Abstract
Proteins of the extracellular matrix often have multiple functions to facilitate complex tasks ranging from signaling to structural support. Here we have focused on the function of one of the matrix proteins expressed in bones and teeth, the matrix adhesion protein ameloblastin (AMBN). Transgenic mice with 5-fold elevated AMBN levels in mandibles suffered from root cementum resorption, delamination, and reduced alveolar bone thickness. AMBN gain of function also resulted in a significant reduction in trabecular bone volume and bone mass dentistry in 42 days postnatal mouse jaws. In an in vitro model of osteoclastogenesis, AMBN modulated osteoclast differentiation from bone marrow derived monocytes (BMMCs), and dramatically increased osteoclast numbers and resorption pits. Furthermore, AMBN more than doubled BMMC adhesion, accelerated cell spreading, and promoted podosome belt and actin ring formation. These effects were associated with elevated ERK1/2 and AKT phosphorylation as well as higher expression of osteoclast activation related genes. Blocking integrin α2β1 and ERK 1/2 pathways alleviated the effects of AMBN on osteoclast differentiation. Together, our data indicate that AMBN increases osteoclast number and differentiation as well as mineralized tissue resorption by regulating cell adhesion and actin cytoskeleton polymerization, initiating integrin-dependent extracellular matrix signaling cascades and enhancing osteoclastogenesis.
Keywords: Osteoclasts, Extracellular matrix, Ameloblastin, Integrin, ERK 1/2
Introduction
Adhesion of cells to extracellular matrices is of fundamental importance for a wide range of cellular functions, including cell differentiation, proliferation, and survival [1]. Among many cell types in which surface adhesion plays a critical role, osteoclasts are arguably some of the most prominent, likely because of the necessity of osteoclast precursors, i.e. circulatory hematopoietic monocytes/macrophages, to attach to specific resorption sites and further differentiate, and because of the proximity of osteoclasts to bone as a requirement for leakage-free delivery of acids and proteases to enable targeted bone resorption [2]. Osteoclast surface attachment is a first and essential step in a cascade of events that continues with osteoclast polarization, formation of podosomes and a sealing zone, and finally secretion of acids and proteases for the resorption of bone. The contact of osteoclasts with the apatitic bone surface activates small GTPases of the Rho family [3,4], which in turn induces osteoclast spreading, podosome formation and the formation of the sealing zone [5-7]. Adhesion of osteoclasts to matrices is aided by integrin receptors (αvβ3, αvβ5, α2β1, and αvβ1) [8], linking osteoclast adhesion with cytoskeletal polarization and activation for bone resorption [9].
From a clinical perspective, changes in osteoclast number or activity have dramatic effects on the life of an organism: Osteoclast hyperactivity results in osteoporosis, while osteoclast defects cause increased bone density in individuals suffering from osteopetrosis. Changes in osteoclast physiology not only affect the entire skeletal system but also the jaws and teeth, where inflammatory or pro-inflammatory cytokines cause root resorption as a result of orthodontic treatment [10,11] or bone resorption and associated tooth loss as a result of periodontal disease [12,13]. The intricate balance between bone resorption and apposition is exemplified during physiological tooth movement where bone resorption at movement-direction alveolar bone walls and bone apposition on the contralateral side plays a role in the continuous maintenance of the attachment apparatus.
The matrix adhesion protein AMBN [14] is one of several extracellular matrix proteins that are significantly upregulated during physiological tooth movement [un-published data] [15,16]. While first discovered in ameloblasts [17], AMBN is also expressed by osteoblasts [18], cementoblasts [19] and Epithelial Rests of Malassez (ERM) in the periodontal ligament [15]. The AMBN protein sequence features a fibronectin interaction site [20] and heparin binding domains [21,22]. In addition, there is a potential α2β1 integrin binding domain and a thrombospondin cell adhesion motif located in rodent ameloblastin proteins [23]. Mutation of AMBN results in detachment of ameloblasts from the developing enamel matrix and a loss of cell polarity in an AMBN null mouse model [14]. The effect of AMBN on cell adhesion has been shown to involve RhoA and cell cycle progression through p27 [24].
To determine the function of AMBN in mineralized tissue homeostasis, we generated an Ambn overexpressing transgenic mouse model and investigated the effect of AMBN on osteoclastogenesis. Based on the importance of surface adhesion on osteoclastogenesis we conducted a series of in vivo and in vitro studies to test whether and how increased AMBN levels would affect osteoclast activity and bone resorption. Our studies shed new light on molecular factors contributing to osteoclastogenesis and explain how the ECM adhesion protein AMBN affects the mineralized state of periodontal tissues.
Materials and methods
Transgene constructs and transgenic mice
The human Keratin 14 (K14) promoter was chosen to drive the Ambn transgene because K14 was expressed in HERS, ERM and bone [25], and the K14 promoter has resulted in substantial overexpression yields in our laboratory [26]. For our studies, two transgenic constructs were generated using a modified pSKII-trans vector in which the K14 promoter, the polyA signal (a generous gift from Dr. Elaine Fuchs, Rockefeller University), the β-Globulin intron, the mouse Ambn coding region, or the LacZ gene were inserted. The β-Globulin intron was used to ensure that the transgenes were properly transcribed. The transgenic fragments were freed from pSKII-K14-Ambn or pSKII-K14-LacZ by digesting the constructs with Sac I and Hind III, gel purified, and microinjected into mouse zygotes [27]. The human K14 promoter-driven Ambn transgenic mice and K14 promoter-driven LacZ transgenic mice were handled in accordance with the UIC Use of Animals in Research Policy.
Genotyping
Genotyping was carried out using tails collected from Ambn or LacZ transgenic heterozygous litters. The tails were lysed in DirectPCR (Tail) buffer (Qiagen, Los Angeles, CA) and PCR amplification was performed using K14 promoter specific primers: 5′GCTTAGCCAGGGTGACAGAG 3′ (forward) and 5′CACAGAGGCGTAAATGCAGA3′ (reverse) [27].
Whole mount X-gal staining and alizarin red staining
For whole mount X-gal staining, mandibles from LacZ transgenic mice at postnatal day 35 were fixed with 4% paraformaldehyde in PBS at 4 °C overnight. The samples were then incubated in the dark with a staining buffer containing 0.05 mM K3Fe(CN)6, 0.05 mM K4Fe(CN)6, 1 mM MgCl2, and 1 mg/ml X-gal at 37 °C for 7 h. For whole mount alizarin red staining, mandibles from wild type (WT) and Ambn transgenic mice at postnatal day 35 were fixed, dehydrated and then stained with saturated alizarin red S (Sigma, St Louis, MO) in 0.5% potassium hydroxide (KOH).
Micro-CT analysis
To visualize mineralized tissues, mandibular tissue blocks were analyzed using microcomputed tomography (micro-CT). For this purpose, 3D X-ray CT images were acquired using a high resolution scanner (Viva CT 40 Scanco Medical AG, Brüttisellen, Switzerland). The micro-CT images were segmented to obtain accurate 3D image data sets.
Scanning electron microscopy
Molars from mandibles of 35-day-old wild-type (WT) and Ambn transgenic (TG) mice were extracted and dehydrated in a series of ethanol, air dried and coated with gold-palladium. Scanning electron micrographs were taken using a JEOL Field Emission SEM (JSM-6320F).
Tissue processing
Mandibles from WT, Ambn or LacZ transgenic mice were dissected and fixed with 10% formalin at 4 °C. For un-decalcified ground sections, tissues were dehydrated, embedded in Technovit 7200 (Exakt Inc., Oklahoma, OK) and prepared into 10 μm sections for subsequent von Kossa staining. For decalcified paraffin sections, mandibles were de-mineralized in EDTA, and processed for paraffin sections. Sections were subjected to H & E staining, Villanueva staining, TRAP staining, in situ hybridization or immunohistochemistry.
In situ hybridization
In situ hybridization analysis was performed using a Digoxingenin (DIG)-labeled probe. Briefly, deparaffinized and rehydrated sections were treated with Proteinase K and then hybridized with a hybridization solution containing DIG-labeled AMBN antisense or sense RNA probe at 65 °C for 16 h. Sections were then washed at high stringency, blocked and incubated with anti-DIG-Alkaline Phosphatase antibody (Roche, Mannheim, Germany). The localization of AMBN mRNA was revealed using the NBT/BCIP substrate.
Immunohistochemistry
Sections were deparaffinized, rehydrated and treated with 6% peroxide and methanol followed by a brief incubation in 10 mM sodium citrate buffer with 0.05% Tween 20 at pH 6.0 for antigen retrieval. After blocking, sections were first incubated with affinity purified anti-AMBN antibody at a dilution of 1:200, and then with anti-rabbit secondary antibody (Abcam, Cambridge, MA) at a dilution of 1:2000. Protein expression was detected with a Histomouse Broad Spectrum AEC kit (Invitrogen, Carlsbad, CA) under a light microscope. To verify AMBN and K14 co-localization, fluorescent immunohistochemistry was performed using rabbit anti-AMBN or mouse anti-K14 antibodies (Abcam). The samples were incubated overnight at 40 °C with primary antibodies and then with secondary FITC-conjugated anti-rabbit IgG antibody or Texas Red-conjugated anti-mouse antibody (Invitrogen). Immunoreactivity was observed under a fluorescence microscope. As a negative control, non-immune rabbit serum was used instead of the primary antibody.
Von Kossa staining
Ground sections were stained with 5% silver nitrate for 1 h under UV exposure and then de-stained with 5% sodium thiosulfate.
TRAP staining
Osteoclasts were visualized using a tartrate resistant acid phosphatase (TRAP) staining procedure. For this procedure, either fixed cells or de-paraffinized sections were incubated in acetate buffered solution containing naphthol AS-MX phosphate, Fast Garnet GBC salt, and tartrate solution (0.67 mol/l) (Sigma, St. Louis/MO) for 60 min, and then counterstained with hematoxylin.
AMBN protein expression and purification
The mouse Ambn coding region was amplified by PCR with a 5′ Nde1 site and a 3′ BamH1 site. The PCR products were inserted in the pET-28 expression vector (Novagen, Madison, WI) which was transformed into BE21 cells. The expression of recombinant mouse AMBN (rmAMBN) was induced with IPTG at a concentration of 1mMol at 32 °C for 4 h. Protein purification was performed used Ni-NTA agarose (Qiagen).
Osteoclast differentiation induction and blocking
Bone marrow cells were isolated from 5-week old CD1 mice by flushing femurs and tibias with α-MEM and cultured in α-MEM with 10% FBS, antibiotics and M-CSF (10 ng/ml, R&D systems, Minneapolis, MN) for 1 day. Non-adherent cells were collected and further cultured for 4 days in the presence of M-CSF (10 ng/ml). Adherent cells were used as bone marrow monocytes/macrophages (BMMCs). To induce osteoclast differentiation, BMMCs were cultured with M-CSF (10 ng/ml) and RANKL (50 ng/ml, R&D Systems) for 5 days. The differentiated BMMCs were subjected to TRAP staining, actin ring formation and bone resorption analyses. To block osteoclast differentiation, an anti-α2β1 integrin antibody (10 μg/ml, Millipore, Billerica, MA) was used to neutralize the integrin receptors. In addition, PI3K inhibitor LY294002 (10 μM, Sigma) and the ERK1/2 inhibitor UO126 (10 μM, Sigma) were applied to inhibit PI3K and MER/ERK pathways.
Cell attachment
Adhesion assays were performed using 35 mm untreated culture dishes (Corning, NY). The dishes were coated with recombinant mouse AMBN (rmAMBN), BSA, or recombinant mouse amelogenin (rmAMEL) proteins at concentrations of 5 or 10 μg/ml at 4 °C overnight, and blocked with 2% denatured BSA at room temperature for 1 h. After washing, 106 of BMMCs were seeded into each dish and incubated at 37 °C for 4 h. Nonadherent cells were removed by washing with PBS and the remaining cells were counted under a microscope.
F-actin staining
BMMCs were placed in rmAMBN coated 35 mm dishes. To elucidate podosome formation, cells were cultured for 12 h. To induce actin ring formation, cells were cultured for 5 days in the presence of M-CSF (10 mg/ml) and RANKL (50 mg/ml). The cultured cells were fixed with 3.7% formalin, permeabilized with 0.1% Triton-100, and then stained with rhodamine — or FITC-conjugated phalloidin (Sigma). Flourescent images were captured under a Leica DMRX fluorescent microscope.
Bone resorption assay
BioCoat Osteologic MultiTest Slides (BD Biosciences, San Jose, CA) were coated with rmAMBN (10 μg/ml). 2×103 of BMMCs were seeded into each well and cultured for 5 days with M-CSF (10 ng/ml) and RANKL (50 ng/ml). The cells were removed with 6% NaOCl and each slice was washed with distilled water, air dried and examined for resorption pits.
Protein extraction and Western blot analysis
Mandibular molar root/bone tissues from the transgenic and control animals at postnatal days 3, 6, 10, 15 and 20 or cultured cells were collected. Equal amounts of protein extracts in a lysis buffer containing 100 mM Tris HCl pH 9.0, 200 mM KCl, 25 mM EGTA, 36 mM MgCl2,2% deoxycholic acid and 10% DTT v/v were subjected to SDS–polyacrylamide gel electrophoresis, and the separated proteins were transferred to a PVDF membrane (Immobilon P®, Millipore). The membrane was incubated with anti-RhoA, pho-AKT, pan AKT, pho-ERK, ERK, β-actin, β-tubulin, NFATc1 or c-Fos primary antibodies (Abcam). Immune complexes were detected with peroxidase-conjugated secondary antibody (Molecular Probes®, Carlsbad, CA) and enhanced by chemiluminescence reagents (Pierce Biotechnology, Rockford, IL). The amounts of protein expression were compared after normalization against β-actin or β-tubulin as an internal calibrator in each lane.
Statistical analysis
Quantitative data were presented as means±SD from three independent experiments and compared with one way ANOVA statistical analysis tests. The difference between groups was considered statistically significant at P<0.05.
Results
AMBN is expressed in Hertwig’s Epithelial Root Sheet (HERS) and in the periodontal ligament of mouse molar teeth
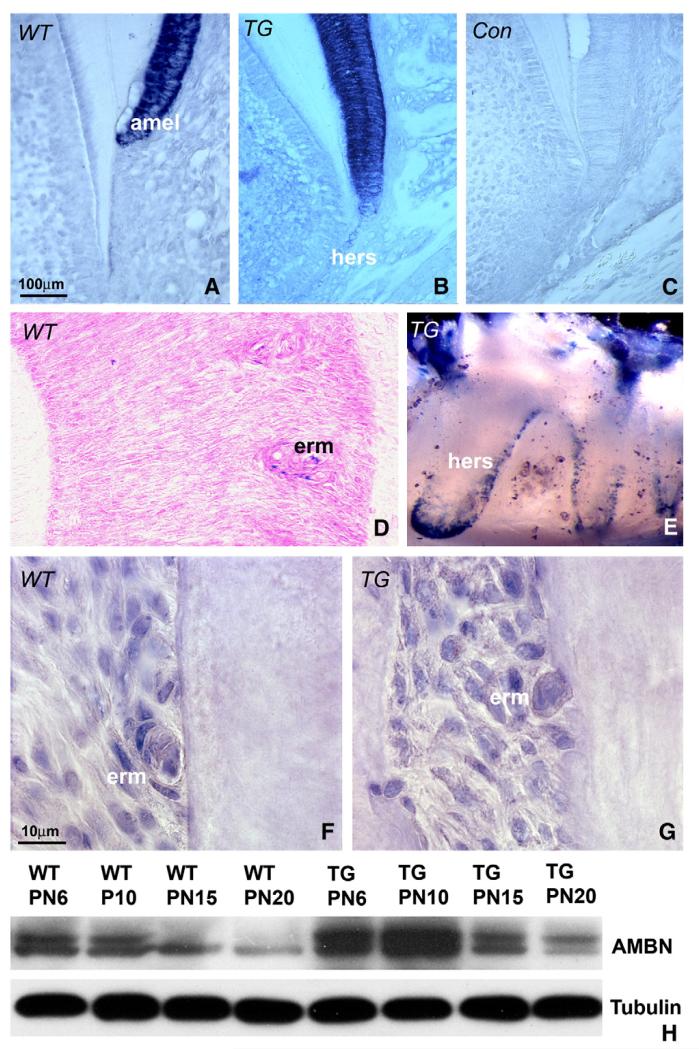
Our interest in AMBN in the periodontium was peaked by a dramatic increase in AMBN expression in the Epithelial Rests of Malassaz (ERM) during tooth movement or after mechanical injury of cementum [15,16]. As a first step to understand AMBN function in the periodontal ligament, the temporo-spatial expression pattern of AMBN was mapped during tooth development. Using in situ hybridization analysis, AMBN mRNA was prominently localized in ameloblasts and HERS (Fig. 1A) as well as ERM (Fig. 1D). ERM were identified as islands of cuboidal cells aligned parallel to the root surface in the midst of elongated periodontal ligament fibroblasts. Lower levels of mRNA were also detected in odontoblasts and dental pulp on postnatal day 12 (Fig. 1). AMBN mRNA was substantially enhanced in Ambn transgenic (TG) mice (Fig. 1B). No signal was detected in the control, in which AMBN sense RNA was used as a probe (Fig. 1C).
Fig. 1.
AMBN expression in postnatal developing molar teeth. (A-D) AMBN mRNA in situ hybridization analysis in mouse molars. Paraffin sections of 12 day postnatal wild-type (A) transgenic (B), and 35 day postnatal WT (D) mouse molars were hybridized against mouse antisense AMBN coding region, and the sense AMBN served as a control (C). (E) X-gal staining of oral epithelial cells and ERM/HERS (blue color) in LacZ transgenic mice. (F and G) Immunohistochemistry analysis of AMBN protein expression in PN 35 mouse molars. Immunoreactivity for the AMBN protein was visible as brown staining in the periodontal ligament and at higher levels in ERM. (H) Changes in AMBN expression in transgenic (TG) and wild-type (WT) molars as revealed by Western blot. Mouse molars from postnatal day 6 (PN6), day 10 (PN10), day 15 (PN15) and day 20 (PN20) were used for analysis. Molecular weights of the expressed AMBN proteins are 55 and 50 kDa. β-Tubulin was applied as a protein loading control. amel = ameloblasts, hers = Hertwig’s Epithelial Root Sheath, erm = epithelial rests of Malassez.
As a second step in our analysis of AMBN expression and function in periodontal tissues AMBN overexpressing transgenic mice driven by the human K14 promoter [27] were generated. To verify overexpression of the K14-driven transgene in HERS and ERM, a K14-LacZ transgenic mouse model was also created. Whole mount X-gal staining demonstrated the localization of the transgene in HERS and ERM cells (Fig. 1E). Immunohistochemistry analysis revealed distinct staining for AMBN protein in ERM cells and surrounding periodontal ligament of postnatal day 35 WT and TG mice, and ERM staining intensity was higher in TG mice than in WT counterparts (Figs. 1F and G).
Our immunohistochemical data indicated that AMBN protein expression in ERM cells was higher than in surrounding periodontal ligament cells, especially in AMBN overexpressing mice. To quantify the expression of AMBN in developing mouse teeth, protein was extracted from molars and surrounding tissues at subsequent stages of periodontal development (postnatal days 6–20). Western blot analysis revealed two bands corresponding to the AMBN protein, one with a higher molecular weight at 55 kDa and a second with a lower molecular weight at 50 kDa (Fig. 1H). The expression of AMBN gradually decreased from 6 dpn to 20 dpn in WT and TG mice (Fig. 1H). Furthermore, densitometry analysis on Western blots revealed that on postnatal day 10, AMBN was five times higher expressed in AMBN overexpressing mice than in WT controls.
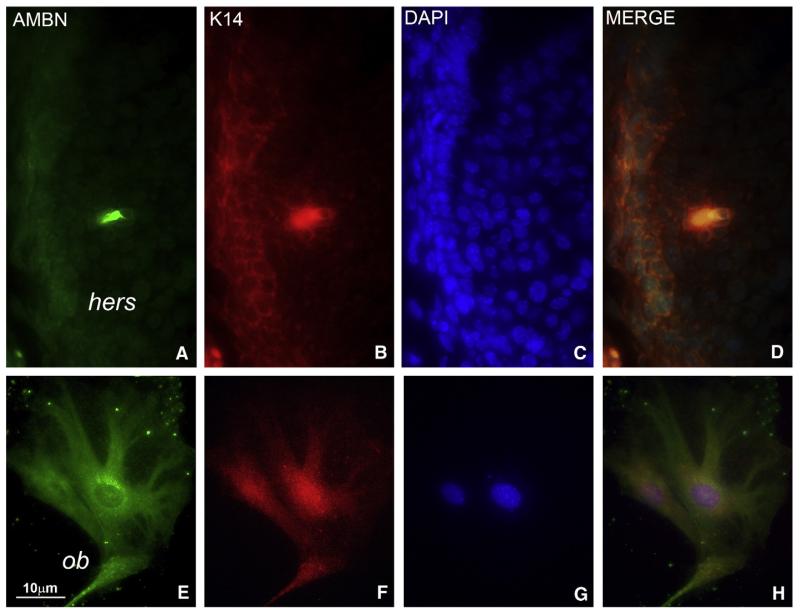
Immunofluorescence-based comparison of AMBN and K14 localization in 10 days postnatal developing tooth roots demonstrated co-localization of both proteins in HERS cells (Figs. 2A–D). There were strings of small patches of overlapping AMBN/K14 labeling along the HERS cell membrane (Fig. 2D). Cultured alveolar bone osteoblasts stained for both AMBN and K14 (Figs. 2E-H). There was an intense intracellular AMBN labeling in the periphery of the nuclear envelope (Fig. 2E).
Fig. 2.
Co-localization of AMBN and Keratin 14 in Hertwig’s Epithelial Root Sheath and in alveolar bone osteoblasts. (A-D) AMBN (A), Keratin 14 (B), and DAPI (C) triple fluorescent staining in Hertwig’s Epithelial Root Sheath (hers). (D) is an overlay image. Hertwig’s root sheath is the bilayered epithelium on the left side of the image. Green fluorescence for AMBN and red fluorescence for K14 were detected in HERS cells (A, B). Note the strong yellow label in patches along the HERS cell membrane indicating presence of both AMBN and K14 (D). (E-H) AMBN (E), Keratin 14 (F), and DAPI (G) triple fluorescent staining in alveolar bone osteoblasts (ob). (H) is an overlay image. Alveolar bone osteoblasts were stained for both AMBN (green label, E) and K14 (red label, F). Note the intense reaction for AMBN in the periphery of the nuclear envelope (E). hers = Hertwig’s Epithelial Root Sheath, ob = alveolar bone osteoblasts.
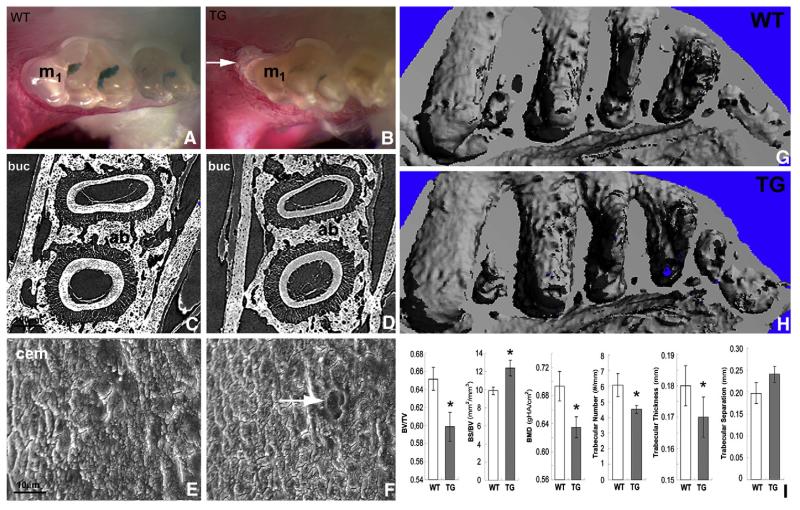
Ambn overexpression induces alveolar bone loss and tooth root resorption
Enamel of Ambn transgenic mice featured a dull surface and yellowish color (Fig. 3B), indicating that our Ambn overexpression model was associated with an enamel defect. Whole mount alizarin red staining revealed that the Ambn overexpressor suffered from significant bone loss at the crest of the alveolar bone as well as an increase in the periodontal ligament thickness (Fig. 3B). Alveolar bone loss was most prominent at the crest of the first root of the first molar (Fig. 3B). Micro-CT analysis revealed a reduction in alveolar bone wall thickness when comparing TG to WT counterparts (Fig. 3D versus Fig. 3C). Scanning electron microscopy revealed a smooth root surface in the control molars (Fig. 3E) compared to a relatively rougher surface with root resorption lacunae on the Ambn over-expressing molar roots (Fig. 3F). Micro-CT 3D-images of the molar-region dentoalveolar complex of wild-type (G) and AMBN transgenic mice (H) revealed widened alveolar crypts, roughened alveolar bone surfaces, and reduced interradicular spaces in AMBN overexpressors (Figs. 3H versus G). Morphometric comparison demonstrated significantly reduced trabecular bone volume (BV/TV), bone mass density (BMD), trabecular number, and trabecular thickness (p<0.05) in AMBN transgenic versus WT mice (Fig. 3I). The approximately 10% reduction in trabecular bone volume and bone mass dentistry as a result of AMBN gain of function was a sizable and significant (p<0.05) effect. In contrast, bone surface/volume ratio (BS/BV) increased slightly and significantly (p<0.05) in AMBN overexpressors, while trabecular separation increased as well, but the difference was not significant (Fig. 3I).
Fig. 3.
Bone loss and root resorption in Ambn over-expressor molars. (A, B) Whole-mount Alizerin red staining of mandibles from Ambn transgenic (TG) and wild-type (WT) mice at 35 days postnatal. First molars are marked M1. Note the significant bone loss at the crest of the alveolar bone (arrow) as well as the increase in periodontal ligament width in AMBN transgenic mice as compared to wild-type controls. (C, D) Micro-CT comparison of root sections of WT and TG mice. (E, F) Scanning electron microscopy images of molar teeth of 35 days old WT and Ambn transgenic mice. The arrow indicates a resorption pit on the TG molar root. (G, H) 3D μCT image reconstruction of the molar-region dentoalveolar complex of wild-type (G) and AMBN transgenic mice at age 42 days postnatal (H). Note the widened alveolar crypts, roughened alveolar bone surfaces, and reduced interradicular spaces in AMBN overexpressors (H versus G). Morphometric comparison between AMBN transgenic and WT mice. The following parameters were compared (3 samples per group, from left to right): trabecular bone volume fraction (BV/TV), surface/volume ratio (BS/BV), bone mass density (BMD), trabecular number, trabecular thickness, and trabecular separation. All but the trabecular separation were significantly different (p<0.05) between both groups. *=p<0.05.
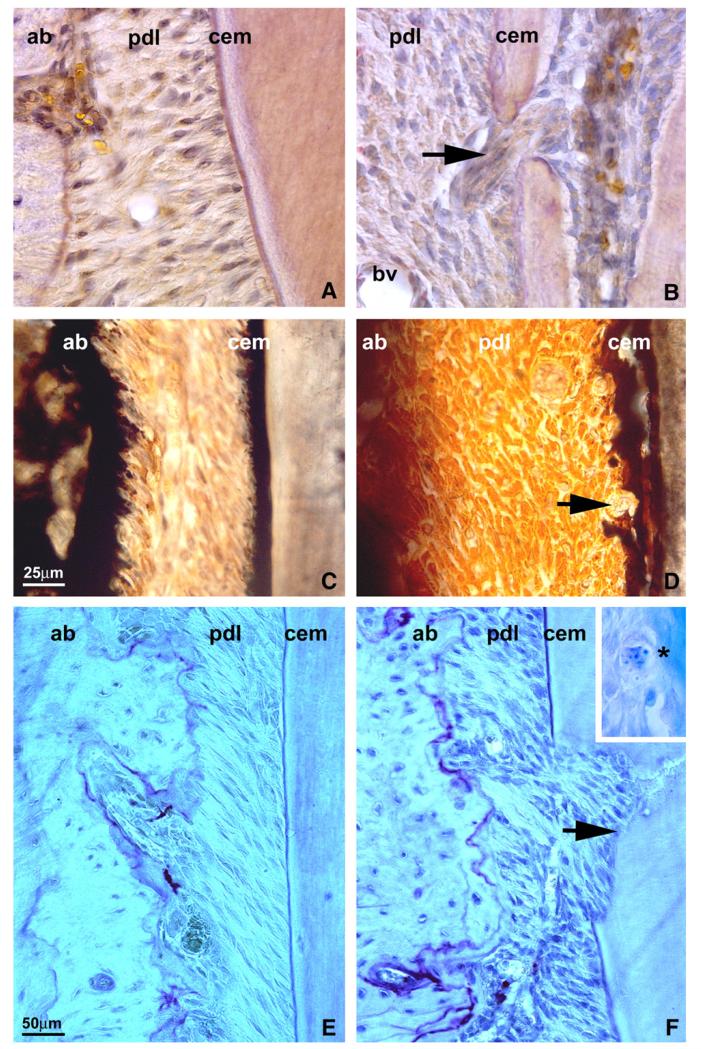
Ambn overexpression is associated with increased osteoclast/cementoclast activity
Villanueva’s Osteochrome staining of ultrathin ground sections demonstrated an acellular cementum layer featuring a homogeneous structural integration with the underlying root dentin in controls (Fig. 4A). A number of layers displayed metachromatic colorations, and transitions occurred gradually between each layer (Fig. 4A). In Ambn over-expressing transgenic mice, the junction between acellular cementum and dentin was disrupted (Fig. 4B). There was an open channel between periodontal ligament and dental pulp with a group of periodontal fibroblasts crossing in between (Fig. 4B). Von Kossa staining of WT mouse molars at PN 35 revealed a dark brown and intact acellular cementum surface (Fig. 4C), while acellular cementum was de-laminated from the root surface in Ambn over-expressing mice (Fig. 4D). In addition, a few giant cells were attached on the surface of the acellular cementum (Fig. 4D). In tandem with increased root resorption, TRAP staining of molar root sections indicated higher osteoclast activity in the periodontal ligament of Ambn overexpressors (Fig. 4F) than in that of WT mice (Fig. 4E). Resorption areas in PN 35 Ambn overexpressors were explained by multiple cementoclasts present on the surfaces of PN 15 Ambn overexpressor molar roots (Fig. 4F, insert), and cementoclasts were absent on WT PN 15 controls (not shown).
Fig. 4.
Changes in osteoclast activity in the periodontal ligament of Ambn overexpressor mice. (A, B) Histological sections of molars from WT (A) and Ambn overexpressor (B) postnatal day 35 mice. The arrow points to a perforated mineralized tissue layer in Ambn TG mice that was sufficiently disrupted to allow periodontal fiber insertions between dental pulp and ligament. (C-D) von Kossa staining of mouse molar roots sectioned through the acellular cementum region of the cervical roots of WT (C) and Ambn overexpressor (D) mice on postnatal day 35. There were de-laminations of acellular cementum in Ambn overexpressing mice (cem) and giant cells on the root surface of Ambn mutant mice (arrow). (E, F) TRAP staining of molar sections of WT (E) and Ambn overexpressor (F) mice. Arrows indicate osteoclast activity. The inset in (F) illustrates the presence of cementoclasts on the root surface of postnatal day 15 Ambn mutant mice (asterisk marking cementoblast and adjacent lacuna of Howship). These were not found in WT mice. ab = alveolar bone, pdl = periodontal ligament, cem = cementum, bv = blood vessel.
AMBN modulates RANKL-stimulated osteoclastogenesis in vitro
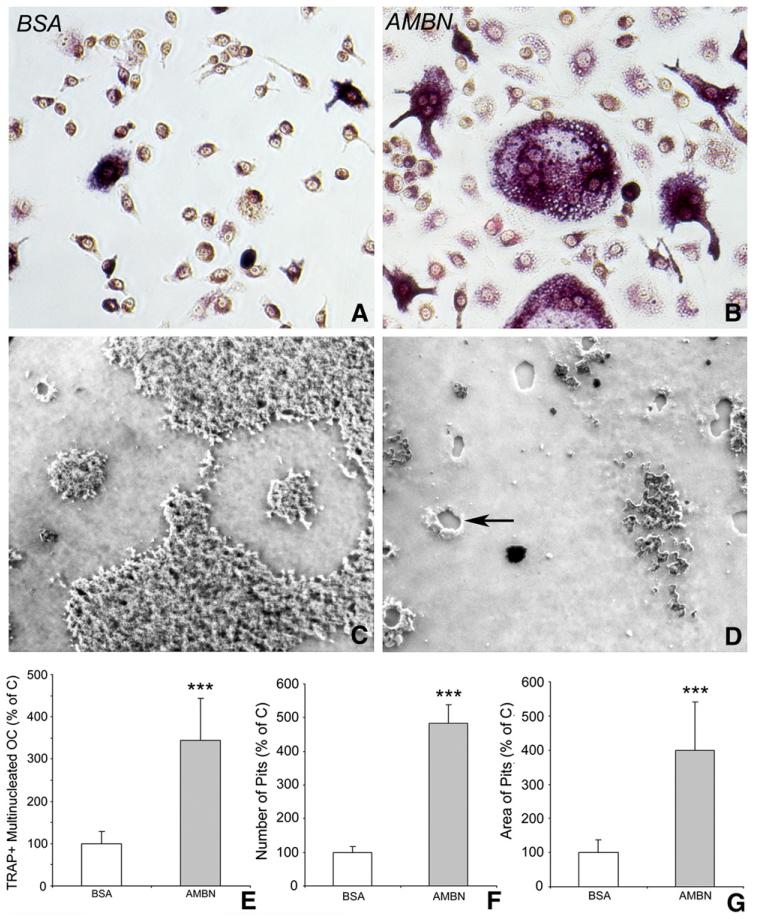
Our findings in Ambn overexpressing mice indicated that increased AMBN levels were associated with enhanced osteoclast activity and mineralized tissue resorption. To determine the role of AMBN on mineralized tissue dynamics in vivo, studies were designed to test AMBN’s function as an extracellular matrix mediator in osteoclasts and their precursors. In these studies, the interaction between cells and AMBN was mimicked in an in vitro model system, in which untreated culture dishes were coated with AMBN at two different concentrations. The effect of AMBN on RANKL induced osteoclast differentiation was evaluated after BMMCs were subjected to BSA- or AMBN-coated cell culture plates or calcium-phosphate substrate, and cultured in the presence of RANKL (50 ng/ml) and CSF-1 (10 ng/ml) for 5 days. The number of multi-nuclear and TRAP-positive osteoclasts was increased by 3.45-fold upon AMBN treatment (Figs. 5A, B and E). In addition to its effect on osteoclast size and number, AMBN also had significant impact on bone resorbing activity as determined by the number and area of resorption pits on Biocoat™-treated culture dishes (Figs. 5C and D), resulting in 4.70-fold higher pit numbers and 4.01-fold increased resorption pit area following AMBN treatment (Figs. 5F and G).
Fig. 5.
AMBN modulation on osteoclast differentiation in vitro. Osteoclast formation was induced from BMMCs cultured on BSA or AMBN coated culture dishes or calcium-phosphate substrate with CSF-1 (10 ng/ml) and RANKL (50 ng/ml) for 5 days. (A and B) TRAP staining showed a pink cytoplasm in multinucleated cells, indicative of mature osteoclasts. (E) Comparison of TRAP-positive/multinucleated cell density between treatment groups. Data are presented as mean±SD for four different cultures. (C and D) Effect of AMBN on the ability of differentiated osteoclasts to form resorption pits. The phase contrast microscopy images showed a comparison between AMBN and BSA treated bone matrix (Biocoat™) subjected to BMMCs and osteoclast induction conditions. Note the presence of resorption pits (arrow) in the AMBN treated group. (F and G) The number and area of resorption pits in the AMBN treatment group are presented as percentage of the control group. ***=p<0.0001.
AMBN enhances BMMS adhesion and induces adhesion-dependent phosphorylation of ERK and AKT
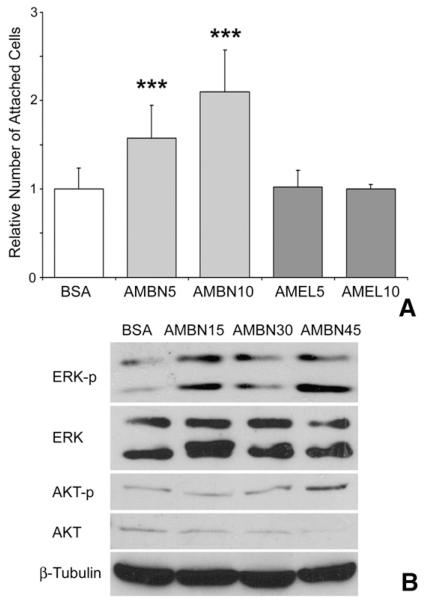
Previous studies have highlighted the role of AMBN as an extracellular adhesion molecule [14]. We therefore hypothesized that AMBN might exert its effects on osteoclastogenesis and mineral homeostasis through cell adhesion related mechanisms. In support of this concept, AMBN treatment significantly enhanced adhesion of both Raw cells (data not shown) and BMMCs (Fig. 6A), and this effect was dosage-dependent (Fig. 6A). Both BSA and recombinant mouse amelogenin (rmAMEL, another enamel matrix protein) were used as controls and did not affect cell adhesion (Fig. 6A).
Fig. 6.
Role of AMBN in BMMC adhesion. BMMCs were seeded onto culture plates that were coated with AMBN or AMEL at two different concentrations, and blocked with BSA. (A) Adhesion of BMMCs on BSA, AMBN or AMEL coated plates. The number of attached cells was counted after 4 h of culture. AMBN5/AMEL5 contained 5 μg/ml and AMBN10/AMEL10 contained 10 μg/ml of the recombinant AMBN or AMEL in the coating solution used to coat the plates. (B) Western blot analysis of the effect of AMBN on adhesion-dependent phosphorylation of key extracellular matrix signaling molecules, ERK and AKT, in BMMCs. The Western blot compares BSA and AMBN treatments after 15 (AMBN15), 30 (AMBN30) and 45 (AMBN45)min of incubation. ***=pb<.0001.
Adhesion of cells to ECM initiates signaling pathways which lead cellular spreading and polarization. Potential intermediaries that might modulate the effect of AMBN on BMMCs include extracellular-signalregulated kinases (ERK) and phosphatidylinositol 3-kinases (PI3K), both of which have been implicated in the regulation of cell adhesion and osteoclastogenesis and act as second messengers [28,29]. To determine whether AMBN affects ERK and PI3K pathways, ERK1/2 and AKT phosphorylation was examined by Western blot. Our study indicated that AMBN treatment enhanced ERK 1/2 and AKT phosphorylation 15 min after cell seeding. The level of ERK1/2 phosphorylation was 1.6-fold and AKT phosphorylation was 2.3-fold higher in the AMBN treated group than in the BSA group (Fig. 6B, p<0.05). Fold increase in ERK1/2 phosphorylation was calculated from the sum of the bands detectable on the Western blot and from three independent experiments.
AMBN regulates osteoclast podosome and actin ring formation and is involved in α2β1 and MER pathways
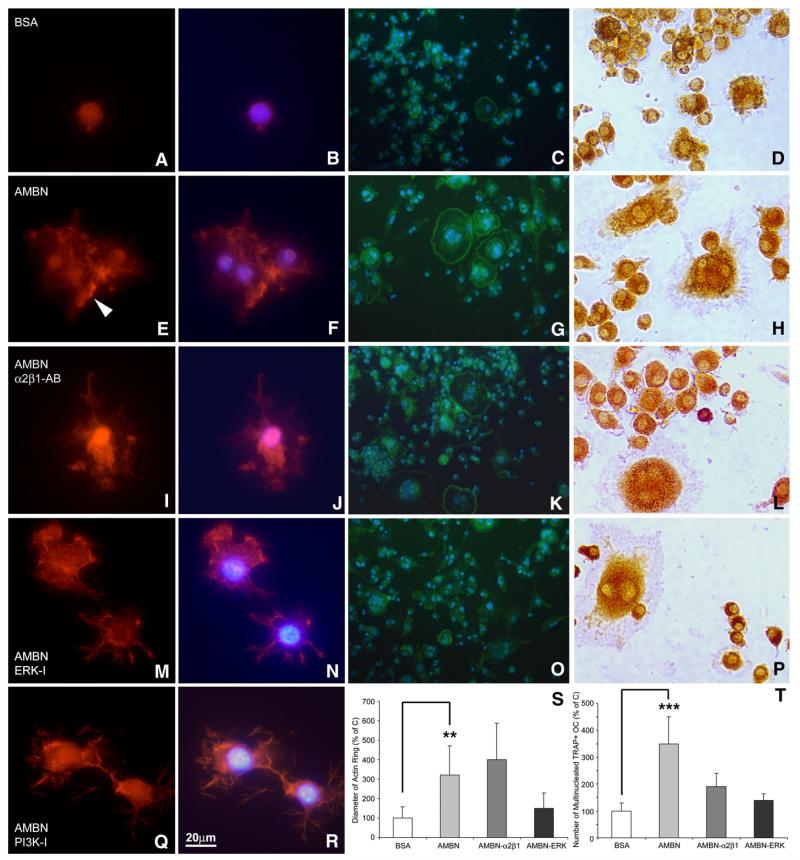
To illustrate cytoskeletal changes as a result of AMBN treatment, BMMCs were cultured in the presence of either AMBN or BSA. Following culture, F-actin was stained using rhodamine-phalloidin. After seeding for 12 h, BMMCs were still rounded in the BSA control culture (Figs. 7A and B), while cells had adhered to and spread on the AMBN-coated substrate (Figs. 7E and F). Individual dot-like structures (podosomes) appeared at the cell periphery of AMBN-treated BMMCs and these podosomes were connected with each other through actin fibers (Figs. 7E and F).
Fig. 7.
Involvement of α2β1 integrins and ERK1/2 in AMBN-modulated formation of podosome and actin ring. (A, B, E, F, I, J, M, N, Q, R) Fluorescent images of BMMCs cultured on dishes treated with either BSA (A and B), AMBN (E and F), AMBN plus α2β1 integrin antibody (I and J), AMBN plus ERK inhibitor (M and N), or AMBN plus PI3K inhibitor (Q and R). Cells were stained with the actin-filament stain rhodamine-phalloidin (A, E, I, M, Q) and nuclei were counterstained with DAPI (B, F, J, N, R). Following AMBN treatment, BMMCs displayed podosome-like structures (arrowhead). (C, G, K, O) FITC-phalloidin staining indicated actin ring formation characteristic of osteoclasts. Multiple nuclei were visualized in osteoclasts with actin rings using DAPI (D, H, L, P), TRAP staining showed a pink cytoplasm in multinucleated cells, indicative of mature osteoclasts. (S and T) Comparison of actin ring diameter and TRAP-positive/multinucleated cell density between treatment groups. Data are presented as mean±SD for four different cultures. **=p<0.001, ***=p<0.0001.
Adhesive interaction between cells and ECM proteins is mediated by cell surface receptor integrins. It has been reported that the rodent AMBN protein contains a potential α2β1 integrin binding domain and a thrombospondin cell adhesion motif [23]. To determine whether integrins participate in AMBN-mediated cell adhesion and subsequent spreading, an anti-α2β1 integrin antibody (10 μg/ml) was used to neutralize the integrin receptors. In addition, we used the ERK1/2 inhibitor U0126 and the PI3K inhibitor LY294002 to inhibit ERK and PI3K pathways to determine their involvement in the effect of AMBN on osteoclasts. In comparison to AMBN-treated BMMCs, the integrin α2β1 antibody did not block BMMC spreading, but reduced the number of podosomes (Figs. 7I and J). Application of the ERK inhibitor U0126 and the PI3K inhibitor LY294002 to AMBN-treated BMMCs resulted in a loss of podosomes and F-actin reorganization (Figs. 7M, N, Q, and R).
Coating of culture substrates with AMBN resulted in a significant 3.3-fold increase in osteoclast actin ring diameter compared to osteoclasts on BSA-coated controls (Figs. 7C and S, p<0.05). In addition, AMBN-treated BMMCs exhibited more and larger sealing zones than BSA-treated cells (Fig. 7G). Treatment with anti-α2β1 integrin antibody in addition to AMBN coating did not affect the actin ring diameter (Figs. 7K and S), while treatment with the ERK-inhibitor U0126 resulted in a reduction of actin ring size compared to the AMBN coated group (Figs. 7O and S). Moreover, the number of multi-nuclear and TRAP-positive osteoclasts was increased by 3.75-fold upon AMBN treatment (Figs. 7H and T, pb0.05). The antibody against integrin α2β1 and the ERK-inhibitor U0126 partially inhibited AMBN-associated osteoclast activation (Figs. 7K, O and T), with multinuclear osteoclast numbers reduced to 53% after anti-α2β1 integrin antibody block and 40% after application of U0126 (Fig. 7T) as compared to the AMBN treated group only.
AMBN upregulates osteoclast activation-related protein expression through α2β1 and MER pathways
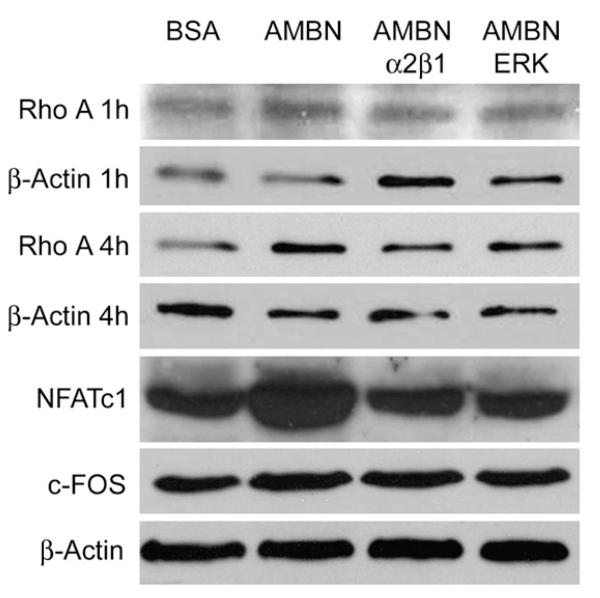
RhoA activity is required to induce integrin clustering and focal complex formation [30,31]. To assess whether AMBN-mediated adhesion of BMMCs stimulated RhoA activity, RhoA expression levels in response to AMBN treatment were examined using Western blot analysis. After 4 h of cell seeding, there was RhoA expression in AMBN-treated cells was 2.2-fold increased compared to BSA-treated cells (Fig. 8, p<0.05). Two inhibitors of adhesion molecules/pathways, α2β1 antibody and UO126, inhibited the effect of AMBN on RhoA expression, with RhoA returning to control levels when AMBN was used in combination with adhesion inhibitory agents (Fig. 8). To determine whether AMBN affects RANKL-stimulated expression of osteoclastogenic regulatory genes, NFATc1 and c-FOS expression was determined using Western blots (Fig. 8). AMBN five times increased NFATc1 protein expression and doubled c-FOS expression as revealed by densitometry (Fig. 8, pb0.01). These results further support our hypothesis that AMBN regulates osteoclast differentiation. In addition, the anti-α2β1 integrin antibody and the ERK-inhibitor UO126 efficiently suppressed AMBN-induced NFATc1 expression (Fig. 8).
Fig. 8.
Effect of AMBN on osteoclast activation-related protein expression. BMMCs were seeded onto BSA or AMBN coated culture dishes and incubated for 1 h (Rho A 1 h), 4 h (Rho A 4 h) or 12 h (NFATc1 and c-FOS) in the presence of M-CSF and RANKL. AMBN function was selectively inhibited with anti-α2β1 integrin antibody or ERK inhibitor. Expression of osteoclast activation-related proteins was detected by Western blot. β-Actin served as loading control.
Discussion
In the present study we have used a number of model systems to examine the relationship between the ECM protein AMBN and osteoclast mediated resorption of mineralized tissues such as bone and cementum. AMBN function was tested using both an overexpressing transgenic mouse line and an in vitro system in which BMMCs were exposed to AMBN surface coatings. For our transgenic approach we took advantage of naturally occurring AMBN expression levels in Hertwig’s Epithelial Root Sheath (HERS) and in the Epithelial Rests of Malassez (ERM), a network of odontogenic epithelial cells remaining in the periodontal ligament after tooth eruption [26,32]. The human Keratin 14 promoter was used to overexpress the AMBN protein in mouse HERS and ERM cells. Keratin 14 is commonly known as an epithelial protein encoded by the KRT14 gene. Here we have further confirmed K14 expression in HERS and ERM by β-gal staining for the K14 LacZ transgene in K14 LacZ transgenic mice using whole mount staining. AMBN and K14 co-localized both in HERS cells and in alveolar bone osteoblasts. In the present study, K14-driven overexpression of AMBN in mouse molars resulted in a robust 5-fold enhancement of AMBN levels compared to controls, confirming the effectiveness of our epithelium-driven AMBN overexpression approach in mouse periodontal tissues. For our in vitro studies, we exploited AMBN’s putative role as an extracellular matrix protein, using surface coating of culture plates as a well established method [33-35] to examine the effect of AMBN on osteoclast attachment and differentiation. Together, these studies indicated that AMBN significantly altered osteoclast activity, both in vivo and in vitro.
In developing tooth roots, AMBN was expressed in HERS, ERM, and PDL, as demonstrated by in situ hybridization and immunoreactions. Our data on AMBN expression in HERS and ERM match previous findings [18,19,36,37]. Evidence of AMBN expression in HERS and ERM contributes to an expanded understanding of a group of proteins previously called “enamel proteins”, including amelogenin, enamelin, AMBN, and others. In addition to ameloblasts and HERS [42], AMBN has now also been identified in pulp cells [37-39], osteoblasts, CD34+ cells and osteoclasts [40,41]. AMBN’s function in bone remodeling involves homing and recruitment of CD34+ cells and commitment to an osteoblast or osteoclast pathway [40,41]. Thus, AMBN function may go beyond its role as cell adhesion modulator in ameloblast differentiation and enamel formation [14,17,42-44], and also include a role in bone formation and periodontal regeneration [41,42,45,46].
Enforced Ambn expression in our mouse model resulted in bone loss and tooth root resorption, and these defects were accompanied by higher osteoclast activity, actin ring and podosome belt formation, and increased number of TRAP positive cells. Enhanced osteoclast activity in our Ambn overexpressors occurred without inflammatory stimuli, indicating that AMBN affects osteoclast activation directly and without inflammatory mediators. The effect of AMBN as it relates to bone, root, and tooth resorption suggests that AMBN activates classic mechanisms of osteoclastogenesis through modulation of attachment and RANKL activation. In support of this concept, our data indicated that AMBN increased BMMC adhesion and multinucleated osteoclast number through integrins and RhoA. Moreover, AMBN also directly enhanced the effect of RANKL and CSF-1 on osteoclastogenesis, suggesting that AMBN has the ability to trigger key mechanisms of osteoclast induction.
Supportive of a potential involvement of integrins in the effect of AMBN on mineralized tissue homeostasis, our study revealed that the AMBN-induced increase in osteoclast count was significantly reduced when antibodies against integrin α2β1 or an inhibitor against ERK-1 were applied to the culture medium. It has been well-documented that αvβ3 and α2β1 are the most prominent integrins expressed in BMMCs and osteoclasts [47]. These integrins mediate osteoclast adhesion to bone matrix proteins and regulate cytoskeleton reorganization [48]. Other integrins αvβ5 and α9β1 are found in osteoclast precursors and regulate osteoclast formation and function [49,50]. Antibodies against integrin α2β1 only resulted in a partial block of osteoclastogenesis in our study, leaving the possibility open that other isoforms or surface receptors may also be involved in the interaction between AMBN protein and BMMCs.
Our study demonstrated that AMBN treatment enhanced RhoA protein expression and resulted in ERK and Akt phosphorylation. Moreover, blocking ERK and PI3K pathways inhibited AMBN-induced formation of podosome and osteoclast differentiation after BMMCs attached on an AMBN-containing substrate, suggesting that that extracellular matrix signaling pathways play a crucial role in mediating the effect of AMBN on osteoclast differentiation. Cell adhesion to the extracellular matrix, especially through integrins, activates various protein kinases such as focal adhesion kinase, mitogen-activated protein kinase, extracellular signal-regulated kinase, and GTPases of the Rho-family [51,52]. These signaling pathways also play distinct roles in osteoclast differentiation and function [53,54]. Cross-talk between pathways and positive/negative feedback loops are therefore likely to be involved in AMBN-mediated osteoclast dynamics.
Further examination of the mechanism by which AMBN regulates osteoclastogenesis illustrated that expression levels of NFATc1, a key transcription factor of osteoclast differentiation, was increased during RANKL-stimulated osteoclast differentiation following AMBN treatment. AMBN addition to BMMCs resulted in increased expression of the osteoclastogenic transcription factor NFATc1 and this effect once more was blocked using the anti-α2β1 integrin antibody and the ERK-1/2 antagonist U0126. NFATc1 plays a pivotal role in osteoclast fusion and activation through regulation of various genes responsible for osteoclast adhesion, migration, acidification and degradation of inorganic and organic matrix [55]. Transcriptional up-regulation of the NFATc1 gene may therefore contribute to enhanced osteoclastogenesis by the AMBN protein. Based on the findings of the present study, we thus propose a model of AMBN-associated osteoclastogenesis by which the AMBN protein enhances BMMC adhesion, induces formation of cell attachment structures, and upregulates Rho A and NFATc1 gene expression, resulting in BMMC differentiation toward an osteoclast lineage through integrin and MEK/ERK pathways.
Conclusion
Albeit first introduced as a protein of the developing enamel matrix, there is increasing evidence that the secretory calcium-binding phosphoprotein (SCPP) family member AMBN is localized in other tissues such as bone, where it plays significant roles associated with mineral metabolism. Here we have explained some of the dramatic effects of AMBN on bone homeostasis by demonstrating that AMBN increases osteoclast number and differentiation as well as mineralized tissue resorption. Specifically, AMBN regulated cell adhesion and actin cytoskeleton polymerization, initiated integrin-dependent extracellular matrix signaling cascades and enhanced osteoclastogenesis. These findings provide a novel mechanism that explains the effect of AMBN on bone mineral metabolism. The effect of AMBN on bone mineral metabolism and cell adhesion might suggest a potential role of AMBN in bone homeostasis and tumor invasion.
Acknowledgments
These studies were supported by generous funding through NIDCR grants DE18057 and DE19155 to XL. Critical suggestions by Dr. Tom Diekwisch, Head of the UIC Department of Oral Biology, are gratefully acknowledged.
References
- [1].Giancotti FG, Rouslahti E. Integrin signaling. Science. 1999;285:1028–32. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- [2].Bilezikian J, Raisz L, Martin TJ. Principles of bone biology. Elsevier; Oxford, UK: 2008. [Google Scholar]
- [3].Saltel F, Destaing O, Bard F, Eichert D, Jurdic P. Apatite-mediated actin dynamics in resorbing osteoclasts. Mol Biol Cell. 2004;15:5231–41. doi: 10.1091/mbc.E04-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jurdic P, Saltel F, Chabadel A, Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur J Cell Biol. 2006;85:195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- [5].Barkalow K, Hartwig JH. Actin cytoskeleton. Setting the pace of cell movement. Curr Biol. 1995;5:1000–2. doi: 10.1016/s0960-9822(95)00200-4. [DOI] [PubMed] [Google Scholar]
- [6].Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, et al. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. 2000;275:11993–2002. doi: 10.1074/jbc.275.16.11993. [DOI] [PubMed] [Google Scholar]
- [7].Bhadriraju K, Yang M, Alom RS, Pirone D, Tan J, Chen CS. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp Cell Res. 2007;313:3616–23. doi: 10.1016/j.yexcr.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nesbitt S, Nesbit A, Helfrich M, Horton M. Biochemical characterization of human osteoclast integrins. Osteoclasts express alpha v beta 3, alpha 2 beta 1, and alpha v beta 1 integrins. J Biol Chem. 1993;268:16737–45. [PubMed] [Google Scholar]
- [9].Duong LT, Lakkakorpi P, Nakamura I, Rodan GA. Integrins and signaling in osteoclast function. Matrix Biol. 2000;19:97–105. doi: 10.1016/s0945-053x(00)00051-2. [DOI] [PubMed] [Google Scholar]
- [10].Ketcham AH. A progress report of an investigation of apical root resorption of vital permanent teeth. Int J Orthod Oral Surg Radiol. 1929;15:310–28. [Google Scholar]
- [11].Hartsfield JK., Jr Pathways in external apical root resorption associated with orthodontia. Orthod Craniofac Res. 2009;12:236–42. doi: 10.1111/j.1601-6343.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McCauley LK, Nohutcu RM. Mediators of periodontal osseous destruction and remodeling: principles and implications for diagnosis and therapy. J Periodontol. 2002;73:1377–91. doi: 10.1902/jop.2002.73.11.1377. [DOI] [PubMed] [Google Scholar]
- [13].Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab Invest. 1976;34:235–49. [PubMed] [Google Scholar]
- [14].Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–83. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hasegawa N, Kawaguchi H, Ogawa T, Uchida T, Kurihara H. Immunohistochemical characteristics of epithelial cell rests of Malassez during cementum repair. J Periodontal Res. 2003;38:51–6. doi: 10.1034/j.1600-0765.2003.01636.x. [DOI] [PubMed] [Google Scholar]
- [16].Talic NF, Evans CA, Daniel JC, Zaki AE. Proliferation of epithelial rests of Malassez during experimental tooth movement. Am J Orthod Dentofacial Orthop. 2003;123:527–33. doi: 10.1067/mod.2003.S0889540602000185. [DOI] [PubMed] [Google Scholar]
- [17].Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J Biol Chem. 1996;271:4431–5. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- [18].Spahr A, Lyngstadaas SP, Slaby I, Pezeshki G. Ameloblastin expression during craniofacial bone formation in rats. Eur J Oral Sci. 2006;114:504–11. doi: 10.1111/j.1600-0722.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- [19].Nunez J, Sanz M, Hoz-Rodriguez L, Zeichner-David M, Arzate H. Human cementoblasts express enamel-associated molecules in vitro and in vivo. J Periodontal Res. 2010;45:809–14. doi: 10.1111/j.1600-0765.2010.01291.x. [DOI] [PubMed] [Google Scholar]
- [20].Beyeler M, Schild C, Lutz R, Chiquet M, Trueb B. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res. 2010;316:1202–12. doi: 10.1016/j.yexcr.2009.12.019. [DOI] [PubMed] [Google Scholar]
- [21].Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, et al. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem. 2009;284:27176–84. doi: 10.1074/jbc.M109.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang X, Diekwisch TGH, Luan X. Structure and function of ameloblastin as an extracellular matrix protein: adhesion, calcium binding, and CD63 interaction in human and mouse. Eur J Oral Sci. 2011;119(Suppl. 1):270–9. doi: 10.1111/j.1600-0722.2011.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fukumoto S, Yamada A, Nonaka K, Yamada Y. Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells Tissues Organs. 2005;181:189–95. doi: 10.1159/000091380. [DOI] [PubMed] [Google Scholar]
- [24].Zhang Y, Zhang X, Lu X, Atwawasuwan P, Luan X. Ameloblastin regulates cell attachment and proliferation through RhoA and p27. Eur J Oral Sci. 2011;119(Suppl. 1):280–5. doi: 10.1111/j.1600-0722.2011.00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kumar A, Singh AK, Gautam AK, Chandra D, Singh D, Changkija B, et al. Identification of kaempferol-regulated proteins in rat calvarial osteoblast during mineralization by proteomics. Proteomics. 2010;10:1730–9. doi: 10.1002/pmic.200900666. [DOI] [PubMed] [Google Scholar]
- [26].Luan X, Ito Y, Diekwisch TG. Evolution and development of Hertwig’s epithelial root sheath. Dev Dyn. 2006;235:1167–80. doi: 10.1002/dvdy.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lu X, Kulkarni A, Gibson C, Luan X, Diekwisch TG. Ameloblasin-rich enamel matrix favors short and randomly orientated apatite crystals. Eur J Oral Sci. 2011;119(Suppl. 1):254–60. doi: 10.1111/j.1600-0722.2011.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pullikuth AK, Catling AD. Extracellular signal-regulated kinase promotes Rho-dependent focal adhesion formation by suppressing p190A RhoGAP. Mol Cell Biol. 2010;30:3233–48. doi: 10.1128/MCB.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lakkakorpi PT, Wesolowski G, Zimolo Z, Rodan GA, Rodan SB. Phosphatidylinositol 3-kinase association with the osteoclast cytoskeleton, and its involvement in osteoclast attachment and spreading. Exp Cell Res. 1997;237:296–306. doi: 10.1006/excr.1997.3797. [DOI] [PubMed] [Google Scholar]
- [30].Martin KH, Slack JK, Boerner SA, Martin CC, Parsons JT. Interin connections map: to infinity and beyond. Science. 2002;296:1652–3. doi: 10.1126/science.296.5573.1652. [DOI] [PubMed] [Google Scholar]
- [31].Vielkind S, Gallagher-Gambarelli M, Gomez M, Hinton HJ, Cantrell DA. Integrin regulation by RhoA in thymocytes. J Immunol. 2005;175:350–7. doi: 10.4049/jimmunol.175.1.350. [DOI] [PubMed] [Google Scholar]
- [32].Hamamoto Y, Nakajima T, Ozawa H. Ultrastructural and histochemical study on the morphogenesis of epithelial rests of Malassez. Arch Histol Cytol. 1989;52:61–70. doi: 10.1679/aohc.52.61. [DOI] [PubMed] [Google Scholar]
- [33].Fujii DK, Massoglia SL, Savion N, Gospodarowicz D. Neurite outgrowth and protein synthesis by PC12 cells as a function of substratum and nerve growth factor. J Neurosci. 1982;2:1157–75. doi: 10.1523/JNEUROSCI.02-08-01157.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kleinman HK, Luckenbill-Edds L, Cannon FW, Sephel GC. Use of extracellular matrix components for cell culture. Anal Biochem. 1987;166:1–13. doi: 10.1016/0003-2697(87)90538-0. [DOI] [PubMed] [Google Scholar]
- [35].Cooke MJ, Phillips SR, Shah DS, Athey D, Lakey JH, Przyborski SA. Enhanced cell attachment using a novel cell culture surface presenting functional domains from extracellular matrix proteins. Cytotechnology. 2008;56:71–9. doi: 10.1007/s10616-007-9119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zeichner-David M, Oishi K, Su Z, Zakartchenko V, Chen LS, Arzate H, et al. Role of Hertwig’s epithelial root sheath cells in tooth root development. Dev Dyn. 2003;228:651–63. doi: 10.1002/dvdy.10404. [DOI] [PubMed] [Google Scholar]
- [37].Fong CD, Cerny R, Hammarstrom L, Slaby I. Sequential expression of an amelin gene in mesenchymal and epithelial cells during odontogenesis in rats. Eur J Oral Sci. 1998;106(Suppl. 1):324–30. doi: 10.1111/j.1600-0722.1998.tb02193.x. [DOI] [PubMed] [Google Scholar]
- [38].Hao J, He G, Narayanan K, Zou B, Lin L, Muni T, et al. Identification of differentially expressed cDNA transcripts from a rat odontoblast cell line. Bone. 2005;37:578–88. doi: 10.1016/j.bone.2005.05.013. [DOI] [PubMed] [Google Scholar]
- [39].Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–70. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- [40].Tamburstuen MV, Reseland JE, Spahr A, Brookes SJ, Kvalheim G, Slaby I, et al. Ameloblastin expression and putative autoregulation in mesenchymal cells suggest a role in early bone formation and repair. Bone. 2010;48:406–13. doi: 10.1016/j.bone.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tamburstuen MV, Reppe S, Spahr A, Sabetrasekh R, Kvalheim G, Slaby I, et al. Ameloblastin promotes bone growth by enhancing proliferation of progenitor cells and by stimulating immunoregulators. Eur J Oral Sci. 2010;118:451–9. doi: 10.1111/j.1600-0722.2010.00760.x. [DOI] [PubMed] [Google Scholar]
- [42].Fong CD, Slaby I, Hammarstrom L. Amelin: an enamel-related protein, transcribed in the cells of epithelial root sheath. J Bone Miner Res. 1996;11:892–8. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- [43].Lee SK, Krebsbach PH, Matsuki Y, Nanci A, Yamada KM, Yamada Y. Ameloblastin expression in rat incisors and human tooth germs. Int J Dev Biol. 1996;40:1141–50. [PubMed] [Google Scholar]
- [44].Lee SK, Kim SM, Lee YJ, Yamada KM, Yamada Y, Chi JG. The structure of the rat ameloblastin gene and its expression in amelogenesis. Mol Cells. 2003;15:216–25. [PubMed] [Google Scholar]
- [45].Kakegawa A, Oida S, Gomi K, Nagano T, Yamakoshi Y, Fukui T, et al. Cytodifferentiation activity of synthetic human enamel sheath protein peptides. J Periodontal Res. 2010;45:643–9. doi: 10.1111/j.1600-0765.2010.01279.x. [DOI] [PubMed] [Google Scholar]
- [46].Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P. Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci. 2006;114(Suppl. 1):244–53. doi: 10.1111/j.1600-0722.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- [47].Teitelbaum SL. Osteoclasts: what do they do and how do they do it? Am J Pathol. 2007;170:427–35. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McHugh KP, Hodivala-Dilke K, Zheng MH, Namba N, Lam J, Novack D, et al. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105:433–40. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Inoue M, Namba N, Chappel J, Teitelbaum SL, Ross FP. Granulocyte macrophagecolony stimulating factor reciprocally regulates alphav-associated integrins on murine osteoclast precursors. Mol Endocrinol. 1998;12:1955–62. doi: 10.1210/mend.12.12.0213. [DOI] [PubMed] [Google Scholar]
- [50].Rao H, Lu G, Kajiya H, Garcia-Palacios V, Kurihara N, Anderson J, et al. Alpha9beta1: a novel osteoclast integrin that regulates osteoclast formation and function. J Bone Miner Res. 2006;21:1657–65. doi: 10.1359/JBMR.060718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–9. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- [52].Howe AK, Juliano RL. Distinct mechanisms mediate the initial and sustained phases of integrin-mediated activation of the Raf/MEK/mitogen-activated protein kinase cascade. J Biol Chem. 1998;273:27268–74. doi: 10.1074/jbc.273.42.27268. [DOI] [PubMed] [Google Scholar]
- [53].Blair HC, Robinson LJ, Zaidi M. Osteoclast signalling pathways. Biochem Biophys Res Commun. 2005;328:728–38. doi: 10.1016/j.bbrc.2004.11.077. [DOI] [PubMed] [Google Scholar]
- [54].Feng X. RANKing intracellular signaling in osteoclasts. IUBMB Life. 2005;57:389–95. doi: 10.1080/15216540500137669. [DOI] [PubMed] [Google Scholar]
- [55].Zhao Q, Wang X, Liu Y, He A, Jia R. NFATc1: functions in osteoclasts. Int J Biochem Cell Biol. 2010;42:576–9. doi: 10.1016/j.biocel.2009.12.018. [DOI] [PubMed] [Google Scholar]