Abstract
The pathogenesis of allergic diseases entails an ineffective tolerogenic immune response towards allergens. Regulatory T cells (TReg) cells play a key role in sustaining immune tolerance to allergens, yet mechanisms by which TReg cells fail to maintain tolerance in allergic diseases are not well understood. We review current concepts and established mechanisms regarding how TReg cells regulate different components of allergen-triggered immune responses to promote and maintain tolerance. We will also discuss more recent advances that emphasize the “dual” functionality of TReg cells in allergic diseases: how TReg cells are essential in promoting tolerance to allergens but also how a pro-allergic inflammatory environment can skew TReg cells towards a pathogenic phenotype that aggravates and perpetuates disease. These advances highlight opportunities for novel therapeutic strategies that aim to re-establish tolerance in chronic allergic diseases by promoting TReg cell and stability function.
Keywords: Asthma, Food Allergy, Regulatory T cells, FOXP3, Interleukin 4, T Helper cells type 2
Introduction
The increased prevalence in allergic diseases has become a major health problem in affluent and rapidly developing societies. Over the last 150 years, accelerated social and environmental changes augured by the industrial revolution, and which profoundly altered patterns of human activity, living arrangements, diet and infections, all came to influence the rise and severity of allergic disorders1. In the USA, food allergy prevalence reaches up to 8% among children and 5% of the adult population whereas 8.6% of children and 7.4% of adults are affected by asthma2,3. The dramatically increased burden of allergic diseases has exacted considerable morbidity on those suffering from those disorders and resulted in substantial financial costs incurred by affected individuals and their healthcare systems. While therapies for allergic diseases have improved over the years with the introduction of agents aimed at combating the inflammatory processes as well as providing symptomatic relief, those therapies have remained for the most non-curative.
Allergic diseases arise in response to normally innocuous environmental agents, including aeroallergens and foods. They involve the participation of components of the innate and adaptive immune responses, such innate lymphoid cells type 2 (ILC2) mast cells, basophils, eosinophils, as well as activated T helper type 2 (Th2) cells and B cells switched to the production of immunoglobulin type E (IgE)4,5. Immune regulatory mechanisms normally operative to maintain tolerance to allergens breakdown for reasons that still remain obscure. The dramatic increase in prevalence of allergic disease during the past decades indicates a strong influence of environmental factors acting on genetically susceptible hosts to promote disease6,7. Emerging studies emphasize the interaction of environmental factors, including diet, antibiotic usage and others, with components of the immune system, affecting their function and modify the outcome of the immune response8. They also support the idea that the commensal bacteria play a central role in the regulation of allergic diseases and that they dynamically interact with host genetic background and environmental factors to promote or disrupt oral tolerance9-11. Genetic and immunological evidence also reinforce the idea of a pivotal role for regulatory T (TReg) cells in promoting tolerance to allergens and preventing allergic disorder12-16. In this review, we will discuss recent advances demonstrating the “dual potential” of TReg cells in allergic diseases; how TReg cells are beneficial in promoting tolerance but also how the pro-allergic environment can derange the TReg cell response to aggravate and perpetuates disease.
Natural and induced Foxp3+ TReg cells
TReg cells were initially described as a population of CD4+ T cells expressing the IL-2 receptor α chain (CD25) and CD45RB, able to protect mice from developing autoimmune diseases17,18. Afterwards, the establishment of TReg cells as a distinct CD4+ T cells sub-population was empowered by the identification of the Forkhead winged helix transcription factor FOXP3 (Forkhead Box 3) as a specific TReg cell maker essential to their function19,20. FOXP3 is required for the differentiation of TReg cells, evidenced by the generation of aberrant TReg cells lacking in regulatory function in mice with loss-of-function mutations in Foxp3 21,22. FOXP3 deficiency results in the development of a multi-organ lymphoproliferative autoimmune disease, referred to as Immune Dysregulation, Polyendocrinopathy, Enteropathy X-linked (IPEX) in human subjects and scurfy in mice12,23-26. Expression of FOXP3 into human and murine conventional CD4+ Foxp3− non-TReg cells by means of retroviral gene transfer, converts naïve T cells into TReg cells19. It is now well established that TReg cells enforce tolerance to both self-antigens and to the “extended-self”, the latter encompassing commensal flora and innocuous environmental antigens such as allergens [Reviewed in 27-30].
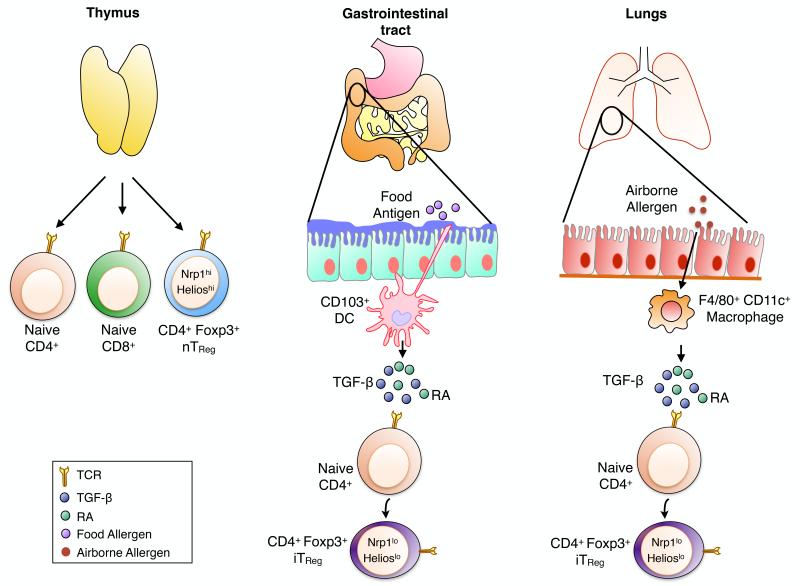
A major population of TReg cells arises in the thymus and is known as CD4+ FOXP3+ “natural” TReg (nTReg, also known as thymus-derived or tTReg) cells, which chiefly mediates tolerance to self-antigens31 (Fig 1). A second population of CD4+ FOXP3+ TReg cells arises extra-thymically in peripheral lymphoid tissues from a pool of naïve conventional CD4+ FOXP3− T cells (Tconv) after exposure to antigens and in the presence of TGF-β [reviewed in32]. These induced TReg (iTReg, also known as peripheral or pTReg) cells are particularly enriched in the gastro-intestinal tract and in the lungs during chronic inflammation, with specificities directed against microbial antigens or environmental allergens33-35 (Fig 1). The generation of iTReg cells at the intestinal mucosa is facilitated by the large abundance of TGF-β and retinoic acid (RA), a vitamin A metabolite, both secreted by the CD103+ CD11c+ dendritic cells (DCs)36-38. In lung tissues, resident macrophages (CD45+ CD11c+ MHC class-IIlow F4/80+) constitutively expressing TGF-β and RA are the main subset of cells driving iTReg cells induction from naïve CD4+ Tconv cells39 (Fig 1). Both FOXP3+ nTReg and iTReg cells subsets play a key function in the maintenance of peripheral tolerance by suppressing reactivity to self-antigens and by containing the amplitude of immune responses to foreign antigens.
Fig 1. Natural and inuced Foxp3+ TReg cells subsets.
The TReg cell pool is composed by two different sub-populations, nTReg and iTReg cells, both expressing the transcription factor Foxp3 crucial for their development and regulatory functions. Foxp3+ Nrp-1high Helioshigh nTReg cells arise in the thymus and mediate tolerance to self- antigens. Foxp3+ Nrp-1low Helioslow iTReg cells, which mediate tolerance to foreign antigens, are induced extra-thymically from naïve CD4+ Foxp3− Tconv cells in the presence of TCR stimulation, TGF-β and RA by either CD103+ DCs at the intestinal mucosa or F4/80+ CD11c+ macrophages at the airways epithelial surfaces.
Because of their different origins, the TCR repertoires of thymic nTReg and peripheral iTReg cells are largely non-overlapping and biased towards self and non-self antigens, respectively 40. However, iTReg cells are known to be less stable than nTReg cells and under inflammatory conditions can lose FOXP3 expression (ex-TReg) and produce cytokines such as IFN-γ and IL-1741,42. This lack of stability can be explained by the methylation status of the conserved non-coding region 2 (CNS2) of the Foxp3 gene. The FOXP3 CNS2 locus, which acts to maintain TReg cell lineage identity under inflammatory conditions, is known to be stably hypomethylated in nTReg whereas it is incompletely demethylated in iTReg cells43-46 .One difficulty for the functional and genetic study of iTReg and nTReg cells is the lack of unique and specific markers allowing the distinction between those two populations and their identification in vivo. nTReg and iTReg cells express similar levels of shared TReg cell markers such FOXP3, CTLA-4, GITR, ICOS, CD103 and CD25. However, many of those markers are also up-regulated by activated CD4+ T cells under inflammatory conditions and their level of expression do not allow the distinction between nTReg and iTReg cells47. The use of Helios and Neuropilin-1 (Nrp-1) has been proposed to specifically discriminate nTReg from iTReg cells, since the expression of those markers is higher in nTReg compared to iTReg cells48-50. While Nrp1 may be upregulated in the context of an inflammatory environment, Helioslow expression has been extensively used as an in vivo marker that distinguishes iTReg from nTReg cells50-52.
In addition to FOXP3+ TReg cells, CD4+ type 1 T regulatory cells (Tr1) represent another subset of TReg cells defined by the expression of IL-10 and the surface marker LAG-3 and CD49b in the face of absent FOXP3 and CD25 expression53. The relationship between FOXP3+ TReg cells and Tr1 cells remains obscure, with both subsets employing common effector pathways including IL-10, TGF-β and CTLA-454. Unlike FOXP3+ TReg cells, Tr1 cells are not uniquely defined by one transcription factor such as FOXP3, but express a number of transcription factors common to other T cell populations including c-MAF, Ahr (Aryl hydrocarbon receptor), and others54 . Many studies that have referred to IL-10 producing TReg cells as Tr1 cells did not discriminate between the two populations by appropriate staining for differentiating markers including FOXP3. In this review, we will focus on FOXP3+ TReg cells as their role in the regulation of allergic disease is far more well defined.
Mechanisms of TReg cells suppression
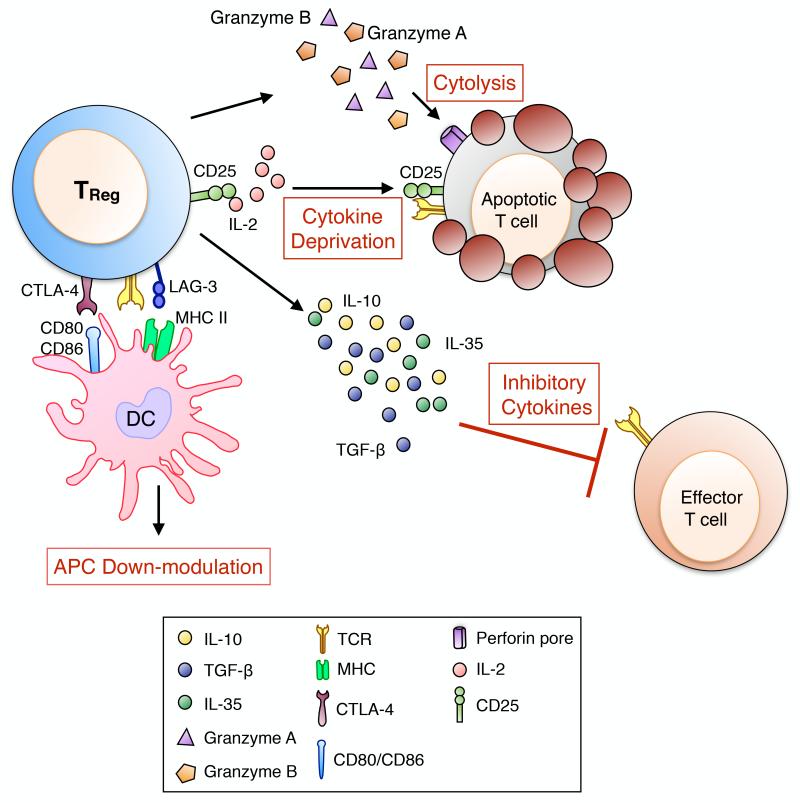
The suppressive functions of TReg cells are essential to control autoimmunity, allergic and inflammatory reactions and responses to infectious agents and tumors. Foxp3+ nTReg and iTReg cells are characterized by a non-overlapping TCR repertoire, resulting in a division of labour where nTReg and iTReg cells regulate immune responses targeting “self” antigens and “non-self” infectious or innocuous agents respectively40,55. TReg cells suppressive function are mediated by multiple mechanisms that involve either the release of inhibitory cytokines (IL-10, TGF-β and IL-35)56-59 and cytolytic molecules (granzymes A and B)60-62, or the down-modulation of antigen presenting cells (Cytotoxic T Lymphocyte Antigen 4 (CTLA-4) and Lymphocyte-activation Gene 3 (LAG-3)63,64, deprivation of trophic cytokines (IL-2 via CD25)65 and modulation metabolic pathways (CD73 and CD39)66 (Fig 2). Expression of select transcription factors and receptors enables TReg cells suppressive functions under inflammatory conditions. GATA-3 expression by TReg cells is triggered by TCR activation and is required to maintain FOXP3 expression and to allow accumulation of TReg cells at inflamed sites67. More recently, it has been demonstrated that Helios expression by TReg cells is key to support their suppressive functions and phenotypic stability during inflammation68. TReg cells functions can also be regulated by endogenous danger signals, or alarmins, released by epithelial cells at the mucosal barrier. Colonic TReg cells express the IL-33 receptor (ST2), allowing them to respond to epithelial cell IL-33 production resulting from tissue damage by amplifying their regulatory functions and restraining intestinal inflammation69.
Fig 2. Mechanisms of Foxp3+ TReg cell-mediated suppression.
Foxp3+ TReg cells mediate tolerance to allergens by diverse suppressive mechanisms. These include T cell cytolysis by a granzyme dependent mechanism, IL-2 deprivation, production of inhibitory cytokines including IL-10, IL-35 and TGF-β capable of blocking the proliferation of Teff cells and down-modulation of antigen presenting cells (APCs) via LAG-3-MHC II and CTLA-4-CD80/CD86 interactions.
TReg cell role in tissue repair
In addition to their immunosuppressive functions and their capacities to restrict the intensity of immune responses, TReg cells can also control non-immunological process such as tissue repair resulting from extensive inflammation. Studies have identified the presence of TReg cells in a wide variety of non-lymphoid organs such as the skin, the intestinal mucosa, the lungs and visceral adipose tissues70. In mice, TReg cells accumulate and remain in skeletal muscles after acute injury and their depletion results in increased muscle damage71. TReg cell production of Amphiregulin (Areg), an epidermal growth factor family member known to promote healing and tissue regeneration, in injured lung and muscle tissues appears to be prevent the tissue damages71,72.
TReg cells and allergic diseases
Allergic diseases reflect a failure to develop tolerance toward a specific allergen, provoking the emergence of an allergen-specific CD4+ T helper type 2 (Th2) cell response, the generation of allergen-specific immunoglobulin E (IgE) and the recruitment of effector cells to the GI tract or lung tissues4,73. In humans, loss-of-function mutations affecting FOXP3 result in the development of IPEX, characterized not only by autoimmunity but also severe allergic inflammation including atopic dermatitis, food allergy, asthma, elevated serum IgE levels and peripheral eosinophilia12,13,74. Foxp3 mutant mice spontaneously exhibit allergic airways inflammation, atopic-dermatitis skin-like disease and elevated serum IgE levels independently of their genetic background75. By using another genetic murine model, the ‘DEREG’ mice which express the diphtheria toxin (DT) receptor under the control of the Foxp3 gene, Hadis et al. have demonstrated that TReg cells depletion in OVA-tolerant DEREG mice was sufficient to break oral tolerance76. Furthermore, in vivo depletion of CD4+ CD25+ T cells in peanut sensitized mice by means of anti-CD25 monoclonal antibody results in impaired oral tolerance development and leads to an heightened allergic77. The central role of TReg cells in oral tolerance development to food allergen has been confirmed in human studies where children who had outgrown milk allergy exhibited higher frequencies of milk protein-specific CD4+ CD25+ TReg cells and where the emergence of allergen-specific TReg cells is highly correlated with a favourable disease outcome78,79.
Among the TReg cell populations, Foxp3+ iTReg cells play an essential role in maintaining tolerance at environmental interfaces, including small and large intestinal and the lung respiratory mucosa32. Allergen-specific iTReg cells are involved in controlling inflammation severity and the IL-4 Th2 cell immune response35. Accordingly, the development of allergic reactions may result from decreased induction and/or impaired function of allergen-specific iTReg cells in genetically allergy-prone subjects. This proposition is supported by a study that took advantage of mice lacking CNS1, an intronic Foxp3 enhancer which contains binding sites for multiple transcription factors such as NFAT and Smad3 and is required for the differentiation of iTReg cells in vivo80,81. Despite decreased iTReg cells expansion, CNS1-deficient mice do not develop a fatal autoimmune lymphoproliferative disease. Nonetheless, with time CNS1-deficient mice develop a pro-allergic phenotype associated with Th2 cell-induced pathologies at mucosal surfaces81. We have recently demonstrated using mice with a gain of function mutation in the IL-4Rα chain (Il4raF709 mice) that heightens susceptibility to oral sensitization that food allergy development is associated with impaired generation and function of allergen-specific TReg cells82. In peanut allergic patients undergoing successful oral immunotherapy (OIT) leading to peanut tolerance induction demonstrated increased numbers of circulating allergen-specific iTReg cells with heighten suppressive capacities and augmented stability, as evidenced by increased demethylation of CpG islands within the FOXP3 gene83. Evidences also point towards a reduced frequency of TReg cells associated with allergic asthma84,85. Compared to healthy controls, frequencies of pulmonary CD4+ CD25high TReg cells in the bronchoalveolar lavage fluid (BALF) were significantly decreased in untreated asthmatic children86. Four weeks of inhaled corticosteroid treatment were sufficient to restore the TReg cells compartment in the blood and the BALF86.
A specific requirement for the cytokines IL-10 and TGF-β1 expressed by TReg cells in the control of allergic responses has emerged. Kearley et al. have demonstrated that adoptively transferred allergen-specific TReg cell suppressive functions during allergic airway inflammation rely on their capacity to induce IL-10 production by CD4+ T cells87. However, subsequent studies by Rubstov et al. employed a genetic approach to show that TReg cell-specific deletion of Il10 promoted allergic airway inflammation; thereby suggesting that TReg cell-derived IL-10 plays a “privileged”, non-redundant role in the induction of immune tolerance in allergic airway inflammation88. IL-10 has immunosuppressive functions and can modulate the activity of key cell subset involved in the allergic reaction such as mast cells82,89, Th2 T cells90, eosinophils and DCs91. The Similar to IL-10, TGF-β1 specifically expressed by TReg cells also appears to play a privileged role in the regulation of allergic responses. In mice, TReg cell-specific deletion of Tgfb1 heightens susceptibility to food allergy (M. NR and T. A. C; unpublished data). The respective role of TReg cell IL-10 and TGF-β1 in the regulation of different allergic responses remains to be fully mapped.
In addition to the defect in allergen-specific iTReg cell induction, aberrations of the TReg cell compartment during allergic disease can also be attributed to a decreased or failure of their suppressive functions. In vitro studies with peripheral TReg (CD4+CD25+) cells isolated from the blood of atopic and non-atopic patients demonstrated that atopic TReg cells can be distinguished from non-atopic TReg cells by decreased capacities to suppress allergen-driven proliferation of effector CD4+ T cells as well as their Th2 cell cytokines secretion92.
TReg cells regulation of the innate immune response in allergic diseases
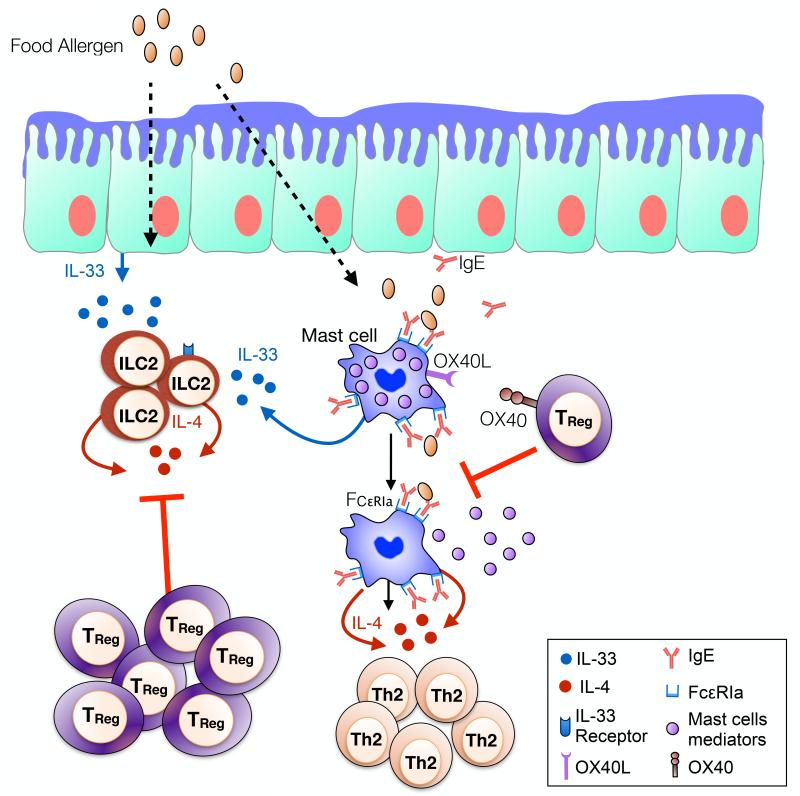
TReg cells can exert their immunosuppressive functions on a broad variety of different cell types, including innate immune cells. Accumulating evidences demonstrate that TReg cells control the immediate hypersensitivity response by acting directly on mast cells and blocking their degranulation (Fig 3)82,89,93. After allergen-sensitization, triggering of mucosal mast cells via the high affinity receptor for IgE (FcεRI) will induce the release of preformed mediators and elicit an IgE-mediated hypersensitity response94. One mechanism by which TReg cells modulate IgE-mediated mucosal mast cells degranulation and decrease mast cells effector mediators release is through the OX40-OX40L pathway93. Direct cell-to-cell contact between OX40 expressed on TReg cells and OX40L on mast cells will lead to increased intracellular levels of cyclic adenosine monophosphate (cAMP) and result in a blockage of extracellular Ca2+ influx and mast cell mediator release inhibition93.
Fig 3. Regulation and suppression of allergic innate immune responses by TReg cells.
TReg cells control innate immune cell subsets involved in promoting allergy. TReg cells block mast cell activation and the release of pre-formed anaphylactic mediators through OX40-OX40L mediated interactions. TReg cells also impede the IL-33-driven ILC2 expansion in the intestinal mucosa and their subsequent IL-4 production. Adapted version from the graphical abstract of ref. 107.
IL-4 production by mast cells is critical in food allergic reaction. IgE interaction with the FcεRI expressed on mast cells act as amplifier of the Th2 cell and IgE responses during allergic sensitization89. Importantly, dysregulated IgE mast cell activation and their subsequent IL-4 production profoundly inhibits allergen iTReg cells generation during allergic processes82. This IL-4 inhibition of iTReg cells generation is mediated through increased intracellular levels of GATA-3 which acts as a FOXP3 inhibitor in early T cell differentiation 95. In contrast, food allergy prone mice that lack the α chain of the high affinity IgE receptor FcεRI (Il4raF709 Fcer1a−/−) were protected from anaphylaxis82. This protection was reflected in decreased mast cell expansion and degranulation and inhibition of the conventional CD4+ cell response, consistent with the key function of IgE/FcεRI axis in mediating not only anaphylaxis but also driving the food allergen-associated cell response 82,89. Moreover, FcεRI deficiency completely corrected the impaired allergen-specific iTreg cell generation82. Similar results were obtained by targeting IgE production (IgE−/− mice or anti-IgE treatment) or by using mast cells ablation genetic murine models 89. The key role of TReg cells in inhibiting mast cell degranulation and their Th2 cell cytokine secretion is thus critical to the prevention of food allergy (Fig 3).
Innate lymphoid cells type 2 (ILC2), a population of mucosal innate cells, are simultaneously characterized by a lack of antigen specificity (absence of T and B cell receptor) and lymphoid traits, as demonstrated by a shared development origin and phenotypic traits with T cells96. ILC2 produce large amounts of Th2 cell cytokines and are linked to allergic disorders such as asthma, chronic rhinosinusitis and atopic dermatitis97-100. In mice, ILC2 can be identified based on the expression of CD25 (IL-2Rα), IL-33R (ST2) and CD127 (IL-7Rα)101 . ILC2 are located in the blood and various organs such as the spleen, the gastrointestinal tract, the liver, the lungs and the lymph nodes102,103. The transcription factor GATA-3 is required for ILC2 differentiation, their stability and Th2 cell cytokines production104,105. Halim et al. demonstrated that ILC2 are required for the development of protease allergen papain-induced airway inflammation as ILC2 deficient mice (RORα−/−) were incapable of mounting an effective Th2 cell immune response and had reduced type 2 lung inflammation102. The critical role of ILC2 in triggering Th2 cell adaptive immune responses involves their production of IL-13, which promotes migration of DC to the draining lymph nodes and enhance the conversion of naïve CD4+ T cells into Th2 cells102
The role of ILC2 in food allergy has been less documented. It appears that IL-13 production by ILC2 enhances allergic mucosal inflammation and promotes IgE-mediated experimental food allergy106. Food allergy development is associated with defective allergen-specific TReg cells induction, consequently resulting in disease promotion82. We have recently demonstrated that increased IL-33 production at the intestinal mucosa during food allergy promotes ILC2 expansion, which further enhances the IgE-mediated food allergic response through their IL-4 production107. ILC2-derived IL-4 inhibits TReg cell response and promotes mast cells activation. Reciprocally, TReg cells block ILC2 expansion and suppress their IL-4 production107(Fig 3). Together, these findings point to the disruption of TReg cell control of mast cells and ILC2 as a key mechanism in the pathogenesis of food allergy. At steady state, TReg cells control both mast cells and ILC2 by restricting their capacity to promote food allergy. Perturbation of this regulatory interaction will subsequently result in a dysregulated pro-allergic innate immune response skewing the immunological balance towards food allergy.
By processing and presenting antigens to naïve T cells, dendritic cells (DCs) are key initiators and master regulators of the allergen-specific immune response. TReg cells also directly act on DCs by down-modulating their surface expression of CD80/CD86 expression and subsequently blocking the generation of an allergen-specific Th2 cell immune response. TReg cell suppression of DCs appears to be mediated via CTLA-4, LAG-3 and Leukocyte Function-Associated antigen-1 (LFA-1)63,64,108. DCs, mostly plasmacytoid DCs (pDCs) have the capacities to prime and naïve T cells and induced their differentiation into IL-10 secreting TReg cells upon ICOS-ICOS ligand interactions109,110.
TReg cells regulation of the adaptive immune response in allergic diseases
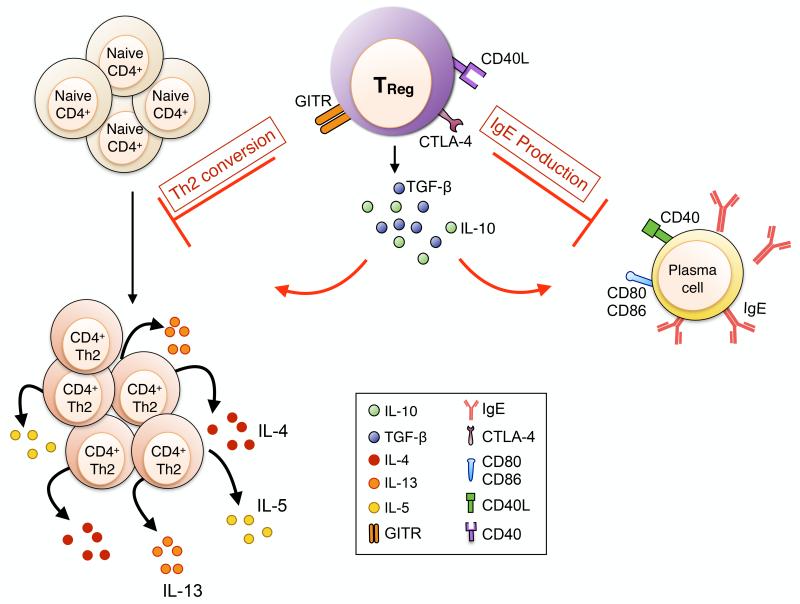
Allergic disorders are characterized by increased dysregulated and aberrant immune responses mediated by the Th2 cell cytokines IL-5, IL-4 and IL-13. TReg cells have also the capacities to regulate allergen-induced adaptive T and B cells responses through diverse mechanisms, either soluble or membrane-bound suppressive molecules (Fig 4). TReg cells express constitutively CTLA-4, a negative co-stimulatory molecule which is essential to their suppressive functions. Mice deficient for CTLA-4 exhibit a lethal multi-organ lymphoproliferative disease111. TReg cell-specific deletion of CTLA-4, by means of crossing Foxp3-Cre with Ctla4fl/fl mice, leads to an autoimmune disease characterized by an increased Th2 cell immune response as evidenced by elevated IL-4 production by CD4+ Foxp3− Tconv cells and increased serum IgE levels91. Ovalbumin (OVA)-specific nTReg cells are efficient in controlling in vitro Th2 cell immune responses and IL-4 production by inhibiting the polarization of naive CD4+ T cells into Th2 cells via a GITR-dependent suppressive mechanisms112. Circulating CD4+ CD25+ TReg cells isolated form the blood of atopic human subjects were also less efficient in vitro than heathy controls CD4+ CD25+ TReg in controlling the Th2 cell cytokines production by effector CD4+ T cells92. Furthermore, frequencies of allergen-specific TReg cells secreting IL-10 with suppressive functions were predominant in peripheral blood mononuclear cells (PBMCs) from healthy subject whereas frequencies of Th2 CD4+ IL-4 secreting T cells frequencies were overrepresented in allergic subjects90. Effector CD4+ IL4+ Th2 cells and suppressive TReg IL-10+ cells are present in both healthy and allergic patients, their ratio frequencies determine either tolerance induction or allergic response development90.
Fig 4. TReg cell mediated suppression of the adaptive allergic immune response.
TReg cells regulate allergen-specific Th2 immune responses and B cell IgE production. GITR stimulation of TReg cells increase their suppressive functions, leading to their blockade of naïve CD4+ Tconv cells conversion into allergen-specific Th2 T cells. TReg cells are also able to control B cells and block their IgE production by a direct CTLA-4 and cell contact-dependent mechanism and through the production of immunosuppressive cytokines such as IL-10.
Through their production of IgE, B cells are essential in the development of allergic immune responses. IgE responses are highly dependent of the immune response Th polarization; the Th2 cell cytokines IL-4 and IL-13 and CD40-CD40L cognate interactions are two signals required to class switching and IgE production by B cells113. In vitro, peripheral allergen-specific TReg cells from healthy subjects repress B cells IgE production by inducing IgG4 class switching114. TReg cells can also exercise their suppressive functions through the release of immunosuppressive cytokines such as IL-10, B cell suppression by TReg cells appears to be cell-to-cell contact mediated and probably occur via CTLA-4 and and TGF-β1114.
Pathogenic TReg cell Th-reprogramming in allergic diseases
An important problem in chronic allergic diseases relates to the mechanisms that enable persistence of inflammation in the face of TReg cell responses115. In the course of regulating Th cell immune responses, TReg cells appropriate partial or “aborted” forms of the transcriptional programs of the target Th cells by expressing their master transcription factors, such as T-bet for Th1 cells and IRF-4 for Th2 cells, and co-opting their function 116,117. Whereas under physiological conditions such partial Th cell programming remains restrained, such restraint is lost under the influence of chronic inflammation leading to pathogenic reprograming of TReg cells into Th cells118,119. In the context of allergic diseases, emerging evidence indicates that a sharply skewed inflammatory environment can overcome the allergen-specific TReg cell regulatory response and redirect those cells towards a pathogenic and pro-inflammatory phenotype (Fig 5 and 6). Recent studies from our laboratory have provided two examples of how allergen-specific TReg cells may acquire T effector (TEff) cell programs and in the process contribute to disease pathogenesis. In the first set of studies, a tyrosine (Y) to phenylalanine (F) mutation at position 709 of the murine IL-4 receptor alpha chain (IL-4Rα) inactivated the receptor’s immunotyrosine inhibitory motif and resulted in augmented activation by IL-4 and IL-13 of the downstream transcription factor STAT6120,121. This mutation, which models human polymorphisms that promote STAT6 activation via the IL-4Rα, imparted on mice heightened susceptibility to allergic diseases, including food allergy and allergic airway inflammation, and reproduced a Th2 cell-high disease “endotype” common in some subjects with those disorders. Importantly, allergen-specific TReg cells became reprogrammed to express a Th2 cell-like phenotype, including IL-4 production, all the while retaining their Foxp3 expression82. Whereas the Th2 cell master transcription factor GATA-3 normally plays a positive role in the accumulation of TReg cells at sites of inflammation and prevents their polarization into Th17 cells, its abnormally expression in TReg cells may contribute to their Th2 cell-like reprogramming under conditions of intense cell polarization67,122,123. The significance of this reprogramming was underlined by the observation that TReg cell-specific deletion of the Il4/Il13 genes restrained the induction of food allergy and allergic airway inflammation in these mice82. Consistent with these results, human food allergic subjects manifest increase expression of Th2 cell cytokines in their circulating allergen-specific TReg cells, indicative of their acquisition of a “Th2-cell like” TEff phenotype 82. Oral immunotherapy was associated with reversal of TH2 cell–like reprogramming of allergen-specific Treg cells, which is coincident with their improved suppression function (data not shown).
Fig 5. Pathogenic “Th2 cell-like” TReg cell reprogramming in food allergy.
Food allergy is characterized by a decreased induction of allergen-specific iTReg cells at the intestinal mucosa. Induced allergen-specific TReg cells in food allergic subjects are prone to acquire a pathogenic skewed “Th2-like” phenotype resulting in increased GATA-3 expression and IL-4 secretion. “Th2 cell-like” iTReg cells are dysfunctional and lacking in suppressor function. They are not able to control the Teff Th2 cell immune response and mast cells expansion, perpetuating in the process the allergic phenotype.
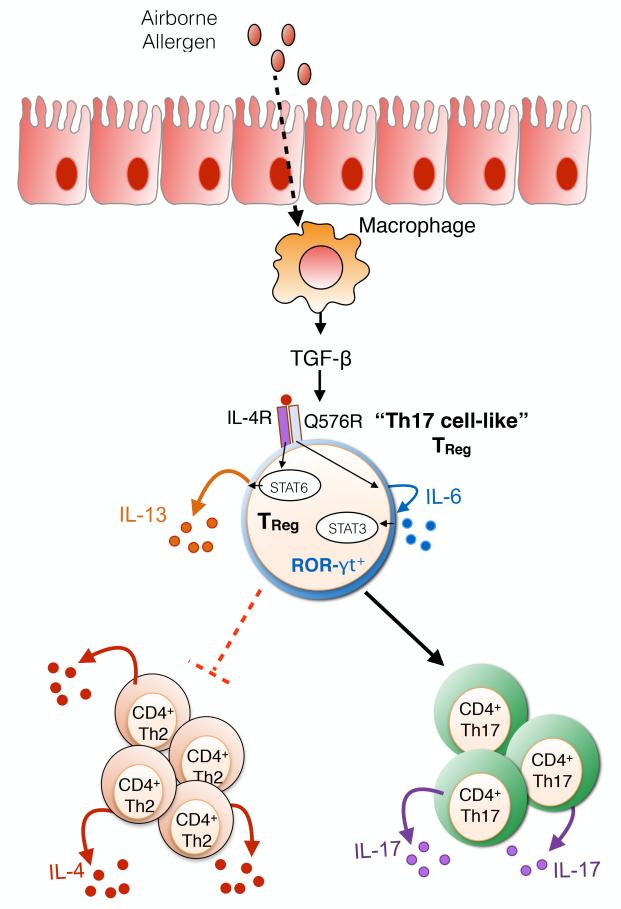
Fig 6. Pathogenic “Th17 cell-like” TReg cell reprogramming by the IL-4Rα-Q576R allele.
Human IL-4Rα-Q576R is associated with increased asthma severity. Signalling through the IL4Rα-Q576R on iTReg cells induces dual activation of STAT6 and STAT3, the latter through an autocrine IL-6 production loop. The IL-6-STAT3 axis promotes pathogenic “Th17 cell-like” TReg cell reprogramming resulting in ROR-γt expression and IL-17 secretion by the reprogrammed TReg cells.
A second example of allergen-specific TReg cell reprogramming came from studies on a human IL-4Rα allele that bears a glutamine (Q) to an arginine (R) substitution at position 576 (IL-4Rα-Q576R)124. This allele is associated with asthma severity, while introduction of the Q576R substitution into the murine IL-4Rα results in exaggerated allergic airway inflammation when the mice are sensitized then challenged in their airways with allergens125-127 (Fig 6). Signalling via IL-4Rα-Q576R allele does not impact the activation by the IL-4R of its dedicated transcription factor STAT6, and Th2 cell responses promoted via the IL-4R are preserved. Nevertheless, in both humans and mice, the Q576R substitution acts to create a novel branch of signalling via IL-4Rα that activates microtubule-associated protein kinases (MAPK), leading to the induction by IL-4 of IL-6 production128 (Fig 6). The newly produced IL-6 destabilizes newly formed allergen-specific TReg cells towards the Th17 cell lineage, thus giving rise to mixed Th2-Th17 cell responses in the context of allergic inflammation128. Inhibition of the capacity of allergen-specific TReg cells to differentiate into Th17 cells, whether by neutralization of IL-6 or by TReg cell-specific deletion of genes encoding IL-6 receptor alpha chain (Il6ra) or the Th17 cell master transcription factor RORγt, reversed the exaggerated allergic inflammation induced by IL-4Rα-Q576Rmice128 (Fig 6).
Microbiome – TReg cell interactions in allergic diseases
Altered environmental exposures early in life may play a critical role in setting in motion the atopic diseases of childhood129. The “hygiene hypothesis” stipulates that increased allergic rates observed over the years result from reduced microbial exposures arising from lifestyle changes, such as family size reduction, use of antibiotics and improved hygiene130. The influence of the intestinal microbiome in tolerance induction and allergies development is becoming more appreciated. The intestinal colonization of neonates starts at birth from the mother’s vaginal flora, as the microbiota composition of vaginally delivered infants is similar to the maternal vagina131. Infants born by cesarian section have a different microbiota composition, mostly derived from maternal skin, and are at increased risks of developing asthma and allergy132. The first months of life are a critical period for the intestinal flora to settle and stabilize as children exhibiting intestinal dysbiosis in this “time-window” are at increased risks of developing asthma133. Exposure to a farm environment and the associated subsequent large diversity of environmental microbial signals reduce the risk of developing allergies133,134. The importance of the microbial flora for allergic diseases development is further emphasized by the observation that Germ Free (GF) mice cannot be tolerized to oral antigens and develop a Th2 cell-biased immune response135. In humans, a polymorphism in the promoter of CD14, a high affinity receptor for bacterial lipopolysaccharide (LPS) and co-receptor of TLR-4, has been associated with the development of atopic disease136. Food allergic responses are also aggravated in TLR4−/− mice or WT mice treated with antibiotics and repopulation of commensal flora in antibiotics treated mice result in reduced allergen-specific IgE and Th2 cell cytokines responses137. These observations highlighted an important function of the intestinal microbial flora and microbial exposure in maintaining and shaping the immune response and inducing protection against the development of atopy138,139.
By using Il4raF709 food allergy-prone mice, we have demonstrated that food allergy is associated with the emergence of an altered intestinal microbial flora10. The microbiome of allergic Il4raF709 mice exhibits decreased relative abundances of members from the Firmicutes phylum and increased abundance of bacteria belonging to the Proteobacteria phylum. Adoptive transfer of allergen-specific TReg cells prevented the development of food allergy in allergic-prone mice as well as the emergence of the food-allergy associated microbial dysbiosis10. Importantly, disease susceptibility can be transferred from allergic Il4raF709 mice to GF mice via transplantation of commensal flora from allergic donors. Allergy susceptibility transfer was associated with increase allergen-specific IgE production and expansion of IL-4 secreting Th2 CD4+ T cells in GF mice reconstituted with the allergic microbial flora10.
The commensal flora can target different immune cell subsets belonging to either the innate or/and adaptive allergic effector responses (Fig 7). In the steady-state, the microbial flora promotes intestinal IgA production by a TReg cell intrinsic MyD88-dependent mechanism that enables the generation of iTReg cells in the gut and their differentiation into T follicular helper cells (TFH)140. Depletion of the commensal flora by antibiotic treatment141 or the use of GF mice11 is associated with the development of Th2 cell-type allergic responses and higher serum IgE levels. Th2 cell-type allergic immune responses were held in check by MyD88-dependent microbial sensing by B cells, which suppressed the IgE responses141. The commensal microbiota also influence the outcome of the allergic response modulating the ILC (Fig 7). Mono-colonization of GF mice with anaerobic bacteria belonging to the Clostridia class blocks and protects from oral allergen sensitization by inducing IL22+ ROR-γt+ ILC at the intestinal mucosa11. ILC2 are expanding in the course of allergic disorders and have a pathogenic function in further promoting disease through their Th2 cell type cytokines secretion102,107. However, how the intestinal or the upper airways microbial flora could affect, regulate or promote the ILC2 immune response needs still to be further investigated.
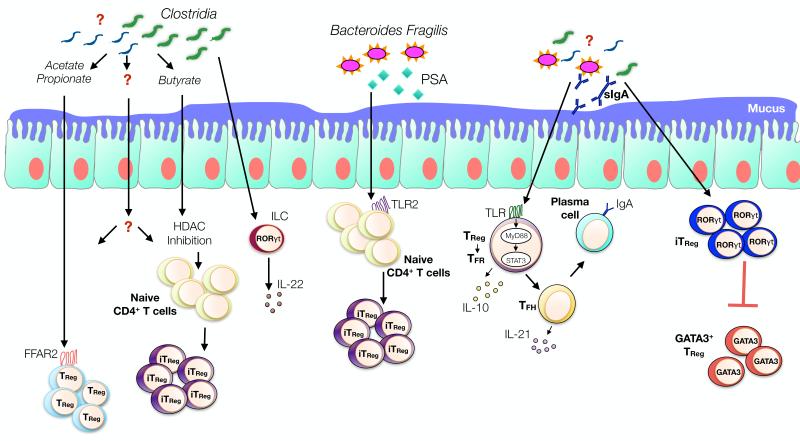
Fig 7. Microbiota-immune cell interactions shape oral tolerance.
Metabolites, such as SFCAs produced by bacterial fermentation of dietary fibers promote the proliferation and de novo induction of iTReg cells through FFAR2 (GPR43) receptor and HDAC inhibition. Clostridial bacterial species promote the production of IL-22 by ROR-γt ILC, reinforcing oral tolerance by decreasing gut permeability and oral allergen uptake. Bacteroides Fragilis production of PSA promotes de novo iTReg cells generation via TLR2 signalling. MyD88/STAT3-sensing by TReg cells enforces oral tolerance by inducing and directing the TFH - TFReg and IgA axis. The microbiota also promote the emergence of ROR-γt expressing iTReg cells. ROR-γt deficiency in TReg cells promotes a Th2 environment and oral allergen sensitization possibly by inducing iTReg cell reprograming into “Th2 cell-like” cells expressing GATA-3.
The microbiota promotes either a tolerant or a pro-Th2 cell type allergic immune response by interacting directly with immune cells and their TLRs or indirectly through the release of microbial products. Polysaccharide A (PSA), produced by the commensal bacterium Bacteroides fragilis, acts by a TLR2-dependent mechanisms to induce the conversion of CD4+ T cells into functional iTReg cells with enhanced suppressive activities and increased IL-10 production142,143. In mice, Clostridial species are among the most abundant Gram-positive bacteria present at the intestinal mucosa. Colonization of GF mice with a mix of Clostridium isolated from either murine or human faeces resulted in a strong induction of iTReg cells in the colonic lamina propia of the reconstituted mice144,145. Through their production of IL-10, Clostridium-induced TReg cells also controlled the systemic IgE production by reducing in vitro the IL-4 production by splenic CD4+ T cells144.
Short chain fatty acids (SCFAs) produced by bacterial fermentation of dietary fibers act on T cells via a G protein coupled receptor (GPR43) and protect mice from intestinal inflammation by expanding the pool of the colonic TReg cells146. SCFAs also promote the generation of intestinal iTReg cells from naïve CD4+ T cells by T-cell intrinsic epigenetic mechanisms147,148. Butyrate, a SCFA known as an histone deacetylase (HDAC) inhibitor, increases Foxp3 protein acetylation conferring thereby increased stability and enhanced suppressive function to de novo generated intestinal iTReg cells148. Accordingly, a high-fiber diet results in modulation of the intestinal flora composition characterized by increased Bacteroidetes and decreased Firmicutes abundances, resulting in increased circulating levels of SCFAs and allergic airways inflammation protection149. More recently, it has been reported that the microbiome and oral antigen promote the induction of iTReg cells expressing RORγt at the intestinal mucosal surfaces150. Specific ablation of RORγt in TReg cells resulted in increased frequencies of GATA-3+ Foxp3+ TReg cells and increased production of IL-4 and IL-13 by CD4+ Foxp3− Tconv cells, leading to the conclusion that the microbiome control the Th2 cell immune response through the expansion of TReg RORγt+ cells and the regulation of DC activation150. Whether “pro-allergic” or “pro-tolerant” bacterial species are linked to and directly affect the generation of those GATA3+ or RORγt+ TReg cells needs to be further investigated. Since atopic disease are associated with a defect in the generation of allergen-specific iTReg cells, it will be of interest to pursue investigation of these pathways to identify potential therapeutic targets to promote allergen-specific tolerance.
Conclusion
A dynamic view of TReg cells in allergic disease is emerging in which they play a central, determining role not only in tolerance induction but also, when destabilized and reprogrammed, in mediating disease pathogenesis, severity and chronicity. Novel approaches to the re-establishment of tolerance are suggested by the results of preclinical models in which reinforcement of iTReg cell stability by interrupting their pathogenic programming may be of therapeutic benefit in these disorders.
What do we know?
TReg cells have a key role in promoting and maintaining tolerance to allergens by regulating both innate and adaptive allergen-triggered immune response.
Allergic diseases are associated with a failure to develop tolerance towards a specific allergen leading to the emergence of a pathogenic Th2 immune response.
A pro-allergic inflammatory environment may skew allergen-specific TReg cells towards a pathogenic phenotype that perpetuates and aggravates disease.
Allergic responses are influenced by the commensal flora, acting in part via TReg cells.
What is still unknown?
Mechanisms by which TReg cells fail to maintain tolerance in allergic diseases are not well understood and require further investigation.
Acknowledgments
This work was supported by the National Institutes of Health (5R01AI065617 and 1R56AI117983), to T.A.C.
Abbreviations
- TReg
Regulatory T cells
- BALF
Bronchoalveolar Lavage Fluid
- ILC
innate lymphoid cells
- Th
T helper
- IgE
Immunoglobulin type E
- Areg
Amphiregulin
- Ahr
Aryl hydrocarbon receptor
- SCFA
Short Chain Fatty Acids
- HDAC
Histone Deacetylase
- TEff
T effector cells
- TConv
Conventional T cells
- DCs
dendritic cells
- APCs
antigen presenting cells
- CTLA-4
Cytotoxic T Lymphocyte Antigen 4
- LAG3
Lymphocyte-Activation Gene 3
- TCR
T cell receptor
- Tr1
Type 1 T regulatory cells
REFERENCES
- 1.Platts-Mills TAE. The allergy epidemics: 1870-2010. J Allergy Clin Immunol. 2015;136:3–13. doi: 10.1016/j.jaci.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. quiz308. [DOI] [PubMed] [Google Scholar]
- 3.Loftus PA, Wise SK. Epidemiology of asthma. Curr Opin Otolaryngol Head Neck Surg. 2016;24:245–9. doi: 10.1097/MOO.0000000000000262. [DOI] [PubMed] [Google Scholar]
- 4.Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2014;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 5.Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol. 2010;125:S116–25. doi: 10.1016/j.jaci.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Sicherer SH. Epidemiology of food allergy. J Allergy Clin Immunol. 2011;127:594–602. doi: 10.1016/j.jaci.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host and Microbe. 2015;17:592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong X, Wang X. Early life precursors, epigenetics, and the development of food allergy. Semin Immunopathol. 2012;34:655–69. doi: 10.1007/s00281-012-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34:671–88. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noval Rivas M, Burton OT, Wise P, Zhang Y-Q, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–12. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. PNAS. 2014;111:13145–50. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, et al. JM2, encoding a fork head–related protein, is mutated in X-linked autoimmunity–allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torgerson TR, Linane A, Moes N, Anover S, Mateo V, Rieux Laucat F, et al. Severe food allergy as a variant of IPEX syndrome caused by a deletion in a noncoding region of the FOXP3 gene. Gastroenterology. 2007;132:1705–17. doi: 10.1053/j.gastro.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 14.Palomares O, Yaman G, Azkur AK, Akkoc T, Akdis M, Akdis CA. Role of Treg in immune regulation of allergic diseases. Eur J Immunol. 2010;40:1232–40. doi: 10.1002/eji.200940045. [DOI] [PubMed] [Google Scholar]
- 15.Jones SM, Burks AW, Dupont C. State of the art on food allergen immunotherapy: oral, sublingual, and epicutaneous. J Allergy Clin Immunol. 2014;133:318–23. doi: 10.1016/j.jaci.2013.12.1040. [DOI] [PubMed] [Google Scholar]
- 16.Palomares O, Martín-Fontecha M, Lauener R, Traidl-Hoffmann C, Cavkaytar O, Akdis M, et al. Regulatory T cells and immune regulation of allergic diseases: roles of IL-10 and TGF-β. Genes Immun. 2014;15:511–20. doi: 10.1038/gene.2014.45. [DOI] [PubMed] [Google Scholar]
- 17.Sakaguchi S, Takahashi T, Nishizuka Y. Study on cellular events in post-thymectomy autoimmune oophoritis in mice. II. Requirement of Lyt-1 cells in normal female mice for the prevention of oophoritis. J Exp Med. 1982;156:1577–86. doi: 10.1084/jem.156.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–64. [PubMed] [Google Scholar]
- 19.Hori S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 20.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 21.Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–68. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 22.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 23.Lyon MF, Peters J, Glenister PH, Ball S, Wright E. The scurfy mouse mutant has previously unrecognized hematological abnormalities and resembles Wiskott-Aldrich syndrome. PNAS. 1990;87:2433–7. doi: 10.1073/pnas.87.7.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 25.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 26.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 27.Alroqi FJ, Chatila TA. T Regulatory Cell Biology in Health and Disease. Curr Allergy Asthma Rep. 2016;16:27–8. doi: 10.1007/s11882-016-0606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatila TA. Regulatory T cells: key players in tolerance and autoimmunity. Endocrinol Metab Clin North Am. 2009;38:265–72. doi: 10.1016/j.ecl.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T Cells: Mechanisms of Differentiation and Function. Annu Rev Immunol. 2012;30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benoist C, Mathis D. Treg cells, life history, and diversity. Cold Spring Harb Perspect Biol. 2012;4:a007021. doi: 10.1101/cshperspect.a007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lio C-WJ, Hsieh C-S. Becoming self-aware: the thymic education of regulatory T cells. Curr Opin Immunol. 2011;23:213–9. doi: 10.1016/j.coi.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilate AM, Lafaille JJ. Induced CD4 +Foxp3 +Regulatory T Cells in Immune Tolerance. Annu Rev Immunol. 2012;30:733–58. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 33.Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille J, Curotto de Lafaille M. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–33. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, Boehmer von H. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–27. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 35.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ Regulatory T Cell-Dependent and -Independent Control of Allergic Inflammation. Immunity. 2008;29:114–26. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Sun C-M, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqui KRR, Powrie F. CD103+ GALT DCs promote Foxp3+ regulatory T cells. Mucosal Immunol. 2008;1:S34–8. doi: 10.1038/mi.2008.43. [DOI] [PubMed] [Google Scholar]
- 38.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soroosh P, Doherty TA, Duan W, Mehta AK, Choi H, Adams YF, et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J Exp Med. 2013;210:775–88. doi: 10.1084/jem.20121849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, et al. A Central Role for Induced Regulatory T Cells in Tolerance Induction in Experimental Colitis. J Immunol. 2009;182:3461–8. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martínez-Llordella M, Ashby M, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–7. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, et al. Molecular Antagonism and Plasticity of Regulatory and Inflammatory T Cell Programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic Control of the foxp3 Locus in Regulatory T Cells. PLoS Biol. 2007;5:0169–78. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkura N, Kitagawa Y, Sakaguchi S. Development and Maintenance of Regulatory T cells. Immunity. 2013;38:414–23. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–63. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 2014;158:734–48. doi: 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrath von MG, Harrison LC. Regulatory Lymphocytes: Antigen-induced regulatory T cells in autoimmunity. Nat Rev Immunol. 2003;3:223–32. doi: 10.1038/nri1029. [DOI] [PubMed] [Google Scholar]
- 48.Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–42. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–22. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–41. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feuerer M, Hill JA, Kretschmer K, Boehmer von H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. PNAS. 2010;107:5919–24. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YC, Bhairavabhotla R, Yoon J, Golding A, Thornton AM, Tran DQ, et al. Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion. Blood. 2012;119:2810–8. doi: 10.1182/blood-2011-09-377895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limón P, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nature Med. 2013;19:739–46. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 54.Gregori S, Goudy KS, Roncarolo MG. The cellular and molecular mechanisms of immuno-suppression by human type 1 regulatory T cells. Front Immunol. 2012;3:30. doi: 10.3389/fimmu.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B, et al. A Requisite Role for Induced Regulatory T Cells in Tolerance Based on Expanding Antigen Receptor Diversity. Immunity. 2011;35:109–22. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitani A, Fuss I, Nakamura K, Kumaki F, Usui T, Strober W. Transforming growth factor (TGF)-beta1-producing regulatory T cells induce Smad-mediated interleukin 10 secretion that facilitates coordinated immunoregulatory activity and amelioration of TGF-beta1-mediated fibrosis. J Exp Med. 2003;198:1179–88. doi: 10.1084/jem.20030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura K, Kitani A, Fuss I, Pedersen A, Harada N, Nawata H, et al. TGF- 1 Plays an Important Role in the Mechanism of CD4+CD25+ Regulatory T Cell Activity in Both Humans and Mice. J Immunol. 2004;172:834–42. doi: 10.4049/jimmunol.172.2.834. [DOI] [PubMed] [Google Scholar]
- 58.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–9. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 59.Collison LW, Chaturvedi V, Henderson AL, Giacomin PR, Guy C, Bankoti J, et al. IL-35-mediated induction of a potent regulatory T cell population. Nat Immunol. 2010;11:1093–101. doi: 10.1038/ni.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McHugh R, Whitters M, A PC, Young D, Skevach EM, Collins M, et al. Immunoregulatory T Cells: Gene Expression Analysis Reveals a Functional Role for the Glucocorticoid-Induced TNF Receptor. Immunity. 2002;16:311–23. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 61.Grossman WJ, Verbsky JW, Barchet W, Colonna M, Atkinson JP, Ley TJ. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting Edge: Contact-Mediated Suppression by CD4+CD25+ Regulatory Cells Involves a Granzyme B-Dependent, Perforin-Independent Mechanism. J Immunol. 2005;174:1783–6. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 63.Read S, Malmström V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang B, Workman C, Lee J, Chew C, Dale BM, Colonna L, et al. Regulatory T cells inhibit dendritic cells by lymphocyte activation gene-3 engagement of MHC class II. J Immunol. 2008;180:5916–26. doi: 10.4049/jimmunol.180.9.5916. [DOI] [PubMed] [Google Scholar]
- 65.Thornton A, Shevach EM. Immunoregulatory T Cells Suppress Polyclonal T Cell Activation In Vitro by Inhibiting Interleukin 2 Production. J Immunol. 1998;188:287–96. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–65. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–15. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H-J, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–9. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schiering C, Krausgruber T, Chomka A, Fröhlich A, Adelmann K, Wohlfert EA, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–8. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14:1007–13. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–95. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–89. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berin CM, Shreffler WG. Mechanisms Underlying Induction of Tolerance to Foods. Immunol Allergy Clin N Am. 2016;36:87–102. doi: 10.1016/j.iac.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Verbsky JW, Chatila TA. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and IPEX-related disorders: an evolving web of heritable autoimmune diseases. Curr Opin Pediatr. 2013;25:708–14. doi: 10.1097/MOP.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin W, Truong N, Grossman WJ, Haribhai D, Williams CB, Wang J, et al. Allergic dysregulation and hyperimmunoglobulinemia E in Foxp3 mutant mice. J Allergy Clin Immunol. 2005;116:1106–15. doi: 10.1016/j.jaci.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 76.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 77.van Wijk F, Wehrens E, Boon L, Pieters R, Knipels LM. CD4+CD25+ T cells regulate the intensity of hypersensitivity responses to peanut, but are not decisive in the induction of oral sensitization. Clin Exp Allergy. 2007;37:572–81. doi: 10.1111/j.1365-2222.2007.02681.x. [DOI] [PubMed] [Google Scholar]
- 78.Karlsson MR. Allergen-responsive CD4+CD25+ Regulatory T Cells in Children who Have Outgrown Cow's Milk Allergy. J Exp Med. 2004;199:1679–88. doi: 10.1084/jem.20032121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shreffler WG, Wanich N, Moloney M, Nowak-Wȩgrzyn A, Sampson HA. Association of allergen-specific regulatory T cells with the onset of clinical tolerance to milk protein. J Allergy Clin Immunol. 2009;123:43–7. doi: 10.1016/j.jaci.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 80.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2007;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 81.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–9. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noval Rivas M, Burton OT, Wise P, Charbonnier L-M, Georgiev P, Oettgen HC, et al. Regulatory T Cell Reprogramming toward a Th2-Cell- like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity. 2015;42:512–23. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Syed A, Garcia MA, Lyu S-C, Bucayu R, Kohli A, Ishida S, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3) J Allergy Clin Immunol. 2014;133:500–10. doi: 10.1016/j.jaci.2013.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mamessier E, Nieves A, Lorec AM, Dupuy P, Pinot D, Pinet C, et al. T-cell activation during exacerbations: a longitudinal study in refractory asthma. Allergy. 2008;63:1202–10. doi: 10.1111/j.1398-9995.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 85.Lloyd CM, Hawrylowicz CM. Regulatory T cells in asthma. Immunity. 2009;31:438–49. doi: 10.1016/j.immuni.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hartl D, Koller B, Mehlhorn AT, Reinhardt D, Nicolai T, Schendel DJ, et al. Quantitative and functional impairment of pulmonary CD4+CD25hi regulatory T cells in pediatric asthma. J Allergy Clin Immunol. 2007;119:1258–66. doi: 10.1016/j.jaci.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 87.Kearley J, Barker JE, Robinson DS, Lloyd CM. Resolution of airway inflammation and hyperreactivity after in vivo transfer of CD4 +CD25 +regulatory T cells is interleukin 10 dependent. J Exp Med. 2005;202:1539–47. doi: 10.1084/jem.20051166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 89.Burton OT, Rivas MN, Zhou JS, Logsdon SL, Darling AR, Koleoglou KJ, et al. Immunoglobulin E Signal Inhibition during Allergen Ingestion Leads to Reversal of Established Food Allergy and Induction of Regulatory T Cells. Immunity. 2014;41:141–51. doi: 10.1016/j.immuni.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akdis M, Verhagen J, Taylor A, Karamloo F, Karagiannidis C, Crameri R, et al. Immune Responses in Healthy and Allergic Individuals Are Characterized by a Fine Balance between Allergen-specific T Regulatory 1 and T Helper 2 Cells. J Exp Med. 2004;199:1567–75. doi: 10.1084/jem.20032058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–5. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 92.Ling EM, Smith T, Nguyen XD, Pridgeon C, Dallman M, Arbery J, et al. Relation of CD4+CD25+ regulatory T-cell suppression of allergen-driven T-cell activation to atopic status and expression of allergic disease. Lancet. 2004;363:608–15. doi: 10.1016/S0140-6736(04)15592-X. [DOI] [PubMed] [Google Scholar]
- 93.Gri G, Piconese S, Frossi B, Manfroi V, Merluzzi S, Tripodo C, et al. CD4+CD25+ Regulatory T Cells Suppress Mast Cell Degranulation and Allergic Responses through OX40-OX40L Interaction. Immunity. 2008;29:771–81. doi: 10.1016/j.immuni.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nature Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mantel P-Y, Kuipers H, Boyman O, Rhyner C, Ouaked N, Rückert B, et al. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eberl G, Colonna M, Di Santo JP, McKenzie ANJ. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132:205–13. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang Y-J, Kim HY, Albacker LA, Baumgarth N, McKenzie ANJ, Smith DE, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–8. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–62. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 100.Roediger B, Kyle R, Yip KH, Sumaria N, Guy TV, Kim BS, et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–73. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 102.Halim TYF, Steer CA, Mathä L, Gold MJ, Martinez-Gonzalez I, McNagny KM, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–35. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Molofsky AB, Van Gool F, Liang H-E, Van Dyken SJ, Nussbaum JC, Lee J, et al. Interleukin-33 and Interferon-γ Counter-Regulate Group 2 Innate Lymphoid Cell Activation during Immune Perturbation. Immunity. 2015;43:161–74. doi: 10.1016/j.immuni.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoyler T, Klose CSN, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–48. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liang H-E, Reinhardt RL, Bando JK, Sullivan BM, Ho I-C, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J-B, Chen C-Y, Liu B, Mugge L, Angkasekwinai P, Facchinetti V, et al. IL-25 and CD4(+) TH2 cells enhance type 2 innate lymphoid cell-derived IL-13 production, which promotes IgE-mediated experimental food allergy. J Allergy Clin Immunol. 2016;137:1216–1225. doi: 10.1016/j.jaci.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Noval Rivas M, Burton OT, Oettgen HC, Chatila TA. IL-4 Production by Group 2 Innate Lymphoid cells Promotes Food Allergy by Blocking Regulatory T cell Function. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2016.02.030. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Onishi Y, Fehervari Z, Yamaguchi T, Sakaguchi S. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. PNAS. 2008;105:10113–8. doi: 10.1073/pnas.0711106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wakkach A, Fournier N, Brun V, Breittmayer J-P, Cottrez F, Groux H. Characterization of dendritic cells that induce tolerance and T regulatory 1 cell differentiation in vivo. Immunity. 2003;18:605–17. doi: 10.1016/s1074-7613(03)00113-4. [DOI] [PubMed] [Google Scholar]
- 110.Ito T, Yang M, Wang Y-H, Lande R, Gregorio J, Perng OA, et al. Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204:105–15. doi: 10.1084/jem.20061660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–8. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 112.Girtsman T, Jaffar Z, Ferrini M, Shaw P, Roberts K. Natural Foxp3(+) regulatory T cells inhibit Th2 polarization but are biased toward suppression of Th17-driven lung inflammation. J Leukoc Biol. 2010;88:537–46. doi: 10.1189/jlb.0110044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Curotto de Lafaille MA, Muriglan S, Sunshine MJ, Lei Y, Kutchukhidze N, Furtado GC, et al. Hyper immunoglobulin E response in mice with monoclonal populations of B and T lymphocytes. J Exp Med. 2001;194:1349–59. doi: 10.1084/jem.194.9.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lim HW, Hillsamer P, Banham AH, Kim CH. Cutting edge: direct suppression of B cells by CD4+ CD25+ regulatory T cells. J Immunol. 2005;175:4180–3. doi: 10.4049/jimmunol.175.7.4180. [DOI] [PubMed] [Google Scholar]
- 115.Krishnamoorthy N, Khare A, Oriss TB, Raundhal M, Morse C, Yarlagadda M, et al. Early infection with respiratory syncytial virus impairs regulatory T cell function and increases susceptibility to allergic asthma. Nature Med. 2012;18:1525–30. doi: 10.1038/nm.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity. 2012;37:501–10. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu T-T, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nature Med. 2014;20:62–8. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 119.Charbonnier L-M, Wang S, Georgiev P, Sefik E, Chatila TA. Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat Immunol. 2015;16:1162–73. doi: 10.1038/ni.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tachdjian R, Khatib Al S, Schwinglshackl A, Sook Kim H, Chen A, Blasioli J, et al. In vivo regulation of the allergic response by the IL-4 receptor α chain immunoreceptor tyrosine-based inhibitory motif. J Allergy Clin Immunol. 2010;125:1128–8. doi: 10.1016/j.jaci.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mathias CB, Hobson SA, Garcia-Lloret M, Lawson G, Poddighe D, Freyschmidt E-J, et al. IgE-mediated systemic anaphylaxis and impaired tolerance to food antigens in mice with enhanced IL-4 receptor signaling. J Allergy Clin Immunol. 2011;127:795–805. e1–6. doi: 10.1016/j.jaci.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13:1010–9. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y, Su MA, Wan YY. An Essential Role of the Transcription Factor GATA-3 for the Function of Regulatory T Cells. Immunity. 2011;35:337–48. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 125.Rosa-Rosa L, Zimmermann N, Bernstein JA, Rothenberg ME, Khurana Hershey GK. The R576 IL-4 receptor alpha allele correlates with asthma severity. J Allergy Clin Immunol. 1999;104:1008–14. doi: 10.1016/s0091-6749(99)70082-5. [DOI] [PubMed] [Google Scholar]
- 126.Tachdjian R, Mathias C, Khatib Al S, Bryce PJ, Kim HS, Blaeser F, et al. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J Exp Med. 2009;206:2191–204. doi: 10.1084/jem.20091480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xia M, Viera-Hutchins L, Garcia-Lloret M, Noval Rivas M, Wise P, McGhee SA, et al. Vehicular exhaust particles promote allergic airway inflammation through an aryl hydrocarbon receptor-notch signaling cascade. J Allergy Clin Immunol. 2015;136:441–53. doi: 10.1016/j.jaci.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Massoud AH, Charbonnier L-M, Lopez D, Pellegrini M, Phipatanakul W, Chatila TA. An asthma-associated IL4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nat Med. 2016;22:1013–22. doi: 10.1038/nm.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heederik D, Mutius von E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 130.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 131.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disesase: meta-analyses. Clin Exp Allergy. 2008;38:634–42. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 133.Braun-Fahrlander C, RIEDLER J, Herz U, EDER W, WASER M, GRIZE L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347:869–77. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 134.Illi S, Illi S, Mutius von E, Erika von Mutius, Lau S, Lau S, et al. Early childhood infectious diseases and the development of asthma up to school age: a birth cohort study. BMJ. 2001;322:390–5. doi: 10.1136/bmj.322.7283.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–45. [PubMed] [Google Scholar]
- 136.Woo JG, Assa’ad A, Heizer AB, Bernstein JA, Hershey GK. The -159 C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol. 2003;112:438–44. doi: 10.1067/mai.2003.1634. [DOI] [PubMed] [Google Scholar]
- 137.Bashir MEH, Louie S, Shi HN, Nagler-Anderson C. Toll-Like Receptor 4 Signaling by Intestinal Microbes Influences Susceptibility to Food Allergy. J Immunol. 2004;172:6978–87. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 138.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–36. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 139.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Sen, Charbonnier L-M, Rivas MN, Georgiev P, Li N, Gerber G, et al. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity. 2015;43:289–303. doi: 10.1016/j.immuni.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nature Med. 2012;18:538–46. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. PNAS. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Round JL, Lee SM, Li J, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 146.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 148.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Med. 2014;20:159–66. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 150.Ohnmacht C, Park J-H, Cording S, Wing JB, Atarashi K, Obata Y, et al. Mucosal Immunology - The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 2015;349:989–93. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]