Abstract
Cardiac cell specification and the genetic determinants that govern this process are highly conserved among Chordates. Recent studies have established the importance of evolutionarily-conserved mechanisms in the study of congenital heart defects and disease, as well as cardiac regeneration. As a basal Chordate, the Ciona model system presents a simple scaffold that recapitulates the basic blueprint of cardiac development in Chordates. Here we will focus on the development and cellular structure of the heart of the ascidian Ciona as compared to other Chordates, principally vertebrates. Comparison of the Ciona model system to heart development in other Chordates presents great potential for dissecting the genetic mechanisms that underlie congenital heart defects and disease at the cellular level and might provide additional insight into potential pathways for therapeutic cardiac regeneration.
Keywords: Ciona, cardiac development, cardiac cell specification, cardiac regeneration
1. Introduction
The animal species comprising the phylum Chordata display very diverse cardiac morphologies and physiology. Cardiac features are highly-adaptive such that diffusely contractile vessels in cephalochordates, simple peristaltic compartments in Tunicates, and multi-chambered pumps with separate high and low pressure circulation in amniotes reflect the evolution and divergence of Chordate species. Yet, across this diversity lies a deeply conserved network of genetic determinants for cardiac cell specification and patterning. Classical examples of key cardiac developmental control genes include orthologs of the transcription-factor-coding genes Nkx2.5, Gata4/5/6, Hand1/2, and Tbx1 that are highly conserved across Chordate species and beyond. These key cardiac gene determinants are thought to form a regulatory kernel, or conserved core subnetwork, that determines cardiac cell specification and differentiation, resulting in conserved cardiac cell types and cellular organization of Chordate hearts [1,2]. Notably, the mechanism of cardiac cell type specification is not only conserved among Chordates, but also determines circulatory pump formation in Drosophila [3], which demonstrates the deep evolutionary origins of this kernel, whereas its widespread conservation presumably results from continuous selective pressure, highlighting its importance for cardiac development in bilateral animals.
The complex events that span from genetic regulation to cellular specification present a puzzle that must be solved in order to fully understand the underlying blueprint of Chordate heart development [4,5]. However, if the cellular and molecular underpinnings of congenital heart defects/disease (CHD) can be studied in simpler Chordates independently of their complex morphogenetic outcomes in higher vertebrates, then solving the riddle of heart development becomes more plausible. Thus, examining the conserved regulatory genetic linkages that lead to cardiac cell specification and differentiation, as well as the resulting cellular outcomes in the hearts of simple Chordates, presents an opportunity to gain insights into the deeply conserved mechanisms of heart development. Comparison of cardiac cell lineages and regulatory mechanisms of heart development across Chordate species elicits many questions. The first of which being, how can the regulatory kernel of genetic programming be so highly conserved and yet produce such great divergence of heart morphologies within the super-phylum Chordata? Furthermore, if cardiac cell specification during heart development and the resulting cardiac cell types that form the heart are also highly conserved in Chordates, then how do some species retain the ability to continuously produce new cardiac cells to replace and regenerate their hearts whereas other species do not?
Here we will focus on the development and cellular structure of the heart of the ascidian Ciona as compared to other Chordates, principally vertebrates. Examining cardiac fate-specification programs and conserved regulatory states of cardiac progenitor cells during Ciona heart development, as compared to vertebrates, presents an opportunity to examine the basic building blocks of the Chordate heart program in a simple model system. Moreover, studies in a simpler model system such as Ciona, which can naturally regenerate cardiac tissue, unlike adult mammals, with a simpler complement of cardiac genes compared to zebrafish, might shed light into the basic cellular and molecular requirements to reactivate the ancestral aspects of the cardiac program in a regenerative context. Thus, a comparison of the Ciona model system to heart development in other Chordates presents great potential for dissecting the genetic mechanisms that underlie congenital heart defects and disease at the cellular level, as well as provides insight into potential pathways for therapeutic cardiac regeneration.
2. Chordate Circulatory Systems: From Divergent Anatomies to Conserved Cytology
2.1. Divergent Cardiac Anatomy and Histology
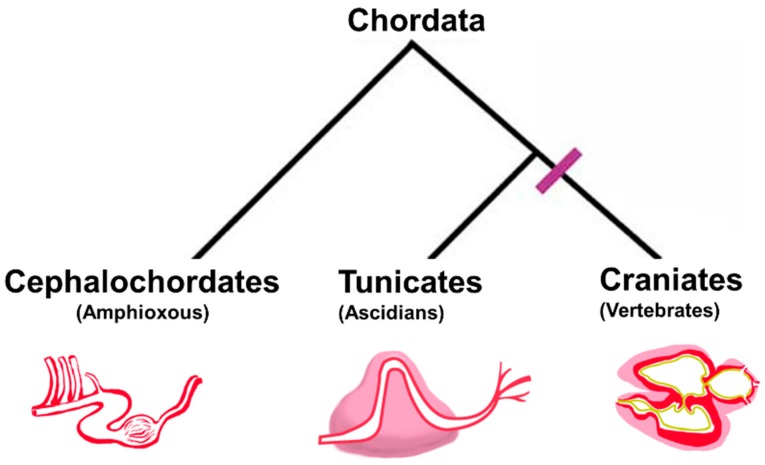
Within the super-phylum Chordata, Cephalochordates, Tunicates, and Craniates are phyla that share common embryonic features including a dorsal nerve cord, a notochord, and a post-anal tail [6]. Ascidians are a class of Tunicates and are primarily soft-bodied filter-feeders that are sessile, which lends to their common name of “sea squirts”. Cephalochordates, commonly known as lancelet or amphioxus, retain Chordate features as adults, and they were previously thought to be more closely related to Craniates. However, phylogenomic studies indicate that Tunicates are the true sister clade to the Craniates, which includes vertebrates [7,8]. Unlike shared embryonic features, the circulatory systems of Chordates vary widely and the final morphogenetic outcomes of heart development in Chordates are highly divergent. The general structure of Chordate hearts and the phylogenetic relationships are represented in Figure 1. Cephalochordates use non-striated muscle cells to power a series of four peristaltic vessels that circulate blood. Within Tunicates, the cardiac anatomy in Ascidians consists of a single-compartment peristaltic pump comprised of a mono-layer of striated muscle myocardium enclosed in a pericardial sac [9]. By contrast, Craniates have many well characterized model systems of heart development and anatomy, which generally display a striated, multi-layered, and trabeculated muscle cell myocardium that is divided into multiple chambers by valves with dedicated inflow and outflow, an endocardium, and enclosed by a pericardium.
Figure 1.
Phylogenetic relationship and general heart structure of Chordate subphyla. Cephalochordates have a series of four peristaltic vessels that serve as a pump. Tunicates have a single-chamber peristaltic pump comprised of a single layer of myocardium (red) surrounded by a pericardium (pink). Vertebrates have at least a two-chambered myocardium comprised of layered cardiac myocytes (red), an endocardium (yellow), valves that separate distinct inflow and outflow chambers and a surrounding pericardium (pink).
Notably, the myocardium of both Cephalochordates and Tunicates, which consist of a monolayer of cardiac myoepithelial cells that lack an endocardium, is also characteristic of other invertebrate blood pumps, such as Drosophila [10]. Interestingly, other invertebrates outside of the super-phylum Chordata also display multi-layered myocardium despite lacking endo- and epi-cardial layers. Species of blue crab and oyster have trabeculae [11]. Whereas cephalopods, which have the most complex circulatory systems as invertebrates, display a closed circulatory system with a heart that has a thickened myocardium that is capable of producing powerful contractions [10,12]. Evidence of thickened myocardium in invertebrates outside the super-phylum Chordata suggests that there are other mechanisms besides endocardial and epicardial signaling that can result in myocardium layering. Thus, comparison of the mechanisms of heart development between all Chordates provides important insights into evolutionary advances; and, there is still much to learn about the genetic and morphometric regulation of heart development across all phyla. However, the fate specification programs and resulting cardiac cell types are most conserved among Chordates. As simple Chordates within the phylum Tunicata, the ascidian species of Ciona provide an excellent model system in which to examine the basic mechanisms of heart development.
Within Tunicates, species of the Ciona genus are among the most studied ascidians and there are multiple species of Ciona that have been characterized, whereas Ciona savignyi and Ciona intestinalis type A provided the first two whole-genome sequences for invaluable comparative analyses [13,14,15,16,17], recent evidence suggests that Ciona intestinalis type A and type B are in fact two separate species. Ciona intestinalis sensu stricto, formerly known as Ciona intestinalis type B, is found in the North Atlantic Ocean and Ciona robusta, formerly known as type A, is found in the Pacific Ocean [18]. In this review, we collectively refer to all of these species as Ciona since they all share the conserved genes, cardiac cell types, and heart structure.
2.2. Cellular Structure of Ciona Heart
Heart formation in Ciona occurs throughout embryonic, larval, juvenile, and adult stages. Larvae develop, hatch, and swim 12 to 18 h post-fertilization, and then attach to a substrate and metamorphose into sessile juveniles with beating hearts within 3–5 days. Juveniles continue to develop into young adults by 10 days post-fertilization, then grow isometrically as adults, mature within 1–3 months and die within 12–18 months [19,20,21,22]. There is a positive correlation between age and body length [23]. Since the embryonic and larval stages retain Chordate features and are more amenable to experimentation, most studies investigating Ciona heart development have focused on the characterization of early development up to the larval stage.
During embryogenesis, Ciona development is highly reproducible, with cell lineages, cell-cell contacts, and successive fate specification events being invariant and designated in part by intrinsic maternal determinants and well stereotyped cell-cell signaling events [24,25,26]. Cell lineage tracing has identified cardiac progenitor cells throughout embryonic development to the larval stage [27,28,29,30,31]. The genetic programs for early cardiac cell fate specification in Ciona share many of the same pathways that govern vertebrate heart development [32,33,34,35]. Cardiac progenitor cells specified during embryogenesis begin to differentiate during metamorphosis in order to form the beating heart [30,36]. Specifically, fate-restricted cardiac precursors arise in swimming larvae from multipotent cardiopharyngeal progenitors that also produce the atrial siphon and body wall muscles (a.k.a. pharyngeal muscles), in a manner analogous to that described in the mouse, where common progenitors of the second heart field cardiomyocytes and branchiomeric head muscles emerge from the primitive streak [37,38].
The main cell types that arise from cardiac progenitor cell differentiation include contractile cardiac myoepithelial cells and an epithelium of pericardial cells. It is not clear how the cardiac progenitor cells in Ciona larvae eventually produce the diverse cell types observed in the post-metamorphic heart. The newly-formed heart can be seen beating 3–5 days post-fertilization in juvenile Ciona (see Video S1). The developmental stages and organ formation up to the juvenile stage of Ciona was reviewed previously [39]. During post-metamorphic development, the Ciona heart is initially round-shaped, then becomes V-shaped about 21 days post-fertilization [39]. There is very little information about the adult Ciona heart in the current literature; however, the most comprehensive resource is a report by R.H. Millar published in 1953 [20]. The adult Ciona heart grows in proportion to the body size of the animal and is a tubular, looped structure that consists of a myocardium with attachments to a pericardium that encloses the myocardium in a fluid-filled cavity (Figure 2). The Ciona circulation is open. The peristaltic contractions of the Ciona heart are rhythmic and directional, even though the direction of the contraction can be reversed (see Video S2).
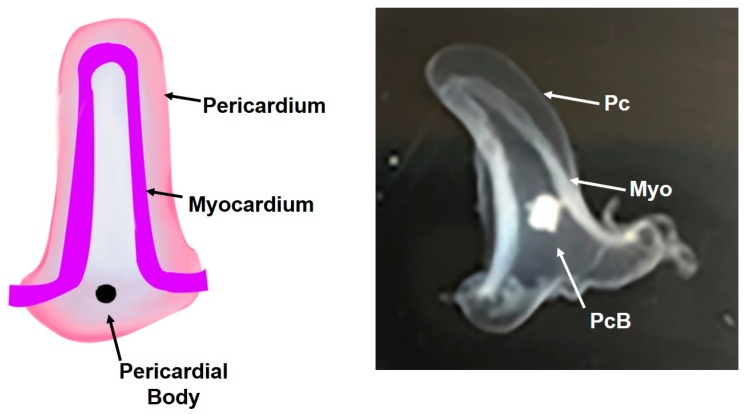
Figure 2.
Adult Ciona Heart Structure. The heart primarily consists of the myocardium (Myo) and the pericardium (Pc). The pericardium encloses the myocardium. As the Ciona ages, a pericardial body (PcB) appears, which consists of decaying cardiac myocytes that are shed from the myocardium.
The cell types that make up the adult Ciona heart are cardiac myocytes; transitional myocardial cells; general and junctional pericardial cells; and cells of the undifferentiated line [9,20,40]. Transitional cardiac myocytes exist at the junction where the pericardium and myocardium meet (Figure 3). These cells are in various stages of differentiation wherein the formation of myofilaments can be observed towards the basal sarcolemma. The pericardium is comprised of a monolayer of non-contractile epithelial cells. The pericardium and myocardium are continuous at the raphe (Figure 3). An acellular extracellular matrix fills the space between the pericardiac and epicardiac walls and becomes more fibrous near the raphe. The cardiac raphe is a structural feature of the adult Ciona heart that occurs at the intersection of the myocardium and the pericardium (Figure 3). As the animal ages, a pericardial body appears from discarded and degenerating cells from the myocardium that resides within the pericardial space. The Ciona myocardium does not have endothelial cells that form an endocardium. However, the surface of cardiac myocytes that face the lumen of the heart is covered by a thin sheet of non-cellular matrix that is continuous with the blood vessels that extend away from the heart. Movat’s pentachrome staining of the Ciona heart shows that this extracellular matrix lining the heart is proteoglycan based. Further studies are necessary to confirm the identity and the role of this extracellular matrix.
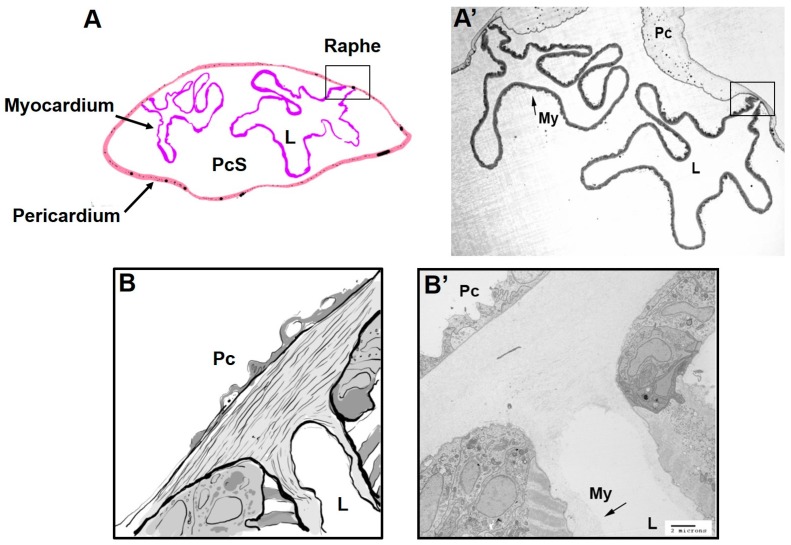
Figure 3.
Relationship between the pericardium and the myocardium. (A) Cartoon representation of transverse section of adult Ciona heart imaged by TEM in A’. The myocardial tube forms a lumen (L) that is surrounded by the pericardial space (PcS). The pericardium (Pc) joins the myocardium (My) at the raphe (boxed region in A and A’); and (B) cartoon representation of raphe as imaged by TEM in B’. Between the cells of the outer pericardium and myocardium, the space is filled with an extracellular matrix that continues into the lumen of the myocardium (arrow, B’). Transitional cardiac myocytes exist where the inner layer of the pericardium transitions to the myocardium. Scale bar = 2 microns.
Cardiac myocytes in Ciona are polarized cells that have nuclei oriented toward the apical pole facing the pericardium and myofilaments on the basal side facing the lumen of the heart (Figure 4). Myofilaments are striated and widely extend away from the central portion of the cell containing the nucleus. Binucleation occurs in ascidian cardiac myocytes, but the rate of occurrence has not been quantified. Ciona cardiac myocytes are in excess of 100 microns in length [20]. The myofilaments make an angle of 60 to 70° with the raphe, which runs parallel to the long axis of the heart [20]. This results in a slight spiral orientation to the myofilaments, which promotes directional contraction of the myocardium in the adult heart (see Video S2).
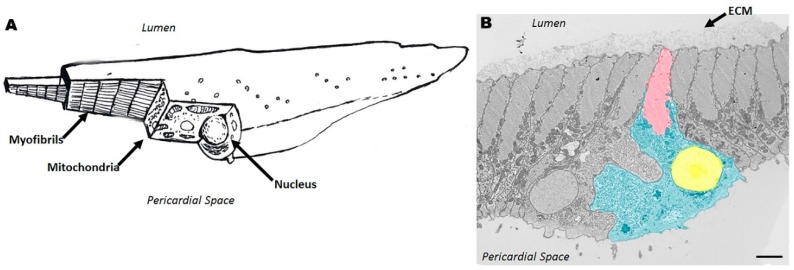
Figure 4.
Cardiac Myocyte in the Ciona myocardium. (A) Schematic of a cardiac myocyte with sectioned areas to show intracellular organization; and (B) false-colored TEM of Ciona myocardium with highlighted cardiac myocyte (red = myofibrils; yellow = nucleus; blue = sarcoplasm). Unknown extracellular matrix is continuous on sarcolemma of cardiac myocytes facing lumen of heart. The nucleus is oriented towards the pericardial space. Scale bar = 2 microns.
3. Development of the Ciona Heart and Parallels with the Vertebrates
Previous studies have shown deep conservation of specific regulatory networks that govern cardiac cell lineage specification [29,34]. The Ciona genome contains single copies of orthologous genes that govern cardiac development, including Gata4/5/6, Nkx2.5, and Hand gene families [41,42,43]. Ciona intestinalis type A/C. robusta has a small genome compared to vertebrates (115.2 Mb) [44]; and Tunicates appear to have diverged before whole genome duplications occurred [7]. Therefore, Ciona retained most of the basal Chordate program for cardiac development without the redundancy of the gene duplications seen in vertebrates.
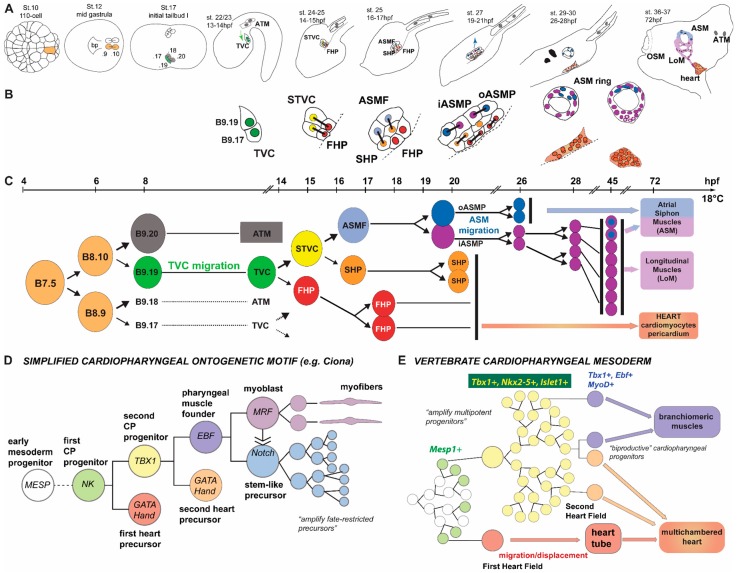
There has been comprehensive mapping of the early heart lineage during ascidian embryogenesis [28,29,30,31,45]. A current summary of known cardiac cell lineage specification and the ontogenetic motif is illustrated in Figure 5. The entire Ciona heart is derived from only two bilateral blastomeres in the gastrulating embryo (St. 12), which express the conserved pre-cardiac specification factor Mesp [28,46]. Likewise, in vertebrates the transcription factor Mesp1 is essential to mesoderm patterning and results in the formation of hematopoietic, skeletal muscle and cardiac cell lineages [37,47,48,49]. Mesp-expressing cells in Ciona divide once in early gastrula embryos to produce equivalent multipotent progenitors referred to as founder cells [50]. The founder cells divide again at the end of gastrulation (St. 22–23), this time asymmetrically, to generate two daughter cell lineages: the anterior tail muscle precursors (ATMs) and the trunk ventral cells (TVCs) [28]. All Ciona heart cells are derived from the TVCs, which are induced by FGF-mediated activation of the Ets1/2 transcription factor. This promotes the TVC-specific activation of conserved cardiac regulators including Gata4/5/6, Nk4/Nkx2-5, and Hand [28,51,52]. Notably, FGF also activates Ets1/2 transcription factors to initiate orthologous cardiac genes and cardiac cell specification in vertebrates [53]. In addition, similar to cardiac progenitor cells in vertebrate heart development, pairs of bilateral clusters of TVCs migrate towards the ventral midline where they meet to form a single cardiac progenitor pool in Ciona [46,51,52,54]. The bilateral fusion of cardiac progenitor cells to form a common progenitor pool is a critical step in vertebrate heart development. Alteration of fusion of cardiac progenitor cells results in cardia bifida phenotypes, whereby bilateral rudimentary hearts develop on either side of the ventral midline. Remarkably, perturbing gene function in the trunk epidermis can produce analogous failure of cardiac progenitors to merge at the midline in Ciona [55]. TVCs are multipotent cardiopharyngeal progenitors that will then divide asymmetrically and along the mediolateral axis to produce small median first heart precursors (FHPs) and large lateral second TVCs (STVCs) [30]. STVCs specifically activate the conserved transcription regulator Tbx1/10 before dividing again asymmetrically and along the mediolateral axis to produce large lateral atrial siphon muscle founder cells (ASMFs) and small median second heart precursors (SHPs; [30,36]). All cells then divide along the anteroposterior axis before the lateral-most atrial siphon muscle precursors (ASMPs) begin to collectively migrate dorsally, towards the atrial siphon placode, where they form a conspicuous ring of ASM precursor cells [30]. Remarkably, ASMFs successively activate the myogenic regulators Ebf and Mrf before the birth of distinct ASMPs, which sets the stage for Notch-mediated lateral inhibition. This process distinguishes between Mrf- undifferentiated stem-cell-like muscle precursors and Mrf+ differentiating myoblasts [31]. Shortly before and during ASMP migration, all cardiopharyngeal cells activate Islet, the sole homolog of the classic vertebrate second heart field marker Islet1 [56,57,58], which becomes restricted and more highly expressed in the migratory ASMPs [30,36].
Figure 5.
Summary of cardiac cell lineage specification in Ciona. (A) Schematic embryos, larvae, and juvenile showing the B7.5-derived cardiopharyngeal lineage, with approximate hours post-fertilization (hpf) and stages according to [59]. Numbers in the left second and third panels are simplified version of cell names according to Conklin (1905) nomenclature and corresponding the (C). Arrows indicate collective TVC (green) and ASMP (blue) migrations. TVC: trunk ventral cells; ATM: anterior tail muscles; STVC: second TVC; FHP: first heart precursor; SHP: second heart precursor; ASMF: atrial siphon muscle founder cells; iASMP: inner atrial siphon muscle precursor; oASMP: outer atrial siphon muscle precursor; LoM: longitudinal body wall muscles (note that they derive from the iASMPs); OSM: oral siphon muscles, derived from the A7.6 lineage [60]; bp: blastopore; and (B) close-up views of cardiopharyngeal lineage cells. Black bars link sister cells (as per the lineage shown in C). White asterisks in the ASM ring at ~72 hours post fertilization indicate iASMP-derived LoM precursors that reactivate Mrf and the body wall muscle differentiation program [31]. Note that the relative contributions of the FHP and SHP to the different parts of the differentiated heart (epicardium, myoepithelium, raphe, and pacemakers) remain elusive; (C) Lineage representation of cadiopharyngeal mesoderm development. Color codes correspond to those used in A and B. Only the progeny of one TVC for one side of the animal is shown, but this pattern is reiterated 4 times in each developing embryo, such that the Ciona equivalent of the first and second heart fields (see B) are composed of distantly-related FHP and SHP. For example, the left second heart field is composed of the anterior/leader and posterior/trailer SHPs, whose last common ancestor is the Mesp+ left B7.5 blastomere; (D) simplified cardiopharyngeal ontogenetic motif as seen in Ciona, illustrating that in ascidian embryos, cell fates are first restricted to a few progenitors, which are secondarily amplified (as seen for both the ASMP, and heart precursors); and (E) schematic representation of equivalent cardiopharyngeal ontogeny in vertebrates, where multipotent progenitors from larger populations in the early embryos, these morphogenetic fields are patterned by cell-cell signaling causing spatially-defined progressive fate restrictions such that not every single multipotent progenitor expresses its full potential. This may explain why a small fraction (~10%) of the bipotent cardiopharyngeal progenitors produce both skeletal head muscles and SFH-derived cardiomyocytes, whereas these populations are easily switched to one fate or another experimentally. The existence of such plastic populations of multipotent progenitors probably fostered the diversification of cardiopharyngeal structures in vertebrates.
These studies have not only provided novel insights into the evolutionary origins of the vertebrate first and second heart fields, but they also lend much insight into the origins of the branchiomeric muscles of the face, jaw and neck [38]. Briefly, branchiomeric muscles and cardiac progenitors share a common origin in the Nkx2-5+, Tbx1+, and Isl1+ anterior splanchnic/pharyngeal mesoderm, which derive from Mesp1+ mesoderm progenitors and has recently been referred to as the cardiopharyngeal field [38,58,61]. Retrospective clonal analyses in the mouse revealed the ontogenetic kinship between distinct head and neck muscles and specific derivatives of the second heart field [62,63]. These studies led to the proposal that the pattern observed in Ciona represents a simplified version of an ancestral/ancient cardiopharyngeal ontogenetic motif, whereby multipotent progenitors progress through transient regulatory states marked successively by Mesp1, Nkx2-5, Tbx1, and Islet1 homologs and produce distinct first and second heart field progenitors, and pharyngeal muscles precursors and associated stem cells (Figure 5); [36,38].
More recent prospective clonal analyses, using state-of-the-art mouse genetic methods, further refined the clonal relationships between distinct cardiac and branchiomeric progenitors. These studies highlighted profound differences between early heart development in the mouse and in ascidians [37,49,64]. For instance, whereas previous analyses indicated that the first heart field (FHF) and second heart field (SHF) emerge from early common pan-cardiac progenitors [65], prospective clonal analyses using Mesp1-rtTA inducible Cre alleles revealed that Mesp1 is activated independently in approximately 250 separate FHF and SHF progenitors that emerge at distinct time points in the primitive streak. These analyses also challenged the view that multipotent cardiovascular progenitors generate cardiomyocytes, endocardium and smooth muscles as all FHF progenitors appeared unipotent. By contrast, a fraction of the SHF progenitors produced either cardiomyocytes and smooth muscles, or cardiomyocytes and endocardial cells, or cardiomyocytes and branchiomeric skeletal muscles [37]. The latter represented ~10% of the total pool of cardiopharyngeal progenitors. Remarkably, this contrasts with the observations that every single one of the four cardiopharyngeal progenitors in ascidians expresses its full potential by generating both first and second heart precursors, and pharyngeal muscle progenitors [30,33,36]. Moreover, studies using explants and perturbations of FGF, BMP, and Wnt signaling in vertebrates demonstrated that cardiopharyngeal progenitors have the potential to generate either cardiomyocytes or skeletal muscles, depending on extrinsic conditions [58,61,66,67,68,69]. Finally, studies using mouse embryonic stem cells showed that forced expression of Mesp1 can promote either skeletal or cardiac muscle differentiation and even generate bipotential cardiopharyngeal progenitors depending upon the culture conditions [70,71]. These observations suggest that, whereas not every single cardiopharyngeal progenitor is actually “multi-productive” in early mouse embryos, there exist plastic populations of multipotent progenitors patterned by cell-cell signaling, which may have fostered the diversification of cardiac and pharyngeal structures during vertebrate evolution [38]. By contrast, the extreme simplification of ascidian cardiopharyngeal development led every single fate choice to be hard-wired, with very little modification in evolution [72], and each one of the four cardiopharyngeal progenitors expresses its full potential in every developing embryo. In terms of cardiopharyngeal clonal dynamics, some of the main differences between ascidians and vertebrates can, thus, be summarized as follows: In vertebrate embryos, multipotent progenitor population are produced and amplified before fate choices occur, whereas in ascidians fate choices occur early and stereotypically within small populations of cells followed by amplification of fate-restricted precursors populations (Figure 5D,E).
While the conserved gene regulatory networks that underlie early cardiac patterning and fate specification in Ciona are increasingly well characterized in embryos and swimming larvae (recently reviewed by [33,34,73]); the subsequent cell fate specification events, behaviors and gene activities are not characterized post-metamorphosis in Ciona juveniles or adults, and this remains an important area for future research.
4. Ciona as a Model for Basic Cellular and Molecular Aspects of Congenital Heart Defects
Many congenital heart defects (CHD) are compatible with life, amenable to surgery and, presumably, arise from complex morphogenetic defects that often impact the derivatives of the second heart field [74]. Whereas terminal cardiac organogenesis is mammalian-specific and without counterpart in simpler invertebrates, morphogenetic defects are often caused by genetic alterations of deeplyyconserved cardiac transcription regulators, which function in early embryonic progenitors. Therefore, it is plausible that understanding the function of conserved transcriptional regulators in the cardiopharyngeal progenitor cells of invertebrate Chordates, like Ciona, will provide novel insights into the immediate cellular and molecular consequences of mutations that cause CHD in humans. For example, TBX1 haploinsufficiency resulting from 22q11.2 deletions is thought to cause cardiac and pharyngeal apparatus defects in the common cardiovelofacial/Di George syndrome [75], and Tbx1 mutations phenocopy 22q11-induced defects in mouse models [76,77,78], whereas the etiology of this syndrome is rather complex, there is mounting evidence that multiple defects arise from alteration of TBX1 function in cardiopharyngeal mesoderm progenitors of early embryos [79,80,81,82]. In this regard, the conserved expression of Tbx1/10 homologs in the second cardiopharyngeal progenitors of ascidians might provide insights into conserved regulatory functions. For instance, Tbx1/10 homologs act upstream of conserved myogenic regulatory factors, such as MyoD, as key determinants of pharyngeal myogenesis in both vertebrates and ascidians [36,83,84,85]. Studies using Ciona first identified a role for Collier/Olf/Ebf (COE) homologs in promoting pharyngeal myogenesis, by acting downstream of Tbx1/10 and upstream of Mrf, and opposing cardiogenesis [30,31]. Notably, COE homologs have since been showed to regulate MyoD expression and myogenesis, including branchiomeric muscle formation, in Xenopus and chick embryos [86,87,88]. We propose that Tbx1/10, Ebf and Mrf homologs are part of an ancient transcriptional network for pharyngeal muscle specification [85], with a role in opposing cardiac development in the cardiopharyngeal lineage. Human EBF homologs may, thus, very well be genetic modifiers of the craniofacial phenotypes observed in 22q11.2 deletion syndrome patients.
Along this line, Tbx1 is thought to delay cardiac differentiation in the mammalian second heart field, allowing the progenitors to proliferate and produce the large amount of cells required to build the right ventricle and outflow tract, among the main SHF derivatives [81,82,89]. Certain defects observed in Tbx1 mutants have been interpreted as failure of the SHF progenitors to proliferate. At the molecular level, Tbx1 interferes with the function of BMP-Smad signaling and the expression of Gata4/5/6 [90,91]. Remarkably, the latter function is conserved in Ciona where Tbx1/10 activity in the multipotent STVC progenitors contributes to delay the re-activation of Gata4/5/6 in the second heart field [36]. This study also showed that Tbx1/10 could activate Ebf and promote a pharyngeal muscle fate in the second heart precursors, if not for the activity of Nk4/Nkx2-5, which inhibits the maintenance of Tbx1/10 expression and directly represses Ebf activation in the SHPs [36]. The latter activities, hinting at cross-regulatory antagonistic interactions between early regulators of the heart and pharyngeal muscle programs, which contribute to their segregation to distinct precursor cells. How many of these relate to CHDs in humans? Mutations in the human homolog NKX2-5 cause various forms of viable CHDs, and Nkx2-5 mutant animals do not entirely lack a heart [92,93,94,95]. Instead, Nkx2-5 probably also functions in SHF progenitors, in part to oppose precocious differentiation [80,95] and either cooperate with or antagonize Tbx1 activities [82,96,97]. Thus, in spite of profound modifications of the complex morphogenetic processes observed between vertebrates and their closest relatives, there is an intriguing possibility for basic cellular and molecular mechanisms to be conserved deeply enough for studies using basal Chordates to illuminate the molecular etiology of congenital heart diseases, as was the case more than 20 years ago when fly genetics first identified the conserved cardiac determinant tinman/Nkx2-5 [3,98], which later proved one of the main loci for CHD-causing mutations [92].
5. Ciona as an Experimental Model for Cardiac Regeneration
Regenerative animal models provide insights into the basic cell biology and unveil novel molecular mechanisms. Cardiac regeneration has been reported in vertebrates, such as newts [99,100], frogs [101] and, most recently, in zebrafish following multiple types of cardiac injury [102,103,104]. Elucidation of the successful models of cardiac regeneration that exist in nature can help illuminate therapeutic avenues toward inducing heart repair. The zebrafish model system has shown regenerative potential of cardiac myocytes via dedifferentiation of pre-existing cardiac myocytes with subsequent proliferation [105,106], whereas murine hearts have been shown to be capable of limited cardiac regeneration via transdifferentiation of epicardial cells into cardiac myocytes [107]. While there are distinct attributes for the use of zebrafish or mice as useful models of regeneration, here we propose Ciona as an additional regenerative model system that can provide further understanding of basic cardiac myocyte biology and the underlying blueprint of heart development and maturation.
Adult mammalian cardiac myocytes are traditionally thought of as, primarily, terminally-differentiated postmitotic cells. However, studies have shown that the adult human heart has a limited capacity for replacement of cardiac myocytes via mitotic events [108,109]. Nonetheless, the rate of replacement of functional cardiac myocytes is insufficient to repair the adult heart after damage, which results in high morbidity following cardiac injury. In order to fully understand the mechanism of cardiac myocyte proliferation during development and the transition to a postmitotic cell, the basic cell biology of cardiac myocytes must be closely examined and compared across species capable of regeneration and those that are not.
Studies of regeneration in Ciona have been conducted over the past 125 years (reviewed in [110]). Recent reports have shown that Ciona have a high capacity to regenerate the neural complex, the oral siphon, and associated organs [111,112]. The regeneration process, including the maintenance of proliferative activity in replacement cells, has been attributed to Notch signaling [113], which is similar to regenerative processes observed in zebrafish cardiac regeneration [114]. The relationship between Notch signaling and cardiac regeneration in Ciona has not been examined; however, it presents an intriguing avenue of future research. In addition, Ciona also presents a model system wherein the effects of aging on tissue repair and regeneration can be addressed. Regeneration in Ciona has been shown to be negatively impacted by age and/or size of the animal (the two being related; [23,111,112,115]). These studies suggest that the regenerative potential might be the greatest during juvenile and young adult stages and declining in adults; however, it is not yet known if an age-related decline in regenerative potential exists in the heart of Ciona. Nonetheless, the decline in regenerative potential with age in the organs that have been studied in Ciona is similar to observations in postnatal and adult cardiac myocytes in mammals. In 2009, a study demonstrated that the adult human heart is capable of replacing cardiac myocytes at a slow rate [108]. This rate of renewal in humans is about 1% at age 25 and declines with age. However, the rate at which mammalian cardiac myocytes are replaced throughout lifespan is controversial [116,117] and the mechanism of replacement is not yet known.
There are several benefits of using Ciona as a model system to study regeneration and rich opportunities for further study. First, there is abundant information about development of Ciona through the larval stages [39,118] and public databases chronicle gene expression throughout the lifespan of Ciona as well as other ascidian species [119,120,121]. In addition, since Ciona have a short life span and can be reared from egg to adult in closed marine systems [122], the entire life cycle of Ciona may be examined. Second, since the Ciona genome has been sequenced and annotated [16], Ciona are amenable to many sophisticated molecular tools, including transgenic lines with specifically-expressed molecular markers [123] that have been used to track regenerative responses [111]. Furthermore, most developmental control genes often reactivated in regeneration are present in Ciona, but in single copies, facilitating functional analyses. Individual adult organs from Ciona can also be cultured as explants in order to manipulate and examine responses in vitro [111]. Whole organs have been removed from Ciona, which then fully regenerated an intact organ [124]; however, this has not been repeated using modern methods. Notably, the adult Ciona heart can be dissected out of the body and survive in culture for up to 10 days (unpublished data, HJEA), which presents an opportunity for further study. Finally, using skeletal muscles as an analogy, detailed knowledge of the early clonal origins of cardiomyocytes opens the possibility to analyze how adult stem cells may emerge from undifferentiated progenitors in swimming larvae. Other Tunicates, such as colonial Ascidians, have the capacity to form cardiac cells as well as other somatic progenitor cells from hemoblasts, which demonstrates wide cellular potential [125]. Other invertebrate regeneration can occur via non-proliferative tissue repair wherein postmitotic cells undergo polyploidization and fusion [126]. Thus, it is evident that regenerative processes and the regulatory mechanisms that mediate the complex state of cells in adult tissue are difficult to decipher; however, the Ciona model system may illuminate possible pathways among Chordates.
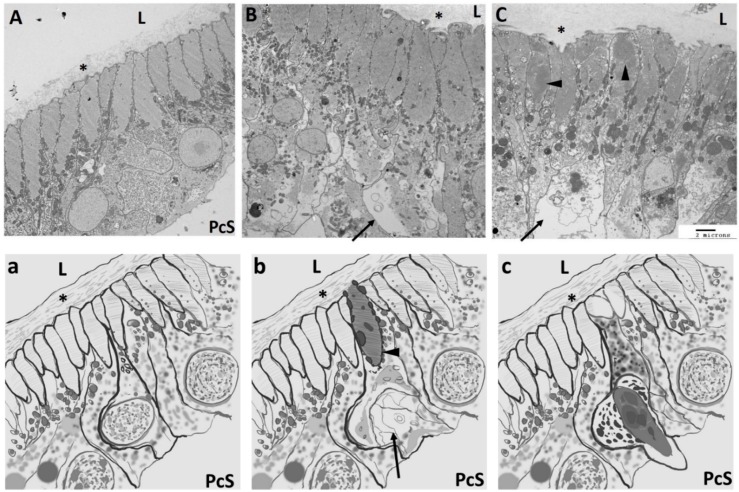
In 1953 Millar described degeneration in the myocardium of Ciona and growth zones of the heart [20]. The process of degeneration begins by thickening of the cardiac myocyte and breakdown of sarcomeric organization. This may be initiated in several neighboring cells. The nuclei lose their sharp outline and the sarcolemma begins to bulge into the pericardial space. The degenerated cells slough off into the pericardial space and collect in the pericardial body. The pericardiac body becomes larger as the Ciona ages (Figure 6). The preliminary data presented in Figure 6 closely resembles Millar’s early depiction wherein sarcomere disassembly and loss of nuclear integrity occurs in injured Ciona myocardium; however, the process of cardiac myocyte replacement in the Ciona myocardium needs further characterization (Figure 6). Notably, this process closely resembles dedifferentiation events that have been well-characterized in the zebrafish model system, which demonstrates that new cardiac myocytes come from existing cardiac myocytes during cardiac regeneration [105,106].
Figure 6.
Degeneration of cardiac myocytes. Panels (A–C) are transmission electron micrographs of Ciona myocardium and panels (a–c) are cartoon depictions of the TEM images to highlight details and provide a proposed model for cardiac myocyte degeneration. (A) Normal Ciona myocardium displays cardiac myocytes with organized myofibrils and thin layer of extracellular matrix on the luminal side (asterisk); (B) 24 h post stress, myofibrils in cardiac myocytes begin to break down, spaces open toward the pericardial space (PcS, arrows), mitochondria migrate toward both poles and enlarge, and the extracellular matrix in the lumen thickens (asterisk); and (C) 48 h post stress, complete breakdown of the myofibrils occurs (arrow heads).
Cardiac myocytes in Ciona share many of the same structural features as mammalian cardiac myocytes, including a complex cytoskeleton with many sarcomeres and tight cell-cell junctions. However, features of mammalian cardiomyocytes such as binucleation and polyploidy might influence the ability of these cells to undergo mitotic division and may reflect the differences in their continued proliferative potential [127]. In regenerative models, such as zebrafish and newts, cardiac myocytes are predominantly mononucleated whereas in mammalian cardiac myocytes binucleation is known to increase postnatally, which coincides with decreased proliferative potential [128,129,130,131]. In addition, sarcomeric organization is much more elaborate in mammalian cardiac myocytes. Given that sarcomeres of cardiac myocytes must be disassembled in order for cell division to occur [132], it is possible that the simpler sarcomeres in Ciona cardiac myocytes are more amenable to break down and, thus, undergo cell division during regeneration. In the zebrafish model system, cardiac myocytes with less organized sarcomeres have higher rates of DNA synthesis and mitosis [105,106]. Furthermore, the extracellular matrix (ECM) of Ciona cardiac myocytes displays intermediate complexity between vertebrate ECM and matrix of non-Chordate invertebrates, such as Drosophila [34]. Notably, the Ciona ECM includes fibronectin [133], which is present only in tunicate and vertebrate genomes, and is known to be an important mediator of cardiac myocyte proliferation in mammals. In mammals, the ECM of cardiac myocytes changes significantly during the neonatal period when the proliferative potential begins to decline [134]. While further studies are required in order to understand the relationship between the regenerative potential of cardiac myocytes and ECM composition, the composition of the ECM in Ciona also needs to be characterized for comparison.
The Ciona heart has been reported to have growth zones at the inflow/outflow regions of the myocardium [9,20]. The growth zone consists of undifferentiated cells that divide rapidly to form transitional cells in which myofilaments begin to appear. Further differentiation towards mature cardiac myocytes results in sarcomeric organization of the myofilaments to form striations and elongation of the cells. The formation of new cardiac myocytes from undifferentiated precursors needs to be validated using modern methods in Ciona. However, this presents an additional possibility of examining transdifferentiation as a mechanism for cardiac regeneration in Ciona. Transdifferentiation has been identified in the zebrafish model system as a mechanism of regeneration [135,136]. Notably, transdifferentiation was not observed in the adult zebrafish heart, which could be an interesting difference between Ciona and zebrafish if both transdifferentiation and dedifferentiation occurs in the adult Ciona myocardium during regeneration. Alternatively, these “undifferentiated cells” could constitute cardiac stem cells that promote the continuous growth of the adult heart, and may also contribute the regeneration. As stated above, Ciona might offer a unique opportunity to study the early developmental origins of cardiac stem cells.
The regenerative potential of the Ciona heart has not yet been completely determined; however, the information collected here provides intriguing possibilities for further studies of the regenerative potential of the Ciona heart and lends the question: Are there similar mechanisms in the adult mammalian heart that can be reactivated in order to stimulate heart regeneration? The field of cardiac biology has learned much from other regenerative model systems such as the newt and the zebrafish. As a basal Chordate with conserved cardiac gene program, the Ciona heart serves as a living scaffold of differentiated cardiac myocytes that can help to further elucidate the blueprint of cardiac regeneration. Cardiac myocytes in Ciona could be pushed toward proliferation and myocardial development using conserved signaling pathway components and/or growth factors. The easily accessible Ciona heart could be used to study effect of blood flow on cardiac myocyte proliferation. The possibilities using modern techniques to further characterize the Ciona heart are vast and these studies may provide important insights into cardiac myocyte biology.
6. Concluding Remarks
Vertebrate heart development is a complex process that requires the coordination of genetic programs, as well as many cellular and morphogenetic events. Studies in comparative genomics have demonstrated that the molecular determinants controlling key features in the development of Chordates are conserved. In order to fully understand how vertebrate features evolved, the genomics, development, and anatomical features of the closest invertebrate relatives provide important insights into the ancestral state of basic biological mechanisms. The Ciona heart displays the basal form of the Chordate heart as well as the basic cardiac genetic program and cell specification events during development and in congenital heart diseases. Moreover, the adult Ciona may serve as a regenerative model system to help further elucidate the differences between organisms that are capable of heart regeneration and those that are not. While the Ciona heart may have some divergent anatomical and histological features, the genetic underpinnings of cardiac cell specification, as well as the basic cellular structures, are deeply conserved. This level of conservation is particularly intriguing considering the regulatory states of cardiac progenitors and the conserved cardiopharyngeal origins of the first and second heart fields. Thus, as a basal Chordate, Ciona can provide important insights into cardiac gene regulation, as well as cardiac myocyte biology. The molecular and cellular impact of CHD-causing mutations on progenitor cells may be modeled in the simpler tunicate Ciona without the complexity of the redundant genome in higher vertebrates. Moreover, Ciona may provide an opportunity to delineate the cellular and molecular requirements for cardiac regeneration with two distinct advantages: a chance to track cellular origins back to early progenitors and the genetic and cellular simplicity. With recent advances in genetic tools (i.e., CRISPR-Cas9; [137,138,139] and culturing methods [122,140] the Ciona model system promises to provide many more important insights into cardiac biology.
Acknowledgments
Funding for this project came from the National Institutes of Health grant 1R15HL104587-01 (H.J.E.A.). Work in the lab of L.C. is funded award R01 HL108643 from the NIH/NHLBI and a Transatlantic Networks of Excellence Program grant from the Leducq Foundation. The authors would like to thank Paul Nguyen for assistance with graphic illustrations and Bob Price for transmission electron microscopy via the University of South Carolina School of Medicine Instrumentation Resource Facility.
Abbreviations
The following abbreviations are used in this manuscript:
| CHD | congenital heart defects/disease |
| Myo | myocardium |
| Pc | pericardium |
| PcB | pericardial body |
| PcS | pericardial space |
| L | lumen |
| TEM | transmission electron microscopy |
| ATMs | anterior tail muscle precursors |
| ATM | anterior tail muscles |
| TVCs | trunk ventral cells |
| FHPs | first heart precursors |
| STVCs | second trunk ventral cells |
| SHPs | second heart precursors |
| ASMFs | atrial siphon muscle founder cells |
| ASMPs | atrial siphon muscle precursors |
| LoM | longitudinal body wall muscles |
| iASMP | inner atrial siphon muscle precursor |
| oASMP | outer atrial siphon |
| OSM | oral siphon muscles |
| ECM | extracellular matrix |
Supplementary Materials
The following are available online at http://www.mdpi.com/2308-3425/3/3/25/s1, Video S1: Juvenile heartbeat, Video S2: Adult heartbeat.
Author Contributions
H.J.E.A. and L.C. equally contributed to writing this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Davidson E.H. Gene Regulatory Networks and the Evolution of Animal Body Plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 2.Olson E.N. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 4.Imai K.S., Levine M., Satoh N., Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava D., Olson E.N. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- 6.Satoh N., Rokhsar D., Nishikawa T. Chordate evolution and the three-phylum system. Proc. Biol. Sci. 2014;281:20141729. doi: 10.1098/rspb.2014.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delsuc F., Brinkmann H., Chourrout D., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 8.Putnam N.H., Butts T., Ferrier D.E.K., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu K., Jr., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 9.Davidson B. Ciona intestinalis as a model for cardiac development. Semin. Cell Dev. Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Pomares J.M., González-Rosa J.M., Muñoz-Chápuli R. Building the vertebrate heart—An evolutionary approach to cardiac development. Int. J. Dev. Biol. 2009;53:1427–1443. doi: 10.1387/ijdb.072409jp. [DOI] [PubMed] [Google Scholar]
- 11.Sedmera D., Thompson R.P. Myocyte proliferation in the developing heart. Dev. Dyn. 2011;240:1322–1334. doi: 10.1002/dvdy.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen B., Wang T., Christoffels V.M., Moorman A.F.M. Evolution and development of the building plan of the vertebrate heart. Biochim. Biophys. Acta. 2013;1833:783–794. doi: 10.1016/j.bbamcr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Vinson J.P., Jaffe D.B., O’Neill K., Karlsson E.K., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., et al. Assembly of polymorphic genomes: Algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cañestro C., Bassham S., Postlethwait J.H. Seeing chordate evolution through the Ciona genome sequence. Genome Biol. 2003;4:208. doi: 10.1186/gb-2003-4-3-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jong H.K., Waterman M.S., Li L.M. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 2007;17:1101–1110. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., et al. The Draft Genome of Ciona intestinalis: Insights into Chordate and Vertebrate Origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 17.Satoh N., Satou Y., Davidson B., Levine M. Ciona intestinalis: An emerging model for whole-genome analyses. Trends Genet. 2003;19:376–381. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 18.Pennati R., Ficetola G.F., Brunetti R., Caicci F., Gasparini F., Griggio F., Sato A., Stach T., Kaul-Strehlow S., Gissi C., et al. Morphological differences between larvae of the Ciona intestinalis species complex: Hints for a valid taxonomic definition of distinct species. PLoS ONE. 2015;10:e0122879. doi: 10.1371/journal.pone.0122879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berrill N.J. The development and growth of Ciona. J. Mar. Biol. Assoc. UK. 1947;26:616–625. doi: 10.1017/S0025315400013825. [DOI] [PubMed] [Google Scholar]
- 20.Millar R.H. L.M.B.C. Memoirs on Typical British Marine Plants and Animals. Liverpool University Press; Liverpool, UK: 1953. Ciona. [Google Scholar]
- 21.Dybern B.I. The Life Cycle of Ciona intestinalis (L.) f. typica in Relation to the Environmental Temperature. Oikos. 1965;16:109–131. doi: 10.2307/3564870. [DOI] [Google Scholar]
- 22.Petersen J.K., Svane I. Larval dispersal in the ascidian Ciona intestinalis (L.). Evidence for a closed population. J. Exp. Mar. Biol. Ecol. 1995;186:89–102. doi: 10.1016/0022-0981(94)00157-9. [DOI] [Google Scholar]
- 23.Jeffery W.R. Siphon regeneration capacity is compromised during aging in the ascidian Ciona intestinalis. Mech. Ageing Dev. 2012;133:629–636. doi: 10.1016/j.mad.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida H. Specification of developmental fates in ascidian embryos: Molecular approach to maternal determinants and signaling molecules. Int. Rev. Cytol. 2002;217:227–276. doi: 10.1016/s0074-7696(02)17016-1. [DOI] [PubMed] [Google Scholar]
- 25.Yamada L., Kobayashi K., Satou Y., Satoh N. Microarray analysis of localization of maternal transcripts in eggs and early embryos of the ascidian, Ciona intestinalis. Dev. Biol. 2005;284:536–550. doi: 10.1016/j.ydbio.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Jeffery W.R. Determinants of cell and positional fate in ascidian embryos. Int. Rev. Cytol. 2001;203:3–62. doi: 10.1016/s0074-7696(01)03003-0. [DOI] [PubMed] [Google Scholar]
- 27.Nishida H. Cell lineage and timing of fate restriction, determination and gene expression in ascidian embryos. Semin. Cell Dev. Biol. 1997;8:359–365. doi: 10.1006/scdb.1997.0160. [DOI] [PubMed] [Google Scholar]
- 28.Satou Y., Imai K.S., Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- 29.Davidson B., Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc. Natl. Acad. Sci. USA. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolfi A., Gainous T.B., Young J.J., Mori A., Levine M., Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razy-Krajka F., Lam K., Wang W., Stolfi A., Joly M., Bonneau R., Christiaen L. Collier/OLF/EBF-Dependent Transcriptional Dynamics Control Pharyngeal Muscle Specification from Primed Cardiopharyngeal Progenitors. Dev. Cell. 2014;29:263–276. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woznica A., Haeussler M., Starobinska E., Jemmett J., Li Y., Mount D., Davidson B. Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis. Dev. Biol. 2012;368:127–139. doi: 10.1016/j.ydbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan N., Razy-Krajka F., Christiaen L. Regulation and evolution of cardiopharyngeal cell identity and behavior: Insights from simple chordates. Curr. Opin. Genet. Dev. 2015;32:119–128. doi: 10.1016/j.gde.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cota C.D., Segade F., Davidson B. Heart genetics in a small package, exploiting the condensed genome of Ciona intestinalis. Brief. Funct. Genom. 2014;13:3–14. doi: 10.1093/bfgp/elt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norton J., Cooley J., Islam A.F.M.T., Cota C.D., Davidson B. Matrix adhesion polarizes heart progenitor induction in the invertebrate chordate Ciona intestinalis. Development. 2013;140:1301–1311. doi: 10.1242/dev.085548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W., Razy-Krajka F., Siu E., Ketcham A., Christiaen L. NK4 Antagonizes Tbx1/10 to Promote Cardiac versus Pharyngeal Muscle Fate in the Ascidian Second Heart Field. PLoS Biol. 2013;11:e1001725. doi: 10.1371/journal.pbio.1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A., et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014;16:829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diogo R., Kelly R.G., Christiaen L., Levine M., Ziermann J.M., Molnar J.L., Noden D.M., Tzahor E. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature. 2015;520:466–473. doi: 10.1038/nature14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiba S., Sasaki A., Nakayama A., Takamura K., Satoh N. Development of Ciona intestinalis juveniles (through 2nd ascidian stage) Zool. Sci. 2004;21:285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- 40.Oliphant L.W. Ph.D. Thesis. University of Washington; Seattle, WA, USA: 1972. Ultrastructure and Mechanism of Contraction of the Ascidian Heart. [Google Scholar]
- 41.Satou Y., Imai K.S., Levine M., Kohara Y., Rokhsar D., Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. I. Genes for bHLH transcription factors. Dev. Genes Evol. 2003;213:213–221. doi: 10.1007/s00427-003-0319-7. [DOI] [PubMed] [Google Scholar]
- 42.Wada S., Tokuoka M., Shoguchi E., Kobayashi K., Di Gregorio A., Spagnuolo A., Branno M., Kohara Y., Rokhsar D., Levine M., et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev. Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- 43.Yamada L., Kobayashi K., Degnan B., Satoh N., Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. IV. Genes for HMG transcriptional regulators, bZip and GATA/Gli/Zic/Snail. Dev. Genes Evol. 2003;213:245–253. doi: 10.1007/s00427-003-0316-x. [DOI] [PubMed] [Google Scholar]
- 44.Satou Y., Mineta K., Ogasawara M., Sasakura Y., Shoguchi E., Ueno K., Yamada L., Matsumoto J., Wasserscheid J., Dewar K., et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirano T., Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi. I. Origin of mesodermal tissues of the juvenile. Dev. Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- 46.Davidson B., Shi W., Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- 47.Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J.I., Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 48.Saga Y., Kitajima S., Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000;10:345–352. doi: 10.1016/S1050-1738(01)00069-X. [DOI] [PubMed] [Google Scholar]
- 49.Devine W.P., Wythe J.D., George M., Koshiba-Takeuchi K., Bruneau B.G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife. 2014;3 doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooley J., Whitaker S., Sweeney S., Fraser S., Davidson B. Cytoskeletal polarity mediates localized induction of the heart progenitor lineage. Nat. Cell Biol. 2011;13:952–957. doi: 10.1038/ncb2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davidson B., Shi W., Beh J., Christiaen L., Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beh J., Shi W., Levine M., Davidson B., Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- 53.Znosko W.A., Yu S., Thomas K., Molina G.A., Li C., Tsang W., Dawid I.B., Moon A.M., Tsang M. Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development. Dev. Biol. 2010;342:11–25. doi: 10.1016/j.ydbio.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christiaen L., Davidson B., Kawashima T., Powell W., Nolla H., Vranizan K., Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 55.Ragkousi K., Beh J., Sweeney S., Starobinska E., Davidson B. A single GATA factor plays discrete, lineage specific roles in ascidian heart development. Dev. Biol. 2011;352:154–163. doi: 10.1016/j.ydbio.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y., Liang X., Najafi N., Cass M., Lin L., Cai C.-L., Chen J., Evans S.M. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nathan E., Monovich A., Tirosh-Finkel L., Harrelson Z., Rousso T., Rinon A., Harel I., Evans S.M., Tzahor E. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135:647–657. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hotta K., Mitsuhara K., Takahashi H., Inaba K., Oka K., Gojobori T., Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- 60.Tokuoka M., Satoh N., Satou Y. A bHLH transcription factor gene, Twist-like 1, is essential for the formation of mesodermal tissues of Ciona juveniles. Dev. Biol. 2005;288:387–396. doi: 10.1016/j.ydbio.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 61.Tirosh-Finkel L., Elhanany H., Rinon A., Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- 62.Lescroart F., Kelly R.G., Le Garrec J.-F., Nicolas J.-F., Meilhac S.M., Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137:3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- 63.Lescroart F., Hamou W., Francou A., Théveniau-Ruissy M., Kelly R.G., Buckingham M. Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc. Natl. Acad. Sci. USA. 2015;112:1446–1451. doi: 10.1073/pnas.1424538112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chabab S., Lescroart F., Rulands S., Mathiah N., Simons B.D., Blanpain C. Uncovering the Number and Clonal Dynamics of Mesp1 Progenitors during Heart Morphogenesis. Cell Rep. 2016;14:1–10. doi: 10.1016/j.celrep.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meilhac S.M., Esner M., Kelly R.G., Nicolas J.-F., Buckingham M.E. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell. 2004;6:685–698. doi: 10.1016/S1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 66.Schultheiss T.M., Xydas S., Lassar A.B. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- 67.Schultheiss T.M., Burch J.B., Lassar A.B. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 68.Tzahor E., Lassar A.B. Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tzahor E., Kempf H., Mootoosamy R.C., Poon A.C., Abzhanov A., Tabin C.J., Dietrich S., Lassar A.B. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 2003;17:3087–3099. doi: 10.1101/gad.1154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan S.S.-K., Shi X., Toyama A., Arpke R.W., Dandapat A., Iacovino M., Kang J., Gengyun L., Hagen H.R., Garry D.J., et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan S.S.-K., Hagen H.R., Swanson S.A., Stewart R., Boll K.A.A., Aho J., Thomson J.A.A., Kyba M. Development of Bipotent Cardiac/Skeletal Myogenic Progenitors from MESP1+ Mesoderm. Stem Cell Rep. 2016;6:26–34. doi: 10.1016/j.stemcr.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stolfi A., Lowe E.K., Racioppi C., Ristoratore F., Brown C.T., Swalla B.J., Christiaen L. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. Elife. 2014;3:e03728. doi: 10.7554/eLife.03728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolkin T., Christiaen L. Development and Evolution of the Ascidian Cardiogenic Mesoderm. Curr. Top. Dev. Biol. 2012;100:107–142. doi: 10.1016/B978-0-12-387786-4.00011-7. [DOI] [PubMed] [Google Scholar]
- 74.Rickert-Sperling S., Kelly R.G. In: Congenital Heart Diseases: The Broken Heart. Driscoll D.J., editor. Springer; Vienna, Austria: 2016. [Google Scholar]
- 75.Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S.I., et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/S0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 76.Lindsay E.A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H.F., Scambler P.J., et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 77.Merscher S., Funke B., Epstein J.A., Heyer J., Puech A., Lu M.M., Xavier R.J., Demay M.B., Russell R.G., Factor S., et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/S0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 78.Jerome L.A., Papaioannou V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Z., Huynh T., Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133:3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prall O.W.J., Menon M.K., Solloway M.J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J.J., Robertson B.R., Chaulet H., et al. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L., Fulcoli F.G., Tang S., Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ. Res. 2009;105:842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu H., Morishima M., Wylie J.N., Schwartz R.J., Bruneau B.G., Lindsay E.A., Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 83.Kelly R.G., Jerome-Majewska L.A., Papaioannou V.E. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum. Mol. Genet. 2004;13:2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- 84.Grifone R., Jarry T., Dandonneau M., Grenier J., Duprez D., Kelly R.G. Properties of branchiomeric and somite-derived muscle development in Tbx1 mutant embryos. Dev. Dyn. 2008;237:3071–3078. doi: 10.1002/dvdy.21718. [DOI] [PubMed] [Google Scholar]
- 85.Tolkin T., Christiaen L. Rewiring of an Ancestral Tbx1/10-Ebf-Mrf Network for Pharyngeal Muscle Specification in Distinct Embryonic Lineages. bioRxiv. 2016 doi: 10.1101/039289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Green Y.S., Vetter M.L. EBF proteins participate in transcriptional regulation of Xenopus muscle development. Dev. Biol. 2011;358:240–250. doi: 10.1016/j.ydbio.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.El-Magd M.A., Saleh A.A., Farrag F., Abd El-Aziz R.M., Ali H.A., Salama M.F. Regulation of chick Ebf1–3 gene expression in the pharyngeal arches, cranial sensory ganglia and placodes. Cells Tissues Organs. 2014;199:278–293. doi: 10.1159/000369880. [DOI] [PubMed] [Google Scholar]
- 88.El-Magd M.A., Allen S., McGonnell I., Otto A., Patel K. Bmp4 regulates chick Ebf2 and Ebf3 gene expression in somite development. Dev. Growth Differ. 2013;55:710–722. doi: 10.1111/dgd.12077. [DOI] [PubMed] [Google Scholar]
- 89.Nevis K., Obregon P., Walsh C., Guner-Ataman B., Burns C.G., Burns C.E. Tbx1 is required for second heart field proliferation in zebrafish. Dev. Dyn. 2013;242:550–559. doi: 10.1002/dvdy.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fulcoli F.G., Huynh T., Scambler P.J., Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS ONE. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Liao J., Aggarwal V.S., Nowotschin S., Bondarev A., Lipner S., Morrow B.E. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev. Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schott J.J., Benson D.W., Basson C.T., Pease W., Silberbach G.M., Moak J.P., Maron B.J., Seidman C.E., Seidman J.G. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 93.Benson D.W., Silberbach G.M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S., Smalls O., Johnson M.C., Watson M.S., Seidman J.G., et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Investig. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lyons I., Parsons L.M., Hartley L., Li R., Andrews J.E., Robb L., Harvey R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 95.Zhang L., Nomura-Kitabayashi A., Sultana N., Cai W., Cai X., Moon A.M., Cai C.-L. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 2014;390:68–79. doi: 10.1016/j.ydbio.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe Y., Zaffran S., Kuroiwa A., Higuchi H., Ogura T., Harvey R.P., Kelly R.G., Buckingham M. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc. Natl. Acad. Sci. USA. 2012;109:18273–18280. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nowotschin S., Liao J., Gage P.J., Epstein J.A., Campione M., Morrow B.E. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 98.Azpiazu N., Frasch M. Tinman and bagpipe: Two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 99.Oberpriller J.O., Oberpriller J.C. Response of the adult newt ventricle to injury. J. Exp. Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 100.Witman N., Murtuza B., Davis B., Arner A., Morrison J.I. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev. Biol. 2011;354:67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 101.Rumyantsev P.P. Autoradiographic study on the synthesis of DNA, RNA, and proteins in normal cardiac muscle cells and those changed by experimental injury. Folia Histochem. Cytochem. 1966;4:397–424. [PubMed] [Google Scholar]
- 102.Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 103.González-Rosa J.M., Martín V., Peralta M., Torres M., Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 104.Wang J., Panáková D., Kikuchi K., Holdway J.E., Gemberling M., Burris J.S., Singh S.P., Dickson A.L., Lin Y.F., Sabeh M.K., et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jopling C., Sleep E., Raya M., Martí M., Raya A., Belmonte J.C.I., Izpisúa Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kikuchi K., Holdway J.E., Werdich A.A., Anderson R.M., Fang Y., Egnaczyk G.F., Evans T., Macrae C.A., Stainier D.Y.R., Poss K.D. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smart N., Bollini S., Dubé K.N., Vieira J.M., Zhou B., Davidson S., Yellon D., Riegler J., Price A.N., Lythgoe M.F., et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Jovinge S., et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M., Wu T.-D., Guerquin-Kern J.-L., Lechene C.P., Lee R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jeffery W.R. Closing the wounds: One hundred and twenty five years of regenerative biology in the ascidian Ciona intestinalis. Genesis. 2014;18:1–18. doi: 10.1002/dvg.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Auger H., Sasakura Y., Joly J.-S.S., Jeffery W.R. Regeneration of oral siphon pigment organs in the ascidian Ciona intestinalis. Dev. Biol. 2010;339:374–389. doi: 10.1016/j.ydbio.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dahlberg C., Auger H., Dupont S., Sasakura Y., Thorndyke M., Joly J.-S. Refining the Ciona intestinalis model of central nervous system regeneration. PLoS ONE. 2009;4:e4458. doi: 10.1371/journal.pone.0004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hamada M., Goricki S., Byerly M.S., Satoh N., Jeffery W.R. Evolution of the chordate regeneration blastema: Differential gene expression and conserved role of notch signaling during siphon regeneration in the ascidian Ciona. Dev. Biol. 2015;405:304–315. doi: 10.1016/j.ydbio.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao L., Borikova A.L., Ben-Yair R., Guner-Ataman B., MacRae C.A., Lee R.T., Burns C.G., Burns C.E. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA. 2014;111:1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jeffery W.R. Regeneration, Stem Cells, and Aging in the Tunicate Ciona: Insights from the Oral Siphon. Int. Rev. Cell Mol. Biol. 2015;319:255–282. doi: 10.1016/bs.ircmb.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 116.Soonpaa M.H., Rubart M., Field L.J. Challenges measuring cardiomyocyte renewal. Biochim. Biophys. Acta. 2013;1833:799–803. doi: 10.1016/j.bbamcr.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirai M., Cattaneo P., Chen J., Evans S.M. Revisiting Preadolescent Cardiomyocyte Proliferation in Mice. Circ. Res. 2016;118:916–919. doi: 10.1161/CIRCRESAHA.115.308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Satoh N. The ascidian tadpole larva: Comparative molecular development and genomics. Nat. Rev. Genet. 2003;4:285–295. doi: 10.1038/nrg1042. [DOI] [PubMed] [Google Scholar]
- 119.Brozovic M., Martin C., Dantec C., Dauga D., Mendez M., Simion P., Percher M., Laporte B., Scornavacca C., Di Gregorio A., et al. ANISEED 2015: A digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 2016;44:D808–D818. doi: 10.1093/nar/gkv966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stolfi A., Christiaen L. Genetic and genomic toolbox of the Chordate Ciona intestinalis. Genetics. 2012;192:55–66. doi: 10.1534/genetics.112.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gilchrist M.J., Sobral D., Khoueiry P., Daian F., Laporte B., Patrushev I., Matsumoto J., Dewar K., Hastings K.E.M., Satou Y., et al. A pipeline for the systematic identification of non-redundant full-ORF cDNAs for polymorphic and evolutionary divergent genomes: Application to the ascidian Ciona intestinalis. Dev. Biol. 2015;404:149–163. doi: 10.1016/j.ydbio.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Joly J.-S., Kano S., Matsuoka T., Auger H., Hirayama K., Satoh N., Awazu S., Legendre L., Sasakura Y. Culture of Ciona intestinalis in closed systems. Dev. Dyn. 2007;236:1832–1840. doi: 10.1002/dvdy.21124. [DOI] [PubMed] [Google Scholar]
- 123.Sasakura Y. Germline transgenesis and insertional mutagenesis in the ascidian Ciona intestinalis. Dev. Dyn. 2007;236:1758–1767. doi: 10.1002/dvdy.21111. [DOI] [PubMed] [Google Scholar]
- 124.Bouchard-Madrelle C. Régéneration de l’ovaire chez une Ascidie simple Ciona intestinalis, L. (Prochordé) Bull. Soc. Zool. Fr. 1966;91:107–114. [Google Scholar]
- 125.Kawamura K., Sunanaga T. Hemoblasts in colonial tunicates: Are they stem cells or tissue-restricted progenitor cells. Dev. Growth Differ. 2010;52:69–76. doi: 10.1111/j.1440-169X.2009.01142.x. [DOI] [PubMed] [Google Scholar]
- 126.Losick V.P., Fox D.T., Spradling A.C. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr. Biol. 2013;23:2224–2232. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aguirre A., Sancho-Martinez I., Izpisua Belmonte J.C. Reprogramming toward heart regeneration: Stem cells and beyond. Cell Stem Cell. 2013;12:275–284. doi: 10.1016/j.stem.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 128.Li F., Wang X., Capasso J.M., Gerdes A.M. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 129.Soonpaa M.H., Kim K.K., Pajak L., Franklin M., Field L.J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 130.Walsh S., Pontén A., Fleischmann B.K., Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo—An analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 2010;86:365–373. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- 131.Mollova M., Bersell K., Walsh S., Savla J., Das L.T., Park S.-Y., Silberstein L.E., dos Remedios C.G., Graham D., Colan S., et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ahuja P., Perriard E., Perriard J.-C., Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J. Cell Sci. 2004;117:3295–3306. doi: 10.1242/jcs.01159. [DOI] [PubMed] [Google Scholar]
- 133.Sasakura Y., Shoguchi E., Takatori N., Wada S., Meinertzhagen I.A., Satou Y., Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. X. Genes for cell junctions and extracellular matrix. Dev. Genes Evol. 2003;213:303–313. doi: 10.1007/s00427-003-0320-1. [DOI] [PubMed] [Google Scholar]
- 134.Ieda M., Tsuchihashi T., Ivey K.N., Ross R.S., Hong T.-T., Shaw R.M., Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D.Y.R., et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;498:497–501. doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Foglia M.J., Poss K.D. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143:729–740. doi: 10.1242/dev.132910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sasaki H., Yoshida K., Hozumi A., Sasakura Y. CRISPR/Cas9-mediated gene knockout in the ascidian Ciona intestinalis. Dev. Growth Differ. 2014;56:499–510. doi: 10.1111/dgd.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stolfi A., Gandhi S., Salek F., Christiaen L. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development. 2014;141:4115–4120. doi: 10.1242/dev.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gandhi S., Christiaen L., Stolfi A. Rational Design and Whole-Genome Predictions of Single Guide RNAs for Efficient CRISPR/Cas9-Mediated Genome Editing in Ciona. Cold Spring Harbor Labs Journals; New York, NY, USA: 2016. [Google Scholar]
- 140.Sasakura Y., Oogai Y., Matsuoka T., Satoh N., Awazu S. Transposon mediated transgenesis in a marine invertebrate chordate: Ciona intestinalis. Genome Biol. 2007;8(Suppl. S1):S3. doi: 10.1186/gb-2007-8-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.