Fig 5 is incorrect. The authors have provided the correct Fig 5 here.
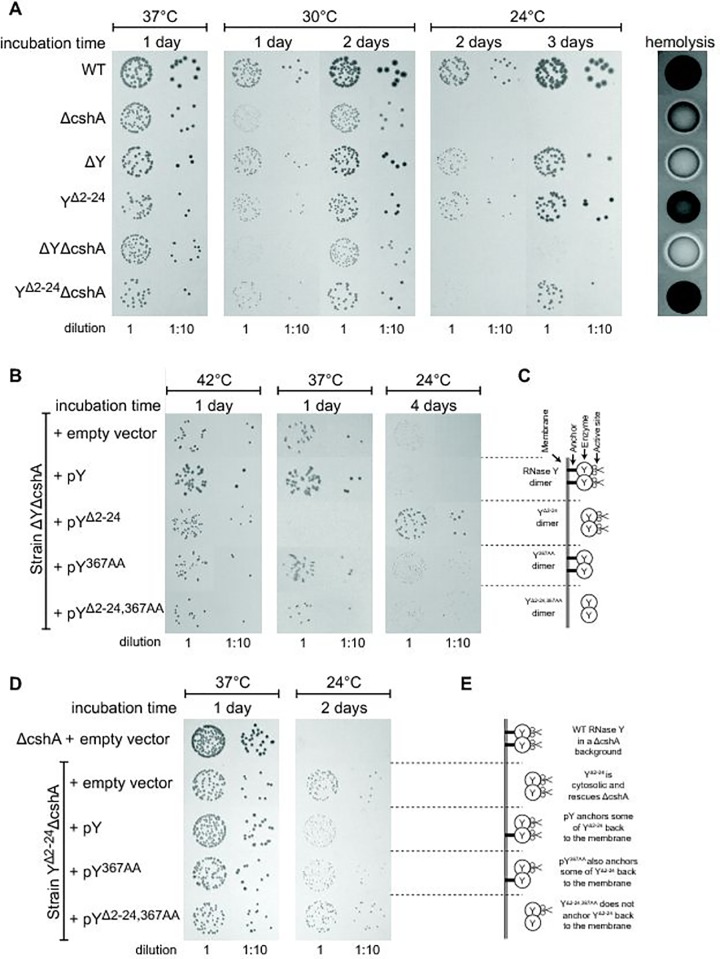
Fig 5. Removal of the membrane anchor enables RNase Y to suppress the phenotypes of a ΔcshA mutant.
(A) In the left panel, over-night cultures of single mutants ΔcshA, ΔY and YΔ2–24 and double mutants ΔYΔcshA and YΔ2–24ΔcshA were diluted, spotted on agar-plates, and incubated at the indicated temperatures and times. In the right panel, over-night cultures were spotted on horse-blood-agar. (B) Transformants of strain ΔYΔcshA with plasmids expressing variants of RNase Y were selected at 42°C, then restreaked and grown over night at 42°C. Finally the cultures were diluted and spotted at the indicated temperatures. ΔYΔcshA with pYΔ2–24 grows significantly better than the other strains at 24°C. (C) Cartoon showing the four versions of RNase Y expressed from the plasmids; wild-type RNase Y (pY), anchorless RNase Y (pYΔ2–24), RNase Y active site mutant (pY367AA), and anchorless RNase Y active site mutant (pYΔ2–24,367AA). (D) The YΔ2–24ΔcshA strain was transformed with the plasmids expressing the wild-type RNase Y, Y367AA or YΔ2–24,367AA. Overnight cultures were diluted, spotted on agar-plates and incubated at the indicated temperatures for the indicated period of time. Both pY and pY367AA inhibit growth at 24°C. (E) Cartoon showing how the wild-type RNase Y and Y367AA can anchor the YΔ2–24 protein back to the membrane, via dimer-formation.
Reference
- 1.Khemici V, Prados J, Linder P, Redder P (2015) Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation. PLoS Genet 11(10): e1005577 doi:10.1371/journal.pgen.1005577 [DOI] [PMC free article] [PubMed] [Google Scholar]