection systems (UVP, Upland, CA, USA) were used for detection.
Luciferase activity assay
For pGL3-SMAD2-3′UTR and pGL3-SMAD7-3′UTR plasmid construction, the SMAD2-3′UTR containing miR-205 binding site and SMAD7-3′UTR containing miR-195 binding site were amplified and cloned into dual-reporter vector pGL3. The PCR primers were designed according to DNA sequence in Gene Bank. KpnI and XhoI were introduced in the primers for inserting pGL3 vector. 3′UTR of SMAD2 (Gene Bank no. NM_005901.5) were as follows: P1, 5′-ATGCGGTACCAGCTTCACCAATCAAG-3′ and P2, 5′-GGTGCTCGAGATAACTACTACTGTTA-3′; 3′UTR of SMAD7 (Gene Bank no. NM_001190821.1) P1, 5′-AATAGGTACCACCGCGTGCGGAGGGGA-3′ and P2, 5′-GGTACTCGAGATACACTGTGTTTGGT-3′.
The pGL3-SMAD3-3′UTR and pGL3-SMAD4-3′UTR plasmids which contained full length of SMAD3 and SMAD4 3′UTR were purchased from iGene Biotechnology, Co. (Shanghai, China). For miR-205 binding detection, U87 cells were transiently co-transfected with pGL3-SMADs-3′UTR and either miR-205 mimics/inhibitor or negative control. For miR-195 binding detection, U87 cells were transiently co-transfected with pGL3-SMADs-3′UTR and either miR-195 mimics/inhibitor or negative control. pGL3-basic vector was used as a negative control. After 12 h, a dual-luciferase reporter assay system was used to measure luciferase activity according to the manufacturer's instructions (Promega).
Co-immunoprecipitation (Co-IP) assays
Co-IP assays were performed using the Co-IP kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer′s protocol. U87 cells were transfected with miR-205 mimic, miR-205 mimic NC, miR-205 inhibitor, miR-205 inhibitor NC, or miR-195 mimic, miR-195 mimic NC, miR-195 inhibitor and miR-195 inhibitor NC, respectively. Total cellular protein extractions from the cells were immunoprecipitated with anti-SMAD4 mAb. Anti-p-SMAD2 mAb was used as the detecting antibody by immunoblotting. The samples were analyzed using the western blotting procedures as described above.
Colony formation assay
Colony formation assay was used to study the effects of miR-205 and miR-195 on cell proliferation. After miRNA mimics, inhibitors and corresponding control transfection, the cells were seeded at a density of 20 cells/well into a 6-well plate and were cultured for 7 days in DMEM with 10% FBS at 37°C with 5% CO2. Colonies that formed were fixed with 4% of paraformaldehyde for 5 min at room temperature and then stained with 0.1% crystal violet for 30 sec. The number of colonies was counted. All the experiments were repeated in triplicate.
Cell invasion assay
Cell migration and invasion assays were performed in 24-well plates using BioCoat™ chambers with 8-µm pore size (BD Biosciences, San Diego, CA, USA). Forty-eight hours post-transfection, 5×104 of U87 cells in 500 µl of serum-free medium were transferred into the upper chamber that were coated with Matrigel™ matrix for the invasion assays. Complete medium (500 µl) with 10% FBS was used as a chemoattractant in the lower chamber. After 48 h of incubation, the cells in the upper chamber were removed, while the invaded cells were fixed with 4% of paraformaldehyde, stained with 0.1% crystal violet and counted. All the experiments were repeated in triplicate.
Cell viability detection
To investigate the effects of miR-205 and miR-195 on glioma cell proliferation, U87 cells were transfected with miRNAs and controls and kept on culturing for 72 h. Viability of transfected U87 cells was determined using CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay kit according to the manufacturer's instructions. Absorbance at optical density (OD) 490 nm was determined using a Thermo Scientific Multiskan GO full wavelength microplate reader.
TGF-β1 has different roles in different cell growth. To further detect the effect of TGF-β1 on U87 cell proliferation, invasion and viability, TGF-β1 combined with or without its inhibitor LY364947 was used to treat U87 cells, the cell proliferation, invasion and viability were determined according to the previous protocol.
Results
TGF-β1 is negatively correlated with miR-205, but positively correlated with miR-195 in patients with brain glioma
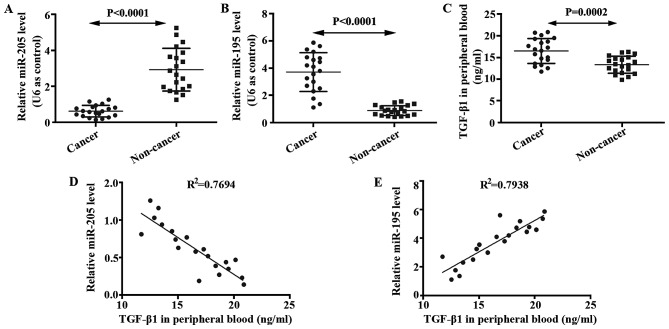
We determined expressions of miR-205 and miR-195 in glioma brain and non-cancer tissues using qRT-PCR. mRNA level of miR-205 in glioma tissue was lower than that in the non-cancer tissue (Fig. 1A). In addition, mRNA level of miR-195 in glioma tissue was higher than that in the non-cancer tissue (Fig. 1B). ELISA was performed to analyze levels of TGF-β1 in the serum of glioma patients and non-cancer volunteers. It was shown that TGF-β1 was enhanced in glioma patients compared with non-cancer volunteers (Fig. 1C). The relationships between miR-205 and TGF-β1, between miR-195 and TGF-β1, were studied by correlation analysis. As shown in Fig. 1D and E, TGF-β1 is negatively correlated with miR-205, but positively correlated with miR-195 in patients with brain glioma.
Figure 1.
Expression of miR-205, miR-195 and TGF-β1 in glioma patients and non-cancer volunteers. (A) Expression of miR-205 in glioma tissue of patients and non-cancer tissue of volunteers was detected by qRT-PCR. (B) Expression of miR-195 in glioma tissue of patients and non-cancer tissue of volunteers was detected by qRT-PCR. (C) Expression of TGF-β1 in serum of patients and non-cancer volunteers was assayed by ELISA. (D) Correlation of miR-205 mRNA levels and TGF-β1 concentration. (E) Correlation of miR-195 mRNA levels and TGF-β1 concentration.
TGF-β1 inhibits miR-205 expression and promotes miR-195 expression in U87 cells
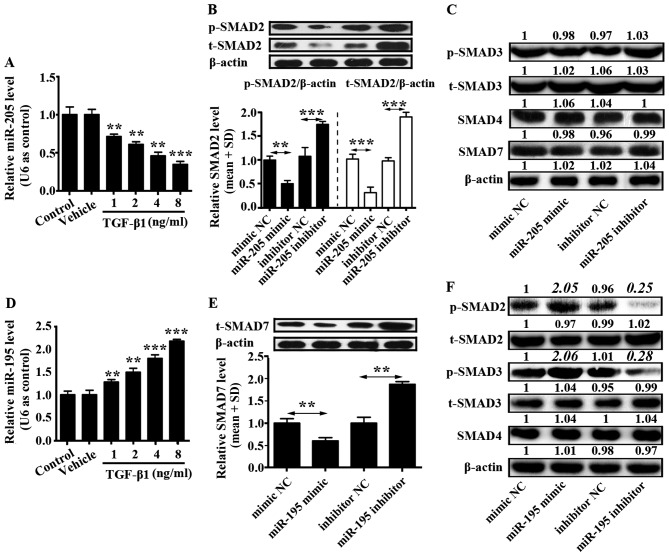
U87 cells (2×105) were seeded into a 6-well plate and cultured in complete DMEM for 12 h. Then, different concentrations of TGF-β1 (1, 2, 4 and 8 ng/ml) were added into U87 cells. Control group was only U87 cells and to vehicle group was added phosphate-buffered saline (PBS) into the U87 cells. The mRNA level of miR-205 was downregulated by TGF-β1 in a concentration-dependent manner, while the mRNA level of miR-195 was upregulated by TGF-β1 in a concentration-dependent manner (Fig. 2A and D). In addition, effects of miR-205 and miR-195 on TGF-β1 signaling pathway were separately examined. U87 cells were transfected with miR-205 and miR-195 mimics, miR-205 and miR-195 inhibitors, or corresponding controls, and western blotting was used to detect the protein level of SMAD2, phosphorylated SMAD2 (p-SMAD2), SMAD3, phosphorylated SMAD3 (p-SMAD3), SMAD4 and SMAD7.
Figure 2.
Relationship between TGF-β1 and miR-205/miR-195. (A) Expression of miR-205 was determined in U87 cells treated with different concentrations of TGF-β1 by qRT-PCR. (B) Expression of SMAD2 was analyzed by western blotting in U87 cells transfected with miR-205 mimic, miR-205 inhibitor or corresponding negative controls. Phosphorylated SMAD2 (p-SMAD2) and total SMAD2 (t-SMAD2) relative protein level was normalized based on β-actin according to the results of western blotting. The black columns represent relative expression of p-SMAD2 and white columns represent relative expression of t-SMAD2. (C) Function of miR-205 mimic, miR-205 inhibitor on SMAD3, 4 and 7 proteins expression in U87 cells. (D) Expression of miR-195 was determined in U87 cells treated with different concentrations of TGF-β1 by qRT-PCR. (E) Expression of SMAD7 was analyzed by western blotting in U87 cells transfected with miR-195 mimic, miR-195 inhibitor or corresponding controls. SMAD7 relative protein level was normalized based on β-actin according to the results of western blotting. (F) Impact of miR-195 mimic, miR-195 inhibitor on SMAD2, 3 and 4 protein expression in U87 cells. All data shown are the means ± SD of three independent experiments; *p<0.05, **p<0.01.
It was demonstrated that miR-205 prohibited SMAD2 and phosphorylation of SMAD2 expression, but did not affect levels of SMAD3, p-SMAD3, SMAD4 and SMAD7 (Fig. 2B and C). miR-205 inhibitor increased expression of SMAD2 and phosphorylation of SMAD2 (Fig. 2B). Otherwise, miR-195 inhibited the expression of SMAD7 which reduced the expression of p-SMAD2 and p-SMAD3. However, there was no influence on the expression of t-SMAD2, t-SMAD3 and SMAD4 (Fig. 2E and F). In addition, miR-195 inhibitor enhanced expression of SMAD7 in U87 cells (Fig. 2E). These findings indicated that TGF-β1 signaling pathway could be activated by miR-195 inhibiting SMAD7 expression, and this pathway could be suppressed by miR-205 inhibiting SMAD2 and phosphorylation of SMAD2 in glioma cells.
SMAD2 is a target of miR-205, and SMAD7 is a target of miR-195 in glioma
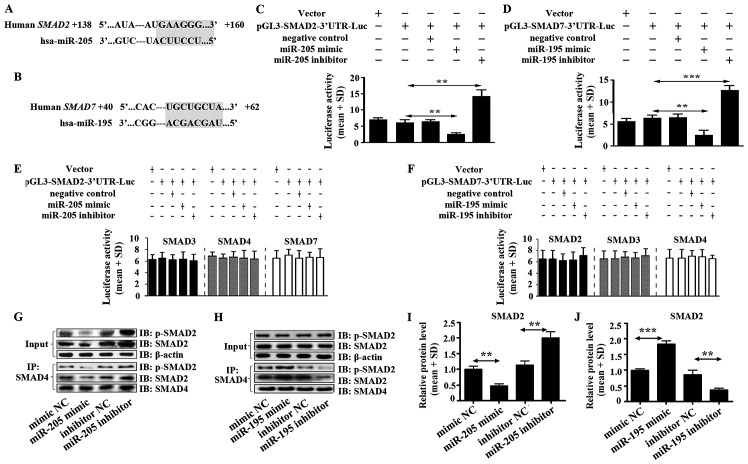
miR-205 and miR-195 were predicted to act with 3′UTR of SMAD2 and 3′UTR of SMAD7, respectively using TargetScan software (Fig. 3A and B). Luciferase activity assay was performed to verify the functions of miR-205 and miR-195 on SMAD2 and SMAD7. We found that miR-205 enormously decreased the luciferase activity of SMAD2 3′UTR in U87 cells (Fig. 3C). In addition, there was no effect on luciferase activity of 3′UTR of SMAD3, SMAD4 and SMAD7 (Fig. 3E). Moreover, high expression of miR-205 significantly inhibited the heteromer formation of SMAD2 and SMAD4 (Fig. 3G and I). This result hinted that miR-205 suppressed TGF-β1 signal pathway in glioma directly by inhibiting the heteromers formation of SMAD2 and SMAD4.
Figure 3.
miR-205 targets SMAD2 and miR-195 targets SMAD7. (A) miR-205 was predicted to target 3′UTR of SMAD2 using TargetScan software. (B) miR-195 was predicted to target 3′UTR of SMAD7 using TargetScan software. (C) The luciferase activity in U87 cells was measured after co-transfection with the indicated SMAD2 3′UTR plasmid and miR-205 mimic, miR-205 inhibitor or corresponding negative control for 12 h. (D) The luciferase activity of U87 cells was measured after co-transfection with the indicated SMAD7 3′UTR plasmid and miR-195 mimic, miR-195 inhibitor or corresponding negative control for 12 h. (E) Effect of miR-205 mimic, miR-205 inhibitor on SMAD3, 4, 7-3′UTR luciferase activity in U87 cells. (F) Influence of miR-195 mimic, miR-195 inhibitor on SMAD2, 3, 4-3′UTR luciferase activity in U87 cells. (G) Heteromer formation of p-SMAD2 and SMAD4 was detected by Co-IP in U87 cells transfected with miR-205 mimic, miR-205 inhibitor or corresponding negative controls. (I) Heteromers of p-SMAD2 and SMAD4 relative expression in miR-205 transfected cells was normalized based on input according to the immunoblotting. (H) Heteromer formation of p-SMAD2 and SMAD4 was detected by Co-IP in U87 cells transfected with miR-195 mimic, miR-195 inhibitor or corresponding negative controls. (J) Heteromers of p-SMAD2 and SMAD4 relative expression in miR-195 transfected cells was normalized based on input according to immunoblotting. All data shown are the means ± SD of three independent experiments; *p<0.05, **p<0.01.
miR-195 inhibited extremely the luciferase activity of SMAD7 3′UTR in U87 cells (Fig. 3D). It was also shown that there was no function of miR-195 on SMAD2, SMAD3 and SMAD4 (Fig. 3F). High expression of miR-195 did not suppress heteromer formation of SMAD2 and SMAD4 but increased heteromer formation (Fig. 3H and J), which suggested that miR-195 could not inhibit TGF-β1 signal pathway but stimulate the pathway by promoting heteromer formation of SMAD2 and SMAD4 in glioma. Mechanism of promoting heteromer formation of SMAD2 and SMAD4 was contributed to miR-195 inhibiting SMAD7 expression (Fig. 2D).
We also evaluated impact of the miR-205 and miR-195 blockade. The luciferase activity assay showed that miR-205 and miR-195 inhibitors enhanced luciferase activity of SMAD2-3′UTR and SMAD7-3′UTR, respectively, in U87 cells. miR-205 inhibitor but not miR-195 inhibitor significantly increased the heteromer formation of SMAD2 and SMAD4 (Fig. 3G–J).
miR-205 inhibits U87 cell proliferation and invasion while miR-195 promotes U87 cell proliferation and invasion
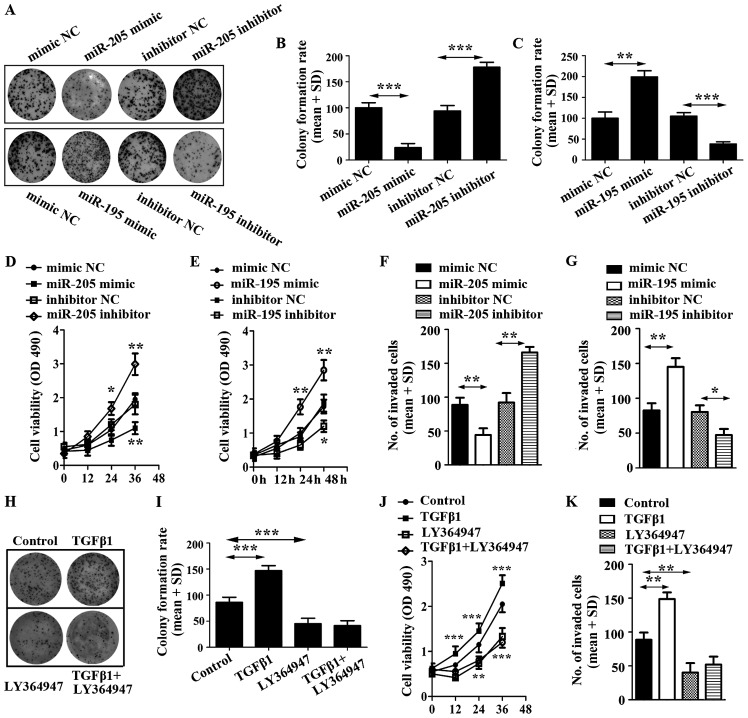
To evaluate the impact of miR-205 and miR-195 on U87 cell proliferation, cell colony and cell viability assays were employed. The results showed that miR-205 decreased U87 cell colonies and viability, but miR-195 increased U87 cell colonies and viability (Fig. 4A–E).
Figure 4.
Effect of miR-205 mimic, miR-205 inhibitor and miR-195 mimic, miR-195 inhibitor on glioma cell proliferation. (A) Proliferation of U87 transfected respectively with miRNA mimic, inhibitor or control was detected and photographed by cell colony formation assay. (B) Relative colony formation rate was calculated in U87 cells transfected with miR-205 mimic, miR-205 inhibitor or corresponding controls. (C) Relative colony formation rate was calculated in U87 cells transfected with miR-195 mimic, miR-195 inhibitor or corresponding controls. (D) Cell viability was detected in U87 cells transfected with miR-205 mimic, miR-205 inhibitor or corresponding controls. (E) Cell viability was detected in U87 cells transfected with miR-195 mimic, miR-195 inhibitor or corresponding controls. (F) Invasive ability of U87 cells transfected with miR-205 mimic, miR-205 inhibitor or corresponding controls was detected by Transwell invasion assay. (G) Invasive ability of U87 cells transfected with miR-195 mimic, miR-195 inhibitor or corresponding controls was detected by Transwell invasion assay. (H) Proliferation of U87 cells treated with TGF-β1 and its inhibitor, LY364947. (I) Relative colony formation rate was calculated in U87 cells treated with TGF-β1 and LY364947. (J) Cell viability was detected in U87 cells stimulated by TGF-β1 and LY364947. (K) Cell invasion was determined in U87 cells stimulated by TGF-β1 and LY364947.
To further explore the potential roles of miR-205 and miR-195 on glioma cells, we still overexpressed miR-205 and miR-195 in U87 cells by transiently transfecting them with miRNAs mimics. We then conducted cell invasion assays, which revealed that miR-205 prohibited the invasive ability of U87 cells (Fig. 4F). In addition, miR-195 displayed a potentiation in the invasive ability of U87 (Fig. 4G).
To corroborate the roles of miR-205 and miR-195 in regulating cell activities, we characterized the effects of miR-205 and miR-195 blockade. We also transfected U87 cells with miR-205 and miR-195 inhibitors, and corresponding controls, respectively. Using cell colony formation assay, cell viability detection and cell invasion assay, it was found that the expression of anti-miR-205 significantly enhanced proliferation and invasion of U87; however, miR-195 inhibitor decreased proliferation and invasion of U87 (Fig. 4)
To identify the effect of TGF-β1 on U87 cells, the cells were stimulated with TGF-β and LY364947. The results suggested that TGF-β1 promoted cell colony formation, increased the viability of U87 cells and enhanced cell invasion. However, LY364947, an inhibitor of TGF-β1, displayed a reverse function on U87 cells and LY364947 efficiently inhibited cell proliferation induced by TGF-β1 (Fig. 4 H–K). The results indicated that inhibitor of TGF-β1 was able to repress the function of miR-195.
Taken together, these observations revealed that miR-205 functioned as a tumor inhibitor and miR-195 was possibly a tumor promoter in U87 cells.
Discussion
Transforming growth factor-β (TGF-β) has been suggested to regulate cellular growth, proliferation and differentiation, and it exhibits a positive or negative function in modulating tumor cells (30). Classical TGF-β signaling is mediated by SMAD proteins. SMAD proteins are divided into three groups: receptor-regulated SMADs including SMAD1, 2, 3, 5 and 8, and common SMAD including SMAD4, inhibitory SMAD including SMAD6 and 7 (31). TGF-β has been demonstrated to be overexpressed in malignant glioma tissues, but undetected in normal brain tissues.
miRNAs have recently been identified to be important regulatory components in TGF-β signal pathway. In the present study, we found that the mRNA level of miR-205 was decreased in glioma patients, and miR-195 and TGF-β1 were both increased in glioma patients. It was revealed that TGF-β1 was negatively correlated with miR-205 mRNA level, but positively correlated with miR-195 mRNA level. In addition, the mRNA level of miR-205 was downregulated by TGF-β1 in a concentration-dependent manner, and the mRNA level of miR-195 was upregulated by TGF-β1 in a concentration-dependent manner.
miRNAs are small non-coding RNAs involved in tumor initiation and progression. Numerous miRNAs can be regulated by TGF-β1 and further influence the downstream of TGF-β signaling. miR-146b is upregulated by TGF-β in a time-dependent manner in intestinal epithelial cells (30). miR-132 is also upregulated by TGF-β in glioma cell line U87 (32). In addition, miR-155, in human fibroblasts, is downregulated by TGF-β. miR-182 can be induced by TGF-β and directly target and suppress the 3′ untranslated regions (3′UTRs) of multiple genes that function as negative regulators of NF-κB, leading to NF-κB hyperactivation and aggressiveness of gliomas (33). Herein, we found that miR-205 and miR-195 were mediated by TGF-β1 in glioma.
Functions of miR-205 in cancers are still vague since it has been found to be upregulated or downregulated targeting different genes (34,35). In various tumors, miR-205 shows an anticancer effect, e.g. prostate cancer, renal cancer and acute lymphoblastic leukemia (36). Otherwise, miR-205 plays a role as an oncogene, such as in nasopharyngeal carcinoma, and cervical cancer (37). In the present study, we found that elevated TGF-β1 induced miR-205 expression in U87 cells. Then, miR-205 targeting SMAD2 protein inhibited heteromer formation of SMAD2 and SMAD4 and reduced angiogenesis and tumor invasion. In contrast, miR-195 was promoted by TGF-β1. miR-195 is related to tumor progression or inhibition. Biological effects of miR-195 remain controversal. miR-195 is reported to increase hepatocellular carcinoma and malignant melanoma (38,39).
In the present study, it was found that miR-195 targeted 3′UTR of SMAD7 and blocked the inhibition of SMAD7. This result is consistent with a study, that miR-195 downregulates the SMAD7 level (40). miR-195 indirectly promoted TGF-β1 signal pathway and enhanced glioma cell invasion and proliferation in U87 cells.
SMAD7 is not only a target gene of TGF-β1 but also target of some inflammatory cytokines, such as IL-1, TNF-α and IFN-γ. It is reported that SMAD7 expression is also induced by EGF, laminar shear stress, UV irradiation and PMA (41). In general, SMAD7 gene expression is controlled by the binding of nuclear active receptor regulated SMAD (R-SMAD) complexes, mainly p-SMAD1-SMAD4 and p-SMAD2-SMAD4 to the SMAD7 promoter region (42). However, in male Sprague-Dawley rats, angiotensin II increased phosphorylation of SMAD2 (p-SMAD2) in aortic protein, but did not affect the level of SMAD7, which suggests that Smad7 expression can be modulated by another mechanism besides Smad2, such as p-SMAD1 (43). In addition, in CCl4-induced liver fibrosis rat model, TGF-β is upregulated and levels of phospho-SMAD2/3 and its nuclear translocation are increased, but SMAD7 is reduced in liver tissue (44). In the present study, miR-205 inhibited SMAD2, which did not influence SMAD7 expression. The result suggested that SMAD7 was probably regulated by another mechanism, than p-SMAD2 binding to SMAD7 promoter in glioma cell line U87.
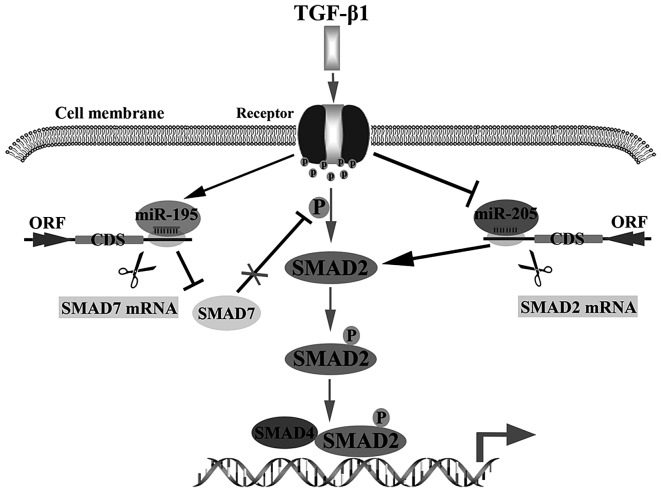
In summary, we concluded that miR-205 could be inhibited by TGF-β1, but miR-195 could be stimulated by TGF-β1. Then, enhancing the activation of TGF-β1 signal pathway via inhibiting miR-205 targeting SMAD2 to reduce degradation of SMAD2 or promoting miR-195 targeting SMAD7 to increase p-SMAD2 in glioma cells (Fig. 5). These findings supply a potential therapeutic target for treating glioma.
Figure 5.
Dual roles of TGF-β1 on glioma development. TGF-β1 induction of miR-195 represses SMAD7 through direct targeting at its 3′UTRs, and TGF-β1 reduction of miR-205 increases SMAD2 via reducing miR-205 targeting SMAD2. The two ways enhance TGF-β1 signal pathway by promoting the heteromer formation of SMAD2 and SMAD4 directly or indirectly.
Abbreviations
- TGF-β
transforming growth factor-β
- UTR
untranslated region
- miRNA
microRNA
- GB
glioblastoma
- FBS
fetal bovine serum
- qRT-PCR
quantitative real-time PCR
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence DA. Transforming growth factor-beta: A general review. Eur Cytokine Netw. 1996;7:363–374. [PubMed] [Google Scholar]
- 3.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-β in cancer. J Pathol. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 4.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/S1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 5.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limón P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8:964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandrow MG, Moses HL. Transforming growth factor beta and cell cycle regulation. Cancer Res. 1995;55:1452–1457. [PubMed] [Google Scholar]
- 8.Wesolowska A, Kwiatkowska A, Slomnicki L, Dembinski M, Master A, Sliwa M, Franciszkiewicz K, Chouaib S, Kaminska B. Microglia-derived TGF-beta as an important regulator of glioblastoma invasion - an inhibition of TGF-beta-dependent effects by shRNA against human TGF-beta type II receptor. Oncogene. 2008;27:918–930. doi: 10.1038/sj.onc.1210683. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop - a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293–35302. doi: 10.1074/jbc.M110.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito A, Suzuki HI, Horie M, Ohshima M, Morishita Y, Abiko Y, Nagase T. An integrated expression profiling reveals target genes of TGF-β and TNF-α possibly mediated by microRNAs in lung cancer cells. PLoS One. 2013;8:e56587. doi: 10.1371/journal.pone.0056587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz LH, Li Y, Chen JS, Muñoz NM, Majumdar A, Chen J, Mishra L. Targeting TGF-β signaling in cancer. Expert Opin Ther Targets. 2013;17:743–760. doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 14.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brower JV, Clark PA, Lyon W, Kuo JS. MicroRNAs in cancer: Glioblastoma and glioblastoma cancer stem cells. Neurochem Int. 2014;77:68–77. doi: 10.1016/j.neuint.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Li L, Li Z, Gong G, Chen P, Liu H, Wang J, Liu Y, Wu X. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol. 2015;137:125–133. doi: 10.1016/j.ygyno.2015.01.531. [DOI] [PubMed] [Google Scholar]
- 19.Cao P, Zhou L, Zhang J, Zheng F, Wang H, Ma D, Tian J. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35:720–728. doi: 10.1002/hed.23011. [DOI] [PubMed] [Google Scholar]
- 20.Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim SW, Kim YT. MicroRNA profiling of a CD133+ spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med Genomics. 2012;5:18. doi: 10.1186/1755-8794-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: Identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7:300–304. doi: 10.4161/rna.7.3.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, Yamamura S, Ueno K, Zaman MS, Singh K, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia LF, Wei SB, Gong K, Gan YH, Yu GY. Prognostic implications of micoRNA miR-195 expression in human tongue squamous cell carcinoma. PLoS One. 2013;8:e56634. doi: 10.1371/journal.pone.0056634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Li Y. Trichostatin A and Tamoxifen inhibit breast cancer cell growth by miR-204 and ERα reducing AKT/mTOR pathway. Biochem Biophys Res Commun. 2015;467:242–247. doi: 10.1016/j.bbrc.2015.09.182. [DOI] [PubMed] [Google Scholar]
- 28.Bai J, Zhu X, Ma J, Wang W. miR-205 regulates A549 cells proliferation by targeting PTEN. Int J Clin Exp Pathol. 2015;8:1175–1183. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Chen H, Fu Y, Ai A, Xue S, Lyu Q, Kuang Y. MiR-195 inhibits proliferation and growth and induces apoptosis of endometrial stromal cells by targeting FKN. Int J Clin Exp Pathol. 2013;6:2824–2834. [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Zhang M, Lönnerdal B. Growth factor TGF-β induces intestinal epithelial cell (IEC-6) differentiation: miR-146b as a regulatory component in the negative feedback loop. Genes Nutr. 2013;8:69–78. doi: 10.1007/s12263-012-0297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gratchev A. TGF-β signalling in tumour associated macrophages. Immunobiology. 2016;S0171–2985:30096–30096. doi: 10.1016/j.imbio.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Wang ZH, Zhang QS, Duan YL, Zhang JL, Li GF, Zheng DL. TGF-β induced miR-132 enhances the activation of TGF-β signaling through inhibiting SMAD7 expression in glioma cells. Biochem Biophys Res Commun. 2015;463:187–192. doi: 10.1016/j.bbrc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C, Wu J, Hu B, Cheng SY, Li M, et al. TGF-β induces miR-182 to sustain NF-κB activation in glioma subsets. J Clin Invest. 2012;122:3563–3578. doi: 10.1172/JCI62339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 35.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 36.Dou L, Li J, Zheng D, Li Y, Gao X, Xu C, Gao L, Wang L, Yu L. MicroRNA-205 downregulates mixed-lineage-AF4 oncogene expression in acute lymphoblastic leukemia. Onco Targets Ther. 2013;6:1153–1160. doi: 10.2147/OTT.S45376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie H, Zhao Y, Caramuta S, Larsson C, Lui WO. miR-205 expression promotes cell proliferation and migration of human cervical cancer cells. PLoS One. 2012;7:e46990. doi: 10.1371/journal.pone.0046990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amer M, Elhefnawi M, El-Ahwany E, Awad AF, Gawad NA, Zada S, Tawab FM. Hsa-miR-195 targets PCMT1 in hepa-tocellular carcinoma that increases tumor life span. Tumour Biol. 2014;35:11301–11309. doi: 10.1007/s13277-014-2445-4. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya A, Schmitz U, Wolkenhauer O, Schönherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene. 2013;32:3175–3183. doi: 10.1038/onc.2012.324. [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Cao S, Liu F, Liu Y. miR-195 plays a role in steroid resistance of ulcerative colitis by targeting Smad7. Biochem J. 2015;471:357–367. doi: 10.1042/BJ20150095. [DOI] [PubMed] [Google Scholar]
- 41.Yan X, Chen YG. Smad7: Not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem J. 2011;434:1–10. doi: 10.1042/BJ20101827. [DOI] [PubMed] [Google Scholar]
- 42.Nicklas D, Saiz L. Computational modelling of Smad-mediated negative feedback and crosstalk in the TGF-β superfamily network. J R Soc Interface. 2013;10:20130363. doi: 10.1098/rsif.2013.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CX, Rhaleb NE, Yang XP, Liao TD, D'Ambrosio MA, Carretero OA. Prevention of aortic fibrosis by N-acetyl-seryl-aspartyl-lysyl-proline in angiotensin II-induced hypertension. Am J Physiol Heart Circ Physiol. 2008;295:H1253–H1261. doi: 10.1152/ajpheart.00481.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang LX, He RH, Yang G, Tan JJ, Zhou L, Meng XM, Huang XR, Lan HY. Asiatic acid inhibits liver fibrosis by blocking TGF-beta/Smad signaling in vivo and in vitro. PLoS One. 2012;7:e31350. doi: 10.1371/journal.pone.0031350. [DOI] [PMC free article] [PubMed] [Google Scholar]