Abstract
Alport syndrome results from mutations in the COL4A5 (X-linked) or COL4A3/COL4A4 (recessive) genes. This study examined 754 previously- unpublished variants in these genes from individuals referred for genetic testing in 12 accredited diagnostic laboratories worldwide, in addition to all published COL4A5, COL4A3 and COL4A4 variants in the LOVD databases. It also determined genotype-phenotype correlations for variants where clinical data were available. Individuals were referred for genetic testing where Alport syndrome was suspected clinically or on biopsy (renal failure, hearing loss, retinopathy, lamellated glomerular basement membrane), variant pathogenicity was assessed using currently-accepted criteria, and variants were examined for gene location, and age at renal failure onset. Results were compared using Fisher’s exact test (DNA Stata). Altogether 754 new DNA variants were identified, an increase of 25%, predominantly in people of European background. Of the 1168 COL4A5 variants, 504 (43%) were missense mutations, 273 (23%) splicing variants, 73 (6%) nonsense mutations, 169 (14%) short deletions and 76 (7%) complex or large deletions. Only 135 of the 432 Gly residues in the collagenous sequence were substituted (31%), which means that fewer than 10% of all possible variants have been identified. Both missense and nonsense mutations in COL4A5 were not randomly distributed but more common at the 70 CpG sequences (p<10−41 and p<0.001 respectively). Gly>Ala substitutions were underrepresented in all three genes (p< 0.0001) probably because of an association with a milder phenotype. The average age at end-stage renal failure was the same for all mutations in COL4A5 (24.4 ±7.8 years), COL4A3 (23.3 ± 9.3) and COL4A4 (25.4 ± 10.3) (COL4A5 and COL4A3, p = 0.45; COL4A5 and COL4A4, p = 0.55; COL4A3 and COL4A4, p = 0.41). For COL4A5, renal failure occurred sooner with non-missense than missense variants (p<0.01). For the COL4A3 and COL4A4 genes, age at renal failure occurred sooner with two non-missense variants (p = 0.08, and p = 0.01 respectively). Thus DNA variant characteristics that predict age at renal failure appeared to be the same for all three Alport genes. Founder mutations (with the pathogenic variant in at least 5 apparently- unrelated individuals) were not necessarily associated with a milder phenotype. This study illustrates the benefits when routine diagnostic laboratories share and analyse their data.
Introduction
Alport syndrome is the commonest cause of inherited renal failure after polycystic kidney disease [1]. It affects at least one in 10,000 individuals and is characterised by progressive kidney failure, hearing loss, and ocular abnormalities [2]. Inheritance is X-linked with COL4A5 mutations [3] in 85% of cases and autosomal recessive with COL4A3 or COL4A4 mutations in most of the others [4,5]. Individuals with heterozygous COL4A3 or COL4A4 mutations usually have Thin basement membrane nephropathy with normal renal function [6,7,8] but some develop renal impairment [9].
The clinical features in Alport syndrome are explained because the COL4A5 and COL4A3/COL4A4 genes code for the collagen IV α5 chain, α3 and α4 chains which form a heterotrimer in the basement membranes of the glomerulus, cochlea and eye [10] (Table 1). Each collagen IV chain has non-collagenous amino and carboxy termini, and an intermediate collagenous domain with Gly-X-Y, with multiple short non-collagenous interruptions [10]. Glycine is found at each third residue in the collagen sequence and is critical for triple helix formation.
Table 1. Characteristics of COL4A5, COL4A3 and COL4A4 genes and the corresponding proteins.
| COL4A51 | COL4A32 | COL4A4 | |
| Gene name | NP_033380.2 | NP_000091.4 | NP_000092.4 |
| Chromosomal location | Xq22 | 2q35-37 | 2q35-37 |
| Family | *COL4A1-like | *COL4A1-like | *COL4A2-like |
| Number of exons | 53 | 52 | 48 |
| Size | 257,623 bp | 150,228 bp | 159,344 bp |
| Total number of amino acids in mature protein | 1665 | 1642 | 1652 |
| Signal peptide | 26 | 28 | 38 |
| Amino terminus | 15 | 14 | 23 |
| Collagenous domain (residues) | 1301 | 1268 | 1284 |
| Number of non-collagenous interrruptions | 22 | 23 | 26 |
| Carboxy terminus (residues) | 229 | 232 | 231 |
*COL4A–like or COL4A2-like genes represent families that have arisen from common progenitors, COL4A1 or COL4A2, and thus have structural similarities
The ‘Expert guidelines on the diagnosis and management of Alport syndrome’ [2] recommend that all individuals with likely Alport syndrome should undergo genetic testing, to confirm the diagnosis and mode of inheritance, and predict the age at renal failure. The Guidelines also recommend cases of suspected Thin basement membrane nephropathy undergo genetic testing when it is important to exclude X-linked Alport syndrome [2].
Mutations in the COL4A5, COL4A3 and COL4A4 genes are mainly nonsense or missense. Genotype-phenotype correlations have only been characterised for X-linked disease [11,12,13]. Nonsense mutations are typically associated with early onset renal failure (before 30), lenticonus and central retinopathy. Deletions and insertions often cause a frameshift, and downstream nonsense variant. Many missense mutations result in a milder phenotype with later onset renal failure without ocular features [11,12,13,14].
The commonest method used currently for mutation detection is whole exome sequencing (WES). The widespread adoption of WES together with the Exome Aggregation Consortium (ExAc collaboration, http://exac.broadinstitute.org/, the ‘1000 genomes’, http://1000genomes.org/ and Hapmap, https://hapmap.ncbi.nlm.nih.gov/, projects), have greatly increased the number of reported pathogenic and normal DNA variants [15].
Collecting all pathogenic and normal variants for each disease-related gene in a publicly-accessible variant database avoids the duplication involved in assessing pathogenicity, and improves the speed and accuracy of testing while limiting cost.
The present study has reviewed known variants in the Alport genes, including 754 recently-submitted from members of the Alport mutation database Consortium to the publicly-accessible LOVD databases, with the aims of further characterising DNA variants, and genotype-phenotype correlations. The LOVD databases continue to be updated.
Methods
Assembly of the variants
DNA variants were extracted from the literature, from other databases (eg ARUP) or from previously-unpublished deidentified submissions direct to the LOVD COL4A5, COL4A3 and COL4A4 database from contributing members of the International Alport Mutation Database Consortium in the UK and Europe, Korea, the US and Australia. Genetic testing from laboratories run by Consortium members was performed after written informed consent had been obtained by the treating clinicians. Submission of variants to the databases was undertaken with IRB approval where this was required (Northern Health HREC, IRB of Seoul National University, CEIC Fundacio Puigvert, Veltishev Pediatric Clinical Research Institute).
In general, individuals had been referred for genetic testing where Alport syndrome was suspected clinically or on renal biopsy (renal failure, hearing loss, retinopathy, lamellated glomerular basement membrane). Clinical data (age, gender, diagnosis and age at onset of renal failure, and, where known, hearing loss and ocular abnormalities) were obtained in a manner conforming with local IRB ethical guidelines according to the Declaration of Helsinki. The testing laboratory deidentified information before submission to the databases.
Variants had been collected by individual laboratories over a ten year period and mutation detection methods varied during this time. All laboratories had used Sanger sequencing at some stage, few had used MLPA, but recently most had used whole exome sequencing. Most diagnostic laboratories confirmed any variant found with WES using Sanger sequencing. All laboratories were accredited by their national board but there was no uniformity in how they confirmed variant pathogenicity. This was most often with consulting the LOVD, and recently, the ExAc databases, and sometimes using an online tool such as Alamut.
Variants were described according to the reference transcripts of COL4A5 (the longer form found in the kidney, LRG_232t2, NM_033380.2), COL4A3 (LRG_230t1, NM_000091.4) and COL4A4 (LRG_231t1, NM_000092.4), and the nomenclature guidelines of the Human Genome Variation Society (HGVS) [16]. Variant descriptions were confirmed with the Mutalyzer programme (http://mutalyzer.nl/) [17]. Pathogenicity assessments were as published or as provided by the submitting laboratory. In general these were based on the HGVS definitions [18]. Variants were not checked independently with any algorithms such as the Human Splicing Finder because their efficacy and accuracy is not confirmed in the genes of interest. Our definition for splicing mutations included the 10 intronic nucleotides immediately adjacent to the exon-intron boundaries. Deep intronic variants were only included where the submitting laboratory believed them to be disease-causing.
Variants, demographic and clinical data were added to the LOVD databases that included all published COL4A5, COL4A3 and COL4A4 variants. Disease-associated variants already recorded in the database but from apparently unrelated families were included to identify the prevalence of Alport syndrome and any common or founder mutations. Mode of inheritance, gender and clinical features were noted where available.
Analysis of the LOVD COL4A5, COL4A3 and COL4A4 databases
Different variant types (deletions, insertions, short deletions, arbitrarily chosen as < than 26 bp, and splicing variants) direct or indirect nonsense mutations, missense variants (resulting in Gly or non-Gly substitutions, pathogenic or not, and synonymous changes) were counted. Each variant was included once only in the analysis.
Large deletions were mapped against the collagen sequence. Missense variants corrected for exon size were compared between exons to determine any increase in variants or gaps.
In addition, COL4A5 missense mutations were tested for a preferential occurrence in CpG-associated codons [19] (especially involving the first G of Gly) using an in-house Python script (www.python.org). CpG sites have an increased tendency for methylcytosine to spontaneously deaminate to thymine. CG>TG and the complementary changes have a 10 times higher mutation rate than normal.
The prevalence of Gly substitutions with each of 8 other amino acids or a stop codon was compared with the expected distribution determined from collagen codon preferences [20].
Age at onset of renal failure
The nature of mutations was correlated with early and late onset renal failure, using the criteria already identified for COL4A5 ‘severe’ variants (copy number variants, frameshifts, nonsense and splice site mutations). Only one age was used for the variant in each family, and for COL4A5 variants, the ages at renal failure in males and females were studied separately. Where several members of the same family or unrelated individuals with the same mutation were described, the youngest age of onset of renal failure was recorded. For COL4A3 and COL4A4, the age at end-stage renal failure in males and females were combined, and the number of severe variants recorded as none, one or two.
Statistical analysis
Results were expressed as mean and SD, and results compared using Fisher’s exact test (DNA Stata). A p value of less than 0.05 was considered significant, and a p values less than 0.10, a trend.
Results
Twelve laboratories involved worldwide in mutation testing for Alport syndrome collaborated to submit 754 novel variants (504 COL4A5, 133 COL4A3, 117 COL4A4 variants) to the LOVD databases. These contributions increased the total number of variants in the databases by 25% (754/2959), to 1168 unique variants for COL4A5 (1951 total), 266 for COL4A3 (518 total) and 268 for COL4A4 (490 total) (Table 2). Most variants were pathogenic, or of unknown significance, from individuals with Alport syndrome or Thin basement membrane nephropathy. Most were from Europeans with COL4A5 variants, or were from Chinese (72 pathogenic, 12 normal), African (10, 8 normal), Hispanic (7 pathogenic) or Indian (2 normal) people.
Table 2. Frequency of variants in COL4A5, COL4A3 and COL4A4 genes in LOVD databases.
| COL4A5 LOVD | COL4A3 LOVD | COL4A4 LOVD | |
|---|---|---|---|
| Total number of variants | 1168 unique (1951 total) | 266 unique (518 total) | 268 unique (490 total) |
| Rearrangements or deletions, copy number variants | 76 (7%) | 0 | 3 (1%) |
| Duplications | 13 (1%) | 3 (1%) | 4 (1%) |
| Insertions | 55 (5%) | 8 (3%) | 8 (3%) |
| Deletions (1-26bp) | 169 (14%) | 31 (12%) | 32 (12%) |
| Indels (with both insertions and deletions) | 16 (1%) | 3 (1%) | 2 (1%) |
| Splicing variants | 273 (152+ and 121-)* (23%) | 25 (15+ and 10-)* (9%) | 52 (25+ and 27-)* (19%) |
| Nonsense codons (direct) | 73 (6%) | 13 (5%) | 13 (5%) |
| Total potential nonsense mutations (direct and downstream) | 329 + 73 = 402 (34%) | 45 + 13 = 47 (18%) | 13 + 49 = 62 (23%) |
| Missense substitutions | 504 (43%) | 136 (51%) | 107 (40%) |
| • Gly | 391 (33%) | 66 (25%) | 46 (17%) |
| • Non-Gly | 113 (10%) | 70 (26%) | 61 (23%) |
| Non-pathogenic changes | 24 | 78 | 65 |
| • Synonymous | 11 | 44 | 28 |
| • Non-Gly missense | 7 | 28 | 14 |
| • Deep intronic | 6 | 6 | 23 |
* For splicing variants, + indicates that the variant is located within the intron immediately adjacent to the 3’ end of the exon; and—indicates that the exon is within the intron immediately adjacent to the 5’ end of the exon.
DNA variants in COL4A5
The 1168 COL4A5 unique variants included 76 complex mutations or deletions of more than 26 nucleotides (7%), 55 insertions (5%), 13 duplications (1%), 169 short deletions (with fewer than 26 nucleotides, 14%), and 16 indels (1%), 73 nonsense mutations (6%), 504 missense variants (43%) and 273 (23%) splicing variants (Table 2). Uncommonly two pathogenic variants were found in the same individual (11/1168, 1%).
Large deletions/indels/rearrangements
There were 25 large deletions. Seven originated in COL4A6 and terminated in exon 1 or the nearby intron of COL4A5. Eight large deletions originated in exons 1–4. Only 9 deletions (fewer than half) had their origins after exon 4, and six at exons 37 to 41.
Direct nonsense mutations
There were 73 (6%) direct nonsense mutations but the insertions/deletions and duplications that result in a frameshift meant that there were potentially 402 variants producing a downstream nonsense change (34%). In addition, 17 direct stop codons resulted from Gly substitutions (4%) which matched the expected prevalence (4%) (Table 3).
Table 3. Observed versus expected likelihood of Glycine substitutions in the COL4A5, COL4A3 and COL4A4 genes in Alport syndrome and Thin basement membrane nephropathy.
Observed number from the LOVD databases; expected number derived from data for collagen I [20].
| Ala | Ser | Cys | Arg | Val | Glu | Asp | Trp | Stop | |
|---|---|---|---|---|---|---|---|---|---|
| Expected (based on residue frequency and collagen coding preferences) | 13% | 18% | 6% | 26% | 10% | 14% | 14% | 1% | 4% |
| COL4A5 observed (n = 391) | 15 (4%) (p less than 0.0001) | 45 (11%) (p = 0.02) | 19 (5%) | 86 (22%) | 64 (16%) | 56 (14%) | 68 (17%) | 3 (1%) | 17 (4%) |
| COL4A3 observed (n = 66) | 0 (0%) | 8 (12%) | 6 (9%) | 23 (35%) | 6 (9%) | 13 (20%) | 8 (12%) | 1 (1%) | 1 (1%) |
| COL4A4 observed (n = 46) | 1 (2%) (p = 0.06) | 7 (15%) | 3 (7%) | 15 (33%) | 4 (9%) | 11 (24%) | 3 (7%) | 1 (2%) | 1 (2%) |
p- not significant for all other comparisons
Missense mutations
There were 504 missense variants (43%) of which 391 (33%) were Gly substitutions, and 113 (10%) non-Gly substitutions. Thus Gly in COL4A5 was substituted three times more often than other amino acids. Glycine can be substituted nine ways (Table 3), and the substitutions with each residue occurred as often as expected except for Ala which was underrepresented (4% compared with 13% expected). Ala is the least destabilising amino acid, and results in a milder clinical phenotype, which may have been overlooked clinically (p<0.0001)[20].
Of the 113 non-Gly substitutions, the most common replacements were of Arg, Asp, Glu, Ser (26, 9.5%), Val (10, 4%) or Ala (7, 2.6%), to Gly.
Only 135 of the 432 Gly in the collagenous sequence were substituted (31%). This was comparable with COL1A1 where fewer than 10% of all variants were calculated to be known (Marini, 2007). Most substitutions were with one amino acid only.
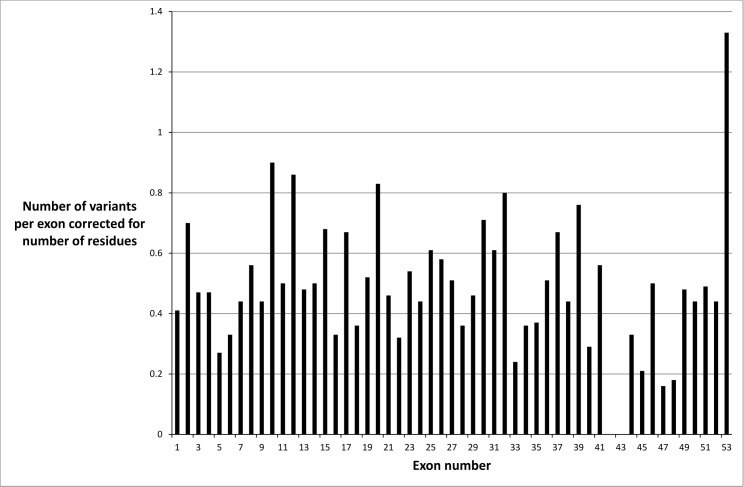
Missense variants were not distributed evenly throughout the COL4A5 gene when exon size was taken into account (Fig 1). There were fewer mutations in exons 47 and 48, which suggested a milder phenotype or an ascertainment bias. It was unlikely that the fewer mutations were due to an embryonic lethal phenotype because the collagen IV α3α4α5 network is mainly expressed after infancy. However exons 47 and 48 correspond to the collagen IV α5 chain before the carboxy terminus, where there is a ‘gap’ for entry of the cleavage enzymes, and the major epithelial binding site on the parallel IV alpha3 chain [21]. Mutations were also underrepresented in exons 42 and 43 probably because these exons are small and not routinely sequenced.
Fig 1. Distribution of missense variants in the COL4A5 gene in each exon corrected for exon size and demonstrating a non-uniform distribution.
CpG site mutations
There were 70 CpG sites among the 5076 nucleotide positions in COL4A5. Of all the mutations causing Alport syndrome, 89 were present at CpG sites which was more often than by chance (p less than10- 41 by Fisher’s exact test). Nonsense mutations were also observed more often at CpG sites. Of the 89 mutations at 70 CpG sites, 27 were nonsense changes at 5 sites (p less than 0.001 by Fisher’s exact test).
Gly substitutions within the collagenous domains were also overrepresented in COL4A5 mutations. Of the 491 missense mutations affecting the 1347 amino acid positions in collagen IV α5 chain, 435 (89%) were substitutions of one of the 449 collagenous domain Gly residues (p<0.001). Similarly, at the CpG sites, 46 missense mutations were observed at 43 CpG sites within the collagenous domains, with 39 resulting in Gly substitutions at 12 Gly positions (p less than 10−7 by Fisher’s exact test).
Splice site mutations
There were 273 splicing variants (23% of total number) with a variant within 10 nucleotides of the intron-exon boundary that was possibly pathogenic. At least one variant was identified for each intron, and there were 152 + (at the 3’ end of the exon) and 121 –(at the 5’ end of the exon) splice variants.
Founder mutations
Two hundred and sixty-five pathogenic variants (23%) were recorded more than once, but only 12 reported at least five times in apparently unrelated families (Table 4). Three were biochemically ‘severe’ mutations being a duplication or nonsense variant. A further three were Gly substitutions. Six resulted in renal failure at less than 30 years of age, and five in a milder phenotype with later onset renal failure.
Table 4. Common Founder mutations in the COL4A5, COL4A3 and COL4A4 genes.
| Variant | Number of reports/families | Ethnicity | Clinical features, Age at onset of renal failure | Reference | |
|---|---|---|---|---|---|
| COL4A5 | Tandem duplication of 35 exons | >160 individuals | French Polynesian | ESRF at 31 years, lenticonus | [28] |
| p.Gly325Arg | Reported 9 times | France, European | ESRF at 22–76 years, hearing loss | [29] | |
| p.Arg373* | Reported 6 times | Italian | ESRF at 15 years, hearing loss | [30] | |
| p.Gly624Asp | Reported at least 5 times | European | ESRF late | [31,32] | |
| p.Gly869Arg | Reported 12 times | Italian | ESRF at 10 years | [30] | |
| p.Ser916Gly | Reported 5 times | US | Not available | [33] | |
| p.Gly953Val | Reported 6 times | France | ESRF at 15 years, hearing loss | [34] | |
| p.Gly1030Ser | Reported 5 times | N European, US | ESRF at 37 years, hearing loss | [31] | |
| p.Arg1569Gln | Reported 7 times | US, France | ESRF at 29 years, hearing loss | [34,35] | |
| p.Leu1655Arg | Reported in 9 families | Western US | ESRF at 40 years, hearing loss | [36] | |
| p.Arg1683Gln | Reported 5 times | Ashkanazi-American | ESRF 40–62 years | [37] | |
| p. Arg1683* | Reported 6 times | British | ESRF at 27 years, hearing loss | [38] | |
| COL4A3 | c.40-63del | One in 183 Ashkenazi | Askhkenazi-American | ARAS | [39] |
| p.Gly43Arg | Reported 5 times | European, US | TBMN, AR AS | [40] | |
| p.Glu162Gly | Reported 8 times | European, US | TBMN, AR AS | [40] | |
| p.Gly695Arg | Reported 5 times | British | Haematuria, TBMN | [41] | |
| p.Gly871Cys | 6 families | Cypriot | Haematuria, TBMN and FSGS | [42,43] | |
| p.Gly1334Glu | 4 families | Cypriot | TBMN, FSGS | [42,43] | |
| p. Gln1495Arg | Reported 5 times | European, British, Cypriot | AR AS, TBMN | [44] | |
| p. 13_22 del LPLLLVLL | 1:183 of Ashkanazi | American Ashkenazi | AR AS | [39] | |
| COL4A4 | p.Gly545Ala | Reported 11 times | Turkish, German | TBMN, AR AS | [45] |
| c.2384-5T>C | Reported 6 times | Italian | TBMN | [44] | |
| p.A880Hisfs69* | Reported 5 times | Australian | AR AS | [46] | |
| p.Gly960Arg | Reported 6 times | Spanish | TBMN | [47] | |
| p.Ser969* | Reported 14 times | British | AR AS with early onset ESRF; TBMN | [48] |
*stop codon; ESRF–end-stage renal failure, FSGS–focal segmental glomerulosclerosis, AR AS–autosomal recessive Alport syndrome, TBMN–thin basement membrane nephropathy
Some of these mutations will have arisen from independent events and others from a single individual, but the distinction can only be made with haplotype analysis. These mutations allow us to examine the consistency of age of onset of renal failure and the effect of modifying genes. More founder mutations are likely but laboratories may not have not reported them because of their ‘lack of novelty’.
Genotype-phenotype correlation
Clinical data were most complete for age at end-stage renal failure. Formal hearing and ophthalmic examinations were rarely reported to the testing laboratory.
Reported mutations were associated more often with a severe phenotype. The age at onset of renal failure was available for 237 COL4A5 mutations (12%) and the mean age was 24.3 years (Table 5). More mutations (162, 68%) were associated with end-stage renal failure before the age of 30 years, than later (75, 32%).
Table 5. Age at end-stage renal failure and severity of mutations with COL4A5, COL4A3 and COL4A4 variants.
| COL4A5 | COL4A3 | COL4A4 | |
|---|---|---|---|
| Total number with end-stage renal failure and all causative mutations identified | 237 | 75 | 48 |
| Age at renal failure for all patients with mutations in this gene (mean, SD, years) | 24.4, 7.8 (23.4 to 25.4) (n = 237) (COL4A5 and COL4A3, p = 0.45; COL4A5 and COL4A4, p = 0.55) | 23.3, 9.3 (20.1 to 26.5) (n = 35) (COL4A3 and COL4A4, p = 0.41). | 25.4, 10.3 (21.3 to 29.6) (n = 26) |
| End-stage renal failure at < 30 years | 162 (68%) | ||
| Number of homozygous mutations | N/A | 16 (21%) | 15 (31%) |
| Number of compound heterozygous mutations | N/A | 59 (79%) | 33 (69%) |
| Age at onset of renal failure | |||
| No severe mutation (mean, SD, years) | 26.7, 8.1 (n = 106) | 24.0, 6.9 (n = 8) | 26.6, 5.0 (n = 9) |
| One severe mutation (mean, SD, years) | 22.5, 7.0 (n = 131) (one compared with none, p less than 0.01) | 20.8, 5.1 (n = 8) | 25.5, 7.8 (n = 4) |
| Two severe mutations (mean, SD, years) | Not applicable | 17.6, 8.5 (n = 14) (two compared with none, p = 0.08) | 21.1, 3.6 (n = 10) (two compared with none, p = 0.01) |
| Direct nonsense mutation | 21.4, 6.4 (16.9 to 25.9) (n = 12) | 17.9, 5.5, (13.3 to 22.5) (n = 8) | |
‘Severe’ mutations were large rearrangements, deletions, insertions or other changes resulting in a nonsense codon.
A severe mutation (rearrangement, large deletion, insertion/deletion, or nonsense change) was associated with a younger average age at onset of renal failure of 22.5 ± 7.0 years (n = 131) than in those without a severe mutation (26.7 ± 8.1 years, n = 106, p less than 0.01).
A direct nonsense mutation, that is where a codon was replaced by a stop signal, was associated with a younger average age at onset of renal failure of 21.4 ± 6.4 years than with other mutations types (p = 0.03).
There was no difference in the age at onset of renal failure in individuals with Gly substitutions with Arg, Glu or Asp compared with other amino acids, nor with non-Gly substitutions (p all greater than 0.05).
COL4A5 mutations were reported in 68 females. Nine of the 23 (39%) who developed renal failure had a direct or indirect nonsense mutation or rearrangement or insertion/deletion, in contrast to 9 of the 45 without renal failure (20%, p = 0.14).
DNA variants in COL4A3 and COL4A4
Many fewer DNA variants have been described for the COL4A3 and COL4A4 genes, presumably because autosomal recessive Alport syndrome is less common, and genetic testing is performed infrequently for Thin basement membrane nephropathy.
DNA variants in the LOVD databases affected the COL4A3 and COL4A4 genes equally frequently (Table 2). The COL4A3 database had 266 unique variants, and COL4A4 had 268. Compound heterozygous variants were more common than homozygous variants for both COL4A3 and COL4A4 (69% and 59% respectively) suggesting lesser consanguinity.
Large deletions/indels/rearrangements
The proportions of duplications, insertions, short deletions and other indels were similar to those found in COL4A5.
Missense
Glycine substitutions were relatively less abundant than for COL4A5 (Table 2). This was unexpected since they are the most common change in X-linked Alport syndrome and other collagen diseases such as osteogenesis imperfecta [20]. The explanation may be that milder disease with heterozygous Gly substitutions was overlooked. Gly to Ala or Ser substitutions were again underrepresented for the COL4A3 and COL4A4 genes.
Too few missense variants were found to determine any mutational hot spots. Non-Gly substitutions were more common than in COL4A5, but their pathogenic significance was often not clear.
Nonsense
The % of direct nonsense mutations was similar to COL4A5 but the total % (of direct and potential indirect nonsense mutations) was fewer because fewer large insertions/deletions were reported.
Splicing
Very few potential splicing mutations were identified within the first 10 nucleotides of the intron-exon boundaries.
Founder mutations
Founder mutations were reported in both COL4A3 and COL4A4 in autosomal recessive Alport syndrome and Thin basement membrane nephropathy (Table 5). For example, the S969X variant in COL4A4 was the commonest variant in British populations, and other variants were reported from Cyprus.
Genotype-phenotype correlations
Clinical features were compared between individuals with autosomal recessive Alport syndrome caused by COL4A3 or COL4A4 pathogenic mutations and those with X-linked Alport syndrome and COL4A5 mutations. The age at onset of renal failure was known for some homozygous or compound heterozygous COL4A3 (n = 35) and COL4A4 (n = 25) pathogenic mutations where both variants were known. Overall the mean age at onset of end-stage renal failure was not different for individuals with COL4A3 (23.2 + 9.3, years, 95% CI 20.1 to 26.5) or COL4A4 mutations (mean 25.4 ±10.3 years, 95% CI 21.3 to 29.6) compared with COL4A5 mutations (24.4 ± 7.8 years, 95% CI 23.4 to 25.4, n = 237) (Table 4).
The age at onset of renal failure was also the same for homozygous and for compound heterozygous COL4A3 and COL4A4 mutations (21.5 ± 7.2, 95%CI 17.2–25.9, n = 13; and 21.8 ± 7.0, 95% CI 19.8 to 23.7, n = 52).
Subjects with two severe mutations (direct or indirect nonsense mutations, insertions/deletions) developed end-stage renal failure at a younger age than those with none (Table 4). Individuals with at least one direct nonsense COL4A3 or COL4A4 mutation also developed renal failure at a younger age than those with none. Thus, the age at onset of renal failure in autosomal recessive Alport syndrome depended on mutation severity using the same criteria as for COL4A5 mutations [11,12]. However this study did not demonstrate any difference in the age at end-stage renal failure due to Gly substitutions with Arg, Glu or Asp compared with other substitutions for COL4A3 and COL4A4.
Twenty COL4A3 or COL4A4 mutations were identified in the LOVD databases in individuals diagnosed clinically with autosomal dominant Alport syndrome. Seven only had developed end-stage renal failure, at a mean age of 42 ± 15.1 years (range 20–60). The others either did not have renal failure or the age at onset was not known.
Biallelic and digenic mutations
There are occasional reports of individuals with two COL4A5 mutations [22,23], and others with autosomal recessive Alport syndrome and mutations in both COL4A3 and COL4A4 with a later age at onset of renal failure [15,24,25].
Discussion
This study confirmed that COL4A5, and COL4A3 and COL4A4 mutations in Alport syndrome all resulted in end-stage renal failure at the same age. Other studies have found that COL4A5 deletions, insertions, and nonsense variants, are associated with early onset renal failure [11,12,13]. This study confirmed that two COL4A3 or COL4A4 mutations resulted in earlier onset renal failure than one or no severe mutations. Nonsense COL4A3 and COL4A4 mutations were also associated with a younger age at renal failure. Cohort studies indicate that hearing loss is common with all mutation types, and that the same mutation types that cause early onset renal failure in X-linked disease are associated with lenticonus and retinopathy [14].
The number of different published COL4A5 variants has increased from 176 in 1997 [26], to 520 by 2010 [27], and 1200 in 2015. Many more pathogenic variants have been reported for COL4A5 than for COL4A3 and COL4A4. There was some bias in the types of mutations reported here (deletions and insertions were more common for COL4A5) probably because of the detection techniques.
The major challenge for the future is to encourage all diagnostic laboratories including those that are commercially-based, to submit their variants to a database, together with accurate pathogenicity assessments and as much clinical information as possible. The classification of some so- called benign variants will be revised. We will understand better genotype-phenotype correlations for heterozygous COL4A3 and COL4A4 mutations in Thin basement membrane nephropathy, and any genetic distinctions from variants that cause autosomal dominant Alport syndrome. In addition we will better understand the consequences of biallelic, triallelic and digenic COL4 mutations [25], and possibly why some heterozygous COL4 mutations are associated with proteinuria.
Increasingly the value of sharing variants and pathogenicity assessments is realised. Some countries allow all variants identified in public laboratories to be shared in public databases without specific assent, and IRBs internationally are finding that the public benefit of sharing outweighs any small adverse risk. Our study exemplifies the advantages of sharing variants in a rare disease.
Acknowledgments
This work was presented in part at the Alport Mutation Updates at Oxford University, January 2014 and at the University of Gottingen, September 2015. We would like to thank the many patients who allowed us to use their data, and the physicians who referred them. We would also like to acknowledge mutations submitted by the ARUP laboratory, Salt Lake City, USA, and by the Hopital Necker–Enfants-malades, Paris, France.
Data Availability
All primary data are available at the LOVD websites for COL4A5, COL4A3 and COL4A4 genes (www:http://www.LOVD.nl).
Funding Statement
JS received seed funding to establish the Alport mutation databases from the US Alport Foundation and the Australian Alport Foundation ($USD 10,000 and $AUD 20,000 respectively). HIC, HGK and EP received funding from the Korean government through the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea ($USD142,000) (HI12C0014).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grunfeld JP, Joly D (1997) [Hereditary kidney diseases in adults]. Rev Prat 47: 1566–1569. [PubMed] [Google Scholar]
- 2.Savige J, Gregory M, Gross O, Kashtan C, Ding J, et al. (2013) Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol 24: 364–375. 10.1681/ASN.2012020148 [DOI] [PubMed] [Google Scholar]
- 3.Barker DF, Hostikka SL, Zhou J, Chow LT, Oliphant AR, et al. (1990) Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248: 1224–1227. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki T, Lemmink HH, Mariyama M, Antignac C, Gubler MC, et al. (1994) Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat Genet 8: 77–81. [DOI] [PubMed] [Google Scholar]
- 5.Feingold J, Bois E, Chompret A, Broyer M, Gubler MC, et al. (1985) Genetic heterogeneity of Alport syndrome. Kidney Int 27: 672–677. [DOI] [PubMed] [Google Scholar]
- 6.Lemmink HH, Nillesen WN, Mochizuki T, Schroder CH, Brunner HG, et al. (1996) Benign familial hematuria due to mutation of the type IV collagen alpha4 gene. J Clin Invest 98: 1114–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzza M, Wang YY, Dagher H, Babon JJ, Cotton RG, et al. (2001) COL4A4 mutation in thin basement membrane disease previously described in Alport syndrome. Kidney Int 60: 480–483. [DOI] [PubMed] [Google Scholar]
- 8.Savige J, Rana K, Tonna S, Buzza M, Dagher H, et al. (2003) Thin basement membrane nephropathy. Kidney Int 64: 1169–1178. [DOI] [PubMed] [Google Scholar]
- 9.van der Loop FT, Heidet L, Timmer ED, van den Bosch BJ, Leinonen A, et al. (2000) Autosomal dominant Alport syndrome caused by a COL4A3 splice site mutation. Kidney Int 58: 1870–1875. [DOI] [PubMed] [Google Scholar]
- 10.Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG (2003) Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Engl J Med 348: 2543–2556. [DOI] [PubMed] [Google Scholar]
- 11.Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M (2002) Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant 17: 1218–1227. [DOI] [PubMed] [Google Scholar]
- 12.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, et al. (2000) X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 11: 649–657. [DOI] [PubMed] [Google Scholar]
- 13.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, et al. (2003) X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a "European Community Alport Syndrome Concerted Action" study. J Am Soc Nephrol 14: 2603–2610. [DOI] [PubMed] [Google Scholar]
- 14.Tan R, Colville D, Wang YY, Rigby L, Savige J (2010) Alport retinopathy results from "severe" COL4A5 mutations and predicts early renal failure. Clin J Am Soc Nephrol 5: 34–38. 10.2215/CJN.01030209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee R, Hoffman M, Cliften P, Seshan S, Liapis H, et al. (2013) Targeted exome sequencing integrated with clinicopathological information reveals novel and rare mutations in atypical, suspected and unknown cases of Alport syndrome or proteinuria. PLoS One 8: e76360 10.1371/journal.pone.0076360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Dunnen JT, Paalman MH (2003) Standardizing mutation nomenclature: why bother? Hum Mutat 22: 181–182. [DOI] [PubMed] [Google Scholar]
- 17.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE (2008) Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat 29: 6–13. [DOI] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schorderet DF, Gartler SM (1992) Analysis of CpG suppression in methylated and nonmethylated species. Proc Natl Acad Sci U S A 89: 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, et al. (2007) Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat 28: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, et al. (2011) Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat 32: 127–143. 10.1002/humu.21401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo C, Van Damme B, Vanrenterghem Y, Devriendt K, Cassiman JJ, et al. (1995) Severe alport phenotype in a woman with two missense mutations in the same COL4A5 gene and preponderant inactivation of the X chromosome carrying the normal allele. J Clin Invest 95: 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammad M, Nanra R, Colville D, Trevillian P, Wang Y, et al. (2014) A female with X-linked Alport syndrome and compound heterozygous COL4A5 mutations. Pediatr Nephrol 29: 481–485. 10.1007/s00467-013-2682-6 [DOI] [PubMed] [Google Scholar]
- 24.Malone AF, Phelan PJ, Hall G, Cetincelik U, Homstad A, et al. (2014) Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int 86: 1253–1259. 10.1038/ki.2014.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mencarelli MA, Heidet L, Storey H, van Geel M, Knebelmann B, et al. (2015) Evidence of digenic inheritance in Alport syndrome. J Med Genet 52: 163–174. 10.1136/jmedgenet-2014-102822 [DOI] [PubMed] [Google Scholar]
- 26.Lemmink HH, Schroder CH, Monnens LA, Smeets HJ (1997) The clinical spectrum of type IV collagen mutations. Hum Mutat 9: 477–499. [DOI] [PubMed] [Google Scholar]
- 27.Crockett DK, Pont-Kingdon G, Gedge F, Sumner K, Seamons R, et al. (2010) The Alport syndrome COL4A5 variant database. Hum Mutat 31: E1652–1657. 10.1002/humu.21312 [DOI] [PubMed] [Google Scholar]
- 28.Arrondel C, Deschenes G, Le Meur Y, Viau A, Cordonnier C, et al. (2004) A large tandem duplication within the COL4A5 gene is responsible for the high prevalence of Alport syndrome in French Polynesia. Kidney Int 65: 2030–2040. [DOI] [PubMed] [Google Scholar]
- 29.Knebelmann B, Deschenes G, Gros F, Hors MC, Grunfeld JP, et al. (1992) Substitution of arginine for glycine 325 in the collagen alpha 5 (IV) chain associated with X-linked Alport syndrome: characterization of the mutation by direct sequencing of PCR-amplified lymphoblast cDNA fragments. Am J Hum Genet 51: 135–142. [PMC free article] [PubMed] [Google Scholar]
- 30.Renieri A, Bruttini M, Galli L, Zanelli P, Neri T, et al. (1996) X-linked Alport syndrome: an SSCP-based mutation survey over all 51 exons of the COL4A5 gene. Am J Hum Genet 58: 1192–1204. [PMC free article] [PubMed] [Google Scholar]
- 31.Martin P, Heiskari N, Zhou J, Leinonen A, Tumelius T, et al. (1998) High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol 9: 2291–2301. [DOI] [PubMed] [Google Scholar]
- 32.Pierides A, Voskarides K, Kkolou M, Hadjigavriel M, Deltas C (2013) X-linked, COL4A5 hypomorphic Alport mutations such as G624D and P628L may only exhibit thin basement membrane nephropathy with microhematuria and late onset kidney failure. Hippokratia 17: 207–213. [PMC free article] [PubMed] [Google Scholar]
- 33.Barker DF, Denison JC, Atkin CL, Gregory MC (2001) Efficient detection of Alport syndrome COL4A5 mutations with multiplex genomic PCR-SSCP. Am J Med Genet 98: 148–160. [DOI] [PubMed] [Google Scholar]
- 34.Knebelmann B, Breillat C, Forestier L, Arrondel C, Jacassier D, et al. (1996) Spectrum of mutations in the COL4A5 collagen gene in X-linked Alport syndrome. Am J Hum Genet 59: 1221–1232. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Gregory MC, Hertz JM, Barker DF, Atkin C, et al. (1993) Mutations in the codon for a conserved arginine-1563 in the COL4A5 collagen gene in Alport syndrome. Kidney Int 43: 722–729. [DOI] [PubMed] [Google Scholar]
- 36.Barker DF, Pruchno CJ, Jiang X, Atkin CL, Stone EM, et al. (1996) A mutation causing Alport syndrome with tardive hearing loss is common in the western United States. Am J Hum Genet 58: 1157–1165. [PMC free article] [PubMed] [Google Scholar]
- 37.Barker DF, Denison JC, Atkin CL, Gregory MC (1997) Common ancestry of three Ashkenazi-American families with Alport syndrome and COL4A5 R1677Q. Hum Genet 99: 681–684. [DOI] [PubMed] [Google Scholar]
- 38.Plant KE, Green PM, Vetrie D, Flinter FA (1999) Detection of mutations in COL4A5 in patients with Alport syndrome. Hum Mutat 13: 124–132. [DOI] [PubMed] [Google Scholar]
- 39.Webb BD, Brandt T, Liu L, Jalas C, Liao J, et al. (2014) A founder mutation in COL4A3 causes autosomal recessive Alport syndrome in the Ashkenazi Jewish population. Clin Genet 86: 155–160. 10.1111/cge.12247 [DOI] [PubMed] [Google Scholar]
- 40.Heidet L, Arrondel C, Forestier L, Cohen-Solal L, Mollet G, et al. (2001) Structure of the human type IV collagen gene COL4A3 and mutations in autosomal Alport syndrome. J Am Soc Nephrol 12: 97–106. [DOI] [PubMed] [Google Scholar]
- 41.Wang YY, Rana K, Tonna S, Lin T, Sin L, et al. (2004) COL4A3 mutations and their clinical consequences in thin basement membrane nephropathy (TBMN). Kidney Int 65: 786–790. [DOI] [PubMed] [Google Scholar]
- 42.Pierides A, Voskarides K, Athanasiou Y, Ioannou K, Damianou L, et al. (2009) Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant 24: 2721–2729. 10.1093/ndt/gfp158 [DOI] [PubMed] [Google Scholar]
- 43.Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, et al. (2007) COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol 18: 3004–3016. [DOI] [PubMed] [Google Scholar]
- 44.Longo I, Porcedda P, Mari F, Giachino D, Meloni I, et al. (2002) COL4A3/COL4A4 mutations: from familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int 61: 1947–1956. [DOI] [PubMed] [Google Scholar]
- 45.Ozen S, Ertoy D, Heidet L, Cohen-Solal L, Ozen H, et al. (2001) Benign familial hematuria associated with a novel COL4A4 mutation. Pediatr Nephrol 16: 874–877. [DOI] [PubMed] [Google Scholar]
- 46.Dagher H, Yan Wang Y, Fassett R, Savige J (2002) Three novel COL4A4 mutations resulting in stop codons and their clinical effects in autosomal recessive Alport syndrome. Hum Mutat 20: 321–322. [DOI] [PubMed] [Google Scholar]
- 47.Badenas C, Praga M, Tazon B, Heidet L, Arrondel C, et al. (2002) Mutations in theCOL4A4 and COL4A3 genes cause familial benign hematuria. J Am Soc Nephrol 13: 1248–1254. [DOI] [PubMed] [Google Scholar]
- 48.Storey H, Savige J, Sivakumar V, Abbs S, Flinter FA (2013) COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J Am Soc Nephrol 24: 1945–1954. 10.1681/ASN.2012100985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All primary data are available at the LOVD websites for COL4A5, COL4A3 and COL4A4 genes (www:http://www.LOVD.nl).