Summary
Many microtubule (MT) functions are mediated by a diverse class of proteins (+TIPs) at growing MT plus ends that control intracellular MT interactions and dynamics, and depend on end-binding proteins (EBs) [1]. Cryoelectron microscopy has recently identified the EB binding site as the interface of four tubulin dimers that undergoes a conformational change in response to β-tubulin GTP hydrolysis [2, 3]. Doublecortin (DCX), a MT-associated protein (MAP) required for neuronal migration during cortical development [4, 5], binds to the same site as EBs [6], and recent in vitro studies proposed DCX localization to growing MT ends independent of EBs [7]. Because this conflicts with observations in neurons [8, 9], and the molecular function of DCX is not well understood, we revisited intracellular DCX dynamics at low expression levels. Here, we report that DCX is not a +TIP in cells, but in contrast is excluded from the EB1 domain. In addition, we find that DCX-MT interactions are highly sensitive to MT geometry. In cells, DCX-binding was greatly reduced at MT segments with high local curvature. Remarkably, this geometry-dependent binding to MTs was completely reversed in the presence of taxanes, which reconciles incompatible observations in cells [9] and in vitro [10]. We propose a model explaining DCX specificity for different MT geometries based on structural changes induced by GTP hydrolysis that decreases the spacing between adjacent tubulin dimers [11]. Our data are consistent with a unique mode of MT interaction in which DCX specifically recognizes this compacted GDP-like MT lattice.
Graphical Abstract
eTOC
Ettinger et al. report that the neuronal migration protein Doublecortin (DCX) is an ‘anti-+TIP’ in cells that is excluded from growing microtubule (MT) ends. MT geometry dependence of DCX-binding to straight MTs without or curved MT segments in the presence of taxanes supports a model in which DCX recognizes the compacted GDP-like MT lattice.
Results and Discussion
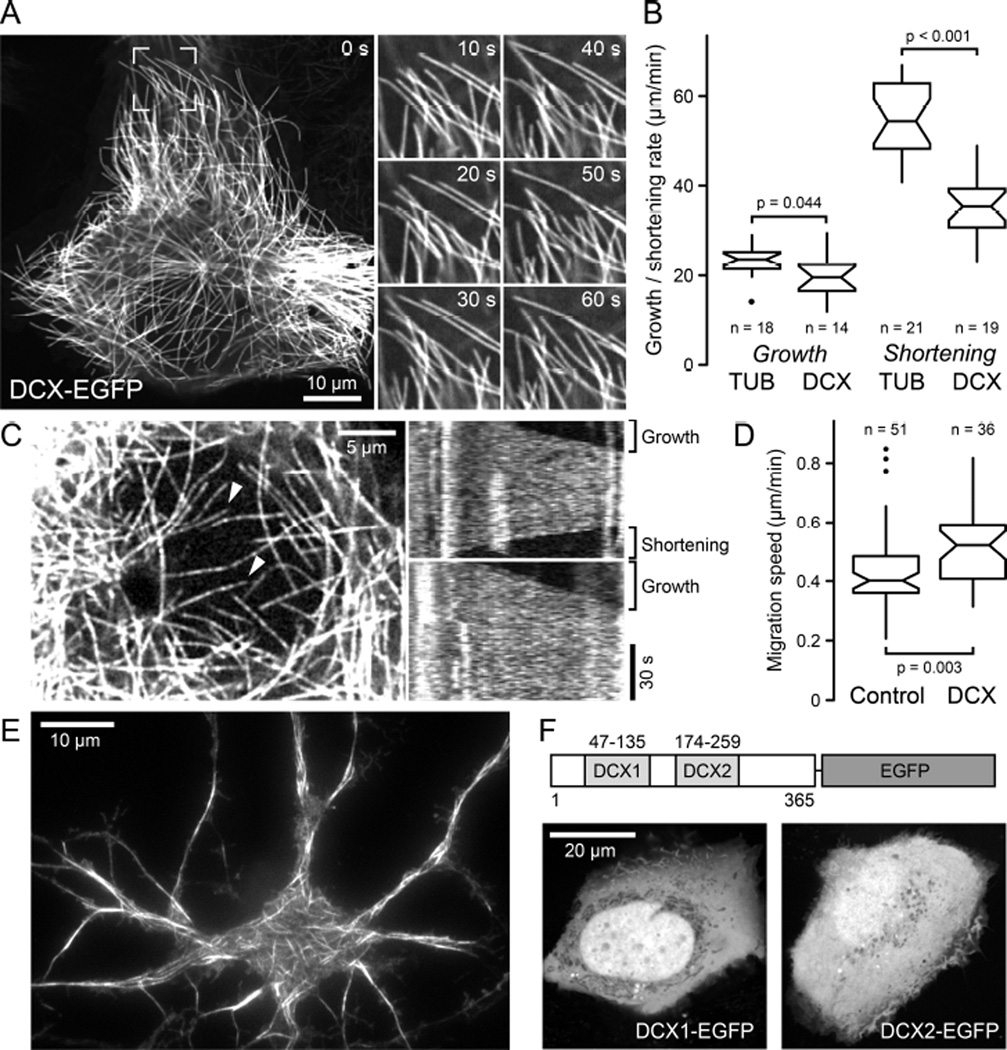
Although DCX has been proposed to bind, stabilize and nucleate microtubules (MTs) [12], early studies on DCX-MT interactions in cells suffered from potential overexpression phenotypes such as massive MT bundling [5]. In contrast, recent studies proposed EB-independent DCX plus-end-tracking at very low concentrations in vitro [7]. Because this is consistent with DCX recognizing the same site between tubulin dimers and between protofilaments that determines EB plus-end-tracking [2, 6], we reexamined intracellular DCX dynamics at low expression levels that are likely similar to endogenous expression in developing neurons. In several stably expressing human cell lines generated by lentivirus transduction, which include non-transformed HaCaT keratinocytes as well as MDA-MB-231 or H1299 cancer cells, DCX-EGFP appeared evenly distributed along individual MTs, and bound MTs with high affinity indicated by the very low level of DCX-EGFP in the cytoplasm (Figure 1A; Movie S1). MT-binding required both DCX domains, and constructs containing only the N- or C-terminal DCX domains showed no MT association in cells (Figure 1F) [13]. At these expression levels, we did not observe noticeable MT bundling and MTs remained dynamic although DCX-EGFP decreased MT shortening rates by ~30% (Figure 1B). Nevertheless, DCX-EGFP did not obviously interfere with interphase or mitotic remodeling of the MT cytoskeleton and did not inhibit cell migration (Figure 1D; Movie S2). At very low expression levels, DCX-EGFP labeling along MTs appeared speckled and the speckle pattern fluctuated rapidly indicating highly dynamic binding kinetics (Figure 1C; Movie S3). Even at these expression levels, DCX-EGFP was never enriched near MT plus ends regardless of whether these MTs were growing or shortening (Figure 1C) demonstrating that even at the lowest observable intracellular concentration, DCX does not display MT plus-end-tracking behavior in cells. Because non-neuronal cells do not normally express DCX, we confirmed these observations in rat embryonic primary neurons infected with the DCX-EGFP lentivirus, and found similar DCX-EGFP localization along MTs and no enrichment at growing MT ends (Figure 1E).
Figure 1. DCX-EGFP associates along MTs and does not accumulate at MT ends in cells.
(A) Localization of lentivirus-transduced DCX-EGFP along MTs in a stably expressing HaCaT cell. Images on the right show the boxed region at higher magnification and indicated time intervals, illustrating MT growth and shortening. See also Movie S1.
(B) MT growth and shortening rates in tubulin-EGFP expressing cells (TUB) compared with DCX-EGFP expressing cells (DCX). n = number of MTs in 5 cells per condition. Notches indicate 90% confidence intervals.
(C) DCX-EGFP dynamics in a low expressing cell. Kymographs of MTs marked with arrowheads from one frame per second time-lapse recording show rapid fluctuations of DCXEGFP intensity indicative of rapid binding kinetics and no indication of plus ends accumulation during phases of growth or shortening. See also Movie S3.
(D) Comparison of HaCaT cell migration speed in control and DCX-EGFP expressing cells (DCX). n = number of cells from 3 experiments.
(E) Total internal reflection microscopy image of live DCX-EGFP lentivirus transduced rat primary neurons showing DCX-EGFP binding along MTs.
(F) HaCaT cells transiently transfected with either N- or C-terminal DCX domains tagged with EGFP.
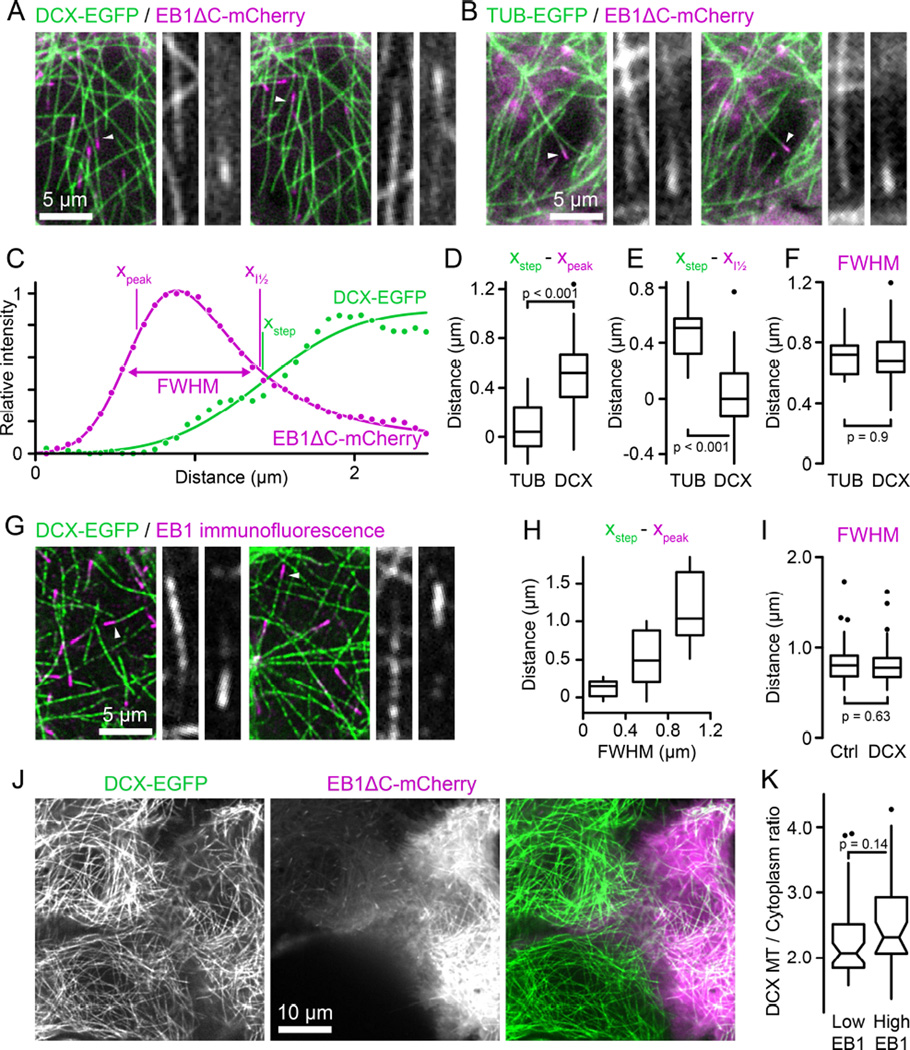
Because DCX recognizes the same site on MTs as EB1 [2, 6], we next asked if DCX and EB1 compete for binding sites near growing MT ends. To this end, we observed the MT-binding domain of EB1 tagged with mCherry (EB1ΔC-mCherry) [14] in DCX-EGFP expressing cells. In images in which both fluorescence channels were acquired simultaneously to eliminate spatial shift between channels due to MT growth or movement, DCX-EGFP appeared to be excluded from the EB1ΔC-mCherry domain near growing MT ends (Figure 2A, Movie S4). To quantitatively compare DCX and EB1 binding, we then fitted fluorescence intensity profiles of fast-growing MTs underneath the nucleus where MTs are relatively sparse and can be traced unambiguously with Gaussian-convolved models of exponentially decaying (EB1) or stepwise (DCX) MT binding [15, 16] (Figure 2C). Calculation of the distance between the start of the underlying exponential decay, xpeak, and the step of the step function, xstep, from these curve fits revealed that DCX binding lags behind EB1 by 530 ± 280 nm (mean ± standard deviation) (Figure 2D). In contrast, in control experiments comparing TUB-EGFP and EB1ΔC-mCherry intensity profiles there was no measurable difference between the EB1 maximum and the MT end (80 ± 220 nm; Figure 2B and 2D). In addition, the half-maximum of DCX-EGFP binding increase, i.e. xstep, coincided with the half-maximum of EB1 decay (Figure 2E), indicating that EB1 and DCX binding are indeed mutually exclusive. To exclude that elevated EB1ΔC-mCherry expression competed with DCX-binding, we stained for EB1 in DCX-EGFP expressing cells, and consistent with our live cell experiments DCX-EGFP did not overlap with endogenous EB1 MT plus end comets in HaCaT (Figure 2G and 2H) and other cell types (Figure S2).
Figure 2. DCX and EB1 bind mutually exclusive domains on MTs.
(A) DCX-EGFP expressing HaCaT cell transiently transfected with EB1ΔC-mCherry. Both channels were acquired simultaneously using an emission beamsplitter and corrected for spatial shift between images. Gray scale panels show the individual DCX-EGFP and EB1ΔC-mCherry channels for the MTs highlighted by arrowheads at higher magnification illustrating that DCX is excluded from the EB1 domain. See also Movie S4.
(B) Tubulin-EGFP expressing HaCaT cell transiently transfected with EB1ΔC-mCherry acquired as described in (A).
(C) EB1ΔC-mCherry and DCX-EGFP intensity profiles corresponding to the right panel shown in (A). Solid lines are curve fits with Gaussian-convolved models as described in the text. xpeak and xstep indicate the positions of the underlying exponential and step functions, respectively. Note that because of the asymmetry of the exponential decay xpeak does not coincide with the maximum of the Gaussian-convolved function. xI½ is the position of the half-maximum of the Gaussian-convolved exponential decay, and FWHM the width of the Gaussian-convolved exponential decay at half-maximum intensity.
(D) Comparison of the distance between the maximum of the EB1ΔC-mCherry exponential decay and the TUB-EGFP or DCX-EGFP half-maximum calculated from the curve fits.
(E) Comparison of the distance between the half-maximum of the EB1ΔC-mCherry decay and the half-maximum of the TUB-EGFP or DCX-EGFP curve fits.
(F) Widths of the EB1ΔC-mCherry comet at half-maximum intensity in TUB-EGFP or DCX-EGFP expressing cells. n = 12 MTs from 5 cells (TUB) and 46 MTs from 11 cells (DCX) in (D) to (F).
(G) Endogenous EB1 staining in EGFP-DCX expressing HaCaT cells. Gray scale panels show the individual DCX-EGFP and EB1 channels for the MTs highlighted by arrowheads at higher magnification illustrating that DCX is excluded from the EB1 domain. See also Figure S1.
(H) Distance between the maximum of the exponential decay of EB1 immunofluorescence and the half-maximum of the DCX-EGFP curve fit as a function of EB1 comet width. n = 45 MTs from 19 cells.
(I) Widths of the EB1 comet at half-maximum intensity in control or DCX-EGFP expressing cells. n = 78 MTs from 14 cells (Ctrl) and 106 MTs from 16 cells (DCX).
(J) DCX-EGFP expressing HaCaT cells expressing different levels of EB1ΔC-mCherry. The cell on the right is expressing high levels resulting in EB1ΔC-mCherry binding along MTs. Yet, there is no obvious difference in DCX-EGFP binding to MTs.
(K) Quantification of relative DCX-EGFP binding to MTs in cells expressing low or high levels of EB1ΔC-mCherry. n = 44 MTs in 8 cells per condition.
In addition, in vitro, DCX-EGFP plus-end-tracking is only observed at very low concentrations (<5 nM) [10], which is ~5-fold below the KD reported for EB1 at MT plus ends [16]. Thus, if DCX competed with EB1 for the same binding site, DCX should bind with higher affinity and displace EB1 from MT plus ends. However, this was not the case and the half maximum width determined from the exponentially-modified Gaussian fits of EB1ΔC-mCherry (Figure 2F) or endogenous EB1 comets (Figure 2I) remained unchanged in the presence of DCX-EGFP. Conversely, DCX-binding along MTs was not reduced in cells expressing high levels of EB1ΔCmCherry at which EB1 begins to bind along MTs (Figure 2J and 2K). Finally, the zone of DCXEGFP exclusion from growing MT ends correlated with EB1 comet length (Figure 2H), indicating that faster growing MTs have a longer zone without DCX at the plus end. Together, these data strongly indicate that in cells DCX and EB1 do not compete for binding, but instead may recognize the same interface between four tubulin dimers in different conformational states. These different MT conformations are most likely defined by different GTP hydrolysis states suggesting that DCX specifically recognizes the GDP-MT lattice. This is indeed consistent with in vitro findings that, in contrast to EB1, DCX shows no increased affinity for MTs assembled in the presence of nucleotide analogs that are believed to mimic either the GTP- or the GDP-Pibound MT conformation [10].
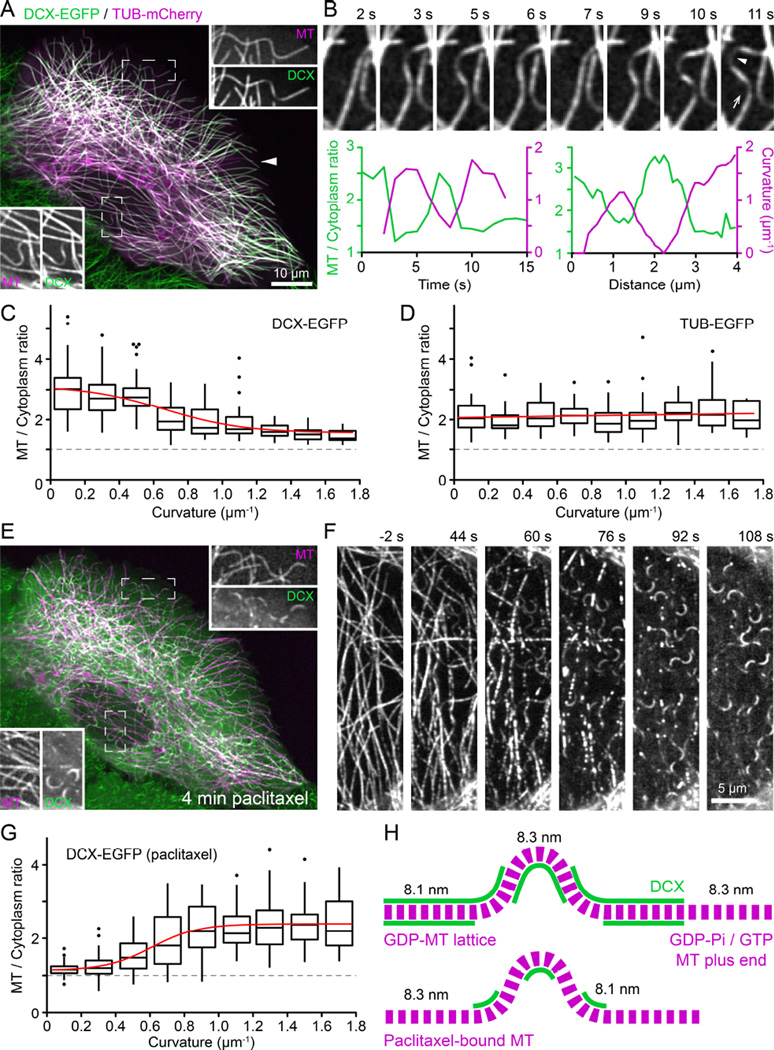
Although at first glance DCX-EGFP appeared to decorate all intracellular MTs evenly, upon closer inspection we noticed that DCX-EGFP fluorescence was strongly reduced on curved MTs (Figure 3A, see also Movie S1). This loss of DCX-EGFP from curved MTs was highly dynamic and reversible: DCX-EGFP dis- and reappearance correlated tightly with MT buckling and straightening, both in time and space (Figure 3B). To quantify this dependence of DCX-MT binding on MT geometry, we analyzed DCX-EGFP fluorescence intensity as a function of local MT curvature of a large number of MT segments, and determined that DCX-EGFP dissociated from MTs with increasing local curvature (Figure 3C). Conversely, as expected, TUB-EGFP fluorescence intensity did not depend on MT curvature (Figure 3D). This preference of DCX for straight MTs was surprising as the opposite was recently reported, namely that DCX specifically recognizes highly curved MTs in vitro [10] even though earlier reports also noted decreased DCX staining at bent MTs in neuronal growth cones [9]. Of note, although the vast majority of straight MTs was DCX-decorated, we observed rare occurrences in which few straight MTs did not bind DCX, which may suggest lattice defects resulting in unusual protofilament number [7, 12].
Figure 3. DCX binds straight but not curved MTs and taxanes reverse this MT geometry preference.
(A) DCX-EGFP expressing HaCaT cell transiently transfected with tubulin-mCherry (TUB). Insets show the individual channels in the boxed regions at higher magnification illustrating decreased DCX-EGFP signal at MT segments with high local curvature. The arrowhead points to an example of a very rare occurrence of straight MTs that are not decorated with DCX-EGFP.
(B) Time-lapse sequence of DCX-EGFP decorated MTs. The graphs show the correlation of DCX-EGFP binding and local MT curvature in time and space. Left: Relative MT-bound DCXEGFP signal and local MT curvature measured at the spot indicated by the arrowhead in the right image. Right: Relative DCX-EGFP signal and curvature profile along the MT in the direction of the arrow.
(C) Relative MT-bound DCX-EGFP as a function of local MT curvature. Box plots show binned data at 0.2 µm−1 curvature intervals. Red line is a sigmoidal fit of the data. The dashed line at aMT to cytoplasm ratio of one represents undetectable MT binding. Outliers are shown as individual data points. n = 308 measurements from 55 MTs in 22 cells.
(D) Relative TUB-EGFP signal as a function of local MT curvature. Box plots show binned data at 0.2 µm−1 curvature intervals. Red line is a linear fit of the data. n = 219 measurements from 50 MTs in 12 cells.
(E) Same cell as in (A) after 4 minutes in 1 µM paclitaxel. Note the drastic redistribution of DCX-EGFP from straight to curved MT segments. Insets show the individual channels in the boxed regions at higher magnification.
(F) Time-lapse sequence of DCX-EGFP redistribution following addition of 1 µM paclitaxel at 0 s. See also Movie S5.
(G) Relative MT-bound DCX-EGFP as a function of local MT curvature in cells treated with 1 µM paclitaxel. Box plots show binned data at 0.2 µm−1 curvature intervals. Red line is a sigmoidal fit of the data. n = 348 measurements from 76 MTs in 11 cells.
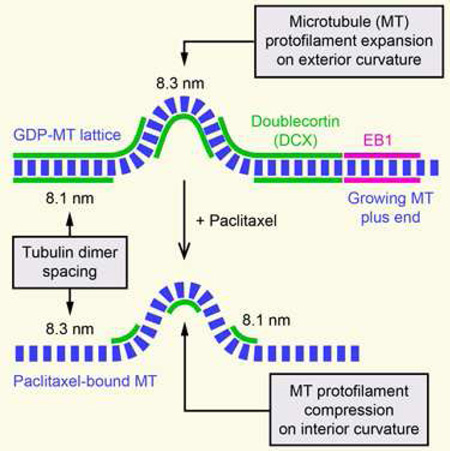
(H) Model explaining observed DCX-EGFP binding to straight and curved MT segments based on tubulin dimer spacing in the GDP-MT lattice (8.1 nm) and in paclitaxel-bound MTs (8.3 nm). Assuming a curvature of 1 µm−1, the outside spacing between tubulin dimers must be 2.5% larger compared with the inside resulting in an expansion of tubulin dimer spacing. The observed DCX-EGFP distribution along MTs in control and paclitaxel-treated cells is thus consistent with DCX recognition of the GDP-like MT lattice conformation. For simplicity, individual protofilaments are not drawn. See also Figure S2.
To reconcile these conflicting results, we considered possible mechanisms by which DCX might recognize MT curvature. Because the loss of DCX-EGFP along curved MT segments was spatially and temporally uniform, it is likely not due to MT lattice defects that would occur in a more anisotropic manner [17]. At a curvature of 1 µm−1, at which DCX-EGFP binding reaches a lower plateau, the angle between adjacent tubulin dimers along a protofilament is less than 0.5° and it is difficult to imagine how such a small deviation from straightness could be recognized. However, assuming tubulin subunits do not normally exchange from within the MT lattice, the number of tubulin dimers per unit length of MT has to be the same along the inside and outside protofilament of a curved MT. At a curve radius of 1 µm and the known MT thickness of 25 nm, tubulin dimer spacing on the outside of the curved MT thus has to be 2.5% larger compared with the inside (Figure S2). Strikingly, this is extremely close to the recently reported structural difference between the GTP-tubulin lattice with a subunit spacing of 8.3 nm and the compacted GDP-tubulin lattice with a subunit spacing of 8.1 nm [11]. This structural change predominantly occurs at the interface between tubulin dimers along a protofilament, which constitutes the DCX binding pocket [6]. It is thus conceivable that DCX is sensitive to this conformational change between adjacent tubulin dimers. Assuming that the 8.1 nm tubulin dimer spacing cannot be compressed any further, the protofilament along the outside of a curved GDP-MT would have to adopt a GTP-like conformation. Therefore, along a curved MT DCX would be expected to remain bound to the inside curvature, but dissociate from the outside consistent with the significant reduction in binding but not total loss of DCX signal that we see in our data (Figure 3H).
Paclitaxel-binding to MTs stabilizes the GTP lattice conformation and reverses lattice compaction following GTP hydrolysis [11, 18]. Thus, we predicted that paclitaxel prevents DCX binding to MTs. Indeed, the effect of paclitaxel addition was rapid and dramatic. Within minutes, concurrent with paclitaxel diffusion into the cell, DCX-EGFP rapidly dissociated from straight MTs (Figure 3F; Movie S5). Depending on paclitaxel concentration, DCX-EGFP appeared to localize to growing MT plus ends for a short period of time, which we interpret as lag between MT polymerization and paclitaxel-binding (Figure 3F; Movie S5). After a few minutes, DCXEGFP remained bound only to MT segments with high local curvature (Figure 3E–3G). Similar to the logic outlined above, compression of the inside of curved taxane-bound MTs in a GTP-like conformation to GDP-like spacing would restore DCX-binding to the inside protofilaments of a curved MT (Figure 3H), which now also fits with the in vitro data in which DCX-binding was tested on paclitaxel-stabilized MTs [10]. Taken together, these data are consistent with the hypothesis that DCX specifically recognizes the compacted tubulin dimer spacing characteristic of the GDP-MT lattice. Because DCX is excluded from the EB1-binding domain near growing MT ends (Figure 2), our data also indirectly suggest that MT lattice compaction occurs after Esite phosphate release as it is thought that EB1 recognizes the GDP-Pi tubulin state following GTP hydrolysis [2, 3, 16].
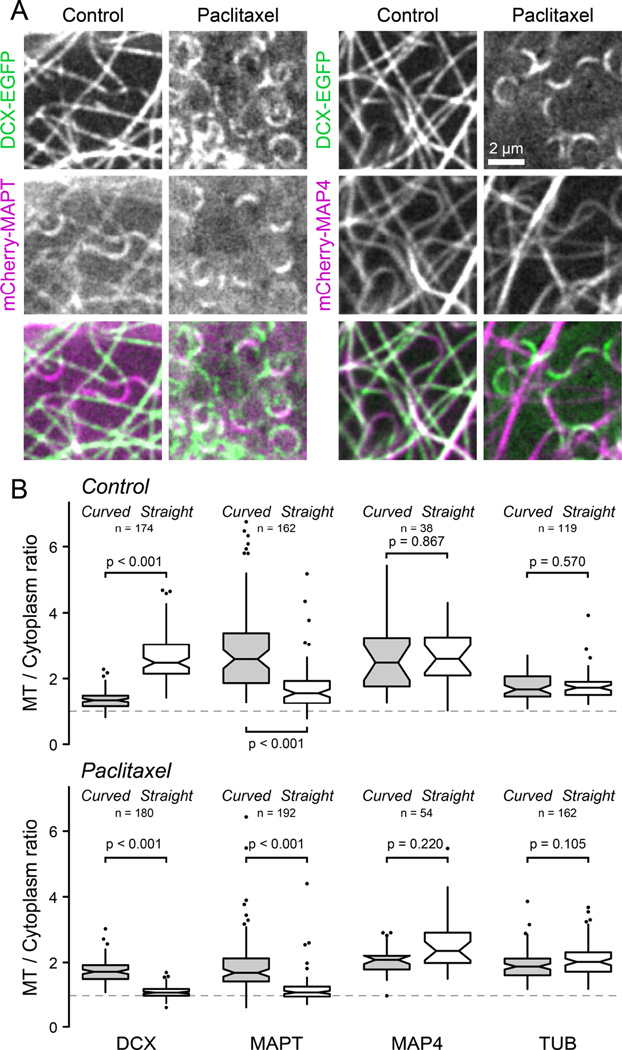
Because this represents a previously unappreciated mode of recognition of MT conformation, we tested the only other MT-associated protein, the neuronal MAPT (tau), for which a similar sensitivity to MT geometry has previously been reported [19]. In contrast to DCX, mCherry-MAPT was enriched at curved MT segments in both control and paclitaxel-treated cells, and paclitaxel decreased MAPT-MT binding along both straight and curved MT segments (Figure 4). This indicates that MAPT recognizes curved MTs through a mechanism that is distinct from DCX and cannot directly rely on inter-tubulin dimer spacing. MAPT and MAP4 belong to a family of structurally related proteins that bind MTs through a repeat array of electrostatic interactions [20]. However, unlike neuronal MAPT, the more ubiquitously expressed MAP4 was completely insensitive to both MT curvature and paclitaxel (Figure 4). Thus, recognition of MT curvature is not an intrinsic property of the classic KXGS MT-binding motif.
Figure 4. Comparison of DCX, MAPT and MAP4 binding to curved and straight MTs.
(A) MTs underneath the nucleus in DCX-EGFP expressing HaCaT cells transiently transfected with mCherry-MAPT (left) or the MT-binding domain of MAP4 fused to mCherry (mCherry-MAP4) before and after addition of 1 µM paclitaxel.
(B) Comparison of the relative MT-bound signal of DCX-EGFP, mCherry-MAPT, mCherry-MAP4 and either EGFP or mCherry-tagged tubulin (TUB) as a control at curved and straight MT segments in the absence (top) or the presence of 1 µM paclitaxel (bottom). The dashed line at a MT to cytoplasm ratio of one represents undetectable MT binding. Outliers are shown as individual data points. n = number of MT measurements from 6–18 cells per condition.
In conclusion, we propose that DCX is the first example of an ‘anti-+TIP’ that specifically recognizes the GDP-MT lattice conformation. Our results are consistent with a recent analysis of MT ultrastructure in the absence and presence of paclitaxel [11], and the simplest explanation for the observed discrimination between straight and curved MT segments is compression or expansion of the DCX binding pocket between adjacent tubulin dimers at a local MT curvature that is typically observed in cells as a result of compressive forces [21]. Similarly, compression of the GTP-like MT lattice at outwardly curved protofilaments in vitro [22] could result in high affinity DCX binding sites anterior to the GDP-Pi EB1 binding platform, explaining DCX plus-end-tracking at very low concentrations in vitro and observed differences between EB1 andDCX dynamics [10]. In vitro, DCX also discriminates between 13 and 14 protofilament MTs [7] indicating that lateral curvature or interactions between adjacent protofilaments can contribute to DCX binding. However, because lateral contacts between protofilaments only minimally differ in different nucleotide conditions [11], we believe it is unlikely that these play a role in the observed DCX binding response to longitudinal curvature of paclitaxel binding. Currently, we cannot explain why DCX plus-end-tracking does not occur in cells. Because our live cell imaging is sufficiently sensitive to observe DCX-EGFP speckles along MTs at low expression levels that likely reflect binding dynamics of few molecules, it seems unlikely that we lack the sensitivity to detect DCX-EGFP on MT tips if it were there. MT polymerization dynamics in cells and in vitro are fundamentally different and it is possible that high affinity DCX binding sites at the outermost tips of growing MTs do not exist or are inaccessible in cells. Alternatively, differences in MT protofilament numbers (13 in cells versus predominantly 14 in GMPCPP-seed nucleated MTs in vitro [23]) and thus lattice geometry, or different posttranslational modifications may explain the observed differences between cells and in vitro. Although we can only speculate what the biological function of this recognition of a specific MT geometry may be, it is possible that DCX-binding stabilizes the compacted GDP-lattice conformation and thus stiffens MTs consistent with qualitative observations that DCX increases the number of straight MTs in neuronal cells [9]. To our knowledge, the sensitivity of DCX interaction with MTs to taxane-induced changes in MT structure is unique, and demonstrates that in vitro MT-binding assays that are predominantly carried out in the presence of paclitaxel have to be interpreted carefully. Finally, DCX and MAPT are both neuronal MAPs. Taxane neuropathy is a widely acknowledged but poorly understood potentially severe side effect of chemotherapy [24], and it will be interesting to evaluate to what extent taxane-induced alterations in DCX and MAPT MT interactions contribute to taxane neurotoxicity.
Experimental Procedures
Human DCX [25] was C-terminally tagged with EGFP and cloned into a lentivirus expression vector. HaCaT cells were cultured, stable lines generated by lentivirus transduction and imaged by spinning disk confocal microscopy essentially as previously described [26–28]. Detailed experimental and image analysis procedures are included in the supplement.
Supplementary Material
Highlights.
Doublecortin (DCX) is excluded from growing microtubule ends in cells
DCX and EB1-binding to microtubules is mutually exclusive but not competitive
DCX-binding is sensitive to taxane-induced microtubule conformation changes
DCX recognizes GDP microtubule lattice-like tubulin dimer spacing
Acknowledgments
We thank Anthony Wynshaw-Boris for the original doublecortin plasmid, Michael Davidson for MAPT and MAP4 constructs, the Parnassus Flow Cytometry Core for help with cell sorting, the Dumont lab for letting us use their Dualview and all members of the HSW-6 community for discussions and comments. This work was supported by National Institutes of Health grants R01 GM079139 and S10 RR26758 to T.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
Conceptualization, A.E. and T.W.; Methodology, A.E. and T.W.; Software, A.E.; Investigation, A.E., J.v.H., S.A.R., and T.W.; Writing – Original Draft, A.E. and T.W..; Writing – Review & Editing, A.E., J.v.H., and T.W..; Funding Acquisition, T.W.; Supervision, T.W.
References
- 1.Kumar P, Wittmann T. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 2012;22:418–428. doi: 10.1016/j.tcb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs Recognize a Nucleotide-Dependent Structural Cap at Growing Microtubule Ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Des PV, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 5.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 6.Fourniol FJ, Sindelar CV, Amigues B, Clare DK, Thomas G, Perderiset M, Francis F, Houdusse A, Moores CA. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J. Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bechstedt S, Brouhard GJ. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev. Cell. 2012;23:181–192. doi: 10.1016/j.devcel.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tint I, Jean D, Baas PW, Black MM. Doublecortin associates with microtubules preferentially in regions of the axon displaying actin-rich protrusive structures. J. Neurosci. 2009;29:10995–11010. doi: 10.1523/JNEUROSCI.3399-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jean DC, Baas PW, Black MM. A novel role for doublecortin and doublecortin-like kinase in regulating growth cone microtubules. Hum. Mol. Genet. 2012;21:5511–5527. doi: 10.1093/hmg/dds395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bechstedt S, Lu K, Brouhard GJ. Doublecortin recognizes the longitudinal curvature of the microtubule end and lattice. Curr. Biol. 2014;24:2366–2375. doi: 10.1016/j.cub.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 11.Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moores CA, Perderiset M, Francis F, Chelly J, Houdusse A, Milligan RA. Mechanism of microtubule stabilization by doublecortin. Mol. Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng Y, Devedjiev Y, Dauter Z, Walsh CA, Otlewski J, Bushweller JH, et al. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- 14.Gierke S, Wittmann T. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr. Biol. 2012;22:753–762. doi: 10.1016/j.cub.2012.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Chimenti MS, Pemble H, Schonichen A, Thompson O, Jacobson MP, Wittmann T. Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of the cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1) J. Biol. Chem. 2012;287:17050–17064. doi: 10.1074/jbc.M111.316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaedel L, John K, Gaillard J, Nachury MV, Blanchoin L, Thery M. Microtubules self-repair in response to mechanical stress. Nat. Mater. 2015;14:1156–1163. doi: 10.1038/nmat4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnal I, Wade RH. How does taxol stabilize microtubules? Curr. Biol. 1995;5:900–908. doi: 10.1016/s0960-9822(95)00180-1. [DOI] [PubMed] [Google Scholar]
- 19.Samsonov A, Yu JZ, Rasenick M, Popov SV. Tau interaction with microtubules in vivo. J. Cell Sci. 2004;117:6129–6141. doi: 10.1242/jcs.01531. [DOI] [PubMed] [Google Scholar]
- 20.Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J. Biol. Chem. 2004;279:6196–6203. doi: 10.1074/jbc.M307261200. [DOI] [PubMed] [Google Scholar]
- 22.Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyman AA, Chretien D, Arnal I, Wade RH. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J. Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JJ, Swain SM. Peripheral neuropathy induced by microtubule-stabilizing agents. J. Clin. Oncol. 2006;24:1633–1642. doi: 10.1200/JCO.2005.04.0543. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 2014;16:558–573. doi: 10.1038/ncb2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stehbens S, Pemble H, Murrow L, Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ettinger A, Wittmann T. Fluorescence live cell imaging. Methods Cell Biol. 2014;123:77–94. doi: 10.1016/B978-0-12-420138-5.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.