SUMMARY
We combine a genome-scale RNAi screen in mouse epiblast stem cells (EpiSCs) with genetic interaction, protein localization, and “protein-level dependency” studies—a systematic technique that uncovers post-transcriptional regulation—to delineate the network of factors that control the expression of Oct4, a key regulator of pluripotency. Our data signify that there are similarities, but also fundamental differences in Oct4 regulation in EpiSCs versus embryonic stem cells (ESCs). Through multiparametric data analyses, we predict that Tox4 is associating with the Paf1C complex, which maintains cell identity in both cell types, and validate that this protein-protein interaction exists in ESCs and EpiSCs. We also identify numerous knockdowns that increase Oct4 expression in EpiSCs, indicating that, in stark contrast to ESCs, Oct4 is under active repressive control in EpiSCs. These studies provide a framework for better understanding pluripotency and for dissecting the molecular events that govern the transition from the pre-implantation to the post-implantation state.
Graphical abstract
INTRODUCTION
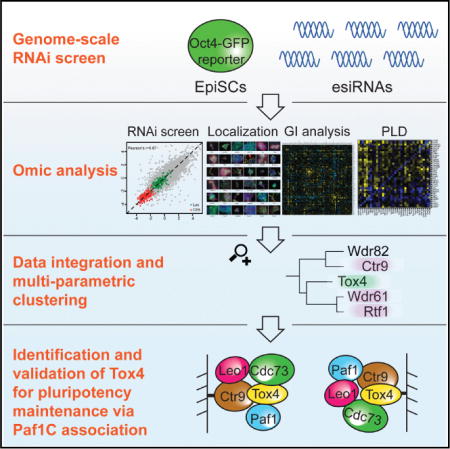
The ability to culture stem cells isolated from early mammalian embryos in vitro, exemplified by mouse embryonic stem cells (ESCs) and the developmentally more advanced epiblast stem cells (EpiSCs), has greatly expanded our knowledge about development and has fueled the field of regenerative medicine (ten Berge et al., 2011). Like ESCs, EpiSCs are pluripotent cells that can give rise to cells of the three germ layers: ectoderm, mesoderm, and endoderm. EpiSCs recapitulate properties of the post-implantation epiblast (Brons et al., 2007; Tesar et al., 2007) and can be propagated indefinitely in vitro. Consequently, they are uniquely positioned to provide insights into the developmental window when somatic and germ cell lineages are first established (Chenoweth et al., 2010). Global gene expression analysis from ESCs and EpiSCs shows that EpiSCs express some of the pluripotency transcription factors (TFs) known to drive self-renewal in ESCs. However, the overall expression pattern of EpiSCs is quite distinct from ESCs (Brons et al., 2007; Tesar et al., 2007). Furthermore, the culture conditions for maintaining the two cell types are different. While ESC self-renewal depends on LIF-Stat3 signaling, EpiSCs require Activin and Fgf signaling for maintaining an undifferentiated state (Brons et al., 2007; Tesar et al., 2007) (Figure 1A).
Figure 1. Genome-Scale RNAi Screen in Mouse EpiSCs.
(A) Illustration of important shared and divergent attributes in ESCs and EpiSCs. Genes for exemplified indicated classes and pathways are shown. The GSK-3 inhibitor (CHIR99021) and MEK inhibitor (PD0325901) that maintain self-renewal of ESCs are indicated.
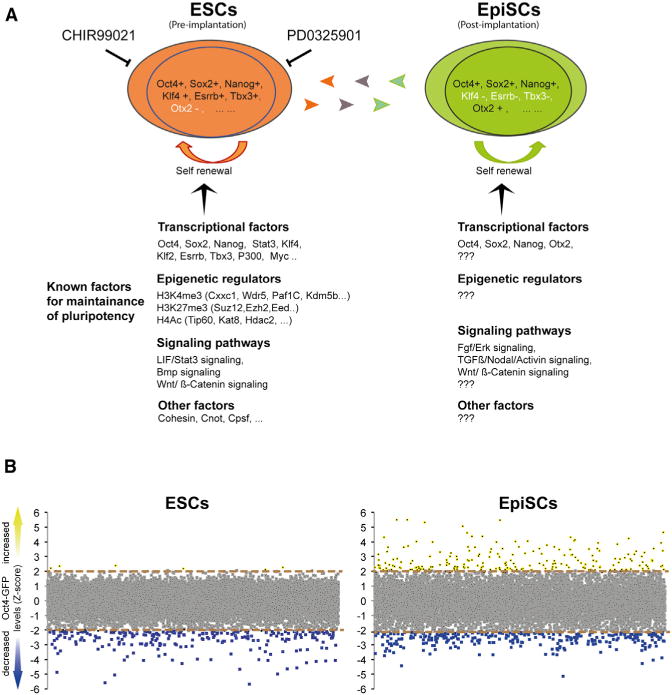
(B) Comparative analysis of the screen results in EpiSCs and in ESCs. The y axis represents the average Z scores for the GFP intensity for each targeted gene. Upregulated (Z score >2) or downregulated (Z score < −2) Oct4 expression is depicted in yellow and blue, respectively. Note the large number of knockdowns that upregulated Oct4 expression in the EpiSCs screen.
ESC pluripotency is regulated by a complex orchestration of TFs, transcriptional coactivators, epigenetic regulators, miRNAs, and extracellular signals. Several TFs, including Oct4 (also called Pou5f1), Nanog, and Sox2, form the core transcriptional circuitry in the maintenance of pluripotent ESCs (Boyer et al., 2005). Oct4, Nanog, and Sox2 are also expressed in EpiSCs. However, many additional TFs that are important for ESC identity and that are vital components of the core circuitry, such as Tbx3 or Esrrb, are not expressed in EpiSCs (Brons et al., 2007; Tesar et al., 2007), signifying the relatedness but also the distinction between EpiSCs and ESCs. Oct4 is critically involved in maintaining pluripotency in both cell types. Accordingly, changing its expression level causes differentiation of EpiSCs (Chenoweth et al., 2010). Hence, Oct4 expression is a valuable reporter for the status of pluripotent cells.
In the past few years, a number of RNA interference (RNAi) screens have been performed in mouse ESCs (reviewed in Ding et al., 2012). Results of these screens have greatly extended our knowledge of ESC biology and have led to the identification of novel components required to maintain ESC identity. Importantly, these screens have also provided new insights into understanding the interplay of multiple pathways of TFs, epigenetic regulators, and cellular signals implicated in ESC pluripotency (Ding et al., 2012).
Much less is known about the genes and pathways that regulate EpiSCs, and comprehensive functional screens in EpiSCs have not been reported. To gain a systematic understanding of the genes associated with EpiSC identity, we performed four large-scale analyses in mouse EpiSCs: a genome-scale RNAi screen, genetic interaction mapping, protein localization, and protein-level dependency (PLD, discussed below). These data define shared and distinguishing factors in naive and primed pluripotent cells and provide insights into the dynamics that accompany the transitions between ESCs and EpiSCs. They also provide one method for translating genome-wide, loss-of-function genetic screens into specific, testable hypotheses about how phenotypes are generated.
RESULTS
Primary RNAi Screen in EpiSCs
Numerous factors and multiple signaling pathways have been recognized to regulate Oct4 expression in ESCs. However, even though it is known that tightly controlled Oct4 levels are also required to maintain EpiSC identity (Chenoweth et al., 2010), much less is known about the factors that influence Oct4 expression in these cells (Hackett and Surani, 2014; Martello and Smith, 2014; Nichols and Smith, 2009) (Figure 1A). To fill this gap, we set out to identify Oct4 regulators in EpiSCs. The mouse EpiSCs line OE7, derived from embryos that express GFP from Oct4 regulatory elements (Ying et al., 2002), was used for the screen. An equivalent Oct4-GFP reporter line (Oct4-GiP) has previously been used to identify Oct4 regulators in ESCs (Ding et al., 2009; Hu et al., 2009). GFP fluorescence intensity reflects the Oct4 levels in individual OE7 EpiSCs and thus can be used as a rapid and accurate readout via fluorescence-activated cell sorting (FACS).
To investigate the feasibility of an RNAi screen in EpiSCs, first we optimized culture conditions and transfection protocols (see Experimental Procedures). To evaluate whether we could accomplish comparable knockdown efficacies in EpiSCs, we transfected OE7 EpiSCs and ESCs with a panel of endoribonuclease prepared (e)siRNAs (Kittler et al., 2007) and measured the knockdown levels by quantitative RT-PCR (RT-qPCR) 24-hr post-transfection. EsiRNAs are enzymatically prepared pools of siRNA-like molecules that have proven efficacy and specificity in ESC and other mammalian cells (Ding et al., 2012; Surendranath et al., 2013). EsiRNAs efficiently and comparably downregulated all tested transcripts in ESCs and EpiSCs (Figure S1A), suggesting that esiRNAs are well suited silencing triggers for RNAi screens in EpiSCs.
Having established a robust esiRNA transfection protocol, we proceeded to execute a similar high-throughput RNAi screen as performed previously in ESCs (Ding et al., 2009). We transfected OE7 EpiSCs in duplicate in 384-well plates and evaluated the reliability of the screen by comparing the scores of each esiRNA. The overall analysis revealed a Pearson correlation of 0.87, suggesting high consistency between the replicates (Figure S1B). To determine thresholds for candidate hit calling, we analyzed all positive (Ctr9 esiRNA) and negative (non-targeting Luc esiRNA) controls and set a Z-score threshold, where 99% Ctr9 esiRNAs scored and 99% of the non-targeting control esiRNAs did not score (Figure S1C).
Screen Comparison between ESCs and EpiSCs
With these criteria, we nominated 467 primary hits that downregulated Oct4 expression in EpiSCs (Figure 1B; Table S1). Many known pluripotency genes that scored in ESC screens scored significantly, including the core circuitry factors Oct4, Nanog, and Sox2 and associated factors such as Tcf3, Sall1, Sall4, and Zfp281, demonstrating the effectiveness of the screen. In addition, multiple components of complexes or genes functioning in the same pathway and that had been identified in ESC screens scored similarly and significantly, including mediator components Med4, Med12, Med14, Med17; Paf1C subunits Ctr9, Rtf1, Wdr61; chromatin remodeling factors Cxxc1, Mll2, Smarcc1, Smarce1, Ep300, Cited1, Jmjd1c, Crebbp; and Wnt pathway component Apc (Table S1). Hence, many genes that are important to sustain Oct4 expression in ESCs are also vital to support Oct4 expression in EpiSCs.
Knockdowns that upregulate Oct4 expression in ESCs were rarely seen among any of the reported RNAi screens (reviewed in Ding et al., 2012), suggesting that repressors for Oct4 expression are rare, or not present in ESCs. In sharp contrast, our EpiSC screen identified 149 esiRNAs that significantly upregulated Oct4 expression (Figure 1B; Table S1). This result likely reflects differences in Oct4 regulation between the two cell types (Yeom et al., 1996) and indicates that Oct4 is under repressive control in EpiSCs.
Validation
To investigate whether GFP fluorescence faithfully reflects Oct4 protein levels, we knocked down five genes (Apc, Med14, Ep300, Wdr82, and Tox4) that downregulated GFP fluorescence and five genes (Brd4, Ctnnb1, Ep400, Smc1a, and Rad21) that upregulated GFP fluorescence in OE7 EpiSCs. At 72 hr post-transfection, Oct4 protein was assessed by quantitative western blot hybridization (Figure S1D). For all ten genes the western blot quantifications of Oct4 were consistent with the FACS-based GFP readouts, suggesting that the GFP measurements faithfully reflect Oct4 expression.
To reduce false positives due to off-target effects, we synthesized 174 independent secondary esiRNAs for the strongest hits (77 for downregulation of Oct4 expression and 97 for upregulation of Oct4 expression). The independent esiRNA transfections validated 53 primary hits (69%) that downregulated Oct4 expression and 54 primary hits (56%) that upregulated Oct4 expression in EpiSCs at high confidence (Figure S1E; Table S2).
Categorization of Validated Genes
To start a more detailed investigation of validated genes, we transfected OE7 EpiSCs and the equivalent Oct4-GFP reporter ESCs (Oct4-Gip; Ying et al., 2002) with the primary and secondary esiRNAs targeting 121 genes (107 validated hits and 14 additional genes from a previous ESCs screen) (Ding et al., 2009) and measured the GFP intensities 72-hr post-transfection in quadruplicates (Table S3). Highly reproducible data were obtained from these experiments with an average correlation coefficient of 0.91 and 0.88 for EpiSCs and ESCs, respectively. Knockdowns were grouped into five categories based on their phenotypes with respect to changes in Oct4 expression in ESCs and EpiSCs (Figure 2A).
Figure 2. Categorization of Validated Oct4 Modulators.
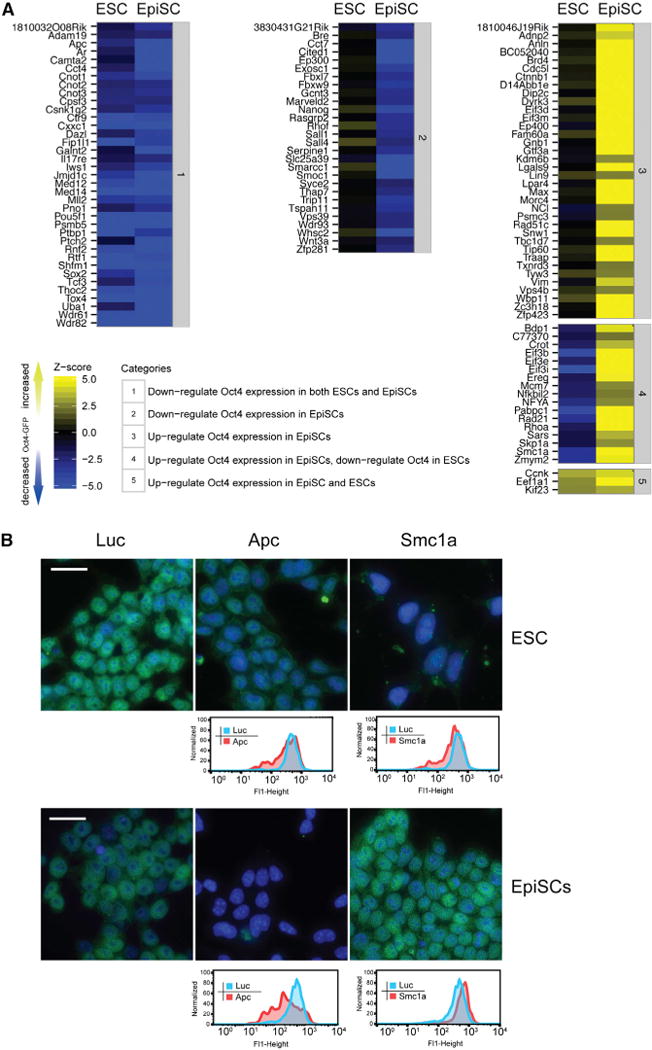
(A) Phenotypic comparison of ESCs and EpiSCs. Indicated categories classify the genes based on their changes of Oct4 expression in ESCs and EpiSCs. Knockdowns that upregulate Oct4 expression are shown in yellow. Knockdowns that downregulate Oct4 expression are shown in blue. The color intensity scales with the intensity of the phenotype.
(B) Examples of knockdowns for different categories. Oct4 expression (green) after transfection with indicated esiRNAs in ESCs and EpiSCs is shown by immunostaining (upper) and quantified by FACS (lower). DAPI was used to stain cell nuclei (blue). Scale bars are 20 μm.
Many genes, including known genes associated with pluripotency, such as Oct4, Sox2, Med12, Med14, Ctr9, Rtf1, Wdr61, Cpsf3, Fip1l1, Cnot1, Cnot2, Cnot3, Tcf3, Cxxc1, Rnf2, and Mll2, clustered in category 1 (Figure 2A), where the knockdown reduced Oct4 levels, signifying that ESCs and EpiSCs share common pathways to maintain pluripotency. However, about 40% of the knockdowns that resulted in reduced Oct4 expression in EpiSCs did not significantly affect its expression in ESCs (Figure 2A, category 2). Hence, these genes represent candidates that are specifically required to maintain Oct4 expression in EpiSCs. Accordingly, this class includes the TFs Sall1, Sall4, and Nanog, all factors that have previously been shown to be dispensable for ESC self-renewal (Chambers et al., 2007; Karantzali et al., 2011; Yuri et al., 2009). Our data indicate that Oct4 expression is more dependent on these factors in EpiSCs.
Fifty-four genes led to an upregulation of Oct4 on knockdown in EpiSCs. Interestingly, components from a recently described Myc-centered network, a module essential to maintain ESC pluripotency that is independent from the Oct4 centered core module, fall into this category (Figure 2A, category 3). Myc, Max, Dmap1, Tip60, Trrap, and Ep400 are tightly interconnected within this protein interaction network and co-occupy a large number of promoters of target genes (Kim et al., 2010). Knockdown of Max, Tip60, Trrap, and Ep400 resulted in marked upregulation of Oct4 expression in EpiSCs, whereas little to no change in Oct4 expression was observed after knockdown in ESCs. These data suggest that, in contrast to the core transcriptional module, which serves as an activator of Oct4 expression in both ESCs and EpiSCs, the Myc module, while important to maintain pluripotency in both cell types (Fazzio et al., 2008; Kim et al., 2010), functions as an Oct4 repressor specifically in EpiSCs.
Possibly the most surprising cluster is category four (Figure 2A). Here, knockdowns led to a reduced expression of Oct4 in ESCs, while knockdowns in EpiSCs increased Oct4 expression. Interestingly, two subunits of the cohesin complex (Rad21 and Smc1a) fall into this category (Figure 2A, category 4, and Figure 2B). Cohesin has a multitude of functions, including a role in transcriptional regulation via chromosome organization (Losada, 2014). Indeed, it was recently shown that cohesin contributes to chromosome organization that supports the pluripotency expression program in ESCs, including the expression of Oct4 (Kagey et al., 2010; Nitzsche et al., 2011). Our data from EpiSCs now suggest that cohesin variably participates in organizing chromatin at the Oct4 locus in ESCs and EpiSCs.
Knockdowns of only three genes showed an upregulation of Oct4 expression in both cell types, reflecting the difference of repressive Oct4 regulation in ESC and EpiSCs (category 5).
Overall, the RNAi screen provides a global view of Oct4 regulators in EpiSCs and allows a comparison to genes influencing Oct4 levels in ESCs.
Generation of an Oct4 Regulator E-MAP
RNAi screens can reveal the function for genes implicated in a particular cellular process, but the interactions between the different genes are usually not identified. Building a genetic epistasis map (E-MAP) by perturbing pairs of genes is a powerful approach for understanding functional relationships between genes, including, but not limited to, physical interactions of their gene products (Roguev et al., 2013). To obtain an E-MAP for 97 identified Oct4 regulators with strong phenotypes in EpiSCs, we dispensed distinct esiRNAs in triplicate each into a 384-well cell culture plate and overlaid these with a single constant esiRNA to produce all pairwise combinations, resulting in 9,409 combinations altogether. We then transfected OE7 EpiSCs with these esiRNA mixtures and quantified GFP signals 72-hr post transfection by FACS (Figure 3A; Table S4). Genetic Interaction scores (GIs) were calculated based on the phenotype strength (GFP intensities) using robust linear fitting to estimate the expected double knockdown phenotypes (Figure S2). GIs were broadly divided into three categories. First, there are synergistic interactions (enhancements) whereby the resulting phenotype is stronger than expected from the phenotypes associated with the single knockdowns. These interactions are typically observed in cases when the two factors act at the same step of a pathway or in parallel (redundant) pathways. Second, there are buffering interactions, where the compound phenotype is weaker than anticipated. These interactions are often observed within linear pathways or between components of protein complexes. Third, there are neutral interactions, where the measured phenotype is close to the expected. Numerous buffering and synergistic GIs were identified (Figure 3A; Table S4), indicating complex connectivity of the factors influencing Oct4 expression in EpiSCs.
Figure 3. Epistasis Map of Oct4 Modulators.
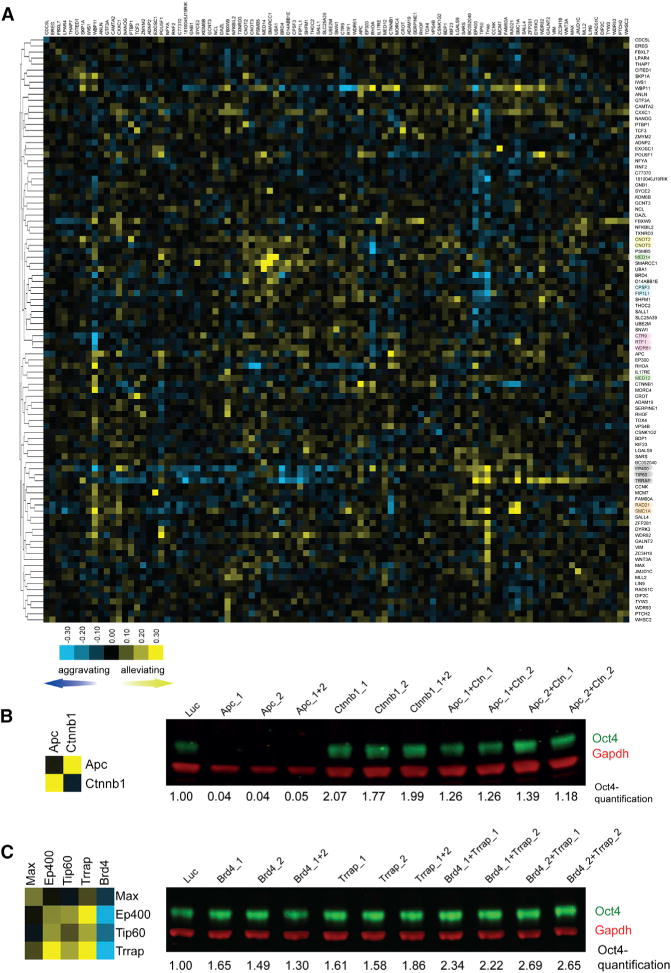
(A) E-MAP profile of Oct4 modulators in EpiSCs. GIs are illustrated as blue squares (negative GIs; synergistic), yellow squares (positive GIs; buffering), and black (neutral GIs). The numbers on the color bar indicate GI scale. Proteins known to form complexes are highlighted with a color code.
(B) Validation of the positive GI between Apc and Ctnnb1 by quantitative western blot. Oct4 protein levels were quantified and normalized to Gapdh after treatment with indicated esiRNA combinations. The non-targeting LUC esiRNA was used as control. The numbers represent fold changes in respect to LUC transfected cells.
(C) Validation of the negative GI between Brd4 and Trrap by quantitative western blot. Oct4 protein levels were quantified and normalized to Gapdh after treatment with indicated esiRNA combinations. The non-targeting LUC esiRNA was used as control. The numbers represent fold changes in respect to LUC transfected cells. Note the synergistic upregulation of Oct4 after co-depletion of Brd4 and Trrap.
To assess reliability of the data, we inspected the clustering of proteins forming core complexes. Proteins from the same complex typically have highly correlated patters of genetic interactions and should therefore cluster together within E-MAPs (Roguev et al., 2013). Indeed, components of known complexes, including members of the Cnot, Cpsf, Paf1, Tip60, and cohesin complexes, tightly clustered together in the E-MAP of Oct4 regulators in EpiSCs (Figure 3A).
Further support for high quality of the data comes from the identification of known GIs. For example, Apc and Ctnnb1 (β-catenin) are both core components of the Wnt signaling pathway. We measured a strong buffering GI score for these two factors (Figure 3B; Table S4), recapitulating the known molecular pathway and genetic interaction of Apc and Ctnnb1 (Bienz, 2002). Double knockdowns with sets of independent esiRNAs for Apc and Ctnnb1 showed that the GI profiles/Oct4 expressions were highly consistent with those from the original esiRNAs (Figure 3B), demonstrating high quality of the data and signifying that the high specificity of esiRNAs (Kittler et al., 2007) minimizes false positives for epistasis experiments (Roguev et al., 2013).
Numerous additional measured GIs have not been described previously and present valuable data to unravel the connectivity of Oct4 modulators. For instance, we observed strong synergistic interactions between bromodomain protein Brd4 and Myc-circuitry components such as Max, Ep400, Tip60, and Trrap (Figure 3C; Table S4). Knockdown of Brd4, Max, Ep400, Tip60, and Trrap alone led to an increase in Oct4 expression, suggesting that these factors act as repressive regulators of Oct4 expression. As seen for other genes encoding proteins forming a complex, the double knockdowns of the Myc-circuitry components among themselves did not further increase expression (e.g., weaker than expected phenotype) and showed a buffering interaction (Figure 3C). In contrast, double knockdowns of Brd4 with Max, Ep400, Tip60, or Trrap led to a further increase (e.g., stronger phenotype) of Oct4 expression (Figure 3C), which manifests itself as a synergistic interaction and points to Brd4 acting independently and in parallel of the Myc circuitry in regulating Oct4 expression in EpiSCs.
Altogether, the epistasis analysis of the selected 97 genes provides a genetic interaction map of Oct4 regulators in EpiSCs. The data should be helpful to decipher connectivity and relations of the genetic network regulating pluripotency.
BAC-TransgeneOmics and PLD
Molecular dissection of biological processes based on RNAi data alone remains difficult. To extend the investigation, we employed BAC transgenes expressing GFP-tagged factors (Hofemeister et al., 2011; Poser et al., 2008). In this approach, candidate genes are GFP tagged on bacterial artificial chromosomes and stably integrated into the genome of mammalian cells in culture. Because the gene remains in its genomic context, BAC tagging provides reliable and reproducible expression of the GFP-tagged protein at levels and patterns matching those of the endogenous counterpart (Poser et al., 2008). From the 97 Oct4 regulators with strong phenotypes, we were able to generate a panel of 36 BAC-tagged ESC and EpiSC lines and employed them for downstream analyses, including subcellular localization (Figure S3).
Technologies that measure global transcript changes after RNAi have been widely employed to understand phenotypes (e.g., Nishiyama et al., 2013). To systematically identify protein level changes after RNAi, we utilized the panel of 36 BAC-tagged EpiSCs and transfected them with an array of 36 corresponding esiRNAs. Based on FACS quantification of the GFP-tagged proteins, protein-level dependencies were calculated from changes in fluorescence intensities compared to control transfected cells (Figure 4A; Table S5). Effective protein depletion was measured for esiRNAs that targeted the corresponding GFP-tagged transcript, validating the efficacy of the employed esiRNAs (Figure 4A; Table S5).
Figure 4. PLD of Oct4 Modulators.
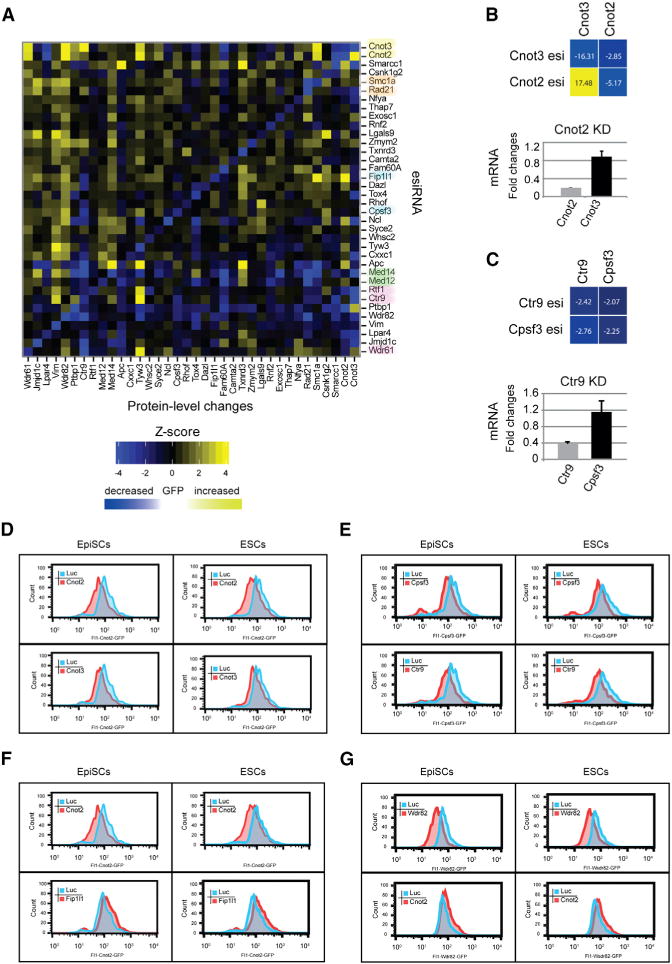
(A) PLD matrix of Oct4 modulators. Protein-level changes are illustrated for each cell line/esiRNA combination with increases marked in yellow and decreases marked in blue. The color intensities scale with the intensity of the changed protein level. Proteins known to form complexes are highlighted with a color code.
(B) PLD between Cnot2 and Cnot3. Depletion of Cnot3 leads to a decrease in Cnot2, whereas depletion of Cnot2 results in an increased in Cnot3 (upper). Quantification of indicated mRNA levels after Cnot2 knockdown are shown in the lower panel. Error bars represent SD from triplicates. Individual Z scores are shown in each square.
(C) PLD between Ctr9 and Cpsf3. Depletion of Ctr9 leads to a decrease in Cpsf3 and vice versa (upper). Quantification of indicated mRNA levels after Ctr9 knockdown is shown in the lower panel. Error bars represent SD from triplicate samples. Individual Z scores are shown in each square.
(D–G) FACS quantification of the expression of BAC-tagged EpiSCs (left) or ESCs (right) after transfection with indicated esiRNAs. The tagged gene is presented in the respective Fl1 channels. The peaks show results after Luc transfection (control; blue) and transfection with indicated esiRNAs (red).
On rare occasions, we observed that protein levels that were not targeted by the esiRNA were altered, indicating that the cells adjust the level of particular proteins, when another protein is present at reduced levels. For 73 cases (5.6%), we observed a significant negative PLD, where the amount of a protein was reduced together with the targeted protein (Figure 4A; Table S5).
We frequently observed reduced protein levels for components of a protein complex when one of the components was knocked down, likely reflecting destabilization of the whole complex when one component was missing (Theis et al., 2009). For 79 cases (6.1%), we detected a significant positive PLD, where the amount of a protein increased when another protein was depleted. However, unlike other protein complexes, Cnot3 protein levels markedly increased on knockdown of Cnot2. Cnot2 and Cnot3 are core components of the Ccr4-Not complex and together with Cnot1 form a tight complex that is important to maintain ESC identity (Zheng et al., 2012). To test whether the increased protein level of Cnot3 after Cnot2 RNAi is due to transcriptional upregulation of Cnot3, we performed RT-qPCR analyses after knockdown of the components. Efficient knockdown was measured for the matching transcripts and esiRNAs, whereas no significant change in transcription was seen for Cnot3 after transfection with esiRNA targeting Cnot2 (Figure 4B). Therefore, the protein level changes cannot be explained by transcriptional changes, indicating that a post-transcriptional mechanism exists that leads to an increase in Cnot3 levels when Cnot2 levels drop.
Many protein dependencies were also discovered for proteins that do not tightly interact with each other. For instance, we observed that the cleavage and polyadenylation specificity factor component Cpsf3 levels decreased significantly when the Paf1C component Ctr9 was depleted (Figure 4C). Again, we checked whether this co-regulation is caused by transcriptional changes and found that transcript levels for Cpsf3 are not significantly altered after Ctr9 RNAi (Figure 4C). Hence, the protein level dependency of Cpsf3 on Ctr9 must be post-transcriptionally regulated.
These data validate the power of the innovative combination between GFP-tagged BAC transgenesis and RNAi, which we term PLD analysis. To investigate whether PLDs in EpiSCs are also observed in ESCs, we selected two negative PLDs (Cnot2-Cnot3, Cpsf3-Ctr9) and two positive PLDs (Cnot2-Fip1l1, Wdr82-Cnot2) and determined the dependencies in ESCs. PLD patterns in ESCs were very similar to those observed in EpiSCs (Figures 4D–4G), suggesting that the adjustment of protein concentrations, at least for the tested cases, is conserved in the two cell types.
Together, our data provide evidence that cells tune the amount of proteins in relation to other proteins and that this tuning is, in the cases examined, controlled by post-transcriptional regulation. Hence, our data represent an additional layer of Oct4 regulation exerted through finely tuned protein levels for Oct4 modulators.
Integration of Omics Data
To integrate the datasets from the RNAi screen, protein localization, E_MAP and PLD analyses, we developed a multiparameter clustering strategy to systematically analyze candidate genes for which all data were available (Figure 5A). We performed hierarchical clustering based on an average of scaled distance matrices calculated for each data source independently, employing a binary distance metric for the localization data and a Euclidean distance metric for all other datasets. A Gower’s coefficient (an average of pairwise distances) was then used to perform hierarchical agglomerative clustering (Kaufman and Rousseeuw, 2009). Components of known complexes closely clustered together in these analyses, suggesting that this composite strategy can be applied to anatomize the relationships of genes and to predict the functions of uncharacterized genes. Integrating multiple datasets is important: inspecting the results from similar calculations where only two datasets were used showed that protein complexes did not cluster as robustly together, although some pairwise combinations appeared to be more reliable than others (Figure S4). Overall, the results indicate that multiparametric clustering is helpful to formulate predictions for proteins that interact and that increasing the number of independent datasets improves the quality of the predictions. Previously we had identified a requirement for Paf1C to maintain Oct4 expression and ESC identity (Ding et al., 2009). Our current screen revealed that Paf1C components are also required for EpiSCs identity (Table S1). The multiparameter clustering analysis placed Tox4 closely together with Paf1C components Ctr9, Rtf1, and Wdr61 (Figure 5A), suggesting that Tox4 could function in a similar fashion. Based on this finding, we selected Tox4 for more detailed functional studies.
Figure 5. Multiparametric Integration of Omics Data.
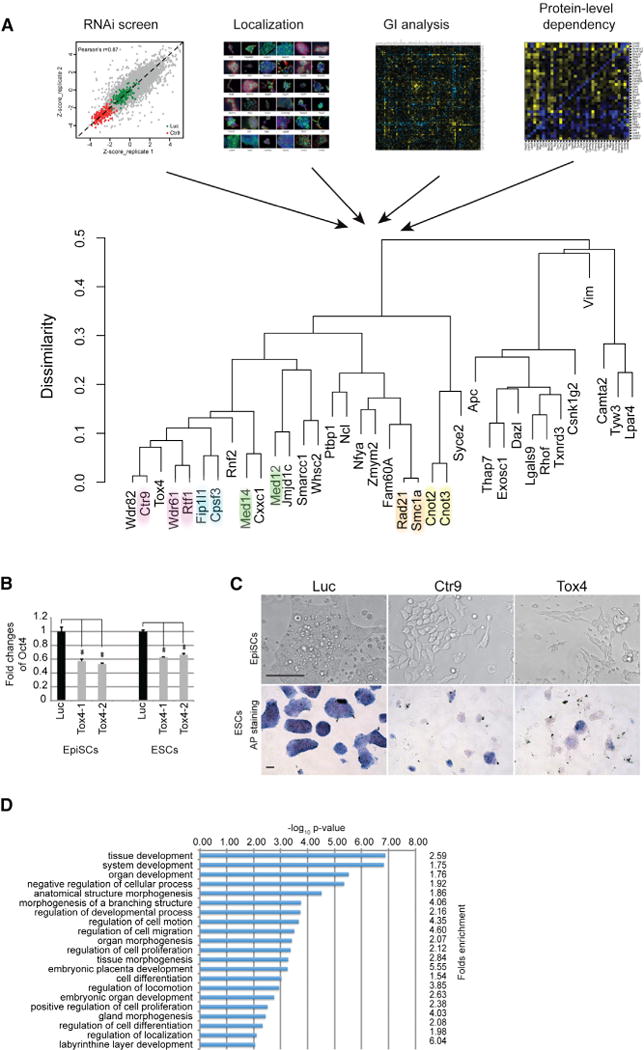
(A) Graphical presentation of hierarchical cluster analysis of indicated Omics data. A binary distance metric was used for the localization data, and a Euclidean distance metric was employed for all other datasets. Components of known protein complexes are highlighted with the same color.
(B) Tox4 depletion decreases Oct4-GFP expression in ESCs and EpiSCs. Results from quantitative FACS analyses using two independent esiRNAs (Tox4-1 and Tox4-2) and the luciferase control esiRNAs (LUC) in Oct4-Gip ESCs and OE7 EpiSCs are shown. Values are means ± SD from triplicate samples. ** indicates significance (p < 0.01) based on the Student’s t test.
(C) Phenotypic analysis of Tox4 knockdown in ESCs and EpiSCs. The upper panel shows brightfield microscopic images of EpiSCs treated with indicated esiRNAs. Note the bigger and flatter cells, loss of cell-to-cell contact, and failure to form tight colonies in the Ctr9- and Tox4-transfected cells. Scale bars are 50 μm. The lower panel shows images of alkaline phosphatase stained ESCs after transfection with indicated esiRNAs.
(D) Tox4 regulates the expression of developmental genes. Gene Ontology term analysis of differentially regulated pathways after Tox4 knockdown is shown. Fold enrichment of genes in each category and p values are presented.
Tox4 Is Required for ESC and EpiSC Identity
Knockdown of Tox4 with two independent esiRNAs resulted in decreased expression of Oct4 in both ESCs and EpiSCs (Figures 5B and S1D). Furthermore, Tox4 and Ctr9 knockdown resulted in colonies with reduced alkaline phosphatase activity, a marker for pluripotency in ESCs and an obvious differentiation phenotype in EpiSCs, which was represented by bigger and flatter cells, loss of cell-to-cell contact and failure to form tight colonies (Figure 5C). Furthermore, transcriptome changes using RNA sequencing (RNA-Seq) after Tox4 knockdown revealed a significant enrichment for genes connected to transcription and development hierarchies, including organogenesis, morphogenesis, pattern specification, neurogenesis, cell differentiation, embryonic development, and cell-fate commitment (Figure 5D; Table S6). Similar results were obtained after Ctr9 knockdown (Ding et al., 2009), consistent with functional overlap of Tox4 and Ctr9. Taken together, these results suggest that Tox4 knockdown promotes differentiation in both cell types, presumably through decreased expression of Oct4.
Tox4 Physically Interacts with Paf1C
To investigate whether functional overlap between Tox4 and Ctr9 is a result of a physical interaction between the two proteins, we employed the BAC-tagged EpiSC lines and WT EpiSCs as controls for co-immunoprecipitation (Co-IP) experiments. After affinity purification with an anti-GFP antibody, prey proteins were eluted and analyzed by western blot hybridization using anti-Gapdh (control), anti-Tox4, and anti-Ctr9 antibodies (Figure 6A). As expected, neither Ctr9 nor Tox4 could be detected in WT EpiSCs IP samples. In contrast, the IP in the Tox4-GFP line prominently pulled down Ctr9 protein, suggesting that these proteins physically interact in EpiSCs. The IP also recovered both tagged and untagged Tox4 protein, indicating that Tox4 is present in a multimeric form. Likewise, Ctr9 and Tox4 were detected in Ctr9-GFP IP samples, confirming physical interaction between Ctr9 and Tox4 in EpiSCs (Figure 6A). To determine whether this interaction can also be detected in ESCs, we repeated the Co-IPs using Tox4 and Ctr9 GFP-tagged ESCs. Results were virtually identical (Figure 6B), suggesting a robust interaction of Tox4 and Ctr9 in pluripotent cells. To further validate the interaction between Tox4 and Ctr9 and to investigate whether Tox4 also interacts with other Paf1C components, we analyzed the Co-IP eluates by mass spectrometry. Consistent with the western blot hybridizations, the proteomic profile of Tox4 IP revealed a prominent interaction with Ctr9. In addition, other core components of the Paf1C, including Paf1, Cdc73, and Leo1, were also readily identified (Figure 6C; Table S7). Furthermore, the Ctr9 IP profile unmasked the canonical Paf1C components Paf1, Leo1, Wdr61, Ctr9, and Cdc73 in addition to Tox4 (Figure 6D; Table S7), demonstrating that Tox4 indeed physically interacts with Paf1C. Together, our proteomic and phenotypic data establishes Tox4 as a new interaction partner of the Paf1 complex in ESCs and EpiSCs that shares functional roles in Oct4 regulation, necessary to maintain ESC and EpiSC identity.
Figure 6. Tox4 Physically Interacts with the Paf1C.
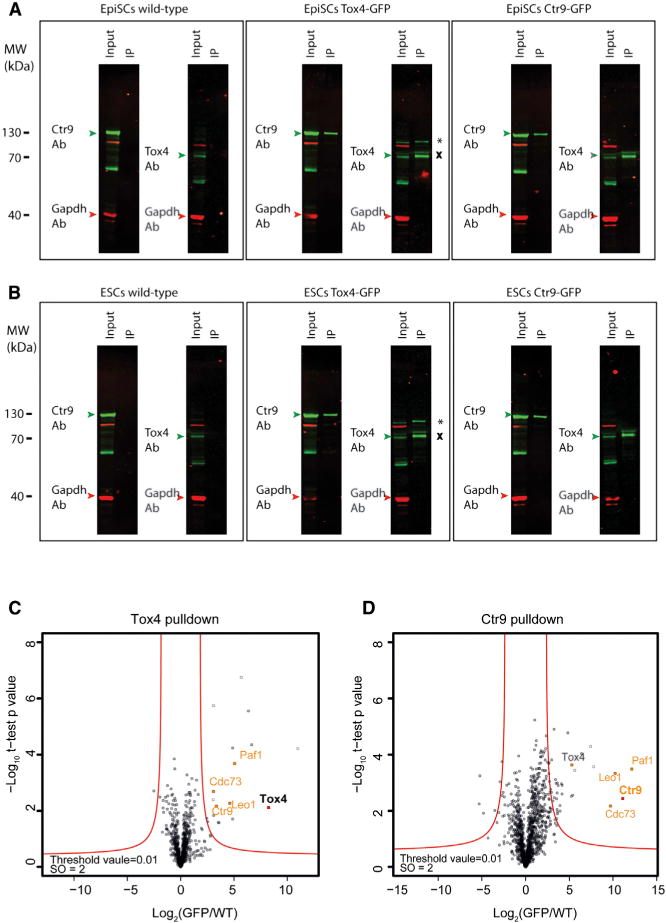
(A and B) Reciprocal Co-IPs of GFP-tagged Ctr9 and Tox4 in EpiSCs (A) and in ESCs (B) are shown. The IP eluates and corresponding input were probed with antibodies against Tox4, Ctr9, and Gapdh. Gapdh served as a reference for Co-IP enrichments. GFP-tagged Tox4 is marked by *, and endogenous Tox4 is marked by×
(C and D) Proteomic profiles of Tox4 and Ctr9 interacting proteins. Results for indicated preys are presented as volcano plots showing (−log10 p value) versus (log2 protein enrichment [GFP/WT]). A hyperbolic curve (red lines, defined by threshold value = 0.01 and S0 = 2) separates specific preyinteracting proteins from background. Significant enrichments of Paf1C core components are presented in orange. Bait proteins are highlighted in bold.
DISCUSSION
While the regulation of pluripotency in ESCs has been studied intensively, the mechanisms that regulate EpiSCs pluripotency are largely unexplored. To close this gap, we present results from a genome-scale RNAi screen for Oct4 regulators in mouse EpiSCs. We observe that knockdown of numerous genes has similar effects on Oct4 expression in ESCs and EpiSCs. However, others have different or even opposite effects, reflecting the divergent regulation of Oct4 in these two cell types. In contrast to ESCs, the Oct4 gene is under repressive control in EpiSCs. This finding argues for a fundamental switch of Oct4 regulation during the transition from pre-implantation to post-implantation. It will be interesting to investigate this switch in the future and relate it to the already known differences in Oct4 enhancer activities in ESCs versus EpiSCs (Tesar et al., 2007; Yeom et al., 1996).
To go beyond a simple description of gene lists, we complement the RNAi screen with localization, genetic interaction, PLD, and proteomic studies and show how meaningful data can be extracted from the various -omics analyses and how testable hypotheses can be formulated from these data. Therefore, our data provide starting points to unravel a variety of mechanisms supporting pluripotency.
The GI investigations should be helpful in resolving how the network of Oct4 modulators governs the pluripotency versus differentiation decision in EpiSCs. For instance, the identified GIs between Brd4 and components of the Myc-module represent an informative epistasis example. Both Brd4 and the Myc-module components Max, Ep400, Tip60, and Trrap are Oct4 repressors in EpiSCs, which synergize to enhance Oct4 expression when depleted together. The Myc-module comprises various members of the NuA4 histone acetyltransferase complex (Kim et al., 2010), possibly implicating histone acetylation for Oct4 repression in EpiSCs. Brd4 is an epigenetic regulator that binds acetylated histones (Dey et al., 2003), providing a possible explanation for the enhanced Oct4 expression. How Brd4 mechanistically synergizes with the Myc-module to repress Oct4 in EpiSCs and how and why this regulation is different in ESCs require further investigation.
PLD mapping reveals unexpected regulation at the protein level. Knowing how the concentrations of key proteins are dependent on the activities of separate genes may provide one key to understanding cellular behavior at a systems level. Protein-level adjustments via transcriptional changes have been studied to some extent, and RNA-Seq is now widely used to investigate transcriptome changes following perturbations (Nishiyama et al., 2013). However, many protein-level adjustments appear to be post-transcriptionally regulated. It will therefore be important to develop appropriate high-throughput methods to investigate processes such as RNA splicing, protein translation, post-translational modifications, and protein degradation to better understand how concentrations of proteins are adjusted in cells. In the meantime, PLD mapping should be helpful to complement other -omics technologies in systems-level analyses of biological processes.
As useful as individual -omics data are to characterize certain biological processes, they often have limited utility when it comes to a deeper understanding of a system. The integration of multidimensional data types is challenging, as is anchoring them in directly testable hypotheses (Cornish and Markowetz, 2014; Flintoft, 2014; Payne, 2012). This study provides one approach to overcoming this hurdle and illuminates new features of pluripotency.
The combined analyses of the employed assays suggested similarity of Tox4 with proteins of the Paf1C. We and others have previously shown that Paf1C has essential roles in maintaining ESC pluripotency and regulating the expression of genes involved in lineage specification, including Oct4 (Ding et al., 2009; Zhang et al., 2013). Our phenotypic and proteomic data indeed classify Tox4 as a new protein required to maintain pluripotency through physical interaction with Paf1C. How exactly Tox4 collaborates with the Paf1C to regulate Oct4 expression and possibly other pluripotency factors at the molecular level will have to be investigated.
The collective analysis of RNAi screening data with protein localization has previously been demonstrated to provide the predictive power to identify new core components of a protein complex (Theis et al., 2009). Our data from this study indicates that this combination is also useful to predict interacting proteins that are not part of the core complex (Figure S4). Hence, the combination of these two datasets seems particularly well suited for multiparametric data analyses, possibly because of the antagonal nature of the two datasets. However, the integration of these data with GI and PLD data improves the prediction (Figure 5A), arguing that complementation of different datasets further enhances the quality of the data. Future studies in this direction should be helpful to extend meta-analysis approaches to various -omics datasets, in particular to refine data mining algorithms by optimizing their ability of detecting true relationships between biological entities.
EXPERIMENTAL PROCEDURES
General Methods
Recombineering, gene targeting, bacterial artificial chromosome (BAC) transgenesis, and Co-IPs using the GFP tag were performed as previously described (Denissov et al., 2014; Poser et al., 2008). Sequences of primers used in this study are provided in Table S8.
Mouse EsiRNA Library
EsiRNAs were synthesized as described previously (Kittler et al., 2005) and normalized to 100 ng/μl in 384-well plates for the genome-scale screen. A complete list of employed esiRNAs is provided in Table S1.
Cell Culture and High-Throughput Screen
ESCs were maintained in Glasgow Minimum Essential Medium (GMEM), supplemented with 10% fetal bovine serum (FBS) and leukemia inhibitory factor (LIF). EpiSCs were cultured in N2B27 medium supplemented with Activin A and Fgf2.
For the genome-scale screen, EpiSCs were reverse-transfected in fibronectin-coated 384-well plates using Lipofectamine 2000 (Invitrogen). GFP fluorescence and cell numbers were measured 72-hr post-transfection using a FACSCalibur (BD biosciences) equipped with a high-throughput sampler (HTS) loader for high-throughput analysis. Full details are provided in the Supplemental Information.
Epistasis Analysis
Pair-wise esiRNA matrix (25 ng esiRNA(A) and 25 ng esiRNA(B) in 10 μl OptiMEM medium) was pipetted onto fibronectin-coated 384-well plates. OE7 EpiSCs were reverse transfected using Lipofectaime 2000 (Invitrogen) in triplicates for each combination. Oct4 expression was quantified 72-hr post-transfection using a FACSCalibur (BD Biosciences) equipped with an HTS loader. Raw data were normalized to the median value of the GFP signal, and the resulting distribution was centered over zero. Phenotypes emerging from individual RNAi knockdowns were defined for each gene as the median of the normalized GFP signal distribution for all gene pairs containing the gene of interest. For each set of double RNAi combinations containing a particular gene, a linear model describing the relationship between the single RNAi phenotypes and the phenotypes observed upon double RNAi knockdown (e.g., D = a*S + b, where D is the phenotype upon double RNAi, S is the computed single RNAi effect, a is the slope of the fit, and b is the y intercept) was computed using a robust linear-fitting algorithm minimizing the effect of the outliers. The y intercepts of these models represent the phenotypes from single RNAi knockdown for each gene and are in very good agreement with the computed single RNAi phenotypes. Genetic interaction scores for each RNAi pairwise combination are defined as the residuals of that fit.
Immunofluorescence
EpiScs and ESCs were fixed with 4% paraformaldehyde in PBS. After permeabilization in 1% Triton X-100/PBS for 30 min, immunostaining was performed using the following primary antibodies: Oct4 (Santa Cruz Biotechnology, Cat# sc-8628, RRID: AB_653551), alpha-Tubulin (Abcam, Cat# ab15246, RRID: AB_301787), and GFP (MPI-CBG, protein facility). Secondary antibodies used were Alexa Fluor 488/594 anti-mouse IgG, anti-rabbit, or anti-goat IgG (Invitrogen). DAPI (Invitrogen) was used for staining the nuclei. Images were taken using a DeltaVision microscopy imaging system (GE Life Sciences).
Co-IP
GFP-tagged protein complexes were isolated by immunoaffinity chromatography using a polyclonal goat anti-GFP antibody that was generated in house as previously described (Poser et al., 2008) and analyzed by mass spectrometry or western hybridization using the following primary antibodies: Ctr9 (Abcam, Cat# ab84487, RRID: AB_1860992), Tox4 (Sigma-Aldrich, Cat# HPA017880, RRID: AB_1845573), and Gapdh (Novus Biologicals, Cat# NB300-221, RRID: AB_10077627). See Supplemental Information for details.
RNA Sequencing
1.5 × 105 EpiSCs cells were reverse transfected with 1,000 ng Luc and Tox4 esiRNAs and 2 μl lipofectamine 2000 in fibronectin-coated 6-well plates. At 72-hr post-transfection, total RNA was isolated using the RNeasy Mini kit (QIAGEN); poly(A) fractions were purified by double selection on oligo(dT) beads, and cDNA was prepared as described previously (Marks et al., 2012).
Sequencing libraries for cDNA samples were prepared using Illumina kits and processed on Illumina HiSeq 2000. Base calling was performed with Illumina CASAVA-1.8.0. RNA-Seq aligner STAR-2.3.0e (Dobin et al., 2013) was used to map sequencing reads to mm10 (Mus musculus, Dec. 2011) genome assembly. Gene expression was calculated as a number of reads per 1 kb of a mappable portion of a gene (evaluated by presence of sequencing reads) and normalized to 1 million reads in a sample Gene ontology (GO) and pathway analyses were performed using DAVID (https://david.abcc.ncifcrf.gov).
Supplementary Material
Highlights.
Oct4, a master pluripotency regulator, is actively repressed in epiblast stem cells
RNAi screen of EpiSCs reveals Oct4 regulators not found in embryonic stem cells
Combining four large-scale techniques predicts that Tox4 interacts with Paf1C
Binding between Tox4 and Paf1C is confirmed in ESCs and EpiSCs
Acknowledgments
We thank Austin Smith for providing the OE7 EpiSC and Oct4-Gip ESC lines. D.C., C.C., A.F.S., and F.B. were supported by the EU FP7 grant SyBoSS (242129). F.B. and L.D. were supported by DFG grant (BU1400/3-1, BU1400/5-1). F.B. was further supported by the DFG grants SFB655 (B5) and the Medical Faculty Carl Gustav Carus. The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (Grant agreement NNF14CC0001). N.K. was supported by grants from NIH (P50 GM081879, R01 GM098101, R01 GM084448, and R01 GM084279) and is a Keck Young Investigator.
Footnotes
ACCESSION NUMBERS
RNA-Seq data can be found on the public server Gene Expression Omnibus (GEO) under the accession number of GEO: GSE62357. Further details are available in Supplemental Information.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2015.08.002.
AUTHOR CONTRIBUTIONS
L.D., M.W., D.C., M.T., G.C., I.P., W.K.C., C.C., M.M., A.F.S., N.K., and F.B. designed and conducted experiments. M.P.-R., A.R., and S.S. performed bioinformatic analysis. L.D., M.P., and F.B. wrote the manuscript.
References
- ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. The subcellular destinations of APC proteins. Nat Rev Mol Cell Biol. 2002;3:328–338. doi: 10.1038/nrm806. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chenoweth JG, McKay RDG, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Cornish AJ, Markowetz F. SANTA: quantifying the functional content of molecular networks. PLoS Comput Biol. 2014;10:e1003808. doi: 10.1371/journal.pcbi.1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Ding L, Poser I, Paszkowski-Rogacz M, Buchholz F. From RNAi screens to molecular function in embryonic stem cells. Stem Cell Rev. 2012;8:32–42. doi: 10.1007/s12015-011-9269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintoft L. Disease genetics: phenome-wide association studies go large. Nat Rev Genet. 2014;15:2. doi: 10.1038/nrg3637. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hofemeister H, Ciotta G, Fu J, Seibert PM, Schulz A, Maresca M, Sarov M, Anastassiadis K, Stewart AF. Recombineering, transfection, Western, IP and ChIP methods for protein tagging via gene targeting or BAC transgenesis. Methods. 2011;53:437–452. doi: 10.1016/j.ymeth.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E, Lekakis V, Ioannou M, Hadjimichael C, Papamatheakis J, Kretsovali A. Sall1 regulates embryonic stem cell differentiation in association with nanog. J Biol Chem. 2011;286:1037–1045. doi: 10.1074/jbc.M110.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis (Google Books) 2009 [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Heninger AK, Franke K, Habermann B, Buchholz F. Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods. 2005;2:779–784. doi: 10.1038/nmeth1005-779. [DOI] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Sharov AA, Piao Y, Amano M, Amano T, Hoang HG, Binder BY, Tapnio R, Bassey U, Malinou JN, et al. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci Rep. 2013;3:1390. doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS ONE. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne PRO. Chapter 1: Biomedical knowledge integration. PLoS Comput Biol. 2012;8:e1002826. doi: 10.1371/journal.pcbi.1002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JRA, Hériché JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendranath V, Theis M, Habermann BH, Buchholz F. Designing efficient and specific endoribonuclease-prepared siRNAs. Methods Mol Biol. 2013;942:193–204. doi: 10.1007/978-1-62703-119-6_11. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger AK, Glatter T, Kruusmaa K, Poser I, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009;28:1453–1465. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hübner K, Schöler HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Yuri S, Fujimura S, Nimura K, Takeda N, Toyooka Y, Fujimura Y, Aburatani H, Ura K, Koseki H, Niwa H, Nishinakamura R. Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells. 2009;27:796–805. doi: 10.1002/stem.14. [DOI] [PubMed] [Google Scholar]
- Zhang K, Haversat JM, Mager J. CTR9/PAF1c regulates molecular lineage identity, histone H3K36 trimethylation and genomic imprinting during preimplantation development. Dev Biol. 2013;383:15–27. doi: 10.1016/j.ydbio.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Dumitru R, Lackford BL, Freudenberg JM, Singh AP, Archer TK, Jothi R, Hu G. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


