Abstract
It has been proposed that miRNAs are involved in the control of telomeres. We test that hypothesis by examining the association between miRNAs and telomere length (TL). Additionally, we evaluate if genetic variation in telomerase reverse transcriptase (TERT) is associated with miRNA expression levels. We use data from a population-based study of colorectal cancer (CRC), where we have previously shown associations between TL and TERT and CRC, to test associations between TL and miRNA expression and TERT and miRNA expression. To gain insight into functions of miRNAs associated with TERT we tested linear associations between miRNAs and their targeted gene mRNAs. An Agilent platform that contained information on over 2000 miRNAs was used. TL was measured using a multiplexed quantitative PCR (qPCR). RNAseq was used to assess gene expression. Our sample consisted of 1152 individuals with SNP data and miRNA expression data; 363 individuals with both TL and miRNA; and 148 individuals with miRNA and mRNA data. Thirty-three miRNAs were directly associated with TL after adjusting for age and sex (false discovery rate (FDR) of 0.05). TERT rs2736118 was associated with differences in miRNA expression between carcinoma and normal colonic mucosa for 75 miRNAs (FDR <0.05). Genes regulated by these miRNAs, as indicated by mRNA/miRNA associations, were associated with major signaling pathways beyond their TL-related functions, including PTEN, and PI3K/AKT signaling. Our data support a direct association between miRNAs and TL; differences in miRNA expression levels by TERT genotype were observed. Based on miRNA and targeted mRNA associations our data suggest that TERT is involved in non-TL-related functions by acting through altered miRNA expression.
Introduction
MicroRNAs (miRNA) are non-coding RNAs that function as gene regulators [1]. As such, they provide a mechanism whereby genes involved in various signaling pathways can be regulated simultaneously [2]. Both aging and cellular senescence are thought to be regulated in part by miRNAs [3, 4]. It also has been suggested that miRNAs control the function of telomeres in cancer [5]. Telomeres, located at the end of chromosomes, shorten with every cell division; when they become too short the cell is unable to divide, leading to the induction of senescence and apoptosis, which are key features of normal cell function, but absent in tumor cells. Telomere length (TL) has been associated with colorectal cancer, with higher risk of disease for both long and short telomeres [6, 7]. Telomerase, a ribonucleoprotein enzyme that is essential for the replication of chromosome ends, is low or non-expressing in normal human somatic cells, but has been shown to be expressed in the majority of tumor cells [8]. Telomerase reverse transcriptase (TERT) is essential for maintenance of telomere length by protecting chromosome ends. TERT has been proposed to regulate miRNAs by regulation of miRNA biogenesis, with diminished primary miRNA expression in TERT-reduced cells [9], and has been shown to be associated with both TL and cancer [6, 10–14]. TERT is thought to be one of the most critical, if not the rate-limiting step, in production of telomerase activity [8, 15]. We have shown previously that TERT rs2853676 was associated with TL and that TL was associated with colorectal cancer [11]. Additionally TERT-CLPTM1L rs2853668 and TERT rs2736118 were associated with colorectal cancer risk in this study population [14].
In this paper we examine the associations between TL and miRNA expression in colorectal tissue. Additionally we test the hypothesis that variation in TERT influences miRNA expression levels. We focus on associations between miRNAs and TERT SNPs previously associated with colorectal cancer and examine miRNA expression in normal colonic mucosa tissue as well as the differential expression of miRNAs between carcinoma and normal colonic mucosa tissue.
Materials and Methods
Study population
The study population consisted of individuals previously enrolled in a study of Diet, Lifestyle, and Colon cancer at the University of Utah and the Kaiser Permanente Medical Research Program (KPMRP) [16] for whom both miRNA from carcinoma and adjacent normal colonic mucosa tissue were available. Study subjects included incident cases of colon cancer between the ages of 30 and 79 who were non-Hispanic white, Hispanic, or African American, and were able to provide a signed informed consent prior to participation in the study. The study was approved by the University of Utah Institutional Review Board for Human Subjects.
miRNA processing
RNA (miRNA) was extracted from formalin-fixed paraffin embedded tissues. We assessed slides and tumor blocks that were prepared over the duration of the study prior to the time of miRNA isolation to determine their suitability. The study pathologist reviewed slides to delineate carcinoma and normal colonic mucosa tissue. Cells were dissected from 1–4 sequential sections on aniline blue stained slides using an H&E slide for reference. Total RNA containing miRNA was extracted, isolated, and purified using the RecoverAll Total Nucleic Acid isolation kit (Ambion); RNA yields were determined using a NanoDrop spectrophotometer. 100 ng total RNA was labeled with Cy3 and hybridized to Agilent Human miRNA Microarray V19.0 and were scanned on an Agilent SureScan microarray scanner model G2600D. The Agilent Human microarray was generated using known miRNA sequence information compiled in the Sanger miRBase database v19.0. The microarray contains probes for 2006 unique human miRNAs. The miRNA array contains 60,000 unique human sequences and averages 30 replicates per probe sequence. Data were extracted from the scanned image using Agilent Feature Extract software v.11.5.1.1. Data were required to pass stringent QC parameters established by Agilent that included tests for excessive background fluorescence, excessive variation among probe sequence replicates on the array, and measures of the total gene signal on the array to assess low signal. If samples failed to meet quality standards for any of these parameters, the sample was re-labeled, hybridized to arrays, and scanned. If a sample failed QC assessment a second time the sample was deemed to be of poor quality and the individual was excluded from down-stream analysis. A 75th percentile scaling was performed to normalize across arrays was done using preprocessCore [17] (www.bioconductor.org) to minimize differences that could be attributed to the array, amount of RNA, location on array, or other factors that could erroneously influence expression measurements. Testing of both reliability and comparative validity were done [18]. We showed a 98% repeatability in the Agilent platform between repeat samples. Additionally, the Agilent platform had fairly good agreement with Nanostring results and 100% agreement in expression values and fold change with qPCR results [19].
RNA-Seq Sequencing Library Preparation and Data Processing
Total RNA was available from 197 carcinoma and normal mucosa pairs. These samples were taken from the study subjects used for miRNA analysis and were extracted, isolated and purified in the same manner as previously described [20]. RNA library construction was done with the Illumina TruSeq Stranded Total RNA Sample Preparation Kit with Ribo-Zero. The samples were then fragmented and primed for cDNA synthesis, adapters were then ligated onto the cDNA, and the resulting samples were then amplified using PCR; the amplified library was then purified using Agencount AMPure XP beads. A more detailed description of the methods can be found in our previous work [21].
Sequencing was done using an Illumina TruSeq v3 single read flow cell and a 50 cycle single-read sequence run was performed on an Illumina HiSeq instrument. Reads were then aligned to a sequence database containing the human genome (build GRCh37/hg19, February 2009 from genome.ucsc.edu) and alignment was performed using novoalign v2.08.01. Python and a pysam library were used to calculate counts for each exon and UTR of the genes using a list of gene coordinates obtained from http://genome.ucsc.edu. We dropped features that were not expressed in our data or for which the expression was missing for the majority of samples. A more detailed description of the methods can be found in our previous work [21].
Telomere Length (TL) Telomere-related Gene Analysis
TL was measured using a multiplexed quantitative PCR (qPCR) method previously described by Cawthon [22]. This method modified earlier qPCR methods in which telomere signals were measured separately from single copy gene signals in order to produce a T/S ratio corresponding to TL, which is proportional to the average telomere length in a cell. Therefore a larger T/S ratio represents a longer telomere length. The multiplexed PCR analysis uses a single dye and measures both the telomere signals and single copy gene signals in the same well. This is achieved by using CG clamps to stabilize the single copy gene giving it a higher melting point. The unique sequence is then amplified during a different cycle than the telomeric sequence, allowing for a single qPCR to determine the T/S ratio [6].
DNA was obtained from immortalized cell lines from study participants for the colon cancer study and from whole blood for the rectal cancer study. To assure the quality of telomere assay for these samples from different DNA origins we evaluated associations with TL separately for cases and controls from each study and as shown previous, results were similar for both study groups and we concluded that DNA source did not significantly alter TL length in this study. Two samples yielded a T/S ratio greater than three and were excluded. These samples were excluded because we concluded, through gel electrophoresis, that the DNA was degraded and therefore the T/S ratio was unreliable. We focused on Single Nucleotide Polymorphisms (SNPs) that were previously associated with TL and/or colon cancer [6, 14]. We analyzed TERT-CLPTM1L rs2853668 which is 5.1kb upstream of TERT and has been associated with colon cancer in GWAS [23]. We also test TERT rs2736118 and TERT rs2853676 with miRNA expression.
Statistical Analysis
Our sample consisted of 1152 individuals with both SNP data and miRNA expression data; 363 individuals with both TL and miRNA expression data in the normal colonic mucosa; and 148 individuals who had both miRNA and mRNA data, and 176 cases in the colon cancer study and 184 cases in the rectal cancer study had both TL and SNP data.
Three major components of the analysis were conducted. First, we evaluated the linear associations between TL and those miRNAs for which at least 20% of the population had expression (N = 817 miRNA) by fitting a linear regression model to the log base 2 transformed expression levels; we adjusted for age and sex. P-values for each regression analysis were generated from the bootstrap method by creating a distribution of 1,000 β coefficients for each miRNA and evaluating H0: β = 0 vs. H1: β≠0 using the boot package in R. We adjusted for multiple comparisons using a false discovery rate (FDR) level of 0.05 [24]. We standardized the slopes generated from the overall dataset in order to compare the results across the miRNA.
Second, we assigned inheritance models for TERT SNPs based on previous findings for colorectal cancer risk using either dominant or recessive models [14]. We compared log base 2 transformed expression levels across the selected genotype models using the significance analysis of microarrays (SAM) technique implemented in the R package siggenes; p-values were based on 1,000 permutations. We utilized an FDR level of 0.05 to determine which TERT SNP/miRNA associations were significant after adjustment for multiple comparisons.
For both the TL/miRNA analysis and the TERT SNP/miRNA analysis, we evaluated miRNA expression in both normal colonic mucosa and for differentially expressed miRNAs (i.e. difference between carcinoma tissue and normal colonic mucosa). Looking at normal colonic mucosa allowed us to evaluate the effect of TL or SNP on overall miRNA expression. However, since an association with normal colonic mucosa could influence baseline level of the miRNA expressed and could potentially miss important changes in miRNA expression between carcinoma and normal colonic mucosa, we also evaluated miRNA differential expression. This allowed us to gain insight into regulation of miRNAs that could be more directly associated with cancer through their influence on carcinoma miRNA expression; by comparing to normal expression we were able to control for differences in tumor subsite.
One of the goals of the study was to identify non-telomere related mechanisms whereby TERT may be operating through altered miRNA expression levels. To accomplish this, we undertook the third part of the analysis, where we used miRTarBase (www.http://mirtarbase.mbc.nctu.edu.tw/) v6.0 (as of 09/15/2015) [25] to identify validated target genes for the significant miRNAs identified in our TERT SNP/miRNA analysis. We calculated the differential mRNA expression as the difference of the log base 2 of the RPKM (Reads per Kilobase per Million) for the carcinoma and normal mucosa tissues for the 4047 target genes identified by miRTarBase. We then evaluated the 6309 combinations of miRNAs with their respective mRNA target genes. We used the bootstrap method as described above to assess any linear relationship between the differential mRNA and miRNA expression levels, adjusting for age and sex.
QIAGEN’s Ingenuity Pathway Analysis (IPA) (QIAGEN Redwood City, www.qiagen.com/ingenuity) [26] was then used to evaluate the genes that were dysregulated in conjunction with the miRNAs. We considered direct relationships from the IPA knowledge base of genes only, limited to experimentally verified and mammalian results only, and considered all mutations, data sources, and tissues. We report canonical pathways that were significant enriched with these genes after adjustment for multiple comparisons.
Results
The average age of study participants from the colon cancer study was slightly older than from the rectal cancer study (Table 1). The distribution of age, sex, and AJCC stage was similar for those individuals included in the smaller TL dataset as with the larger SNP dataset.
Table 1. Description of study population (includes cases only).
| Colon | Rectal | ||||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Telomere Length (TL)1(T/S ratio) | 1.17 | 0.55 | 1.20 | 0.29 | |
| Age (years) | 65.3 | 9.1 | 61.9 | 10.9 | |
| N | % | N | % | ||
| Sex | Male | 380 | 55.7 | 280 | 59.6 |
| Female | 302 | 44.3 | 190 | 40.4 | |
| AJCC Stage | 1 | 167 | 24.7 | 213 | 45.8 |
| 2 | 220 | 32.6 | 88 | 18.9 | |
| 3 | 222 | 32.9 | 136 | 29.2 | |
| 4 | 66 | 9.8 | 28 | 6.0 | |
| TERT (rs2736118)2 | AA | 341 | 51.5 | 237 | 53.1 |
| AG/GG | 321 | 48.5 | 209 | 46.9 | |
| TERT (rs2853668) | GG/GT | 631 | 93.3 | 445 | 94.7 |
| TT | 45 | 6.7 | 25 | 5.3 | |
| TERT (rs2853676) | CC | 348 | 53.0 | 155 | 58.5 |
| CT/TT | 309 | 47.0 | 110 | 41.5 | |
1 179 individuals in the colon cancer study and 184 individuals in the rectal cancer study had TL, miRNA and SNP data.
2 53 individuals with AA genotype had RNAseq data; 50 individuals with the AG/GG genotype had RNAseq; 148 individuals had both miRNA and mRNA (RNAseq) data.
Assessment of the linear relationship between TL and miRNA expression levels in for the 363 individuals with both TL and miRNA from normal colorectal mucosa showed that 34 miRNAs were associated with TL after adjustment for age and sex (Table 2). Thirty-three of the miRNAs were directly associated with TL, in that TL increased as miRNA expression level increased. Only miR-487b was inversely associated with TL, showing lower levels of miR-487b expression with longer TL length.
Table 2. Associations between TL (T/S Ratio) and miRNA expression levels adjusted for age (FDR<0.05).
| miRNA | Mean Expression | Beta Coefficient1 | P-value2 | Q-Value |
|---|---|---|---|---|
| hsa-miR-1185-2-3p | 6.81 | 0.17 | < .0001 | < .0001 |
| hsa-miR-1207-5p | 10.65 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-1226-5p | 5.5 | 0.16 | 0.002 | 0.0481 |
| hsa-miR-1229-5p | 9.07 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-1247-3p | 4.64 | 0.12 | 0.002 | 0.0481 |
| hsa-miR-134 | 7.84 | 0.16 | < .0001 | < .0001 |
| hsa-miR-3141 | 7.4 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-3158-5p | 4.82 | 0.16 | < .0001 | < .0001 |
| hsa-miR-3162-5p | 11.19 | 0.15 | 0.002 | 0.0481 |
| hsa-miR-3194-5p | 7 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-3196 | 10.19 | 0.18 | < .0001 | < .0001 |
| hsa-miR-3200-5p | 4.75 | 0.17 | 0.002 | 0.0481 |
| hsa-miR-3937 | 5.8 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-4253 | 5.29 | 0.18 | < .0001 | < .0001 |
| hsa-miR-4459 | 14.38 | 0.15 | < .0001 | < .0001 |
| hsa-miR-4496 | 5.74 | 0.17 | < .0001 | < .0001 |
| hsa-miR-4497 | 11.57 | 0.17 | 0.002 | 0.0481 |
| hsa-miR-4530 | 12.75 | 0.15 | < .0001 | < .0001 |
| hsa-miR-4535 | 5.93 | 0.15 | < .0001 | < .0001 |
| hsa-miR-4673 | 6.3 | 0.16 | 0.002 | 0.0481 |
| hsa-miR-4689 | 6.64 | 0.16 | 0.002 | 0.0481 |
| hsa-miR-4739 | 9.71 | 0.18 | < .0001 | < .0001 |
| hsa-miR-487b | 1.6 | -0.17 | < .0001 | < .0001 |
| hsa-miR-5006-5p | 9.36 | 0.15 | < .0001 | < .0001 |
| hsa-miR-5088 | 5.64 | 0.15 | 0.002 | 0.0481 |
| hsa-miR-5195-3p | 7.14 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-575 | 9.18 | 0.15 | < .0001 | < .0001 |
| hsa-miR-601 | 5.31 | 0.15 | < .0001 | < .0001 |
| hsa-miR-6076 | 7.84 | 0.14 | 0.002 | 0.0481 |
| hsa-miR-662 | 5.32 | 0.16 | 0.002 | 0.0481 |
| hsa-miR-718 | 6.59 | 0.18 | < .0001 | < .0001 |
| hsa-miR-762 | 10.72 | 0.15 | 0.002 | 0.0481 |
| hsa-miR-887 | 5.74 | 0.2 | < .0001 | < .0001 |
| hsa-miR-939-5p | 9.06 | 0.14 | < .0001 | < .0001 |
1Beta coefficients are from a linear regression of TL on miRNA expression after adjustment for age, sex, and study
Evaluation of differences in miRNA expression levels between carcinoma tissue and normal colorectal mucosa showed that TERT rs2736118 was associated with differential miRNA expression based on genotype (Table 3). TERT rs2736118 was associated with 75 miRNAs with an FDR of <0.05. Individuals with the AA (homozygous dominant) genotype were more likely to have greater miRNA expression in the tumor than in the normal colorectal mucosa (61% of the time) while individuals with the AG/GG genotypes were more likely to have greater miRNA expression in the normal colorectal mucosa (84% of the time). Neither TERT-CLPTMIL rs2853668 nor TERT rs2853676 significantly altered miRNA expression in normal colonic mucosa or miRNA expression between carcinoma and normal colorectal mucosa.
Table 3. Associations between TERT rs2736118 and miRNA differential expression1 with an FDR of <0.05.
| AA (N = 556) | AG/GG (N = 508) | ||||||
|---|---|---|---|---|---|---|---|
| miRNA | Mean Tumor Expression | Mean Normal Expression | Mean Difference in Expression (T-N) | Mean Tumor Expression | Mean Normal Expression | Mean Difference in Expression (T-N) | Q-values |
| hsa-miR-1203 | 0.9040 | 1.2164 | -0.3124 | 0.7861 | 1.3040 | -0.5179 | 0.0044 |
| hsa-miR-1237-5p | 2.7048 | 2.8009 | -0.0961 | 2.5824 | 2.8954 | -0.313 | 0.0003 |
| hsa-miR-125b-1-3p | 3.3566 | 3.3173 | 0.0393 | 3.2864 | 3.3280 | -0.0416 | 0.0069 |
| hsa-miR-1266 | 0.9616 | 0.7167 | 0.2449 | 0.8296 | 0.7950 | 0.0345 | 0.0061 |
| hsa-miR-1276 | 1.1104 | 1.0311 | 0.0793 | 0.9951 | 1.1146 | -0.1195 | 0.0083 |
| hsa-miR-1295b-3p | 1.0382 | 1.0006 | 0.0376 | 0.9074 | 1.0876 | -0.1803 | 0.0055 |
| hsa-miR-1323 | 2.2143 | 2.1357 | 0.0786 | 2.0477 | 2.2264 | -0.1788 | 0.0011 |
| hsa-miR-1470 | 2.0574 | 2.3573 | -0.2999 | 1.8649 | 2.4017 | -0.5368 | 0.0008 |
| hsa-miR-184 | 0.9277 | 0.9309 | -0.0032 | 0.7507 | 1.0149 | -0.2641 | 0.0011 |
| hsa-miR-206 | 2.8032 | 3.1652 | -0.3621 | 2.6350 | 3.2131 | -0.5781 | 0.0109 |
| hsa-miR-302c-5p | 1.2712 | 0.9455 | 0.3257 | 1.1561 | 1.0997 | 0.0564 | 0.0001 |
| hsa-miR-3122 | 0.8753 | 0.8425 | 0.0328 | 0.7107 | 0.8545 | -0.1437 | 0.0151 |
| hsa-miR-3131 | 4.0127 | 3.8855 | 0.1272 | 3.9049 | 3.9703 | -0.0654 | 0.0001 |
| hsa-miR-3150b-3p | 1.0041 | 0.8180 | 0.1861 | 0.8778 | 0.8914 | -0.0136 | 0.0100 |
| hsa-miR-3161 | 3.8361 | 3.9214 | -0.0852 | 3.8044 | 3.9553 | -0.1509 | 0.0080 |
| hsa-miR-3177-5p | 1.9296 | 2.0896 | -0.16 | 1.7230 | 2.1565 | -0.4335 | 0.0003 |
| hsa-miR-3181 | 0.7395 | 1.0470 | -0.3075 | 0.6513 | 1.1449 | -0.4936 | 0.0155 |
| hsa-miR-3186-3p | 0.7814 | 0.6846 | 0.0968 | 0.6631 | 0.7670 | -0.1039 | 0.0046 |
| hsa-miR-3189-5p | 2.1073 | 2.1018 | 0.0055 | 2.0026 | 2.2128 | -0.2102 | 0.0087 |
| hsa-miR-339-3p | 1.9875 | 1.8349 | 0.1527 | 1.8464 | 1.8830 | -0.0366 | 0.0050 |
| hsa-miR-34c-3p | 2.3869 | 2.3467 | 0.0401 | 2.3206 | 2.4220 | -0.1014 | 0.0152 |
| hsa-miR-3615 | 0.7203 | 0.9008 | -0.1804 | 0.6091 | 0.9742 | -0.3651 | 0.0071 |
| hsa-miR-3660 | 1.9127 | 1.9104 | 0.0022 | 1.7900 | 1.9813 | -0.1913 | 0.0158 |
| hsa-miR-3679-3p | 0.4062 | 0.3906 | 0.0156 | 0.2972 | 0.4282 | -0.131 | 0.0011 |
| hsa-miR-3680-3p | 2.9284 | 2.6281 | 0.3003 | 2.7893 | 2.6970 | 0.0923 | 0.0008 |
| hsa-miR-378e | 1.0146 | 0.8963 | 0.1183 | 0.8816 | 0.9639 | -0.0824 | 0.0037 |
| hsa-miR-3922-5p | 0.6227 | 0.4820 | 0.1406 | 0.5307 | 0.5691 | -0.0384 | 0.0031 |
| hsa-miR-4300 | 0.8662 | 0.7173 | 0.1488 | 0.7436 | 0.7864 | -0.0428 | 0.0022 |
| hsa-miR-4303 | 0.8538 | 0.9330 | -0.0792 | 0.7200 | 0.9955 | -0.2755 | 0.0138 |
| hsa-miR-4421 | 0.5460 | 0.7538 | -0.2078 | 0.4893 | 0.8849 | -0.3956 | 0.0113 |
| hsa-miR-4436a | 2.0374 | 1.9462 | 0.0911 | 1.9109 | 2.0796 | -0.1688 | 0.0039 |
| hsa-miR-4444 | 3.2323 | 2.9691 | 0.2632 | 3.0676 | 3.0396 | 0.028 | 0.0011 |
| hsa-miR-4450 | 1.8216 | 1.6023 | 0.2192 | 1.7353 | 1.7504 | -0.0151 | 0.0072 |
| hsa-miR-4461 | 1.9050 | 1.7542 | 0.1507 | 1.7372 | 1.8575 | -0.1203 | 0.0014 |
| hsa-miR-4479 | 0.7581 | 0.7816 | -0.0235 | 0.6689 | 0.8772 | -0.2083 | 0.0158 |
| hsa-miR-4489 | 3.5472 | 3.6088 | -0.0616 | 3.4330 | 3.6676 | -0.2347 | 0.0135 |
| hsa-miR-4510 | 2.4625 | 2.5053 | -0.0429 | 2.3292 | 2.5568 | -0.2276 | 0.0064 |
| hsa-miR-4519 | 3.0204 | 2.9033 | 0.1171 | 2.8279 | 2.9748 | -0.1469 | 0.0004 |
| hsa-miR-4522 | 3.4239 | 3.6634 | -0.2395 | 3.2646 | 3.6946 | -0.4300 | 0.0033 |
| hsa-miR-4526 | 1.8260 | 1.8567 | -0.0307 | 1.6271 | 1.8806 | -0.2535 | 0.0013 |
| hsa-miR-4638-5p | 1.4805 | 1.5457 | -0.0652 | 1.3145 | 1.6703 | -0.3559 | 0.0014 |
| hsa-miR-4654 | 1.3087 | 1.0013 | 0.3074 | 1.1610 | 1.1001 | 0.0609 | 0.0022 |
| hsa-miR-4657 | 1.5101 | 1.0805 | 0.4295 | 1.3891 | 1.1962 | 0.1929 | 0.0032 |
| hsa-miR-4659b-3p | 1.7201 | 1.9028 | -0.1827 | 1.5847 | 2.0117 | -0.4271 | 0.0081 |
| hsa-miR-4660 | 2.0659 | 1.8028 | 0.2631 | 1.9383 | 1.9375 | 0.0008 | 0.0043 |
| hsa-miR-4674 | 2.5960 | 2.6555 | -0.0594 | 2.4912 | 2.7185 | -0.2273 | 0.0029 |
| hsa-miR-4676-5p | 1.0025 | 0.8863 | 0.1162 | 0.9258 | 1.0054 | -0.0796 | 0.0127 |
| hsa-miR-4682 | 0.7889 | 0.6533 | 0.1356 | 0.6994 | 0.7349 | -0.0355 | 0.0114 |
| hsa-miR-4684-3p | 1.1204 | 1.1684 | -0.048 | 1.0038 | 1.2391 | -0.2353 | 0.0083 |
| hsa-miR-4715-5p | 1.6858 | 1.6206 | 0.0652 | 1.4839 | 1.7073 | -0.2233 | 0.0007 |
| hsa-miR-4717-3p | 2.0039 | 1.7400 | 0.2639 | 1.7938 | 1.7920 | 0.0018 | 0.0001 |
| hsa-miR-4731-3p | 0.7447 | 0.6289 | 0.1158 | 0.6076 | 0.7042 | -0.0966 | 0.0011 |
| hsa-miR-4732-5p | 4.1588 | 3.8034 | 0.3555 | 4.1413 | 3.9066 | 0.2347 | 0.0089 |
| hsa-miR-4748 | 2.3375 | 2.2502 | 0.0874 | 2.1273 | 2.2557 | -0.1285 | 0.0084 |
| hsa-miR-500a-3p | 1.5768 | 1.4419 | 0.1348 | 1.4121 | 1.5348 | -0.1227 | 0.0006 |
| hsa-miR-516b-5p | 2.1095 | 2.0461 | 0.0634 | 1.9679 | 2.2117 | -0.2438 | 0.0005 |
| hsa-miR-518c-5p | 0.7122 | 0.8026 | -0.0904 | 0.6082 | 0.8813 | -0.2731 | 0.0101 |
| hsa-miR-5195-5p | 2.8941 | 2.6472 | 0.2469 | 2.7678 | 2.7966 | -0.0287 | 0.0020 |
| hsa-miR-519e-5p | 1.0583 | 1.0823 | -0.024 | 0.9249 | 1.2156 | -0.2907 | 0.0003 |
| hsa-miR-520e | 3.2490 | 3.6373 | -0.3883 | 3.1534 | 3.7298 | -0.5763 | 0.0068 |
| hsa-miR-525-5p | 0.8990 | 0.7384 | 0.1606 | 0.7517 | 0.8720 | -0.1203 | 0.0000 |
| hsa-miR-550a-5p | 2.9657 | 3.0505 | -0.0848 | 2.8758 | 3.0853 | -0.2096 | 0.0097 |
| hsa-miR-551b-5p | 0.8574 | 0.6666 | 0.1908 | 0.7783 | 0.7348 | 0.0436 | 0.0109 |
| hsa-miR-5572 | 2.7054 | 2.6641 | 0.0413 | 2.4570 | 2.6748 | -0.2178 | 0.0010 |
| hsa-miR-5584-5p | 0.4473 | 0.4448 | 0.0024 | 0.3820 | 0.5262 | -0.1442 | 0.0066 |
| hsa-miR-566 | 1.7370 | 1.8000 | -0.0631 | 1.6106 | 1.8912 | -0.2806 | 0.0019 |
| hsa-miR-583 | 2.7128 | 1.5976 | 1.1152 | 2.6492 | 1.7677 | 0.8814 | 0.0065 |
| hsa-miR-616-3p | 0.8733 | 0.5788 | 0.2945 | 0.8258 | 0.6369 | 0.1889 | 0.0157 |
| hsa-miR-639 | 3.0031 | 3.1208 | -0.1177 | 2.9192 | 3.1285 | -0.2093 | 0.0072 |
| hsa-miR-6509-5p | 3.6249 | 3.6661 | -0.0411 | 3.5866 | 3.7262 | -0.1396 | 0.0104 |
| hsa-miR-658 | 0.5168 | 0.6255 | -0.1087 | 0.4250 | 0.6472 | -0.2221 | 0.0055 |
| hsa-miR-659-3p | 3.4147 | 3.2518 | 0.1629 | 3.2092 | 3.3117 | -0.1025 | 0.0003 |
| hsa-miR-6716-5p | 2.2443 | 2.1112 | 0.1331 | 2.1058 | 2.1845 | -0.0787 | 0.0017 |
| hsa-miR-6718-5p | 2.9358 | 2.7755 | 0.1604 | 2.8682 | 2.8986 | -0.0304 | 0.0008 |
| hsa-miR-708-5p | 3.1203 | 3.2337 | -0.1133 | 3.0296 | 3.3207 | -0.2911 | 0.0122 |
1Expression based on log2 transformed data
The 75 miRNAs dysregulated by TERT rs2736118 were associated with 4047 genes and 6309 miRNA/mRNA associations according to miRTarBase. To determine which of these genes might be associated with mRNA regulation in colonic tissue, we assessed each miRNA with its mRNA gene target in colonic carcinoma tissue and colonic normal mucosa using a linear regression. We identified 573 miRNA/mRNA associations p-value of <0.05 prior to adjustment for multiple comparisons. Of these, 150 significant miRNA/mRNA target associations remained when the FDR was 0.05 and 270 miRNA/mRNA associations remained with the FDR of <0.1 (Table 4). The miRNAs and their targeted genes shown in Table 4 also indicate the direct of the association based on the beta coefficient. Of note is that the associations were limited to 13 miRNAs. All but five of these associations showed inverse associations between the miRNA and the mRNA.
Table 4. Regression of differential miRNA expression and differential mRNA expression (FDR<0.05).
| miRNA | Gene | Beta | FDR Q |
|---|---|---|---|
| hsa-miR-3161 | WIZ | -0.24 | 0.006 |
| hsa-miR-3161 | SLC25A40 | -0.27 | 0.006 |
| hsa-miR-3161 | EFNB2 | -0.11 | 0.006 |
| hsa-miR-3161 | TRIM5 | -0.28 | 0.006 |
| hsa-miR-3161 | PPIG | -0.24 | 0.006 |
| hsa-miR-3161 | RELA | -0.2 | 0.006 |
| hsa-miR-3161 | CSTF2T | -0.27 | 0.006 |
| hsa-miR-3161 | SRGAP1 | -0.31 | 0.006 |
| hsa-miR-3161 | HMGN2 | -0.26 | 0.006 |
| hsa-miR-3161 | NUP50 | -0.23 | 0.009 |
| hsa-miR-3161 | TXNDC12 | -0.25 | 0.009 |
| hsa-miR-3161 | BCL2L11 | -0.2 | 0.009 |
| hsa-miR-3161 | RYBP | -0.25 | 0.009 |
| hsa-miR-3161 | TMEM41A | -0.2 | 0.009 |
| hsa-miR-3161 | PRDX3 | -0.29 | 0.009 |
| hsa-miR-3161 | WIPF2 | -0.23 | 0.009 |
| hsa-miR-3161 | TMEM30B | -0.31 | 0.009 |
| hsa-miR-3161 | CTNND1 | -0.22 | 0.009 |
| hsa-miR-3161 | VMP1 | -0.19 | 0.012 |
| hsa-miR-3161 | TRIP11 | -0.26 | 0.012 |
| hsa-miR-3161 | INVS | -0.24 | 0.012 |
| hsa-miR-3161 | GEMIN6 | -0.17 | 0.012 |
| hsa-miR-3161 | EOGT | -0.27 | 0.012 |
| hsa-miR-3161 | UBXN2A | -0.2 | 0.012 |
| hsa-miR-3161 | NUPL2 | -0.18 | 0.016 |
| hsa-miR-3161 | PCMTD1 | -0.16 | 0.016 |
| hsa-miR-3161 | TOM1L2 | -0.21 | 0.016 |
| hsa-miR-3161 | PTP4A1 | -0.18 | 0.017 |
| hsa-miR-3161 | QKI | -0.23 | 0.017 |
| hsa-miR-3161 | SHFM1 | -0.17 | 0.017 |
| hsa-miR-3161 | THRB | -0.18 | 0.017 |
| hsa-miR-3161 | SREK1 | -0.21 | 0.017 |
| hsa-miR-3161 | ACTBL2 | 0.15 | 0.017 |
| hsa-miR-3161 | CCDC43 | -0.24 | 0.017 |
| hsa-miR-3161 | PSMC2 | -0.15 | 0.02 |
| hsa-miR-3161 | ARMC10 | -0.18 | 0.02 |
| hsa-miR-3161 | SPOP | -0.27 | 0.023 |
| hsa-miR-3161 | HIF1AN | -0.22 | 0.023 |
| hsa-miR-3161 | ZNF706 | -0.14 | 0.026 |
| hsa-miR-3161 | ZNF100 | -0.21 | 0.028 |
| hsa-miR-3161 | MCTS1 | -0.17 | 0.031 |
| hsa-miR-3161 | ZNF431 | -0.17 | 0.034 |
| hsa-miR-3161 | ABHD2 | -0.07 | 0.036 |
| hsa-miR-3161 | PDIK1L | -0.21 | 0.036 |
| hsa-miR-3161 | ELOVL5 | -0.12 | 0.041 |
| hsa-miR-3161 | BTC | -0.22 | 0.041 |
| hsa-miR-3161 | NUP37 | -0.09 | 0.045 |
| hsa-miR-3161 | TPCN2 | -0.19 | 0.045 |
| hsa-miR-3161 | LRRC40 | -0.25 | 0.048 |
| hsa-miR-3161 | DSE | -0.19 | 0.052 |
| hsa-miR-3161 | PPIL1 | -0.14 | 0.055 |
| hsa-miR-3161 | VSNL1 | -0.19 | 0.057 |
| hsa-miR-3161 | STARD7 | -0.33 | < .001 |
| hsa-miR-3161 | SRP54 | -0.3 | < .001 |
| hsa-miR-3161 | RBM3 | -0.22 | < .001 |
| hsa-miR-3161 | DGKH | -0.15 | < .001 |
| hsa-miR-3161 | MON1B | -0.3 | < .001 |
| hsa-miR-3161 | RPS6KB1 | -0.32 | < .001 |
| hsa-miR-3161 | EIF4G2 | -0.24 | < .001 |
| hsa-miR-3161 | TRMT5 | -0.3 | < .001 |
| hsa-miR-3161 | ZBED3 | -0.27 | < .001 |
| hsa-miR-3161 | SLC38A2 | -0.24 | < .001 |
| hsa-miR-3161 | SETDB2 | -0.27 | < .001 |
| hsa-miR-3161 | ABI2 | -0.28 | < .001 |
| hsa-miR-3161 | POU2F1 | -0.27 | < .001 |
| hsa-miR-3161 | ANP32E | -0.25 | < .001 |
| hsa-miR-3161 | LBR | -0.27 | < .001 |
| hsa-miR-3161 | MBNL1 | -0.32 | < .001 |
| hsa-miR-3161 | SREK1IP1 | -0.24 | < .001 |
| hsa-miR-3161 | CBR1 | -0.25 | < .001 |
| hsa-miR-3161 | DIP2A | -0.25 | < .001 |
| hsa-miR-3161 | UBE2Q1 | -0.3 | < .001 |
| hsa-miR-3161 | SRSF2 | -0.27 | < .001 |
| hsa-miR-3161 | ZNF367 | -0.21 | < .001 |
| hsa-miR-3161 | NUDT21 | -0.31 | < .001 |
| hsa-miR-3161 | CDK1 | -0.19 | < .001 |
| hsa-miR-3161 | TOMM20 | -0.26 | < .001 |
| hsa-miR-3161 | NR2C2 | -0.24 | < .001 |
| hsa-miR-3161 | LRRC57 | -0.23 | < .001 |
| hsa-miR-3161 | NGRN | -0.25 | < .001 |
| hsa-miR-3161 | PTAR1 | -0.28 | < .001 |
| hsa-miR-3177-5p | PRRG4 | -0.18 | < .001 |
| hsa-miR-3181 | FAM3C | -0.18 | 0.008 |
| hsa-miR-3181 | ATP5E | -0.13 | 0.061 |
| hsa-miR-3181 | KLHDC3 | -0.17 | 0.061 |
| hsa-miR-3615 | KHSRP | -0.17 | 0.008 |
| hsa-miR-3615 | NIPA2 | -0.25 | 0.008 |
| hsa-miR-3615 | ZYG11B | -0.18 | 0.008 |
| hsa-miR-3615 | HIST1H1B | -0.09 | 0.008 |
| hsa-miR-3615 | PPP6R1 | -0.2 | 0.011 |
| hsa-miR-3615 | WNT4 | -0.2 | 0.011 |
| hsa-miR-3615 | CCNDBP1 | -0.27 | 0.011 |
| hsa-miR-3615 | CTC1 | -0.21 | 0.011 |
| hsa-miR-3615 | VASP | -0.19 | 0.013 |
| hsa-miR-3615 | ARF6 | -0.21 | 0.013 |
| hsa-miR-3615 | LRCH3 | -0.19 | 0.013 |
| hsa-miR-3615 | ZBTB7A | -0.21 | 0.021 |
| hsa-miR-3615 | SRM | -0.1 | 0.027 |
| hsa-miR-3615 | OPA3 | -0.21 | 0.042 |
| hsa-miR-3615 | WDR12 | -0.14 | 0.042 |
| hsa-miR-3615 | SPRY1 | -0.13 | 0.047 |
| hsa-miR-3615 | ATP9A | -0.09 | < .001 |
| hsa-miR-3615 | RPS8 | -0.18 | < .001 |
| hsa-miR-3615 | ALDOA | -0.22 | < .001 |
| hsa-miR-3615 | COX20 | -0.29 | < .001 |
| hsa-miR-3615 | BMPR2 | -0.22 | < .001 |
| hsa-miR-378e | TGFB2 | -0.14 | 0.052 |
| hsa-miR-4510 | CDH7 | 0.25 | < .001 |
| hsa-miR-4638-5p | GINM1 | -0.16 | 0.073 |
| hsa-miR-4638-5p | OGFOD1 | -0.15 | 0.073 |
| hsa-miR-4638-5p | ALDH2 | -0.17 | 0.073 |
| hsa-miR-4638-5p | FEM1A | -0.19 | 0.073 |
| hsa-miR-4638-5p | CNBP | -0.25 | 0.073 |
| hsa-miR-4638-5p | RSBN1L | -0.17 | 0.073 |
| hsa-miR-4638-5p | FAM120AOS | -0.19 | 0.073 |
| hsa-miR-4638-5p | HMGB1 | -0.15 | 0.073 |
| hsa-miR-4638-5p | CAPZA2 | -0.14 | 0.073 |
| hsa-miR-4638-5p | PEX26 | -0.2 | 0.073 |
| hsa-miR-4638-5p | PI4K2B | -0.18 | 0.077 |
| hsa-miR-4638-5p | RAB10 | -0.14 | 0.077 |
| hsa-miR-4638-5p | RRP36 | -0.14 | 0.077 |
| hsa-miR-4638-5p | OXA1L | -0.22 | 0.077 |
| hsa-miR-4638-5p | ZBTB43 | -0.21 | 0.077 |
| hsa-miR-4638-5p | FGFR1OP2 | -0.19 | 0.079 |
| hsa-miR-4638-5p | HSD17B12 | -0.14 | 0.079 |
| hsa-miR-4638-5p | SGPP2 | -0.22 | 0.079 |
| hsa-miR-4638-5p | ERAP2 | -0.24 | 0.079 |
| hsa-miR-4638-5p | ZNF652 | -0.14 | 0.079 |
| hsa-miR-4638-5p | GABPB1 | -0.19 | 0.081 |
| hsa-miR-4638-5p | CIAO1 | -0.1 | 0.081 |
| hsa-miR-4638-5p | MTMR10 | -0.15 | 0.081 |
| hsa-miR-4638-5p | KCMF1 | -0.12 | 0.081 |
| hsa-miR-4638-5p | ZNF485 | -0.17 | 0.081 |
| hsa-miR-4638-5p | GNPNAT1 | -0.16 | 0.087 |
| hsa-miR-4638-5p | YIPF5 | -0.16 | 0.087 |
| hsa-miR-4638-5p | SNRPD1 | -0.21 | 0.087 |
| hsa-miR-4638-5p | TSPAN6 | -0.12 | 0.092 |
| hsa-miR-4638-5p | ZNF207 | -0.09 | 0.092 |
| hsa-miR-4638-5p | MRI1 | -0.13 | 0.092 |
| hsa-miR-4638-5p | MAPK1 | -0.15 | 0.092 |
| hsa-miR-4638-5p | RBM23 | -0.13 | 0.092 |
| hsa-miR-4638-5p | NUP93 | -0.13 | 0.092 |
| hsa-miR-4638-5p | RPL34 | -0.11 | 0.092 |
| hsa-miR-4638-5p | GNB4 | -0.14 | 0.092 |
| hsa-miR-4638-5p | RNF11 | -0.16 | 0.092 |
| hsa-miR-4638-5p | RAB9A | -0.18 | 0.092 |
| hsa-miR-4638-5p | IRF1 | -0.18 | 0.092 |
| hsa-miR-4638-5p | TEP1 | -0.13 | 0.092 |
| hsa-miR-4638-5p | SCO1 | -0.19 | 0.092 |
| hsa-miR-4638-5p | CDK4 | -0.09 | 0.092 |
| hsa-miR-4638-5p | UGGT1 | -0.13 | 0.092 |
| hsa-miR-4638-5p | PRPF4 | -0.17 | 0.092 |
| hsa-miR-4638-5p | PNPT1 | -0.11 | 0.092 |
| hsa-miR-4638-5p | ENSA | -0.11 | 0.092 |
| hsa-miR-4638-5p | CNKSR3 | -0.14 | 0.092 |
| hsa-miR-4638-5p | FOXK1 | -0.09 | 0.092 |
| hsa-miR-4638-5p | TRMT10B | -0.18 | 0.092 |
| hsa-miR-4638-5p | MELK | -0.16 | 0.092 |
| hsa-miR-4638-5p | ZNF561 | -0.16 | 0.092 |
| hsa-miR-4638-5p | KIAA1919 | -0.13 | 0.092 |
| hsa-miR-4638-5p | CCS | -0.14 | 0.092 |
| hsa-miR-4638-5p | GK5 | -0.13 | 0.092 |
| hsa-miR-4638-5p | ZNF682 | -0.18 | 0.092 |
| hsa-miR-4638-5p | SF3B1 | -0.08 | 0.096 |
| hsa-miR-4638-5p | TNFAIP8L1 | -0.12 | 0.096 |
| hsa-miR-4638-5p | PSMB5 | -0.14 | 0.097 |
| hsa-miR-4638-5p | MCM4 | -0.12 | 0.097 |
| hsa-miR-4638-5p | SMG1 | -0.1 | 0.097 |
| hsa-miR-4638-5p | LRIG2 | -0.13 | 0.097 |
| hsa-miR-4638-5p | OPTN | -0.22 | < .001 |
| hsa-miR-4638-5p | ZNF655 | -0.17 | < .001 |
| hsa-miR-4660 | CTBS | -0.2 | < .001 |
| hsa-miR-4731-3p | HSP90AB1 | -0.16 | 0.015 |
| hsa-miR-4731-3p | IKZF4 | -0.22 | 0.015 |
| hsa-miR-4731-3p | SPTLC2 | -0.24 | 0.023 |
| hsa-miR-4731-3p | N4BP1 | -0.24 | 0.023 |
| hsa-miR-4731-3p | NRF1 | -0.25 | 0.023 |
| hsa-miR-4731-3p | SLC38A2 | -0.21 | 0.023 |
| hsa-miR-4731-3p | STAT5A | -0.2 | 0.027 |
| hsa-miR-4731-3p | CHEK1 | -0.22 | 0.027 |
| hsa-miR-4731-3p | MFSD6 | -0.34 | 0.027 |
| hsa-miR-4731-3p | PPP2R5E | -0.25 | 0.027 |
| hsa-miR-4731-3p | TMED10 | -0.24 | 0.033 |
| hsa-miR-4731-3p | ZNF678 | -0.27 | 0.033 |
| hsa-miR-4731-3p | HFE | -0.22 | 0.034 |
| hsa-miR-4731-3p | TJP1 | -0.25 | 0.034 |
| hsa-miR-4731-3p | TFCP2 | -0.17 | 0.034 |
| hsa-miR-4731-3p | ZIC5 | -0.2 | 0.034 |
| hsa-miR-4731-3p | MYO10 | -0.16 | 0.034 |
| hsa-miR-4731-3p | ASB6 | -0.21 | 0.034 |
| hsa-miR-4731-3p | PIP4K2A | -0.17 | 0.034 |
| hsa-miR-4731-3p | SRSF2 | -0.21 | 0.034 |
| hsa-miR-4731-3p | ZBTB40 | -0.21 | 0.034 |
| hsa-miR-4731-3p | TPM2 | -0.24 | 0.034 |
| hsa-miR-4731-3p | SRSF1 | -0.19 | 0.038 |
| hsa-miR-4731-3p | PTPN3 | -0.21 | 0.041 |
| hsa-miR-4731-3p | ANKS6 | -0.22 | 0.041 |
| hsa-miR-4731-3p | PTMA | -0.17 | 0.055 |
| hsa-miR-4731-3p | SREK1 | -0.19 | 0.058 |
| hsa-miR-4731-3p | TRAM1 | -0.17 | 0.059 |
| hsa-miR-4731-3p | GALNT7 | -0.18 | 0.059 |
| hsa-miR-4731-3p | QKI | -0.16 | 0.059 |
| hsa-miR-4731-3p | SKIL | -0.17 | 0.059 |
| hsa-miR-4731-3p | GATAD2B | -0.22 | 0.059 |
| hsa-miR-4731-3p | ANKRD40 | -0.16 | 0.059 |
| hsa-miR-4731-3p | TAF1D | -0.13 | 0.059 |
| hsa-miR-4731-3p | VCPIP1 | -0.19 | 0.059 |
| hsa-miR-4731-3p | CCDC43 | -0.17 | 0.059 |
| hsa-miR-4731-3p | DIDO1 | -0.17 | 0.06 |
| hsa-miR-4731-3p | KCNC4 | -0.18 | 0.06 |
| hsa-miR-4731-3p | MAP3K9 | -0.18 | 0.068 |
| hsa-miR-4731-3p | EIF5A2 | -0.19 | 0.068 |
| hsa-miR-4731-3p | ATP9A | -0.13 | 0.074 |
| hsa-miR-4731-3p | SHOC2 | -0.21 | 0.074 |
| hsa-miR-4731-3p | SLC1A4 | -0.12 | 0.074 |
| hsa-miR-4731-3p | GAN | -0.16 | 0.074 |
| hsa-miR-4731-3p | SFXN5 | -0.17 | 0.074 |
| hsa-miR-4731-3p | ZNF432 | -0.15 | 0.074 |
| hsa-miR-4731-3p | ZNF585A | -0.1 | 0.076 |
| hsa-miR-4731-3p | ACBD5 | -0.3 | < .001 |
| hsa-miR-4731-3p | SOS1 | -0.28 | < .001 |
| hsa-miR-4731-3p | FBXL5 | -0.26 | < .001 |
| hsa-miR-4731-3p | NDUFA5 | -0.31 | < .001 |
| hsa-miR-4731-3p | CLSTN3 | -0.24 | < .001 |
| hsa-miR-4731-3p | THYN1 | -0.26 | < .001 |
| hsa-miR-4731-3p | RABGEF1 | -0.26 | < .001 |
| hsa-miR-4731-3p | MYO6 | -0.33 | < .001 |
| hsa-miR-4731-3p | TLK1 | -0.25 | < .001 |
| hsa-miR-4731-3p | LYN | -0.24 | < .001 |
| hsa-miR-500a-3p | C18orf25 | -0.24 | < .001 |
| hsa-miR-520e | CDIPT | -0.29 | 0.075 |
| hsa-miR-520e | WWC1 | -0.23 | 0.075 |
| hsa-miR-520e | PDPK1 | -0.3 | 0.075 |
| hsa-miR-520e | ZNF12 | -0.37 | 0.075 |
| hsa-miR-520e | KBTBD6 | -0.29 | 0.075 |
| hsa-miR-520e | ZBTB33 | -0.23 | 0.075 |
| hsa-miR-520e | TSPAN6 | -0.24 | 0.094 |
| hsa-miR-520e | TFAP4 | -0.22 | 0.094 |
| hsa-miR-520e | SUGP1 | -0.3 | 0.094 |
| hsa-miR-520e | SERINC1 | -0.22 | 0.094 |
| hsa-miR-520e | NUDT3 | -0.29 | 0.094 |
| hsa-miR-520e | KCNJ8 | -0.17 | 0.094 |
| hsa-miR-520e | ZNFX1 | -0.29 | 0.094 |
| hsa-miR-520e | ABI2 | -0.33 | 0.094 |
| hsa-miR-520e | EFCAB11 | -0.22 | 0.094 |
| hsa-miR-520e | TM4SF5 | -0.28 | 0.094 |
| hsa-miR-520e | ZNF7 | -0.24 | 0.094 |
| hsa-miR-520e | HSPA13 | -0.25 | 0.094 |
| hsa-miR-520e | NEK8 | -0.27 | 0.094 |
| hsa-miR-520e | ITGA2 | -0.29 | 0.094 |
| hsa-miR-520e | FOXK1 | -0.27 | 0.094 |
| hsa-miR-520e | CYB5A | -0.23 | 0.094 |
| hsa-miR-520e | BLCAP | -0.34 | 0.094 |
| hsa-miR-520e | BSCL2 | -0.26 | 0.094 |
| hsa-miR-520e | KBTBD2 | -0.35 | 0.094 |
| hsa-miR-520e | ZMAT3 | -0.17 | 0.094 |
| hsa-miR-520e | ARL10 | -0.23 | 0.094 |
| hsa-miR-520e | CAPZA2 | -0.31 | 0.094 |
| hsa-miR-520e | ZNF805 | -0.28 | 0.094 |
| hsa-miR-520e | SLC35F6 | -0.26 | 0.094 |
| hsa-miR-520e | ZNF174 | -0.37 | < .001 |
| hsa-miR-520e | SLC6A4 | -0.36 | < .001 |
| hsa-miR-520e | BMP8B | -0.28 | < .001 |
| hsa-miR-520e | ZNF426 | -0.31 | < .001 |
| hsa-miR-520e | ABHD15 | -0.35 | < .001 |
| hsa-miR-525-5p | KCNH1 | 0.21 | < .001 |
| hsa-miR-583 | NKD1 | 0.08 | 0.068 |
| hsa-miR-583 | SIGLEC9 | -0.25 | < .001 |
| hsa-miR-583 | KIF2C | 0.2 | < .001 |
| hsa-miR-583 | PDZRN4 | -0.25 | < .001 |
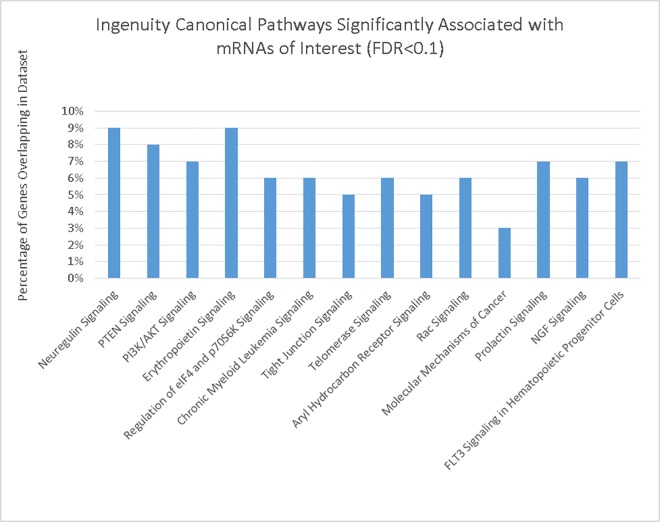
Using IPA to construct pathways enriched with the 270 genes with an FDR of <0.1, we identified 14 pathways (Fig 1). Although less than 10% of genes in each pathway were included in our gene list, these pathways were significantly enriched by our genes. The enriched pathways included: neuregulin signaling, PTEN signaling, PI3K/AKT signaling, erythropoietin signaling, regulation of EIF4 and p706SK signaling, chronic myeloid leukemia signaling, tight junction signaling, telomerase signaling, aryl hydrocarbon receptor signaling, Rac signaling, molecular mechanisms of cancer, prolactin signaling NGF signaling, and FLT3 signaling in hematopoietic progenitor cells.
Fig 1. IPA canonical pathways associated with 270 genes targeted by miRNAs associated with TERT.
Discussion
These data suggest a direct association between miRNAs and TL as well as between TERT rs2736118 and miRNA differential expression between carcinoma tissue and normal colonic mucosa. While TERT expression is low in most somatic cells, endothelial cells represent an established exception and associations observed for differential expression between carcinoma and normal mucosa is a likely extension of that observation. Our results add support to the hypothesis that miRNAs are associated with TL and that genetic variation in telomere-related genes influence miRNA expression levels. Given the number of miRNAs altered by TERT rs2736118, our ability to show associations between altered miRNAs and their targeted mRNA, and the pathways represented by these target genes, we believe that our data support the hypothesis that TERT has functions beyond that of regulating TL. Focusing only on those miRNAs that were statistically associated with target mRNA expression in colon tissue, we identified 14 canonical pathways that were associated significantly with these TERT-regulated miRNAs after adjustment for multiple comparison, 13 of which were not associated with telomerase.
There have been few reports of miRNAs associated with TL that have looked at actual miRNA expression levels in conjunction with TL; our sample size is by far the largest study to date to examine miRNA expression with TL. One study with only two samples compared shortened telomeres to intact telomeres and showed that 47 miRNAs had over a twofold difference in expression levels [27]. MiR-155, miR-138, and miR-34a,b,c also have been hypothesized as being associated with TL [3, 5]; none of these miRNAs were associated in a linear fashion with TL in our study. The miRNAs associated with TL in our data for the most part increased TL with increased miRNA expression. Differences in association between our study and others may in part be from the methods of analysis. We assessed a linear association, while other studies have often used categorical variables and looked at either short or long TL, which could account for some of the differences observed.
A limitation of the study is that we are unable to confirm our observed associations between TL and miRNAs in bio-functional studies; we encourage others to do so. However, support for our findings come from several sources. First, we have previously shown that 26 of the 34 miRNAs associated with TL were differentially expressed between colorectal carcinoma and normal colonic mucosa and 28 of the 34 were dysregulated between carcinoma and adenomatous polyp tissue; three miRNAs were dysregulated between normal colonic mucosa and adenomatous polyp tissue [18]. Interestingly, the only miRNA that was directly associated with TL, miR-487b, was the only miRNA that was not associated with dysregulation between carcinoma, adenoma, and/or normal mucosa. Additionally, using bioinformatics tools to identify the target genes for the dysregulated miRNAs associated with TL, we observed that 12 of these miRNAs associated with TL target genes (summary in Table 5) are components of the alternative lengthening of telomeres pathway (ALT) and Shelterin [28]. Shelterin is a conserved protein component on telomeres that serves as a functional framework of telomere chromatin pathways. ALT is a telomerase-independent pathway of lengthening telomeres in human cells. Given that there is no overlap between TERT SNP-associated miRNAs and TL-associated miRNAs, and that a large proportion of these miRNAs target genes in the ALT pathway, it is possible that miRNAs regulate TL through this alternative mechanism. As TL has been shown to be associated with CRC, the associations between TL and miRNA expression may unique clinical significance.
Table 5. Summary of miRNAs related to telomere length and TERT rs2736118 differentially expressed in colorectal cancer.
| Differentially Expressed: | ||||
|---|---|---|---|---|
| miRNA | T—N1 | T—AD | AD—N | ALT Gene(s) Targeted |
| Telomere Length | ||||
| hsa-miR-1185-2-3p | x | |||
| hsa-miR-1207-5p | x | x | CBX5, TERT, TP53 | |
| hsa-miR-1226-5p | x | x | CBX5, RAD18 | |
| hsa-miR-1229-5p | x | x | ||
| hsa-miR-1247-3p | x | FANCA, RAD51L3-RFFL, SMC5 | ||
| hsa-miR-134 | x | x | ||
| hsa-miR-3141 | x | |||
| hsa-miR-3158-5p | x | x | ||
| hsa-miR-3162-5p | x | x | ||
| hsa-miR-3194-5p | x | x | ||
| hsa-miR-3196 | x | x | ||
| hsa-miR-3200-5p | x | x | ||
| hsa-miR-3937 | FANCA, CBX5 | |||
| hsa-miR-4253 | x | |||
| hsa-miR-4459 | x | x | TERF2, ERCC1, HSP90B1, SMC5 | |
| hsa-miR-4496 | x | RAD50, RAD51B, RAP1B | ||
| hsa-miR-4497 | x | x | FANCA | |
| hsa-miR-4530 | x | x | ||
| hsa-miR-4535 | x | x | ||
| hsa-miR-4673 | x | x | XRCC3 | |
| hsa-miR-4689 | CDKN1A, RAD18, RAD50, XRCC3 | |||
| hsa-miR-4739 | x | x | ||
| hsa-miR-487b | ||||
| hsa-miR-5006-5p | x | x | ||
| hsa-miR-5088 | x | x | ||
| hsa-miR-5195-3p | x | x | ||
| hsa-miR-575 | x | x | x | RAD51 |
| hsa-miR-601 | x | x | ||
| hsa-miR-6076 | x | x | ||
| hsa-miR-662 | x | x | ||
| hsa-miR-718 | x | x | ||
| hsa-miR-762 | x | x | CDK2, RAD54L2 | |
| hsa-miR-887 | x | x | ||
| hsa-miR-939-5p | x | x | CDKN1A | |
| TERT rs2736118 | ||||
| hsa-miR-1203 | x | x | ||
| hsa-miR-1237-5p | x | |||
| hsa-miR-125b-1-3p | ||||
| hsa-miR-1266 | ||||
| hsa-miR-1276 | ||||
| hsa-miR-1295b-3p | WRN | |||
| hsa-miR-1323 | ||||
| hsa-miR-1470 | x | |||
| hsa-miR-184 | ||||
| hsa-miR-206 | x | x | HSP90B1 | |
| hsa-miR-302c-5p | CBX4 | |||
| hsa-miR-3122 | PARP2 | |||
| hsa-miR-3131 | ||||
| hsa-miR-3150b-3p | CDK2, CDKN1A, RAD18, RAD50, XRCC3 | |||
| hsa-miR-3161 | x | |||
| hsa-miR-3177-5p | x | |||
| hsa-miR-3181 | x | |||
| hsa-miR-3186-3p | ||||
| hsa-miR-3189-5p | ||||
| hsa-miR-339-3p | ||||
| hsa-miR-34c-3p | ||||
| hsa-miR-3615 | x | |||
| hsa-miR-3660 | ||||
| hsa-miR-3679-3p | x | x | ||
| hsa-miR-3680-3p | CBX4 | |||
| hsa-miR-378e | RAP1B | |||
| hsa-miR-3922-5p | ||||
| hsa-miR-4300 | ||||
| hsa-miR-4303 | ||||
| hsa-miR-4421 | x | |||
| hsa-miR-4436a | ||||
| hsa-miR-4444 | ||||
| hsa-miR-4450 | ||||
| hsa-miR-4461 | ||||
| hsa-miR-4479 | x | |||
| hsa-miR-4489 | ||||
| hsa-miR-4510 | x | x | TP53, MRE11A | |
| hsa-miR-4519 | ||||
| hsa-miR-4522 | x | PARP2 | ||
| hsa-miR-4526 | ||||
| hsa-miR-4638-5p | ||||
| hsa-miR-4654 | ||||
| hsa-miR-4657 | x | x | ||
| hsa-miR-4659b-3p | CDKN1A | |||
| hsa-miR-4660 | ||||
| hsa-miR-4674 | x | |||
| hsa-miR-4676-5p | RAD51 | |||
| hsa-miR-4682 | CDKN1A, TP53 | |||
| hsa-miR-4684-3p | NR2F2 | |||
| hsa-miR-4715-5p | CBX4 | |||
| hsa-miR-4717-3p | WRN | |||
| hsa-miR-4731-3p | ||||
| hsa-miR-4732-5p | x | x | ||
| hsa-miR-4748 | WRN | |||
| hsa-miR-500a-3p | ||||
| hsa-miR-516b-5p | ||||
| hsa-miR-518c-5p | ||||
| hsa-miR-5195-5p | ||||
| hsa-miR-519e-5p | ||||
| hsa-miR-520e | x | x | CBX5, FANCA, NR2F2 | |
| hsa-miR-525-5p | ||||
| hsa-miR-550a-5p | x | RAD51 | ||
| hsa-miR-551b-5p | ERCC1 | |||
| hsa-miR-5572 | ||||
| hsa-miR-5584-5p | CDKN1A, RAD50, RAD54L2 | |||
| hsa-miR-566 | CBX5 | |||
| hsa-miR-583 | x | x | CBX5, CDKN1A | |
| hsa-miR-616-3p | x | x | ||
| hsa-miR-639 | x | CDKN1A | ||
| hsa-miR-6509-5p | ||||
| hsa-miR-658 | x | HSP90B1, SMC5 | ||
| hsa-miR-659-3p | ||||
| hsa-miR-6716-5p | CBX5 | |||
| hsa-miR-6718-5p | x | x | ||
| hsa-miR-708-5p | x | TOP2A | ||
1’T’ signifies ‘Tumor tissue’, ‘N’ signifies ‘Normal colonic mucosa’, and ‘AD’ signifies ‘Adenoma tissue’.
We also observed that genetic variation in TERT, which we have previously reported as being associated with TL and cancer [6, 14], was associated with miRNA expression levels. Many of these miRNAs have been shown to be dysregulated in CRC [18] and 26 of these miRNAs target ALT pathway genes (Table 5). One of the pathways enriched by genes targeted by dysregulated miRNAs is ‘telomerase signaling’. Since telomerase is generally not expressed in non-tumor somatic tissue but is expressed in the majority of tumors, it is logical that TERT rs2836118 was not associated with miRNA expression in normal colonic mucosa, but only in differential miRNA expression between carcinoma and normal colorectal mucosa. Telomerase is often reactivated by TERT upregulation, which gives tumor cells their immortal quality, and this is thought to be a key step in the adenoma-carcinoma transition in human tumorigenesis [29]. As stated previously, we identified miRNAs that were differentially expressed between normal colonic mucosa and adenomatous polyp tissues as well as between adenoma and carcinoma tissues, which show the changes in miRNA expression as tissue progresses from normal to adenomatous to cancerous [18]. Both hsa-miR-520e and hsa-miR-583 were differentially expressed between normal colonic mucosa and adenomatous polyp tissue [18]; in this study we show that they also regulate mRNA expression. These two miRNAs regulated 40 genes, including ITGA2, BMP8B, and PDPK1, which were found in numerous pathways in IPA, including PTEN Signaling, BMP signaling pathway, and Basal Cell Carcinoma Signaling. Hsa-miR-520e was associated with CBX5, FANCA and NR2F2, and hsa-miR-583 was associated with CBX5 and CDKN1A, all of which are part of the ALT pathway. We also identified six miRNAs that were downregulated with the AG/GG (heterozygous-homozygous recessive) genotype of rs2736118 that we previously found were dysregulated between adenomatous and carcinoma tissue; these miRNAs were: hsa-miR-206, hsa-miR-4510, hsa-miR-4674, hsa-miR-639, hsa-miR-6718-5p, and hsa-miR-708-5p. This supports the hypothesis that TERT regulates miRNA expression and this influences the formation of adenomatous tissue and contributes to tumorigenesis.
The association of TERT rs2736118 genotype with miRNA expression levels may provide additional insight into pathways in which TERT is involved. We therefore examined genes and pathways regulated by the miRNAs that were influenced by TERT rs2736118. We observed that genetic variation in TERT was associated with dysregulation of 75 miRNAs, which in turn could influence translation and function of over 6000 genes. While many genes are regulated by these miRNAs, it is not clear which of these are related to colorectal cancer. Evaluation of over 6000 genes leads to a very non-specific assessment of potential functionality. To better focus on functionality we attempted to cluster genes together into what could be potentially a more common function and we also assessed seed regions. Using a cluster analysis approach, we still had over fifty pathways that also lacked specificity to colorectal cancer. Focusing on seed regions, we were left with 73 seed regions and did not minimize the potential number of genes that could be associated with TERT. Thus, we took an approach that focused only on those genes where the gene targeted by the miRNA was linearly associated with the corresponding mRNA expression in colorectal cancer. This reduced the number of genes and pathways considerably and was specific to colorectal cancer. However, while we gained specificity, we undoubtedly were limited in the target genes we assessed. We believe that the pathway information obtained is meaningful, but most certainly not complete. These data do however support the hypothesis that TERT has non-TL-related pathways [29, 30] that may be acting through altered miRNA expression.
In addition to its established role in telomere maintenance, it has been proposed that TERT operates through other pathways. Further examples of TERT involvement in non-TL related pathways include both NFκB complex and the Wnt/β-catenin pathways, which have been hypothesized as being telomerase-targeted pathways as both are pathways related to inflammation [5]; these pathways were both represented by our target genes in IPA although they are not shown in Fig 1. Most notably, the NFκB-signaling pathway activation has been suggested as being related to TERT [31]. TERT levels also have been associated with differential expression of genes involved in development/morphogenesis, signal transduction, and cell adhesion-signaling pathways [32, 33]. Our data also suggest that associations between TERT and colorectal cancer, possibly mediated by miRNAs, involve numerous pathways other than telomerase. These pathways involve tumor suppressor genes (such as PTEN signaling), apoptosis (PI3K/AKT signaling, NGF signaling, and FLT3 signaling), angiogenesis (Erythropoietin signaling), immune regulation and response (Prolactin signaling and Aryl Hydrocarbon Receptor), cell migration and proliferation (Neuregulin signaling), and growth factor activity (NGF signaling and EIF4 and p70S6K signaling). Another enriched pathway, Tight Junction Signaling, is important in cellular communication and facilities crosstalk across several of these signaling pathways including PI3K/AKT and PTEN [34].
Additionally, specific genes such as STAT5A, MELK, MAPK1 and MAP3K9, RELA (part of the NFKB1 complex), CDK1, tumor necrosis factor genes, IRF1, and TGFβ-related genes were associated with miRNAs differentially expressed by TERT genotype. A number of translocations associated with cancer involve the Ig heavy chain locus on chromosome 14. For example t(8;14) brings c-MYC from chromosome 8 to the IgH gene locus causing an overexpression of c-MYC in Burkitt lymphoma. A t(18;14) translocation moving Bcl-2 from chromosome 18 to the IgH locus on 14 is involved in follicular lymphoma. Also CCND1 is involved in a t(11;14) translocation bringing cyclin d1 to the IgH resulting in cell cycle dysregulation in Mantle cell lymphoma. Studies suggest that these cancers are associated with telomere length [35–38] as well as with miRNA levels [39]. We show that TERT regulation of miRNAs alters Bcl-2 and an enriched pathway associated with the dysregulated genes is chronic myeloid leukemia, which has overlapping genes with some of the lymphomas previously studied. Our data support the hypothesis that TERT is involved in disease processes that include non-TL-related pathways.
One of the major strengths of our study was the availability of miRNA expression data, SNP data, and mRNA data on a large population. We have previously analyzed these SNPs with both TL and colon cancer, which helps facilitate the interpretation of our data. Additionally, we applied a FDR to guide our interpretation of meaningful results. Larger samples would have enabled us to split the sample and replicate our findings with a second data set. While we are unable to replicate these findings in a second data set, we encourage others with similar data to undertake such analysis. There are many available tools to assist in identifying verified mRNA targets for miRNA. In this study we used several bioinformatics tools to better understand function of these miRNAs, however all bioinformatics tools have limitations as to the extent of the knowledge and the specificity of the information to specific miRNAs. To have a more focused assessment of pathways, we restricted our bioinformatics assessment to only those targeted genes significantly associated with miRNA. Although functionality data of specific miRNA and TERT associations are not available or feasible from data available to us, we believe that the information provided gives direction for more laboratory-based studies that can directly test these associations.
Conclusions
In conclusion, our data suggest that miRNAs are involved in regulating TL. TERT appears to influence carcinoma/normal mucosa differential expression. Given the number of miRNAs associated with TL, TERT rs2736118 and ALT genes, our data also support the hypothesis that telomere-related genes, in addition to affecting TL, impact non-TL-related functions that can importantly influence cancer risk.
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We would like to acknowledge Dr. Bette Caan and the Kaiser Permanente Medical Research Program for sample contributions, Brett Milash and the Bioinformatics Shared Resource of the Huntsman Cancer Institute and University of Utah for miRNA and mRNA bioinformatics data processing, Sandra Edwards for the data collection and management, Wade Samowitz for slide review, and Erica Wolff and Michael Hoffman for miRNA processing.
Abbreviations
- CRC
Colorectal cancer
- FDR
False discovery rate
- IPA
Ingenuity pathway analysis
- FPMRP
Kaiser Permanente Medical Research Program
- mRNA
messenger RNA
- miRNA
microRNA
- PTEN
Phosphate and tensin homolog
- PI3K
Phosphatidylinositide 3-kinases
- AKT
Protein kinase B
- QC
Quality Control
- qPCR
Quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- SNP
Single nucleotide polymorphism
- TERT
Telomerase reverse transcriptase
- TL
Telomere length
- T/S ratio
Telomere-to-single copy gene
Data Availability
Data are available upon request to Dr. Slattery who may be contacted at marty.slattery@hsc.utah.edu. Dr. Slattery will work with individuals who request data to obtain necessary data transfer agreements and MOU.
Funding Statement
This study was supported by NCI grants CA163683, CA48998 and CA61757 from the National Cancer Institute at the National Institutes of Health. NCI was not involved in any of the work described in this manuscript.
References
- 1.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7(17):2643–6. . [DOI] [PubMed] [Google Scholar]
- 2.Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792(4):341–52. 10.1016/j.bbadis.2009.02.003 . [DOI] [PubMed] [Google Scholar]
- 3.Harries LW. MicroRNAs as Mediators of the Ageing Process. Genes (Basel). 2014;5(3):656–70. 10.3390/genes5030656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacio LN, Jarstfer MB. MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PLoS One. 2010;5(9). 10.1371/journal.pone.0012519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberto Dinami EP, Rosanna Sestito, Roberta Benetti, Stefan Schoeftner. microRNAs control the function of telomeres in cancer. RNA & Disease. 2014. Epub e292. [Google Scholar]
- 6.Pellatt AJ, Wolff RK, Lundgreen A, Cawthon R, Slattery ML. Genetic and lifestyle influence on telomere length and subsequent risk of colon cancer in a case control study. Int J Mol Epidemiol Genet. 2012;3(3):184–94. Epub 2012/10/11. [PMC free article] [PubMed] [Google Scholar]
- 7.Boardman LA LK, Seo S, Johnson RA, Vanderboom RJ, Kimmel GW, Cunningham JM, Gangon RE,. Engelman CD,. Riegert-Johnson DL, Potter J, Haile R, Buchanan D, Jenkins MA, Rider DN, Thibodeau SN, Petersen GM, Skinner HG. The association of telomere length with colorectal cancer differs by the age of cancer onset. Clinical and Translational Gastroenterology. 2014. (online);e52 10.1038/ctg.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Hum Mol Genet. 2001;10(7):677–85. . [DOI] [PubMed] [Google Scholar]
- 9.Lassmann T, Maida Y, Tomaru Y, Yasukawa M, Ando Y, Kojima M, et al. Telomerase reverse transcriptase regulates microRNAs. Int J Mol Sci. 2015;16(1):1192–208. 10.3390/ijms16011192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellatt AJ, Wolff RK, Torres-Mejia G, John EM, Herrick JS, Lundgreen A, et al. Telomere length, telomere-related genes, and breast cancer risk: The breast cancer health disparities study. Genes, chromosomes & cancer. 2013;52(7):595–609. Epub 2013/05/01. 10.1002/gcc.22056 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellatt AJ, Wolff RK, Lundgreen A, Cawthon R, Slattery ML. Genetic and lifestyle influence on telomere length and subsequent risk of colon cancer in a case control study. Int J Mol Epidemiol Genet. 2012;3(3):184–94. [PMC free article] [PubMed] [Google Scholar]
- 12.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131(2):217–34. 10.1007/s00439-011-1055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao JL, Yuan P, Abuduwufuer A, Lv W, Yang YH, Hu J. Association between the TERT Genetic Polymorphism rs2853676 and Cancer Risk: Meta-Analysis of 76,108 Cases and 134,215 Controls. PLoS One. 2015;10(6):e0128829 10.1371/journal.pone.0128829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellatt AJ, Wolff RK, Herrick J, Lundgreen A, Slattery ML. TERT's role in colorectal carcinogenesis. Molecular carcinogenesis. 2013;52(7):507–13. 10.1002/mc.21885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–9. . [DOI] [PubMed] [Google Scholar]
- 16.Slattery ML, Potter J, Caan B, Edwards S, Coates A, Ma KN, et al. Energy balance and colon cancer—beyond physical activity. Cancer research. 1997;57(1):75–80. . [PubMed] [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. Epub 2003/01/23. . [DOI] [PubMed] [Google Scholar]
- 18.Slattery ML, Herrick JS, Pellatt DF, Stevens JR, Mullany LE, Wolff E, et al. MicroRNA profiles in colorectal carcinomas, adenomas, and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016. 10.1093/carcin/bgv249 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellatt DF, Stevens JR, Wolff RK, Mullany LE, Herrick JS, Samowitz W, et al. Expression Profiles of miRNA Subsets Distinguish Human Colorectal Carcinoma and Normal Colonic Mucosa. Clin Transl Gastroenterol. 2016;7:e152 10.1038/ctg.2016.11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slattery ML, Herrick JS, Mullany LE, Valeri N, Stevens J, Caan BJ, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137(2):428–38. 10.1002/ijc.29384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slattery ML, Pellatt DF, Mullany LE, Wolff RK, Herrick JS. Gene expression in colon cancer: A focus on tumor site and molecular phenotype. Genes, chromosomes & cancer. 2015;54(9):527–41. 10.1002/gcc.22265 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21 Epub 2009/01/09. gkn1027 [pii] 10.1093/nar/gkn1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2011. Epub 2011/07/16. 10.1007/s00439-011-1055-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini YH, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 25.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(Database issue):D78–85. 10.1093/nar/gkt1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.QIAGEN's Ingenuity Pathway Analysis [Internet]. 2014. Available from: http://www.qioagen.com/ingenuity.
- 27.Uziel O, Yosef N, Sharan R, Ruppin E, Kupiec M, Kushnir M, et al. The effects of telomere shortening on cancer cells: a network model of proteomic and microRNA analysis. Genomics. 2015;105(1):5–16. 10.1016/j.ygeno.2014.10.013 . [DOI] [PubMed] [Google Scholar]
- 28.Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem. 2011;149(1):5–14. 10.1093/jb/mvq119 . [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74(6):1639–44. 10.1158/0008-5472.CAN-13-3568 . [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, et al. Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol. 2012;14(12):1270–81. Epub 2012/11/20. 10.1038/ncb2621 . [DOI] [PubMed] [Google Scholar]
- 31.Ding D, Xi P, Zhou J, Wang M, Cong YS. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-kappaB-dependent transcription. FASEB J. 2013;27(11):4375–83. 10.1096/fj.13-230904 . [DOI] [PubMed] [Google Scholar]
- 32.Lee J, Sung YH, Cheong C, Choi YS, Jeon HK, Sun W, et al. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene. 2008;27(26):3754–60. 10.1038/sj.onc.1211037 . [DOI] [PubMed] [Google Scholar]
- 33.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4(1):e10 10.1371/journal.pgen.0040010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778(3):729–56. 10.1016/j.bbamem.2007.08.018 . [DOI] [PubMed] [Google Scholar]
- 35.Ladetto M, Compagno M, Ricca I, Pagano M, Rocci A, Astolfi M, et al. Telomere length correlates with histopathogenesis according to the germinal center in mature B-cell lymphoproliferative disorders. Blood. 2004;103(12):4644–9. 10.1182/blood-2003-12-4412 . [DOI] [PubMed] [Google Scholar]
- 36.Cottliar AS, Panero J, Pedrazzini E, Noriega MF, Narbaitz M, Rodriguez A, et al. Analysis of telomere length in mantle cell lymphoma. Eur J Haematol. 2009;83(5):433–8. 10.1111/j.1600-0609.2009.01313.x . [DOI] [PubMed] [Google Scholar]
- 37.Jebaraj BM, Kienle D, Lechel A, Mertens D, Heuberger M, Ott G, et al. Telomere length in mantle cell lymphoma. Blood. 2013;121(7):1184–7. 10.1182/blood-2012-08-452649 . [DOI] [PubMed] [Google Scholar]
- 38.Hosnijeh FS, Matullo G, Russo A, Guarrera S, Modica F, Nieters A, et al. Prediagnostic telomere length and risk of B-cell lymphoma-Results from the EPIC cohort study. Int J Cancer. 2014;135(12):2910–7. 10.1002/ijc.28934 . [DOI] [PubMed] [Google Scholar]
- 39.Robaina MC, Mazzoccoli L, Arruda VO, Reis FR, Apa AG, de Rezende LM, et al. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp Mol Pathol. 2015;98(2):200–7. 10.1016/j.yexmp.2015.03.006 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon request to Dr. Slattery who may be contacted at marty.slattery@hsc.utah.edu. Dr. Slattery will work with individuals who request data to obtain necessary data transfer agreements and MOU.