Abstract
Mutagenesis screening is a powerful forward genetic approach that has been successfully applied in lower-model organisms to discover genetic factors for biological processes. This phenotype-based approach has yet to be established in vertebrates for probing major human diseases, largely because of the complexity of colony management. Herein, we report a rapid strategy for identifying genetic modifiers of cardiomyopathy (CM). Based on the application of doxorubicin stress to zebrafish insertional cardiac (ZIC) mutants, we identified 4 candidate CM-modifying genes, of which 3 have been linked previously to CM. The long isoform of DnaJ (Hsp40) homolog, subfamily B, member 6b (dnajb6b(L)) was identified as a CM susceptibility gene, supported by identification of rare variants in its human ortholog DNAJB6 from CM patients. Mechanistic studies indicated that the deleterious, loss-of-function modifying effects of dnajb6b(L) can be ameliorated by inhibition of ER stress. In contrast, overexpression of dnajb6(L) exerts cardioprotective effects on both fish and mouse CM models. Together, our findings establish a mutagenesis screening strategy that is scalable for systematic identification of genetic modifiers of CM, feasible to suggest therapeutic targets, and expandable to other major human diseases.
By conducting a mutagenesis-based modifier screening in adult zebrafish, DNAJB6 is identified as a new susceptibility gene in addition to 3 known cardiomyopathy genes.
Introduction
Cardiomyopathy (CM) is a disease of the heart muscle that can ultimately lead to heart failure. Despite the identification of more than 100 CM susceptibility genes, it is estimated that about one-half of human inherited CMs, particularly dilated CM (DCM), remain genetically elusive (1, 2). Moreover, modifier genes that contribute to phenotypic variation in individual patients are largely unknown. Quantitative trait loci (QTL) mapping and GWAS have been explored in rodents, leading to the identification of modifiers such as Tnni3k and Abcc6 (3, 4). Mutagenesis screening is a powerful forward genetic approach that has been applied to lower-model organisms, such as Drosophila, to screen genetic candidates for heart failure (5). However, this phenotype-based approach has yet to be established in vertebrates for probing CM specifically, largely because of the high complexity of colony management.
DNAJB6 belongs to the J protein family and functions as a molecular chaperon that facilitates protein folding and protein assembly (6–8). In humans, there are two DNAJB6 protein isoforms that result from alternative splicing of the same gene (9). While the first 6 exons encode the short cytoplasmic DNAJB6(S), the inclusion of the additional 2 exons at the 3′-end of the cDNA encodes the long nucleic DNAJB6(L). Mutations in DNAJB6(S) were reported to cause limb-girdle muscular dystrophy type 1D (LGMD1D), and DNAJB6(S) has been proposed to ameliorate protein aggregation–induced cytotoxicity in neurodegenerative diseases via its antiaggregation function (10, 11). In contrast to the more widely studied DNAJB6(S), functions of DNAJB6(L) remains less investigated (12, 13).
In this study, we report a rapid mutagenesis screening strategy in the vertebrate zebrafish for identification of CM modifying genes. We identified DNAJB6(L) as a genetic modifier and/or susceptibility gene for human CM, supported by its predominant expression in the cardiac tissues and identification of rare variants in human CM patients. Through both loss-of-function and gain-of-function studies, our results suggest a function of DNAJB6(L) as an ER stress inhibitor, which can be targeted to achieve therapeutic benefits for CM.
Results
Leveraging a gene-breaking transposon–based (GBT-based) insertional mutagenesis screening platform in the scalable zebrafish (Danio rerio) model, we sought to develop a forward genetic approach for discovering CM genes and/or modifiers in a vertebrate (14). By checking cardiac monomeric red fluorescent protein (mRFP) expression in 609 GBT lines, we were able to identify 44 Zebrafish Insertional Cardiac (ZIC) mutants (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.88797DS1) (15). Having shown cardiac remodeling in adult zebrafish heart upon stress imposed by either anemia or doxorubicin (DOX) injection (16, 17), we applied DOX stress to these heterozygous ZIC mutants. By using fish survival as a quantifiable index, we demonstrated that GBT0002/sorbs2b, GBT0136/ano5a, and GBT0411/dnajb6b(L) accelerated DOX-induced fish death, and GBT0419/rxraa reduced mortality rates (Figure 1 and Supplemental Figure 1). ANO5 is a known human DCM gene, and studies in mouse and cell culture have previously linked Sorbs2b to CM (18, 19). Consistent with a salutary modifying effect of GBT419/rxraa, activation of retinoid acid signaling has been shown to attenuate CM in a neonatal cardiomyocyte culture model and a mouse model of diabetes-induced CM (20, 21). Identification of known CM genes demonstrates the effectiveness of this mutagenesis screening approach.
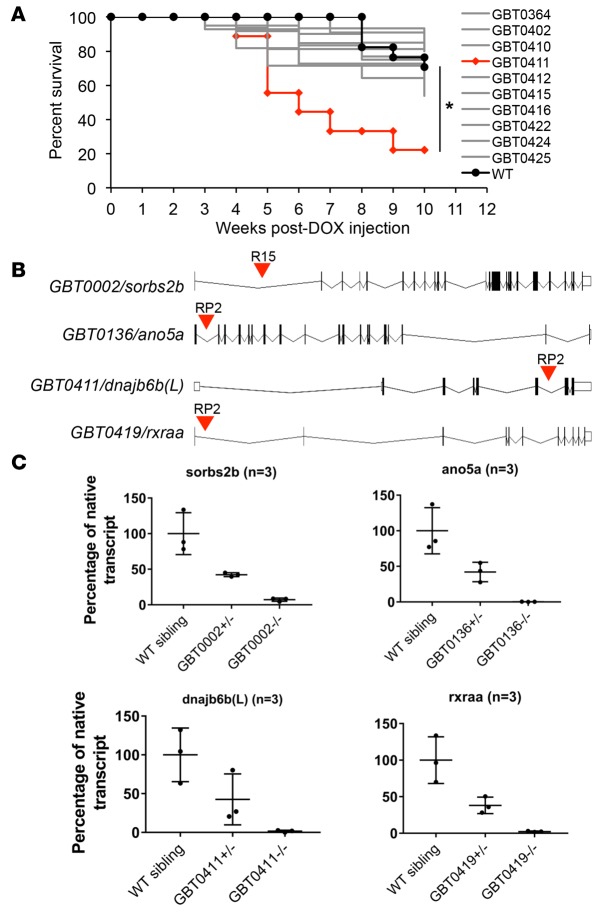
Figure 1. Mutagenesis-based modifier screen identified 4 gene-breaking transposon (GBT) mutants that modified doxorubicin-induced (DOX-induced) adult zebrafish death.
(A) Kaplan-Meier survival curves of adult WT and GBT heterozygous zebrafish injected with a single bolus of 20 μg/gram body mass (gbm) DOX (n = 10–18). GBT0411 mutant was identified that exacerbated DOX-induced fish death. GBT0411 vs. WT, *P < 0.05, log-rank test. (B) The insertional positions of RP2 transposon element in the 4 candidate cardiomyopathy-modifying GBT mutants. (C) qPCR analysis of the percent native transcripts of the tagged genes remaining in the adult fish heart of designated GBT heterozygous and homozygous mutants, respectively. Values represent mean ±SEM.
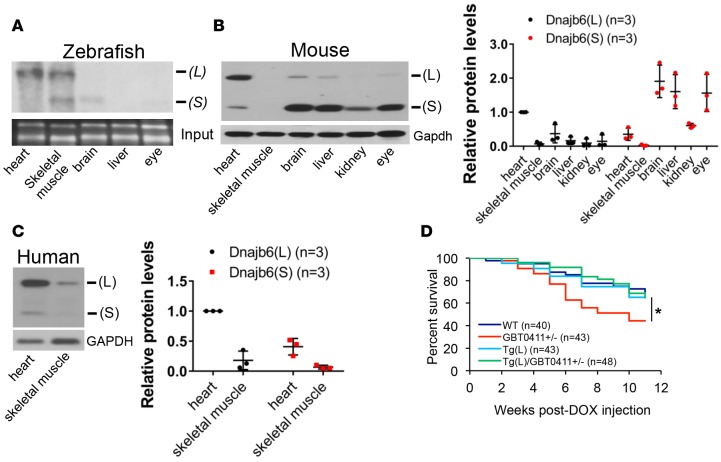
GBT0411/dnajb6b(L) was further investigated as a potential new CM susceptibility gene or disease modifier. GBT0411 homozygous mutants demonstrated cardiomegaly at 1 year, whereas both heterozygous and homozygous GBT0411 mutants accelerated DOX-induced fish death at only 3 months of age (Figure 1A and Supplemental Figure 2A) (15). At the molecular level, GBT0411 specifically disrupted the long isoform of DnaJ (Hsp40) homolog, subfamily B, member 6b (dnajb6b(L)) (Figure 1, B and C). In contrast to a mainly brain-enriched expression of mouse dnajb6 short isoform (dnajb6(S)) (7), zebrafish dnajb6b(L) and both its human and mouse orthologs exhibit previously unrecognized strong cardiac expression (Figure 2, A–C, and Supplemental Figure 2, B–D). Cardiomyocyte-specific overexpression of zebrafish dnajb6b(L) rescued the deleterious modifying effects seen in GBT0411 mutants (Figure 2D and Supplemental Figure 3), supporting a loss-of-function mechanism for the deleterious modifying effects of dnajb6b(L).
Figure 2. The GBT0411 mutant was tagged to the dnajb6b(L) isoform that predominantly expresses in the heart.
(A) Northern blot analysis of dnajb6b(S) and dnajb6b(L) isoforms in indicated 3-month-old adult fish tissues demonstrated a predominant expression of dnajb6b(L) in the fish heart and skeletal muscle. (B) Western blot analysis (left) and quantification (right) of Dnajb6(S) and Dnajb6(L) isoforms expression in 1-year-old WT C57BL/6 mouse tissues indicated a predominant expression of Dnajb6(L) in the mouse heart. Values are presented relative to heart expression. Data represent mean ±SEM. (C) Western blot analysis (left) and quantification (right) of DNAJB6(L) and DNAJB6(S) protein expression in human heart and skeletal muscle lysates. Values are presented relative to heart expression. Data represent mean ±SEM. (D) Kaplan-Meier survival curves of indicated fish injected with a single bolus of 20 μg/gbm doxorubicin (DOX). Cardiomyocyte-specific overexpression of zebrafish dnajb6b(L) rescued DOX-induced fish death in the GBT0411 heterozygous mutant. *P < 0.05, log-rank test. Comparison of Tg(cmlc2:dnajb6b(L)-EGFP); GBT0411+/– vs. GBT0411+/–.
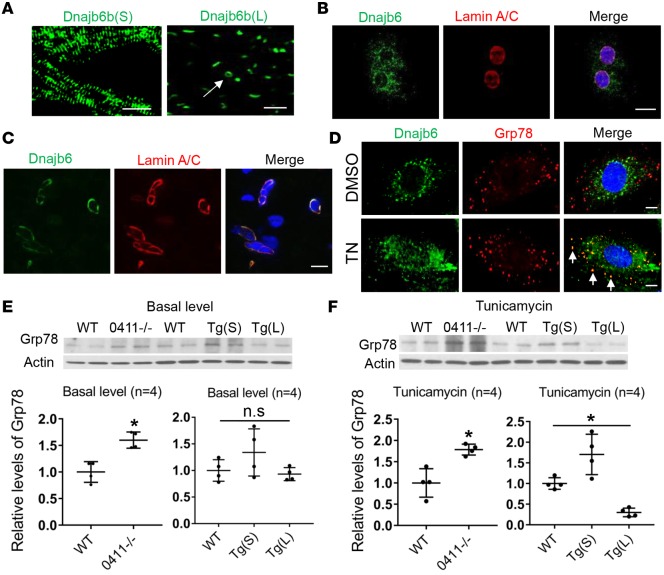
Human DNAJB6(L) is reported to localize to the nuclei (12). We noted perinuclear expression of the zebrafish Dnajb6b(L)-EGFP fusion protein (Figure 3A). In both HL-1 cells and mouse heart tissues, the Dnajb6 endogenous protein colocalized well with the nuclear envelope protein Lamin A/C (Figure 3, B and C). Dnajb6 could also translocate to the ER on treatment with the ER stress inducer tunicamycin (TN) in an H9c2 cell line, as indicated by partial colocalization with the ER resident protein called glucose-regulated protein 78 (Grp78), a well-established molecular marker for ER stress response (Figure 3D) (22). Consistent with the function of Dnajb6 in the ER stress response, dnajb6b(L) transcripts are upregulated upon TN treatment (Supplemental Figure 4). In the GBT0411 mutant with disrupted dnajb6b(L), the ER stress level was elevated both at basal level and upon TN treatment, as indicated by Grp78 protein levels (Figure 3, E and F). In contrast, a suppressed ER stress level was observed in hearts of transgenic dnajb6b(L) fish upon TN treatment but not of transgenic dnajb6b(S) fish. Together, these data characterize a function of dnajb6b(L) as an ER stress inhibitor.
Figure 3. Dnajb6b(L) localizes to the nuclear envelope and functions as an ER stress inhibitor.
(A) EGFP reporter imaged from Tg(cmlc2:dnajb6b(S)-EGFP) fish heart (left) indicated sarcomeric localization of Dnajb6b(S). EGFP reporter from the Tg(cmlc2:dnajb6b(L)-EGFP) fish heart (right) indicated a primary nuclear localization and an occasional perinuclear localization (arrow) for the Dnajb6b(L). Scale bars: 10 μm. (B and C) Costaining of anti-Dnajb6 with the nuclear envelope protein anti-Lamin A/C antibodies in the HL-1 cell line (B) and in sections of WT C57BL/6 mice heart (C) merged with DAPI (blue), indicating Dnajb6 localization to the nuclear envelope. Scale bar: 10 μm. (D) Costaining of anti-Dnajb6 with the ER-resident protein glucose-regulated protein (Grp78) antibody indicated a partial colocalization (arrows) in the ER of the H9c2 cell line after tunicamycin (TN) treatment (1 μg/mL for 24 hours). Scale bar: 10 μm. (E and F) Western blot (upper) and quantification (lower) analysis of Grp78, an ER stress marker, in the GBT0411–/– mutant, Tg(cmlc2:dnajb6b(S)-EGFP), Tg(cmlc2:dnajb6b(L)-EGFP), and WT sibling fish hearts at both basal level (E) and upon TN treatment (F). Values in E and F are expressed relative to WT controls. Data represent mean ±SEM; 1-way ANOVA comparisons, *P < 0.05.
Response to ER stress, also called unfolded protein response, is commonly known as a condition when the ER lumen is overloaded with unfolded or misfolded proteins. It has been implicated as a pathologic event in CM and heart failure (23). We tested the hypothesis that ER stress is a major pathologic event in DOX-induced CM and could serve as a target to ameliorate the deleterious modifying effects of GBT0411. Indeed, we detected late onset activation of ER stress in DOX-injected zebrafish (Figure 4A). Whereas ER stress is further activated in the GBT0411 mutant, its activation is suppressed by the cardiac-specific dnajb6b(L) transgene (Figure 4B). Consistent with the cardioprotective effects of sodium phenylbutyrate (PBA), a chemical chaperone that inhibits ER stress in DOX-induced cardiac dysfunction in mice (24), we found that PBA treatment could decrease the fish mortality rate in both WT and GBT0411 mutant after DOX injection (Figure 4C).
Figure 4. Inhibition of ER stress protected against DOX-induced zebrafish death.

(A) Western blot (upper panel) and quantification (lower panel) analysis of glucose-regulated protein (Grp78) levels in adult WT fish heart at indicated time points after a single bolus of 20 μg/gram body mass (gbm) doxorubicin (DOX) injection. DOX injection results in elevation of Grp78 protein since 7 days after injection. Values are expressed relative to WT controls. Data represent mean ±SEM; *P < 0.05, 1-way ANOVA comparisons. (B) Western blot (upper) and quantification (lower) analysis of Grp78 protein levels at 2 months after DOX injection in indicated mutants and/or double-transgenic fish hearts. Grp78 protein level was further elevated in the GBT0411 mutant fish heart after DOX injection. Cardiomyocyte-specific overexpression of dnajb6b(L) inhibited the elevated ER stress in the GBT0411 mutant fish heart at 2 months after DOX injection. Values are expressed relative to WT controls. Data represent mean ±SEM; 1-way ANOVA comparisons, *P < 0.05. (C) Kaplan-Meier survival curves of indicated adult fish injected with a single bolus of 20 μg/gbm DOX with or without incubation with sodium phenylbutyrate (PBA), a chemical chaperone of ER stress inhibitor (250 μM, 10 hours/day, 5 days/week for the entire experiment). PBA administration protected against DOX-induced fish death in both WT and the GBT0411 mutant. *P < 0.05, log-rank test.
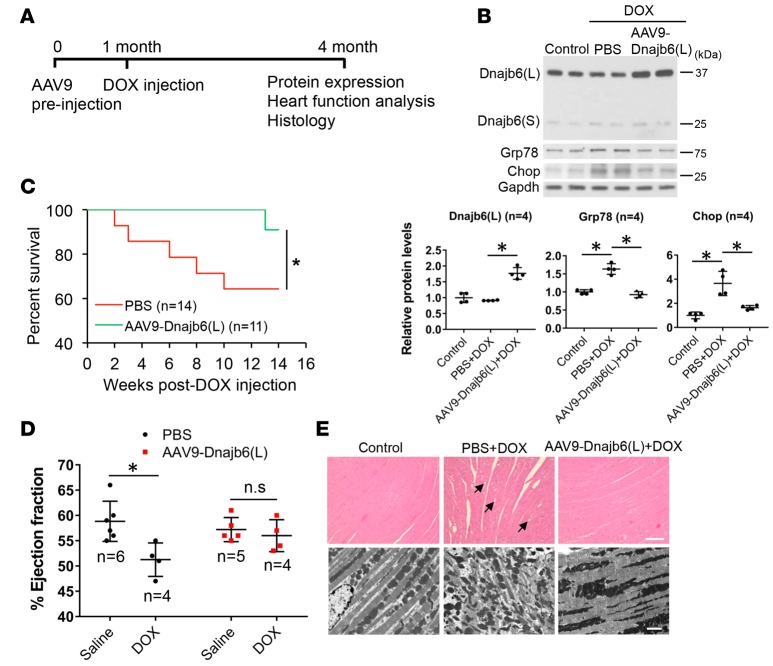
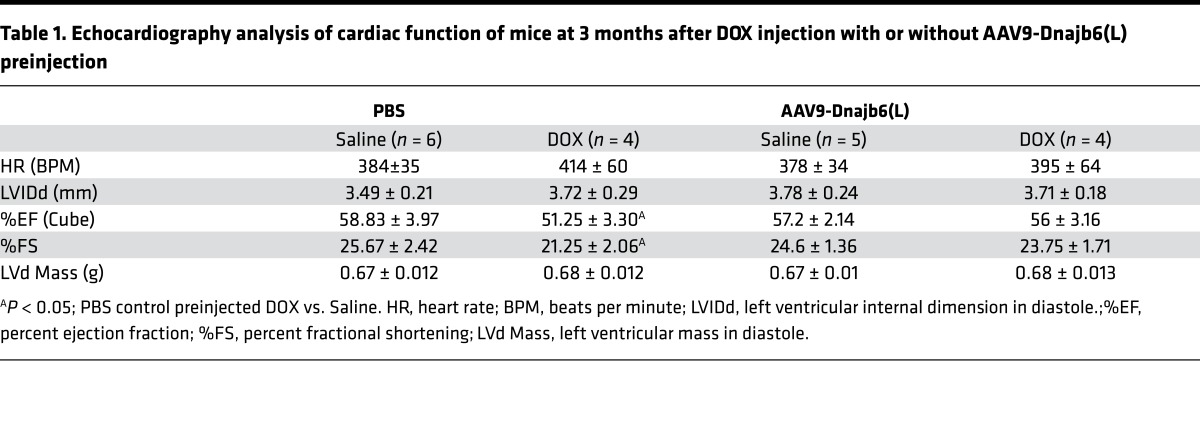
The identification of Dnajb6b(L) as a negative regulator of ER stress and the therapeutic effects of ER stress inhibition through PBA treatment prompted us to test Dnajb6b(L) as a potential therapeutic target for CM. This hypothesis was supported by our observation in zebrafish, where overexpression of dnajb6b(L) in cardiomyocytes reduced the mortality rate in anemia-induced CM (Supplemental Figure 5). To test the cardioprotective function of Dnajb6(L) in mammals, we used the adeno-associated virus serotype 9 (AAV9) medicated gene delivery system (25). Robust ectopic expression of Dnajb6(L) in mouse heart was detected (Figure 5, A and B). To assess the impact on CM development, DOX at 12 mg/kg body weight was i.p. injected 1 month after AAV9-Dnajb6(L) preinjection. The PBS-injected mice showed a severe mortality rate, with the surviving animals showing markedly reduced cardiac function at 3 months after DOX injection. By comparison, the majority of the mice preinjected with AAV9-Dnajb6b(L) survived with well-preserved heart function (Figure 5, C and D, and Table 1). Persistent elevation of ER stress was noted in hearts from PBS but not from AAV9-Dnajb6(L) preinjected mice (Figure 5B). Pathologic events such as cardiomyocyte vacuolization and loss of myofibril structural integrity found in PBS-injected mice were largely absent in the AAV9-Dnajb6b(L) preinjected mice (Figure 5E). Together, these data suggest that Dnajb6(L) overexpression is cardioprotective on DOX-induced CM in mammals.
Figure 5. Overexpression of Dnajb6(L) in cardiomyocytes protected against DOX-induced cardiomyopathy in mouse.
(A) Schematics of adeno-associated virus serotype-9–Dnajb6(L) (AAV9-Dnajb6[L]) and doxorubicin (DOX) injection protocol in mice. (B) Western blot (upper) and quantification (lower) analysis of Dnajb6, glucose-regulated protein 78 (Grp78), and C/EBP homologous protein (CHOP) protein expression levels at 3 months after 5 × 1011 genomic copy of AAV9-Dnajb6(L) virus preinjection with or without injection of 12 mg/kg body weight of DOX. Values are expressed as relative unit to the GAPDH input. Data represent mean ±SEM. *P < 0.05, 1-way ANOVA comparisons. (C) Kaplan-Meier survival curves of mice preinjected with AAV9-Dnajb6(L) or PBS control and then injected with DOX. Overexpression of Dnajb6(L) through AAV9-mediated gene delivery protected against DOX-induced mice death. Data represent mean ±SEM. *P < 0.05, log-rank test. (D) Percent ejection fraction of mice at 3 months after DOX injection with or without AAV9-Dnajb6(L) preinjection. AAV9-Dnajb6(L) virus preinjected mice had preserved cardiac function. Data represent mean ±SEM. *P < 0.05, 2-way ANOVA comparisons. (E) H&E staining (upper panel) and transmission electron microscopic analysis (lower panel) of mouse myocardium at 3 months after DOX injection with or without AAV9-Dnajb6(L) preinjection. Arrows indicate cytoplasmic vacuoles. The images are representative of n = 3 per treatment. Scale bars: 100 μm (upper) and 2 μm (lower).
Table 1. Echocardiography analysis of cardiac function of mice at 3 months after DOX injection with or without AAV9-Dnajb6(L) preinjection.
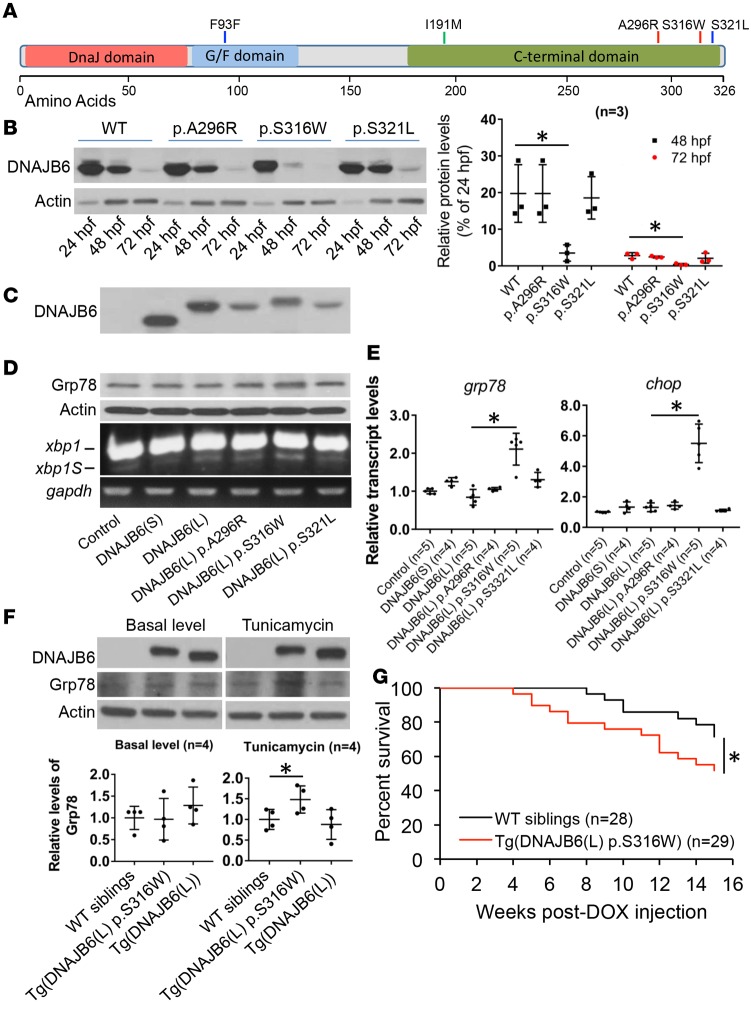
To seek direct evidence for DNAJB6 as a genetic factor for CM, we performed genetic mutational analysis of the translated sequence of DNAJB6 in DNA of human CM patients for possible disease association. We identified 2 unreported missense variants in 214 Mayo Clinic patients with idiopathic DCM, 2 rare variants (1 missense and 1 cryptic splice-site variant) in 86 Mayo Clinic patients with hypertrophic CM (HCM), and 1 unreported missense variant in 17 DCM patients and 8 HCM patients from Shanghai First People’s Hospital, China, cumulatively resulting in identification of 5 rare variants in 5 unique CM patients (Figure 6A and Table 2). We pursued the 3 variants located in the C-terminal domain, which solely affect the DNAJB6(L) isoform — 2 identified in patients with DCM (A296R and S316W) and 1 in a patient with HCM (S321L). In vivo functional analysis in zebrafish embryonic system revealed that the DNAJB6(L)-S316W variant exhibited faster protein turnover and a slower migration rate than WT DNAJB6(L), while the p.A296R and p.S321L variants showed no difference (Figure 6, B and C). Transient overexpression of DNAJB6(L)-S316W, but not the DNAJB6(L) WT and p.A296R and p.S321L variants, led to activation of Grp78 protein and transcriptional activation of ER stress markers (Figure 6, D and E) (22, 26). Similarly, stable overexpression of DNAJB6(L)-S316W in a transgenic line in adult zebrafish resulted in elevated ER stress upon ER stress inducer TN treatment and accelerated the transgenic fish mortality rate after DOX injection (Figure 6, F and G). Together, the rare sequence variants identified in human patients implicate DNAJB6(L) as a CM susceptibility gene, supported by in vivo evidence of aberrant ER stress activation for the DCM-associated S316W variant.
Figure 6. Ectopic expression of the human DNAJB6(L)-S316W variant activated ER stress and accelerated DOX-induced zebrafish death.
(A) Illustration of DNAJB6 rare variants identified from patients with dilated cardiomyopathy (DCM) at Mayo Clinic (red line), patients with hypertrophic cardiomyopathy (HCM) at Mayo Clinic (blue line), and patients with DCM from China (green line). (B) Western blot (left) and quantification (right) analysis of protein levels of DNAJB6(L) WT and the 3 variants in zebrafish embryos injected with corresponding mRNA at one-cell stage. hpf, hours postfertilization. Values are expressed as relative percentage to 24 hpf. Data represent mean ±SEM. *P < 0.05. One-way ANOVA comparisons. (C) Western blot analysis of DNAJB6(S), DNAJB6(L) WT, and the 3 variants expressed in zebrafish embryos injected with corresponding mRNAs at 24 hpf. The DNAJB6(L)-S316W variant showed a slower migration rate compared with DNAJB6(L) WT protein. (D) Transient overexpression of DNAJB6(L)-S316W, but not other variants in zebrafish embryos, led to mild activation of Grp78 protein and transcriptional elevation of spliced X-box binding protein (xbp1S) at 48 hpf. (E) qPCR analysis of grp78 and chop, 2 ER stress molecular markers, in indicated mRNA-injected embryos at 48 hpf. Values are expressed as relative units to uninjected controls. Data represent mean ±SEM. *P < 0.05. vOne-way ANOVA comparisons. (F) Western blot (upper panel) and quantification (lower panel) analysis of DNAJB6(L) and Grp78 proteins in stable transgenic fish heart of Tg(ttna:DNAJB6(L)), Tg(ttna:DNAJB(L) p.S316W), and WT sibling controls at both basal level and at 6 hours after i.p. tunicamycin injection of 1 μg/g body weight. Grp78 protein was elevated in the Tg(ttna:DNAJB6(L) p.S316W) but not Tg(ttna:DNAJB6(L)) transgenic fish heart upon tunicamycin (TN) treatment. Values are expressed as relative units to WT siblings. Data represent mean ±SEM. *P < 0.05. One-way ANOVA comparisons. (G) Kaplan-Meier survival curves of Tg(ttna:DNAJB6(L) p.S316W) transgenic fish and WT siblings after a single bolus of 20 μg/gbm doxorubicin (DOX) injection. *P < 0.05, log-rank test.
Table 2. Summary of DNAJB6 variants identified in human cardiomyopathy patients (n = 325).
Discussion
In summary, our data identified DNAJB6(L) as a cardiac isoform that might function as a CM modifier. In contrast, previous reports mostly focused on the functional study of DNAJB6(S) in neurodegenerative disease and LGMD1D (8, 10, 11), while the function of DNAJB6B(L) was less investigated (12, 13). Our present study further suggested that DNAJB6(L), but not DNAJB6(S), might function as an ER stress inhibitor. The differential subcellular localization of these 2 isoforms must contribute to their different effects. Because these 2 isoforms share their N-terminal sequences, it is plausible that the C-terminal amino acids unique to either isoform are key regions that govern their different effects. Our studies in the heart strongly suggest that these functions of DNAJB6(L) need to be investigated in diseases affecting other organs, including neurological disorders and muscular dystrophy.
Our mechanistic studies of DNAJB6(L) have clear clinical implications. First, ER stress inhibition might be an effective strategy to protect the rare case of a patient with DNAJB6-mediated DCM or, more importantly and more commonly, cancer patients treated with DOX. Because the drug PBA is approved by the US FDA (27), future studies to repurpose PBA to these patient groups are warranted. An immediate future study will be to generate S316W transgenic or knock-in mice, which will allow us to test whether its susceptibility to DOX-induced cardio toxicity is conserved in mammals and whether PBA administration is cardioprotective. Second, our results that AAV9-mediated overexpression of Dnajb6(L) exhibited a cardioprotective effect on mice indicated that DNAJB6(L) could be further pursued as a therapeutic target for human CM and heart failure.
Taken together, we present a rapid mutagenesis screening strategy in an in vivo vertebrate animal model for effective identification of genetic modifiers and potential therapeutic targets for human CM. Of note, our current screen focuses on DOX-induced CM because of its efficiency and throughput for an initial mutagenesis screening method. An obvious future direction is to determine whether genetic modifiers and therapeutic strategies identified herein are applicable to other types of CM. Nevertheless, because this approach is scalable to cover the entire genome with reasonable cost, we anticipated that our strategy will facilitate the generation of a comprehensive genetic landscape of CM, serving to advance the capacity to interpret individual genomes for diagnosis, prognosis, and individualized therapy for CM.
Methods
Animals.
Zebrafish (Danio rerio) WIK were maintained under a 14 hours light/10 hours dark cycle at 28.5°C. WT C57BL/6 male mice (age, 8 weeks) were purchased from the Jackson Laboratory and maintained on a 12-hour light-dark cycle and were inspected daily.
Expansion of ZIC mutant collection and DOX modifier screen.
We previously developed an expression-based strategy to enrich ZIC mutants from GBT lines (15). In the present study, we expanded the ZIC mutant collection by identifying GBT lines with visible mRFP signal in the heart during either embryonic or adult stages. To examine cardiac expression during embryonic stages, we anesthetized embryos 2–5 days postfertilization (dpf) using 0.16 mg/ml tricaine (Argent Chemical Laboratories) to arrest the heartbeat. mRFP signal was observed using a Zeiss microscope. To examine cardiac expression at adult stages, fish 3–18 months of age were anesthetized with tricaine (Argent Chemical Laboratories). Hearts were dissected and observed under a Zeiss microscope. To perform the DOX screen, 10–25 fish for each ZIC mutant at 3–12 months of age were i.p. injected with a single dose at 20 μg/g body weight of DOX dissolved in 5 μl Hank’s buffer using a NanoFil 10-μl syringe (World Precision Instruments Inc). Fish survival was monitored daily for 2–4 months after DOX injection.
Northern blotting.
Northern blot hybridization was performed with the NorthernMax-Gly kit (Ambion) following the manufacturer’s instructions. Briefly, approximately 1 μg of total RNA was isolated from zebrafish hearts (6 months old) or C57BL/6 male mice hearts (8 weeks old) using the TRIzol reagent (Thermo Fisher Scientific). 32P-labeled RNA probes that bind to the N-terminal common region of both dnajb6(S) and dnajb6(L) isoforms were synthesized from 100-ng fish or mouse cDNA fragments using a random primed DNA labeling kit (Roche Diagnostics). The primers for amplifying the zebrafish dnajb6b cDNA are forward 5′ GGATTATTACCACATCCTTGGTG 3′ and reverse 5′ GCTGTCCAGTCGGAGTAACTGC 3′. The primers for amplifying the mouse dnajb6 cDNA are forward 5′ ACCATAACAAGCACCATTTGAG 3′ and reverse 5′ AACAATAGTCAAACTGACGAGG 3′.
Generation of transgenic fish lines.
Zebrafish dnajb6b(S) and dnajb6b(L) cDNAs were amplified with total RNA isolated from zebrafish heart tissue through reverse transcriptase PCR (RT-PCR) using 2 primer pairs: forward 5′ TAGCGGATCCGCCACCATGGAGGAGGATTATTACCACATCCTTGGTG 3′ and reverse 5′ AGCTCTCGAGAACCACACCCCAATCCGGGGAGTGAGTCTT 3′ and forward 5′ TAGCGGATCCGCCACCATGGAGGAGGATTATTACCACATCCTTGGTG 3′ and reverse 5 AGCTCTCGAGCATTTGCTGTCCAGTCGGAGTAACTGC 3′. Human DNAJB6(L) WT and DNAJB6(L) p.S316W cDNAs were amplified through PCR using the forward primer 5′ ATCGGGATCCGCCACCATGGTGGATTACTATGAAGTTCTAGG 3′ and reverse primer 5′ ACTGCTCGAGCAGTGATTGCCTTTGGTCGACTTCTT 3′ using human pCS2-DNAJB6(L) WT or pCS2-DNAJB6(L) p. S316W as a template, respectively.
The resulting PCR fragments were digested with BamH I and XhoI and then cloned into pENTRI1a to generate pENTRI-dnajb6b(S), pENTRI-dnajb6b(L), pENTRI-DNAJB6(L) WT, and pENTRI-DNAJB6(L) p.S316W. The final constructs for fish dnajb6b(S) and dnajb6b(L) were generated through a recombination reaction among p5E-cmlc2 (15), pENTRI1a with corresponding cDNAs, p3E-EGFP-polyA, and pDestTol2pA destination vector that is facilitated by the Gateway LR clonase II plus enzyme (Thermo Fisher Scientific). For the human DNAJB6(L) WT and p.S316W variant, we used the p5E-ttna plasmid containing ttna enhancer. Subsequently, 50 ng of the resulting final plasmid was coinjected with 100 ng of transposase mRNA into single-cell–staged WT WIK embryos to generate stable transgenic lines.
Cell culture and antibody staining.
The HL-1 myocytes, a cardiac muscle cell line derived from adult mouse atrium, were maintained in Complete Claycomb Medium (Sigma Aldrich) supplemented with 100 μM norepinephrine, 4 mM L-glutamine (Sigma Aldrich), and 10% FBS (Sigma Aldrich). The myocytes were cultured in flasks coated with 12.5 μg/ml fibronectin and 0.02% gelatin (both Sigma Aldrich). The H9c2 cells, rat heart–derived embryonic myocytes (American Type Culture Collection), were cultured in DMEM (Thermo Fisher Scientific) with 4 mM/l glutamine, 4.5 g/l glucose, 1.0 mM sodium pyruvate, and 10% FBS at 37°C in 5% CO2. For antibody staining in HL-1 and H9c2 cells, the cultured cells with or without TN treatment (1 μg/ml for 24 hours) were subjected to antibody staining using previously described methods (16). Briefly, cells were fixed in 4% PBS-buffered formaldehyde for 10 minutes, followed by incubation with the primary antibodies anti-DNAJB6 (1:200, Novus Biologicals LLC, catalog H00010049-M01), anti-Lamin A/C (1:200, Santa Cruz Biotechnology Inc., catalog sc-6215), and anti-GRP78 (1:200, Novus Biologicals LLC, catalog NBP-06274) for 2 hours at room temperature. Next, samples were washed in PBS and incubated with secondary antibodies for 30 minutes at room temperature, followed by PBS wash. For immunostaining in the mouse heart tissue, the left ventricle of 8-week-old WT C57BL/6 mice hearts were dissected, and frozen 8-μm sections were fixed with ice cold acetone followed by the staining protocol described above. The anti-Dnajb6 (1:100, Santa Cruz Biotechnology Inc., catalog sc-104204) and anti-Lamin A/C (1:200, Cell Signaling Technolgoy, catalog 4777) were used. All images were captured using a Zeiss Axioplan II microscope equipped with ApoTome and AxioVision software (Carl Zeiss Microscopy).
Western blotting.
Human heart and skeletal muscle tissue lysates were purchased from Novus Biologicals LLC. Tissues harvested from overdose anesthesia–treated mice or zebrafish were transferred immediately to RIPA buffer (Sigma Aldrich) supplemented with complete protease inhibitor cocktail (Roche Diagnostics) and homogenized using a Bullet Blender tissue homogenizer (Next Advance Inc.). Resultant protein extracts were subjected to Western blotting using a standard protocol. The following primary antibodies were used: anti-actin (1:4,000, Santa Cruz Biotechnology Inc., catalog sc-1615), anti-DNAJB6 (1:6,000, Abcam, catalog ab198995 ), anti-GAPDH (1:8,000, Santa Cruz Biotechnology Inc., catalog sc-25778), anti-GRP78 (1:2,000, Novus Biologicals LLC, catalog NBP-06274), anti-Chop (1:1,000, Abcam, catalog ab11419), and an anti-GFP antibody (1:10,000, Torrey Pines Biolabs Inc., catalog TP401).
Quantitative PCR.
To determine the relative expression level of ER stress molecular markers grp78 and chop, we performed quantitative PCR (qPCR) as previously described (15). gapdh was used as an internal control to normalize expression levels. The following primers were used: grp78 forward 5′ TCAGCGTCAGGCCACTAA 3′, grp78 reverse 5 GTCAGCAGAGACACGTCAAA 3′; chop forward 5′ CGGTTCCCGACACATCA 3′, and chop reverse 5′ CACTTTCCTTTCATTCTCCTGTT 3′. qPCR was also carried out to determine the transcriptional induction of dnajb6b gene on treatment with the ER stress inducer TN. dnajb6a was also included. The primers for dnajb6b(L) were forward 5′ ACACAACCGATACCTCCGAAACGT 3′ and reverse 5′ GCTTCTTTTTGGTGTCTGACTCCTTC 3′; for dnajb6b(S), forward primer 5′AAGGAGCAGTTACTCCGACTGG 3′ and reverse primer 5′ AGACTTCACCCTGTTTACCTCTGC 3′; and for the dnajb6a gene, forward primer 5′ CACATTCCGCAATCCAGAG 3′ and reverse 5′ AAATCCCGCACCAAAAGG 3′. To quantify the percent-native transcript of a tagged gene in corresponding GBT mutants, each heart was dissected from adult zebrafish at 3–6 months of age. qPCR was performed using a previously described method (15). The following primers were used: GBT0002/sorbs2b forward 5′ AACTTCTCCAGTGCCAAGCAAAGC 3′ and reverse 5′ CCAGGCATTAGGACTGAAAGTAGGCA 3′; GBT0136/ano5a forward 5′ GGCACTGGAGATCAGATCAC 3′ and reverse 5′AAGTTGAAACGTCTCCTTATCC 3′; GBT0411/dnajb6b(L) forward 5′ CCTCCATTTCCACATCCACCA 3′ and reverse 5′ CCTTTTCTGAAGGGTTCTGGGC 3′; and GBT0419/rxraa forward 5′ TGGAAACCAAACCTTTCTTGTC 3′ and reverse 5′ CTGTAGAAAGCCTGCGTTG 3′.
Whole-mount in situ hybridization.
Digoxigenin-labeled probes were generated from PCR products using T7 promoter–tagged primer pairs. The dnajb6b(S) and dnajb6b(L) isoform-specific primers were used: dnajb6b(S) forward 5′ CAGTGAATCGCAGAGGTAAACAG 3′ and reverse 5′ GGTAATACGACTCACTATAGGGTGCAGCATTCAAAACAGGTG 3′; and dnajb6b(L) forward 5′ ACACAACCGATACCTCCGAAACGT 3′ and reverse 5′ GGTAATACGACTCACTATAGG GAACCACACCCCAATCCGGGGAGTGAGTCTT 3′. Whole-mount in situ hybridization procedures were subsequently performed as described previously (15).
Mouse AAV9–Dnajb6(S) and (L) virus and DOX injection.
To produce mouse AAV9-Dnajb6(L) virus, total RNA was extracted from C57BL/6 mice heart tissue using TRIzol reagent (Thermo Fisher Scientific). After purification with the RNeasy Plus kits (QIAGEN), 1 μg resultant total RNA was used to synthesize cDNAs with a SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher Scientific). Mouse Dnajb6(L) cDNAs were PCR amplified using Expand High-Fidelity PCR System (Roche Diagnostics) with the primers Dnajb6(L) forward 5′ ATCGGGATCCGCCACCATGGTGGATTACTATGAAGTGCTGGGCG 3′ and reverse 5′ ACTGGGATCC CTAATGGTTCCCTTTGGTCGACTTCTTCTTC 3′. The resultant RT-PCR products were subcloned into the pAAV-MCS vector backbone (Stratagene Corp.) after BamH I enzyme digestion. The cDNA fragment was under the transcriptional control of the cytomegalovirus promoter and hGH polyA sequence. Clones with correct insertional direction were identified by direct sequencing. The production of recombinant AAV9 was carried out as described previously (25). Briefly, AAV vectors were generated by packaging AAV2-based recombinant genomes in AAV9 capsids. The production of these pseudotyped AAV2/9 vectors (AAV9) was performed by triple transfection using an adenovirus-free system (Agilent Technologies). Next, 5 × 1011 genomic copy of virus diluted in 100 μl PBS was injected into mice via tail vein. One month after the AAV9 injection, the first echocardiography was conducted to determine the baseline before DOX injection, and DOX at a dose of 12 mg/kg body weight was dissolved in 200 μl saline and i.p. injected into C57/B6 male mice (8 weeks old) using a 28-gauge 1/2-inch insulin syringe (BD Biosciences). Three months after the DOX and 4 months after the AAV9 injection, the second echocardiography was carried out. Heart tissues were then harvested for subsequent histology and protein expression analysis.
Echocardiography in mice.
Mice were anesthetized under light isoflurane (0.5%–1.0% v/v) administered via a nose cone. Echocardiography gel was placed on the shaved chest, and the mouse heart was imaged with a 13-MHz probe using 2-dimensional echocardiography (GE Healthcare). All measurements were made by an operator to whom the study groups were masked.
H&E staining.
To harvest tissues from mice, ventilation was ceased and mice were euthanized by administration of high-dose (5%) isoflurane anesthesia. Dissected mice heart left ventricles were immediately fixed in 4% PBS buffered formaldehyde overnight at 4°C and sent to the Mayo Clinic Histology Core Laboratory for sample process and H&E staining.
Electron microscopy.
For transmission electron microscopy analysis, the left ventricle apexes of dissected mice were fixed immediately in Trump’s solution (4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer [pH 7.2]) at room temperature for 1 hour, followed by overnight incubation at 4°C. Fixed samples were subsequently processed and imaged at the Mayo Clinic Electron Microscopy Core Facility using a Philips CM10 transmission electron microscope.
Identification of DNAJB6 variants in human patients with idiopathic DCM from Mayo Clinic.
Patients were clinically evaluated at Mayo Clinic, and phenotypic characterization of idiopathic DCM was based on echocardiographic measurements. Diagnostic criteria for DCM were left ventricular diastolic dimension, systolic dimension, or both; >95th percentile indexed for body surface area; and left ventricular ejection fraction <50%. Genomic DNA was isolated from peripheral blood white cells or saliva. For DNAJB6 mutation scanning and Sanger sequencing, genomic DNA samples from 214 unrelated probands with DCM were used. PCR primer pairs were designed to amplify the 9 translated exons of DNAJB6 (NM_058246.3). PCR products from 6 of the 9 translated exons (exons 2–7) were analyzed with denaturing high-performance liquid chromatography (DHPLC) heteroduplex analysis (WAVE DHPLC System; Transgenomic Inc.). Chromatograms of amplified and heteroduplexed products were analyzed against the WT pattern. Abnormal traces were Sanger sequenced to determine the precise genetic variants. Because of special interest in the DNAJB6 long isoform and its isoform-specific exons, all DCM probands underwent Sanger sequencing for exons 8–10.
Identification of DNAJB6 variants in human patients with HCM from Mayo Clinic.
Patients were clinically evaluated at Mayo Clinic, and phenotypic characterization of HCM in patients and family members was based on echocardiographic measurements. Diagnostic criterion for HCM was a left ventricular wall thickness >13 mm in the absence of any other conditions that could cause cardiac hypertrophy, such as hypertension or aortic stenosis. Genomic DNA was isolated from peripheral blood white cells. For DNAJB6 mutation scanning and Sanger sequencing, we used genomic DNA samples from 86 unrelated probands with heretofore genetically elusive HCM having been analyzed previously for mutations in the 10 most common HCM susceptibility genes. For mutation scanning and Sanger sequencing, PCR primer pairs were designed to amplify the 9 translated exons of DNAJB6 (NM_058246.3). PCR products from 6 of the 9 translated exons were analyzed by DHPLC (WAVE DHPLC System; Transgenomic Inc.). Chromatograms of amplified and heteroduplexed products were analyzed against the WT pattern. Abnormal traces were Sanger sequenced to determine the precise genetic variants.
Identification of a DNAJB6 variant in human patients with idiopathic DCM and HCM from China.
Seventeen DCM patients, 8 HCM patients, and 23 controls were recruited from the Han Chinese population. The diagnosis of DCM was based on echocardiographic measurements according to WHO criteria. Enrichment of DNAJB6 genomic sequences was performed with genomic DNA templates using the Access Array System (Fluidigm) for Ion Torrent PGM (personal genome machine) according to the manufacturer’s instructions. Each sample DNA was combined with primer pairs in a microfluidic chip. The chip was loaded with PCR reagents and transferred to a thermocycler. Sample barcodes and sequence indexes and sequencing adaptors were attached with a second-step PCR reaction. Deep sequencing was conducted using Ion OneTouch 200 (Thermo Fisher Scientific) reagent kits, Ion PGM Sequencing 200 Kit (Thermo Fisher Scientific), and 318 scale chips. We use the Ion Torrent Suite 3.2 package for sequence analysis and variant calling. Primers were trimmed and variants called with a variant frequency threshold at 25%. The Integrative Genomics Viewer was used for visualization. To validate candidate variants, the targeted region was amplified by PCR using a 96-well GeneAmp PCR System 9700 (Applied Biosystems). GRCh37/hg19 was used as the reference sequence. From each sample, 10 ng DNA was used as a template, together with the 2X Taq PCR Master Mix (Lifefeng Biotech). Sequencing was conducted using a 3730xl DNA Analyzer (Applied Biosystems), a capillary-based Sanger sequencing machine.
Frequency of all 5 identified variants in the population were obtained from online variant and exome databases, such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), 1000 genome project (http://browser.1000genomes.org/index.html), Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), and Exome Aggregation Consortium (http://exac.broadinstitute.org/).
Transient embryonic expression of human DNAJB6(S) and DNAJB6(L) WT and 3 DNAJB6(L) variants.
DNAJB6(S) cDNA was PCR amplified from pEGFP-N1-MRJ(S)-GFP (12) using Expand High-Fidelity PCR System (Roche Diagnostics). The following primers were used: DNAJB6(S/L) forward 5′ ACTGGGATCCGCC ACCATGGTGGATTACTATGAAGTTCTAGG 3′ and DNAJB6(S) reverse 5′ACTGGGATC CTTGTTATCCAAGCGCAGCAGCTGCTC 3′. DNAJB6(L) cDNA was PCR amplified from MGC human DNAJB6 sequence–verified cDNA (Dharmacon, GE Healthcare; catalog MHS6278-202827593) as a template. The sequences of the primers are DNAJB6(S/L) forward 5′ ACTGGGATCCGCC ACCATGGTGGATTACTATGAAGTTCTAGG 3′ and DNAJB6(L) reverse 5′ ACTGGGATCGTGATTGCCTTTGGTCGACTTCTT 3′. The resultant PCR products were digested with BamH I and cloned into the pCS2 + GFP (28) vector to generate pCS2-DNAJB6(S) and pCS2-DNAJB6(L), respectively. GeneArt Site-Directed Mutagenesis System (Invitrogen, catalog A13282) was used to generate 3 DNAJB6(L) variants — DNAJB6(L) p. A296R, DNAJB6(L) p.S316W, and DNAJB6(L) p.S321L — using the pCS2-DNAJB6(L) as a template. The following primers were used: forward 5′ GCCCTGGGACCCC CTCAGGTCCGCAGCAGGATTG 3′ and reverse 5′ CAATCCTGCTGCGGACCTGAGGGGGTCCCAGGGC 3′ for pCS2-DNAJB6(L) p.A296R; forward 5′ AAGCAGAGAGAGGAGTGGAAGAAGAAGAAGTCG 3′ and reverse 5′ CGACTTCTTCTTCTTCCACTCCTCTCTCTGCTT 3′ for pCS2-DNAJB6(L) p.S316W; and forward 5′ TCGAAGAAGAAGAAGTTGACCAAAGGCAATCAC 3′ and reverse 5′ GTGATTGCCTTTGGTCAACTTCTTCTTCTTCGA 3′ for pCS2-DNAJB6(L) p.S321L. The pCS2-based plasmids were linearized by NotI digestion, and mRNAs were produced using mMESSAGE mMACHINE Kit (Thermo Fisher Scientific). About 100 pg synthesized mRNA was injected into single-cell staged WIK embryos.
Statistics.
Student’s t test was used for comparisons between two groups. One-way or 2-way ANOVA was used for assessment differences among multiple group comparisons. Log-rank test was used to determine the difference for animal survivals. P values less than 0.05 were considered significant.
Study approval.
All animal study procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press. 2011.). and were approved by the Mayo Clinic Institutional Animal Care and Use Committee. All patients from Mayo Clinic provided written informed consent under a research protocol approved by the Mayo Clinic IRB prior to inclusion in the study. All patients from China provided written informed consent under a research protocol approved by the ethics committee of Shanghai First People’s Hospital affiliated to Shanghai Jiao Tong University.
Author contributions
YD and XX conceived the project, designed research, analyzed the data, and wrote the paper; YD performed the experiments. PAL, RSS, and TMO identified variants from DCM patients at Mayo Clinic. JMB and MJA identified variants from HCM patients at Mayo Clinic. JC, YJ, LZ, XH, JW, and YS identified a variant from DCM patients in China. YHS and XM produced the data for Supplemental Figure 4. XL, SCE, MMR, and TMO made intellectual contributions and assisted in editing the manuscript.
Supplementary Material
Acknowledgments
We thank Beninio T. Gore and Kashia B. Stragey for managing the zebrafish facility, Ronald H. May for murine echocardiography, and Rebecca A. Nace for murine AAV9 tail vein injection. This work was supported in part by a Scientist Development Grant from the US American Heart Association (14SDG18160021 to YD), the US NIH R01 grants HL 81753 and HL 107304 to XX, HL071225 to TMO, HG 006431 and GM63904 to SCE, the Windland Smith Rice Sudden Death Genomics Research Program and the Tsai Family HCM Research Fund to JMB and MJA, and the Mayo Foundation to XX.
Footnotes
PAL and XM’s present address is Mayo Graduate School, Mayo Clinic College of Medicine, Rochester, Minnesota, USA.
YHS’s present address is Department of Molecular, Cell, and Cancer Biology, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
Conflict of interest: MJA is a consultant for Boston Scientific, Gilead Sciences, Medtronic, and St. Jude Medical. MJA and Mayo Clinic receive sales-based royalties from Transgenomic and their FAMILION-LQTS and FAMILION-CPVT genetic tests.
Reference information:JCI Insight. 2016;1(14):e88797. doi:10.1172/jci.insight.88797.
Contributor Information
Yonghe Ding, Email: ding.yonghe@mayo.edu.
Yu-Huan Shih, Email: yu-huan.shih@umassmed.edu.
Xiao Ma, Email: ma.xiao@mayo.edu.
Jianhua Chen, Email: chenjhv@hotmail.com.
Yiwen Jiang, Email: yiwenjiang931208@126.com.
Liqun Zhao, Email: zhaoliq8570@hotmail.com.
Xinyang Hu, Email: hxy0507@hotmail.com.
Jianan Wang, Email: wja@zju.edu.cn.
Yongyong Shi, Email: shiyongyong@sjtu.edu.cn.
Michael J. Ackerman, Email: ackerman.michael@mayo.edu.
Xueying Lin, Email: lin.xueying@mayo.edu.
Xiaolei Xu, Email: xu.xiaolei@mayo.edu.
References
- 1.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12(11):655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60(8):705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler FC, et al. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009;5(9):e88797. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rau CD, et al. Mapping genetic contributions to cardiac pathology induced by Beta-adrenergic stimulation in mice. Circ Cardiovasc Genet. 2015;8(1):40–49. doi: 10.1161/CIRCGENETICS.113.000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res. 2010;106(7):1233–1243. doi: 10.1161/CIRCRESAHA.109.213785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang JZ, Zhou H, Zhu M, Li SH, Li XJ, Sung CH. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J Biol Chem. 2002;277(22):19831–19838. doi: 10.1074/jbc.M109613200. [DOI] [PubMed] [Google Scholar]
- 8.Hageman J, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37(3):355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Hanai R, Mashima K. Characterization of two isoforms of a human DnaJ homologue, HSJ2. Mol Biol Rep. 2003;30(3):149–153. doi: 10.1023/A:1024916223616. [DOI] [PubMed] [Google Scholar]
- 10.Kakkar V, et al. The S/T-Rich Motif in the DNAJB6 Chaperone Delays Polyglutamine Aggregation and the Onset of Disease in a Mouse Model. Mol Cell. doi: 10.1016/j.molcel.2016.03.017. [published online ahead of print April 12, 2016]. [DOI] [PubMed] [Google Scholar]
- 11.Sarparanta J, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44(4):450–455. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra A, et al. Large isoform of MRJ (DNAJB6) reduces malignant activity of breast cancer. Breast Cancer Res. 2008;10(2):e88797. doi: 10.1186/bcr1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu VZ, et al. Nuclear Localization of DNAJB6 Is Associated With Survival of Patients With Esophageal Cancer and Reduces AKT Signaling and Proliferation of Cancer Cells. Gastroenterology. 2015;149(7):1825–1836.e5. doi: 10.1053/j.gastro.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Clark KJ, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8(6):506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Y, et al. Trapping cardiac recessive mutants via expression-based insertional mutagenesis screening. Circ Res. 2013;112(4):606–617. doi: 10.1161/CIRCRESAHA.112.300603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y, et al. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109(6):658–669. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, et al. Cardiac hypertrophy involves both myocyte hypertrophy and hyperplasia in anemic zebrafish. PLoS One. 2009;4(8):e88797. doi: 10.1371/journal.pone.0006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bang C, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124(5):2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahbi K, et al. Dilated cardiomyopathy in patients with mutations in anoctamin 5. Int J Cardiol. 2013;168(1):76–79. doi: 10.1016/j.ijcard.2012.09.070. [DOI] [PubMed] [Google Scholar]
- 20.Rassaf T, Kelm M. Protection from diabetic cardiomyopathy - putative role of the retinoid receptor-mediated signaling. J Mol Cell Cardiol. 2013;59:179–180. doi: 10.1016/j.yjmcc.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Zhou MD, Sucov HM, Evans RM, Chien KR. Retinoid-dependent pathways suppress myocardial cell hypertrophy. Proc Natl Acad Sci U S A. 1995;92(16):7391–7395. doi: 10.1073/pnas.92.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 23.Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107(9):1071–1082. doi: 10.1161/CIRCRESAHA.110.227819. [DOI] [PubMed] [Google Scholar]
- 24.Fu HY, et al. Chemical Endoplasmic Reticulum Chaperone Alleviates Doxorubicin-Induced Cardiac Dysfunction. Circ Res. 2016;118(5):798–809. doi: 10.1161/CIRCRESAHA.115.307604. [DOI] [PubMed] [Google Scholar]
- 25.Moulay G, et al. Cardiac AAV9 Gene Delivery Strategies in Adult Canines: Assessment by Long-term Serial SPECT Imaging of Sodium Iodide Symporter Expression. Mol Ther. 2015;23(7):1211–1221. doi: 10.1038/mt.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 27.Iannitti T, Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drugs R D. 2011;11(3):227–249. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Xu X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: an inhibitory mechanism of ventricle-specific gene expression. Dev Dyn. 2009;238(6):1564–1573. doi: 10.1002/dvdy.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.