Abstract
The periplasm of Gram-negative bacteria includes a variety of molecular chaperones that shepherd the folding and targeting of secreted proteins. A central player of this quality control network is DegP, a protease also suggested to have a chaperone function. We serendipitously discovered that production of the Bordetella pertussis autotransporter virulence protein pertactin is lethal in Escherichia coli ΔdegP strains. We investigated specific contributions of DegP to secretion of pertactin as a model system to test the functions of DegP in vivo. The DegP chaperone activity was sufficient to restore growth during pertactin production. This chaperone dependency could be relieved by changing the pertactin signal sequence: an E. coli signal sequence leading to co-translational inner membrane (IM) translocation was sufficient to suppress lethality in the absence of DegP, whereas an E. coli post-translational signal sequence was sufficient to recapitulate the lethal phenotype. These results identify a novel connection between the DegP chaperone and the mechanism used to translocate a protein across the IM. Lethality coincided with loss of periplasmic proteins, soluble σE, and proteins regulated by this essential stress response. These results suggest post-translational IM translocation can lead to the formation of toxic periplasmic folding intermediates, which DegP can suppress.
Introduction
Although all bacterial proteins are synthesized in the cytoplasm, many are subsequently secreted to the cell surface, where they function to sense and respond to the cellular environment. In Escherichia coli, approximately 10% of proteins contain an N-terminal signal sequence for translocation across the inner membrane (IM) to the periplasm [1]. Most bacterial secreted proteins, including those used for virulence, enter the periplasm through the primary export channel SecYEG [2–4], either co- or post-translationally. While the rate of co-translational translocation is limited by the rate of protein synthesis [3,4], post-translational translocation can occur ~10-fold faster [5,6]. Most secreted proteins in E. coli are thought to be translocated post-translationally [2,7]. The effects of changing the IM translation mechanism on downstream steps in the periplasm are poorly understood, but there is some evidence that the IM translocation mechanism can affect the conformation of the secreted protein after its translocation [5,8,9], which could lead to misfolding and cell envelope stress.
In the periplasm, secreted proteins encounter several abundant chaperones that comprise an efficient quality control network [10–12]. Periplasmic chaperones often have overlapping substrate specificities [13], which has made it challenging to dissect contributions of individual chaperones to specific aspects of protein quality control. For example, the periplasmic chaperone DegP can function as either a chaperone or a protease [14–16]. The main substrates of the DegP protease are OM β-barrel proteins (OMPs) [17], but it has been more difficult to identify its contributions as a chaperone. As a result, most of our current understanding of the DegP chaperone activity is from in vitro studies [18–20]. During heat-shock, E. coli ΔdegP is lethal, but growth can be rescued by production of a chaperone-only DegP mutant, suggesting that DegP may function as a chaperone for OMPs during their assembly in the OM [21,22]. However, recent studies have shown that while assembly-defective OMPs are sequestered by DegP during heat shock, DegP does not promote OM assembly of OMPs and functions primarily as a protease in E. coli [17].
We serendipitously discovered that DegP is required during production of a B. pertussis autotransporter (AT) virulence protein in E. coli. AT proteins are the largest family of virulence proteins secreted from Gram-negative pathogens, including E. coli [23]. The AT secretion mechanism consists of three discrete steps (Fig 1A): (i) The N-terminal AT signal sequence mediates translocation of the AT precursor across the IM, (ii) the C-terminal β-barrel domain folds and inserts into the outer membrane (OM), where it assists with (iii) translocation of the central AT passenger across the OM to the cell surface, assisted by the β-barrel assembly machinery (BAM) and the translocation and assembly module (TAM) [24–31]. After OM translocation, the AT passenger is typically cleaved from its β-barrel [32]. Much work has been devoted to understanding the OM translocation portion of AT secretion, but details of earlier steps, including IM translocation, are less well understood. We and others have shown that the AT passenger remains in an unstable, non-native conformation while it resides in the periplasm [33–36]. This unstable, non-native conformation is required for the passenger to remain compatible with OM translocation [24,29,37,38]. ATs therefore represent unique model substrates to dissect how the E. coli periplasmic chaperone network handles the accumulation of unstable, unfolded substrates during AT secretion.
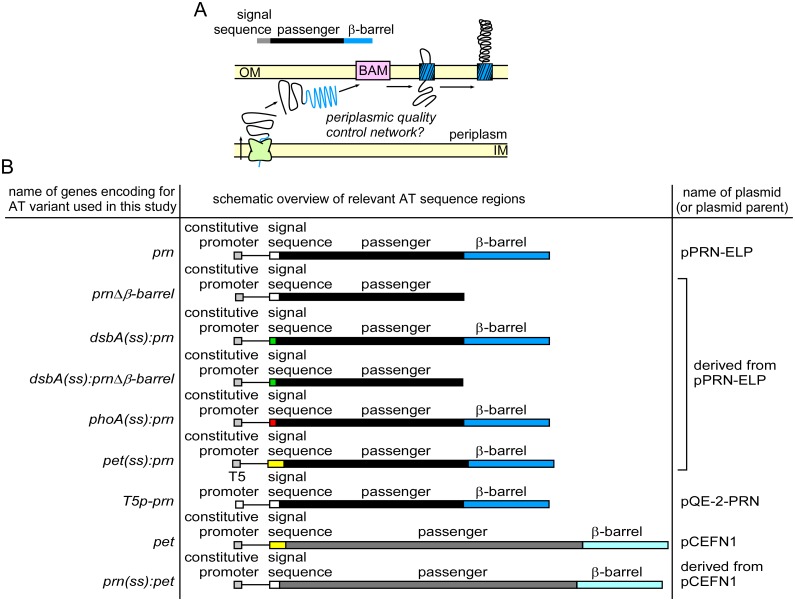
Fig 1. The autotransporter (AT) secretion pathway.
(A) Schematic overview of AT secretion mechanism. The passenger domain remains in a non-native, unfolded conformation during transit across the periplasm. (B) Constructs used in this study. The lengths of the AT variants are drawn to scale and include annotations of promoters and regions encoding annotated domains.
We found that the DegP chaperone activity is required during IM translocation of the B. pertussis pertactin AT passenger in E. coli. We used this model system to investigate the basis for DegP dependent growth and found that changing the AT IM translocation mechanism from post- to co-translational was sufficient to relieve the DegP chaperone requirement. An analysis of the proteome revealed that failure of the σE periplasmic stress response was sufficient to explain lethality. These results provide unique insights into the periplasmic protein quality control network and bacterial cell death triggered by early steps in secretion of an extracellular protein.
Materials and Methods
Construction of plasmids encoding autotransporter (AT) variants and summary of E. coli strains
The genotypes of the AT variants used in this study and the plasmids used to express these genes are summarized in Fig 1B. A plasmid to express the pertactin passenger alone was described previously as pPERPLC01; the protein product accumulates as inclusion bodies in the E. coli cytosol [39]. Plasmid pCEFN1 was used to express pet under the control of its endogenous promoter [40]. All plasmids were constructed using standard molecular biology techniques. All primers were purchased from IDT. E. coli DH5α (Invitrogen) was used for cloning procedures.
Plasmid pPRN-ELP, to express prn from a constitutive tac promoter, was generated from its parents pP.93WT [29] and pPERPLC01 [39]. Three point mutations present in pP.93WT were removed by ligating a portion of the gene encoding the passenger region from pPERPLC01 into pP.93WT. To do this, both plasmids were digested using at the unique KpnI and RsrII sites and the encompassing region from pPERPLC01 with the correct sequence was ligated in pP.93WT. As a result, prn differs from the gene encoded by pP.93WT in three positions (resulting in three amino acid substitutions: D262E, P264L, S332P). All prn gene variants used in this study were in this background (D262E, P264L, S332P).
Construct T5p-prn was made by cloning the prn sequence in pQE-2 (Qiagen), resulting in plasmid pQE-2-PRN. First, a BamHI site was introduced in pQE-2 by site-directed mutagenesis using primers pQE2 BamHI SDM FW and pQE2 BamHI SDM RV (S1 Fig), resulting in the plasmid pQE-2-BamHI. The BamHI site was located downstream of the region encoding an N-terminal His-tag in pQE-2 to ensure that the pertactin gene would not have a His-tag. The coding sequence of pertactin was amplified from prn using primers PCR amp P.93 BamHI FW and PCR amp P.93 HindIII RV (S1 Fig) and ligated in pQE-2-BamHI via the BamHI and HindIII sites, resulting in plasmid pQE-2-PRN.
Signal sequences of ATs were exchanged using MEGAWHOP PCR [41] and the primers listed in S1 Fig. A pertactin deletion variant corresponding to residues 631–910 lacked the region coding for the C-terminal β-barrel domain of pertactin. This mutant was generated by introducing a stop codon in the corresponding parent plasmid using primers N631Stop FW and N631Stop RV (S1 Fig).
E. coli KS303 (F- ΔlacX74, galE, galK, phoA ΔPvuII, StrR lpp) and KS476 (KS303 degP41::km) are derivatives of MC1000, described previously [38,42]. Both strains were cultured at 30°C. The E. coli chaperone deletion strain series are derivatives of BW30270, described previously [43]. The following deletion strains from this series were used for colony forming unit (CFU) measurements as described below: JB4 (fkbA::cm), JB117 (skp::kan), and JB8 (degP::kan).
Growth curves
All cultures were grown at 30°C. To compare growth of different strains in parallel, cultures were first normalized during inoculation from overnight cultures as follows: The overnight culture with the highest optical density (OD600) was inoculated at a 1:50 dilution and the remaining cultures were normalized to have the same starting OD600. After growth to log phase (~OD600 = 0.4), fresh cultures were once again started to ensure that all cultures started at a similar growth phase. For the second inoculation, the equivalent of a 1:50 dilution from a culture corresponding to OD600 = 2.5 was used. The growth curve after this second inoculation was recorded.
Cell viability assay
To quantify the phenotypic effects of ATs on E. coli growth in different genetic backgrounds, the viability of E. coli (KS303 and KS476) was quantified after plasmid transformations. In each transformation reaction, 1 μL of plasmid DNA from a 50 ng μL-1 stock was added to 50 μL of chemically competent E. coli. In some cases, a plasmid encoding a variant of degP (degP S210A) expressed from a trc promoter (pCS10) [16] was co-transformed. The gene product from this degP variant is a protease deficient version of DegP. After heat shock transformation for 45 s at 42°C, 200 μL of SOC medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) was added and cells were allowed to recover for 1 h at 30°C. Transformation reactions were diluted in SOC medium and plated on LB-agar supplemented with appropriate antibiotics (100 μg mL-1 ampicillin (Amp) for single transformations with plasmids encoding AT variants, plus 100 μg mL-1 and 35 μg mL-1 chloramphenicol (Cam) for co-transformations with pCS10).
Mass spectrometry
E. coli KS303 or E. coli KS476 transformed with pQE-2-PRN (Fig 1B) were grown at 30°C. For each sample, 200 mL LB medium supplemented with 100 μg mL-1 Amp was inoculated with an overnight culture at a 1:50 dilution. Expression of prn was induced with 0.5 mM IPTG at the indicated OD600, and cells were pelleted at the indicated times. Cell pellets were lysed using a bead beater (BioSpec) in 50 mM Tris pH 8.0, 1% TritonX-100, 150 mM KCl, 1mM EDTA, 1mM PMSF, and clarified by centrifugation at 12,000 x g for 12 min.
Total protein were quantified using Micro BCA (Pierce) according to the manufacturer’s instructions, and 100 μg of each lysate was precipitated and digested with trypsin using the FASP protocol as described previously [44–46]. All reagents were purchased from Sigma unless otherwise indicated. Briefly, Tris(2-carboxyethyl)phosphine-reduced and iodoacetamide alkylated samples were buffer-exchanged using filtration concentrators (Amicon; 10,000 MW cutoff) with five washes of 8 M urea in 100 mM Tris pH 8.0, then exchanged with two washes of 50 mM ammonium bicarbonate pH 8.0. One μg of trypsin (Promega) was added to each sample and digested at 37°C for 8 h. Peptides were recovered from the filtration concentrator by spinning and a second spin with addition of 100 μl of 500 mM NaCl and quenched with addition of 5% trifluoroacetic acid. Peptide mixtures were desalted using C18 SPE spin columns (Protea Sciences), dried and resuspended in 0.2% formic acid for LC/MS/MS analysis.
LC/MS/MS was performed as described previously [44]. Briefly, 750 ng of each sample was analyzed in duplicate and separated over a 90 min gradient using 3–40% water, 0.1% formic acid—acetonitrile, 0.1% formic acid (Honeywell Burdick & Jackson) using a 100 μm x 100 mm C18BEH column (Waters) running at 900 nL min-1. MS/MS was performed on an LTQ Velos Orbitrap mass spectrometer running a TOP12 method.
Proteomic data analysis
MS/MS files were searched using the Andromeda search engine [47] against the current E. coli FASTA database supplemented with laboratory contaminants and the sequence of pertactin (B. pertussis) produced in these strains. Deamidation of NQ, oxidation of M, carboxyamidomethyl-C, and N-terminal acetylation were considered as modifications. The peptide-spectral matches false-discovery rate (FDR) was set to 0.01%. Protein FDR was set to 0.02% using target-decoy false discovery calculation [48].
Protein quantification was performed using label-free-quantification based on extracted ion chromatograms. MaxLFQ within MaxQuant v 1.5.2.8 software was used for peak integration and normalization [49]. Suggested settings were used for integration. Two or greater quantified peptides per protein were required and normalization against total protein and corrected mass spectrometry response was performed. Deletion of DegP was readily verified within these quantifications (panel A in S2 Fig). Quantitative data is given as the log2 transformed fold-change versus the E. coli KS303 strain after 2 h of pertactin induction. Error bars are plotted as standard error (∑/mean) for each label-free measurement. RAW data files, search results, search parameters and databases were deposited into MassIVE and are available for download at massive.ucsd.edu with the identifier MSV000080095 or ftp://MSV000080095@massive.ucsd.edu.
To estimate abundance of periplasmic proteins, the 1000 most abundant proteins across all conditions were analyzed further. Out of this set, 39 proteins have ‘periplasm’ in their name. These proteins were used as a proxy for all periplasmic proteins. The quantity of these periplasmic proteins during death (5 h post induction, KS476) was compared to the control condition (2 h time point, KS303, no induction). Out of all 39 periplasmic proteins identified in this way, six did not change significantly or increased and one had ambiguous quantification. The remaining 32 proteins (82% of total) decreased by at least two-fold during death.
Measuring accumulation and secretion of AT variants
Accumulation and secretion of AT variants in vivo was verified by measuring the appearance and proteolytic processing of the AT precursor in cell lysates using western blotting, as described previously [24,25,29,32,50]. E. coli were transformed with plasmids encoding the indicated genes and grown overnight. The equivalent of 1 mL of cells at OD600 = 0.5 were pelleted for 5 min at 8,000 x g and resuspended in 50 μL SDS loading buffer. For cells where variants of the Pet passenger were produced, a sample of the growth medium was collected to quantify the secreted passenger. After removing bacteria by centrifugation, the spent growth medium was filtered through a 0.22 μm filter and diluted 15-fold with SDS loading buffer. WCL and spent media samples were boiled for 5 min, separated by SDS-PAGE and analyzed by western blotting as described below.
Western blotting
Samples for western blot analysis were separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) (Bio-Rad) membrane as follows. The membrane was wet in methanol, and then both the membrane and the SDS-PAGE gel were equilibrated in western transfer buffer (WTB, 25 mM Tris base, 192 mM glycine, 20% (v/v) methanol) for 10–15 min. The gel was transferred to the PVDF membrane at 4°C for 2 h at 70 V. After transfer, the membrane was equilibrated for 5 min in 20 mL Tris buffered saline Tween-20 (TBST) consisting of 20 mM Tris HCl pH 7.5, 150 mM NaCl and 0.5% (v/v) Tween-20 supplemented with 5% (w/v) powdered dry milk (PDM). The primary antibody was added to the TBST/5% PDM solution and incubated while shaking. The membrane was washed with 20 mL TBST three times to remove unbound antibody followed by incubation with the secondary antibody in 20 mL TBST/5% PDM. Then, the membrane was incubated for 30 min with an alkaline phosphatase (AP)-conjugated secondary antibody and washed three times with 20 mL TBST to remove unbound antibody. The membrane was rinsed with ddH2O five times and developed by incubation with 10 mL alkaline phosphatase (AP) buffer (100 mM Tris HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2) containing 66 μL of nitro blue tetrazolium and 33 μL of 5-bromo-4-chloro-3-indoyl-phosphate (Promega). The blot was rinsed with water to stop the reaction.
The primary antibodies were a polyclonal rabbit anti-pertactin antibody (1:7,000 dilution) [29], a polyclonal rabbit anti-Pet antibody (1:7,000 dilution) [40] or a polyclonal rabbit anti-MBP/DegP antibody (1:15,000 dilution) [51]. The secondary antibody was an AP-coupled goat anti-rabbit IgG antibody (Novus Biologicals, Littleton, CO, 1:10,000 dilution).
Results
Constitutive expression of prn, but not pet, is lethal in the absence of DegP
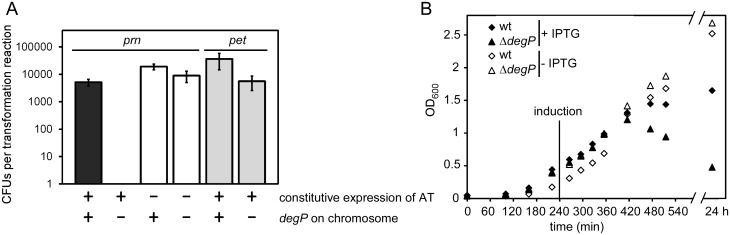
To systematically investigate the effects of deleting degP during AT expression, we compared E. coli growth during the synthesis of two dissimilar ATs, Bordetella pertussis pertactin (gene name: prn) [24,29,39] and Pet, an AT from the serine protease autotransporters of Enterobacteriaceae (SPATE) subfamily [24,40,52] (Fig 1B). Surprisingly, expression of prn in the absence of DegP was lethal: transformation of a degP null strain with a plasmid to constitutively express prn yielded no colonies (Fig 2A). Likewise, induction of prn expression in the degP deletion background resulted in a dramatic decrease in cell density (OD600; Fig 2B), a characteristic phenotype of cell death. Such conditional synthetic lethality has not previously been observed during AT expression in the absence of DegP. Past studies have indicated DegP deletion has no significant effect on E. coli growth, with or without AT expression [38]. Consistent with these studies, we observed that transforming a plasmid to constitutively express pet did not affect viability of E. coli in the absence of DegP (Fig 2A).
Fig 2. Expression of prn in an E. coli degP deletion strain is lethal.
(A) Viability of the E. coli degP null strain versus its parent strain was assessed after transforming plasmids that constitutively express AT variants. Viability was measured by counting colony forming units (CFUs); see Methods. Error bars are standard deviations from three biological repetitions. (B) Growth of the E. coli degP null strain versus its parent strain, with or without IPTG-induction of prn expression. Shown are representative growth curves.
The DegP chaperone function is required for E. coli survival during pertactin production and secretion
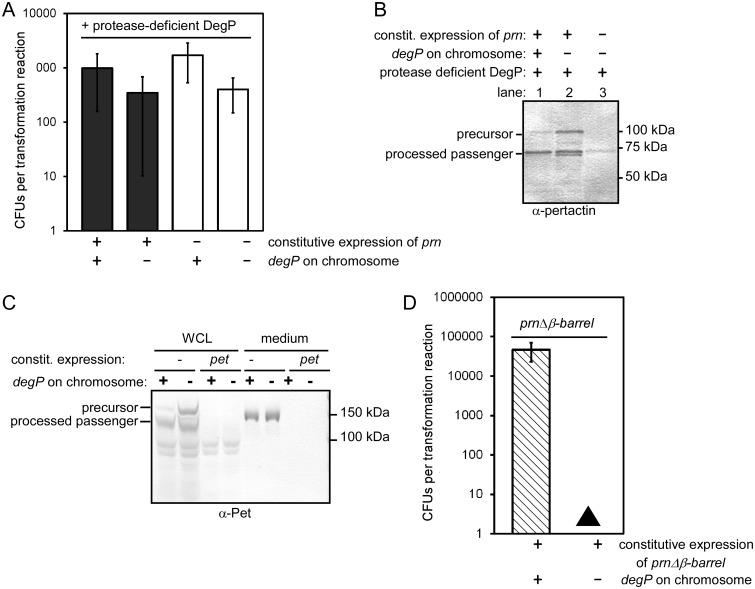
To dissect the specific contributions of the chaperone vs. protease function of DegP to the observed lethality, we first determined whether the DegP chaperone activity alone was sufficient to rescue the lethal phenotype. We transformed a plasmid to constitutively express prn in the E. coli degP deletion strain as before, but also co-transformed a plasmid encoding a protease-deficient version of DegP that retains the DegP chaperone activity (DegP S210A) [16]. Supplying the DegP chaperone function was sufficient to restore E. coli growth during prn expression (Fig 3A).
Fig 3. Differential requirement of the DegP protease and chaperone activity during expression of model ATs.
(A) E. coli viability as a function of degP, with or without constitutive expression of prn. To test if the DegP chaperone alone is sufficient to restore viability, a protease deficient version of DegP [16] was produced by co-transforming a plasmid expressing degP S210A. (B) Expression of prn results in production of the precursor and processing of the passenger (samples shown here were collected from cells described in (A). The role of the DegP protease for pertactin accumulation and secretion was assessed by expressing prn when only the DegP chaperone was present, versus the DegP chaperone plus the chromosomal wild type DegP. In the presence of the DegP protease activity, less pertactin precursor accumulates (compare lanes 1 vs. 2). (C) Wild type pet was expressed overnight in E. coli with or without a chromosomal copy of degP (same samples as grey bars in Fig 2A). In the absence of DegP, more full-length precursor accumulates in whole cell lysates (WCL). The yield of secreted Pet in the media after overnight production was not significantly altered with or without DegP. (D) After transforming prnΔβ-barrel in the E. coli degP null strain, very small colonies appeared after prolonged incubation (black triangle). (A, D) Error bars are standard deviations from at least three biological repetitions.
We next asked whether supplying the DegP chaperone rescues pertactin secretion, in addition to enabling E. coli growth. We and others have previously shown that pertactin processing is a reliable assay for secretion across the OM; secretion-impaired mutants of ATs remain unprocessed in the periplasm [24,25,28,29,53,54]. We observed that the processed pertactin passenger accumulated to similar levels when the DegP chaperone variant was produced (Fig 3B), demonstrating that the DegP chaperone is sufficient to restore growth and allow pertactin secretion in E. coli.
Next, we tested the role of the DegP protease during AT secretion in our model systems. We asked whether the pertactin precursor is also a substrate for the DegP protease while critically requiring the DegP chaperone. We constitutively expressed prn in E. coli strains with or without degP on the chromosome. To avoid the lethal phenotype described above, we also supplied the DegP chaperone activity by producing the protease deficient DegP variant from a plasmid in both strains (Fig 3B). We observed that the processed pertactin passenger accumulated to similar levels with or without the DegP protease activity (Fig 3B), indicating that the DegP protease activity was not crucial for pertactin secretion or cell viability. However, the pertactin precursor accumulated to higher levels when the protease activity of DegP was omitted (Fig 3B, compare lanes 1 and 2), indicating that DegP degraded a portion of the pertactin precursor pool. We tested the DegP protease activity during expression of pet as well (Fig 3C). As seen for pertactin, absence of the DegP protease resulted in an increase in the Pet precursor pool, but no detectable changes for passenger secretion efficiency. Taken together, these results demonstrate that DegP, when present, degrades a portion of both the pertactin and Pet precursors. This result is consistent with previous reports of DegP degradation of AT secretion intermediates in the periplasm [38,54,55].
We next tested whether prn expression was dependent on other E. coli chaperones besides DegP. To do this, we turned to an existing series of E. coli chaperone deletion strains [43]. We transformed the degP deletion strain from this series (JB8) with a plasmid constitutively expressing prn and confirmed that, as for the degP deletion strain KS476, transformation with the prn plasmid results in lethality (panel A in S3 Fig). In contrast, strains lacking FkpA (JB4; fkpA::cm) or Skp (JB117; skp::kan) were viable after transformation (panel A in S3 Fig) and led to efficient secretion of pertactin (panel B in S3 Fig). These results demonstrate that despite the known overlapping substrate specificity amongst periplasmic chaperones [13], lethality upon prn expression in E. coli is related to a specific requirement for the DegP chaperone activity.
Lethality in the absence of DegP occurs prior to pertactin OM translocation
Previous reports suggested that DegP contributes a quality control function during AT OM translocation [37,56,57]. To test whether the lethality of prn expression in the absence of DegP is related to pertactin OM translocation, versus an earlier step in secretion, we constructed prnΔβ-barrel (Fig 1B). Because the product of this construct lacks the C-terminal OM β-barrel, it will neither integrate into the OM nor will the passenger translocate across the OM [58]. If lethality was related to pertactin β-barrel OM assembly or OM translocation, deleting this domain should reverse the lethal phenotype. However, expression of prnΔβ-barrel in the degP deletion strain resulted in a severely toxic phenotype (Fig 3D), yielding only extremely small colonies after prolonged incubation (see Table 1 for a comparison of relevant phenotypes). The severe growth defect of prnΔβ-barrel in the absence of DegP indicates that DegP plays a crucial role in early steps of pertactin secretion, prior to OM translocation.
Table 1. Summary of phenotypes observed upon AT expression in the E. coli degP deletion strain.
| prn | phoA(ss):prn | prnΔβ-barrel | pet(ss):prn | prn(ss):pet | dsbA(ss):prnΔβ-barrel | dsbA(ss):prn | pet | |
|---|---|---|---|---|---|---|---|---|
| Cell growth | - | - | + | ++ | +++ | +++ | +++ | +++ |
| Precursor accumulation | N/A | N/A | ++ | ++ | + | +++ | +++ | ++++ |
| Passenger secretion | N/A | N/A | N/A | ++ | + | N/A | +++ | +++ |
A phenotype similar to expression in the wild type E. coli parent strain was scored as (+++), whereas absence of the phenotype in comparison to the parent strain was scored as (-). Note that precursor accumulation of wild type Pet is increased in the degP deletion strain versus the parent (++++). See Fig 1B for details on these constructs.
Fusion of pertactin to a co-translational signal sequence is sufficient to suppress DegP chaperone dependency
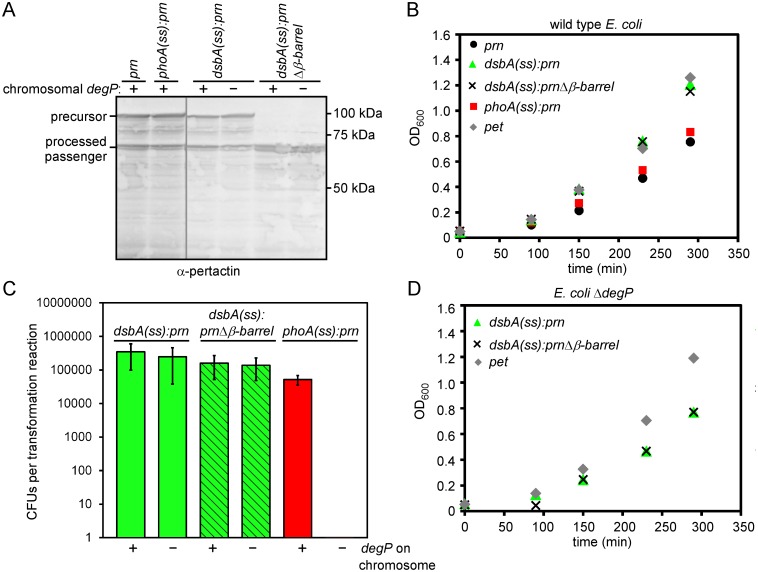
We next sought to identify what features of pertactin, but not Pet, trigger lethality in the degP deletion background during early secretion steps. A notable difference lies within the signal sequences: the Pet signal sequence has an N-terminal extension specific to the SPATE sub-family of AT proteins [59,60]. Although the precise function of this N-terminal signal sequence extension remains unclear, it has been proposed to slow down the rate of IM translocation [60–65]. To systematically test the effects of altering the IM translocation mechanism on DegP-dependent prn expression, we replaced the pertactin signal sequence with a well-characterized signal sequence from dsbA or phoA, to generate dsbA(ss):prn and phoA(ss):prn (Fig 1B). DsbA is known to be translocated co-translationally across the IM [66], whereas PhoA IM translocation is mainly post-translational [5,67]. Both of these E. coli signal sequences are routinely used to target proteins to the periplasm [67–69]. As expected, neither of these signal sequences altered pertactin production or secretion in the parent E. coli strain (Fig 4A). Likewise, we observed no significant changes in growth rates (Fig 4B). However, we found that replacing the pertactin signal sequence with the co-translational signal sequence of dsbA was sufficient to bypass the lethality of expressing prn in the degP deletion strain and restore pertactin secretion (Fig 4). In contrast, fusion to the PhoA signal sequence was lethal (Fig 4C). The lethality of phoA(ss):prn in in the degP deletion background indicates that post-translational translocation of pertactin across the IM is sufficient to recapitulate the conditional synthetic lethality observed for pertactin in the absence of DegP. Importantly, both dsbA and phoA are endogenous to E. coli. Recapitulation of both the lethal and viable phenotypes when these signal sequences were fused to pertactin confirms that the phenotype is directly linked to co- vs. post-translational IM translocation in E. coli.
Fig 4. The co-translational DsbA signal sequence, but not the post-translational PhoA signal sequence, bypasses lethality.
(A) Production and secretion of pertactin chimera constructs in the E. coli degP null strain versus its parent. Bacteria were grown to stationary phase and protein production was analyzed by western blotting of whole cell lysates. Successful secretion of the pertactin passenger is measured by appearance of the processed passenger [24,25,29,32,50]. Two non-adjacent portions of the same blot are shown (grey line). (B) Expression of signal sequence chimera constructs does not dramatically affect growth of wild type E. coli. (C) Plasmids to express dsbA(ss):prn and phoA(ss):prn under control of a constitutive promoter were transformed in the E. coli degP null mutant versus its parent as described in Fig 2A. Error bars are standard deviations from at least three biological replicates. (D) Expression of dsbA(ss):prn, dsbA(ss):prnΔβ-barrel and pet does not affect growth in the degP null strain. (B, D) Shown are representative growth curves.
The extended Pet signal sequence can partially rescue lethality of pertactin in the absence of degP
The dsbA(ss):prn and phoA(ss):prn results indicate that the nature of the signal sequence or the resulting differences in IM translocation mechanism largely determined the DegP dependence, leading us to closely examine the differences in the Pet and pertactin signal sequences. Our current understanding of AT IM targeting is based on studies of the SPATE sub-family of AT proteins, all of which share the N-terminal signal sequence extension described above [59,60]. A survey of the literature revealed that DegP is dispensable during production of ATs with extended signal sequences, including IcsA [56,57,70], Hbp [34,38,54,55,64], EspP, Pet and other SPATEs [37], as well as the two-partner secretion (TPS) protein FHA [71,72]. Furthermore, the SPATE signal sequence extension appears to reduce the rate of post-translational IM translocation and/or release of the AT from the IM [60–65]. We hypothesized that, like the co-translational DsbA signal sequence, the extended SPATE signal sequence could similarly reduce DegP dependence.
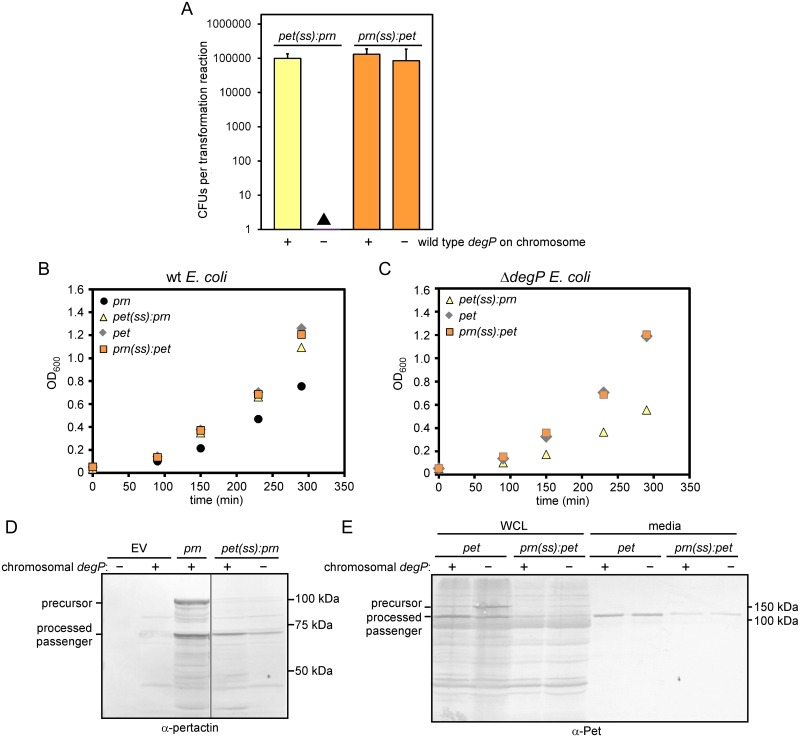
To test whether the extended signal sequence from Pet can rescue lethality in pertactin, we replaced the region of prn encoding the signal sequence with the region encoding the Pet signal sequence (resulting in construct pet(ss):prn; Fig 1B) and transformed the plasmid expressing this chimera into the degP deletion strain to assess viability. We observed small colonies after prolonged incubation, but these did not appear within the timeframe required to qualify as CFUs in our assay (Fig 5A). Note that, in contrast, we did not detect any colonies, even very small ones, when prn was transformed into the degP deletion strain (Fig 2A). We hypothesized that the colonies resulting from transformations with the plasmid harboring pet(ss):prn might be viable, albeit with a strong growth defect. We tested growth of these small colonies in liquid culture. As a control, we transformed the E. coli parent strain with the plasmid to express pet(ss):prn and no growth defect was observed in the E. coli parent (Fig 5B). As hypothesized, we were able to grow the degP deletion strain transformed with a plasmid to express pet(ss):prn in culture (Fig 5C), albeit at a slow growth rate (see also Table 1). These results suggest that fusing the Pet signal sequence to pertactin improves viability of the degP deletion strain, albeit to a limited extent. To test directly if pertactin secretion was also rescued in this chimera context, we measured pertactin accumulation and secretion as described above. Indeed, mature pertactin accumulated in the degP deletion strain, although the secretion yield for the chimera was reduced (Fig 5D). The overall slower growth rate (Fig 5C) likely contributes to this reduced secretion yield. Taken together, these results indicate that the extended Pet signal sequence can partially rescue the lethal phenotype of pertactin in E. coli, although significant toxicity remains.
Fig 5. The Pet signal sequence partially rescues lethality of pertactin in the degP null strain.
(A) The degP null strain and its parent were transformed with plasmids to express pet(ss):prn or prn(ss):pet as described in Fig 2A. Error bars are standard deviations from at least three biological replicates. Filled triangle: After transforming a plasmid to express pet(ss):prn in the degP deletion strain, very small colonies appeared after prolonged incubation. (B) Growth of E. coli with a chromosomal copy of degP is not significantly altered when AT signal sequence chimera variants are produced. (C) Expression of pet and prn(ss):pet does not affect growth in the degP deletion background, whereas expression of pet(ss):prn reduces the growth rate. (D) The chimera pet(ss):prn was expressed and the chimera protein was secreted in E. coli regardless of the presence or absence of DegP, but the secretion yield was reduced in the degP deletion strain. Two non-adjacent portions of the same blot are shown (grey line). (E) Expression of prn(ss):pet resulted in accumulation of the chimera protein at reduced levels in E. coli. Likewise, secretion of the Pet passenger to the medium was reduced, regardless of the presence or absence of DegP. (B, C) Shown are representative growth curves.
We next tested the reverse hypothesis: whether fusion of the pertactin signal sequence to Pet was sufficient to induce toxicity in the degP null strain. We transformed a plasmid encoding prn(ss):pet (Fig 1B) in the degP deletion strain and observed cell growth similar to cells transformed with wild type pet (Fig 5A and 5C). In contrast to wild type Pet, there was a dramatic reduction of Pet precursor accumulation in cell lysates and mature Pet passenger in the media (Fig 5E). A similar reduction was also observed when the chimera was produced in the E. coli parent strain. Taken together, these results indicate that E. coli growth is not DegP-sensitive when Pet is produced with a pertactin signal sequence. Reduced accumulation of the chimera may permit viability.
Loss of periplasmic proteins during prn-induced cell death
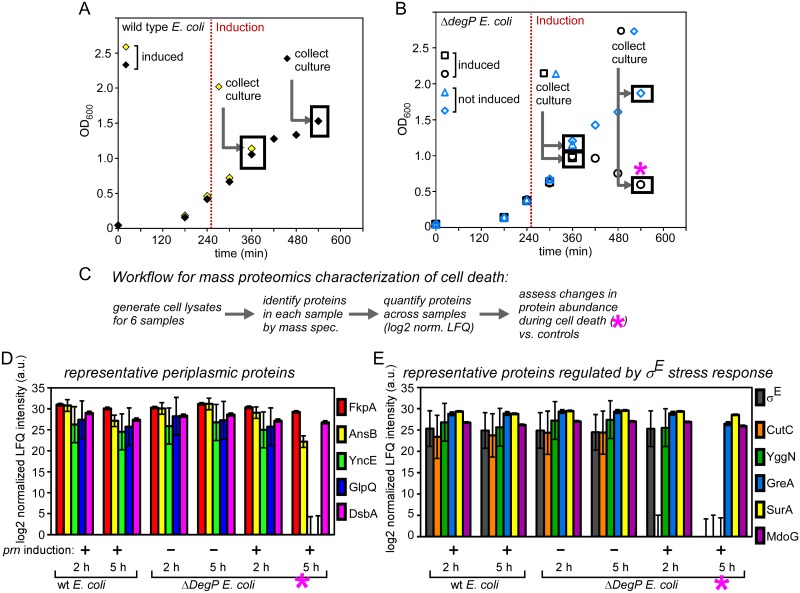
The results above demonstrate that in the absence of DegP, post-translational IM-translocation of pertactin is lethal. This finding was unexpected: There is no precedent in the literature for a link between DegP dependence, the IM translocation mechanism and cell death. To broadly explore the underlying cell death mechanism, we used quantitative mass spectrometry to measure changes in protein abundance across the proteome upon induction of this lethal phenotype. Defects in growth upon induction of prn in the degP deletion strain were first detectable 2 h post-induction, indicating that this represents an early stage of cell death (Fig 2B). At 5 h post induction, the OD600 had decreased significantly, indicating lethality. To capture both early and advanced stages of cell death, we analyzed changes to the proteome at both 2 h and 5 h after prn induction. Cells were grown as described previously (Fig 2B), and we collected samples for proteomic analysis at the chosen time points (Fig 6A and 6B). We compared prn induction in the degP deletion strain versus its parent strain and also included a control condition where prn expression was not induced in the degP deletion strain. Together, we collected samples in these three different conditions at 2 h and 5 h post induction, resulting in six samples overall (see Fig 6A and 6B for details).
Fig 6. Analysis of proteome-wide changes during pertactin-induced death of E. coli ΔdegP.
(A, B) Expression of prn was induced in the presence (A) or absence (B) of a chromosomal copy of degP and cells were collected at 2 and 5 h post induction for mass spectrometry. For wild type E. coli, two samples were collected for mass spectrometry analysis (2 h and 5 h post prn induction). For the degP deletion strain, a total of four samples were collected (2 h and 5 h post prn induction and uninduced controls at the same time points). To express prn, construct T5p-prn was used and expression was induced in mid-log phase (see also Fig 2B). (C) Workflow to generate dataset for unbiased analysis of the proteome during E. coli death. (D) Quantification of representative periplasmic proteins across all experimental conditions indicates that overall periplasmic proteins decrease during death (last condition, see pink star in (B, C)). (E) Quantification of proteins in the σE stress response pathway reveals a loss of σE during death (last condition, pink star). Note that the σE protein itself along with CutC and YggN are undetectable in the last condition. Error bars correspond to the standard error (∑/mean) for each label-free measurement.
Quantification of the entire proteome across our experimental conditions was performed using normalized label-free-quantification (LFQ), where measured peak areas from peptides specific to each protein were integrated, averaged and normalized [45,49]. Overall, 1897 proteins were quantitatively described across all mass spectrometry runs, representing approximately 50% of the total E. coli proteome (see Fig 6C and Methods for details).
Our results above indicated that lethality was caused by a periplasmic defect, motivating us to investigate changes to the periplasmic proteome during cell death. Of the most abundant periplasmic proteins across our experimental conditions, 82% (32 proteins) were ≥2-fold less abundant (or absent entirely) in the degP deletion strain at 5 h post prn induction. Representative examples are shown in Fig 6D. The striking reduction in the abundance of most periplasmic proteins suggests severe stress within the cell envelope. In contrast, we detected no loss of outer membrane porins (including OmpC, LamB and BamA) during prn expression in the absence of degP, and similarly stable levels for cytosolic markers including σ70 and DnaK (panel B in S2 Fig). These results are consistent with a model where expression of prn in the absence of degP disrupts protein homeostasis the periplasm.
Collapse of the σE stress response during prn-induced cell death
The loss of periplasmic proteins as cells approached death led us to hypothesize that dysfunction of one or both of the two major periplasmic stress responses, σE and/or Cpx [73], might trigger lethality. We first investigated the σE stress response, in which the alternative sigma factor σE regulates expression of downstream genes [74–77]. Surprisingly, the σE protein itself was undetectable in the degP deletion strain 5 h after the induction of prn, but was detected in all other control conditions (Fig 6E). From this we conclude that the σE pathway is inactivated during death. Loss of σE could arise due to its degradation and/or reduced production. Alternatively, the σE protein could be sequestered at the IM where it becomes more difficult to detect using mass spectrometry. Indeed, when the σE pathway is inactivated the σE protein is sequestered at the IM [78]. Either of these scenarios is consistent with our data and would result in inactivation of the σE pathway. Since cytosolic σE is an essential component of E. coli [79], its absence is sufficient to explain the lethality of prn expression in the absence of DegP [80].
To test further for the activity of the σE response in our experimental conditions, we quantified 25 σE-dependent gene products [74] detected in our dataset. We found that at least 13 σE-dependent gene products are less abundant (≥2-fold) or undetectable in the degP deletion strain upon induction of prn (representative examples are shown in Fig 6E). These results indicate that collapse of the σE response contributes to lethality when pertactin accumulates in the periplasm in the absence of degP.
Next we investigated the Cpx pathway, which leads to degP upregulation upon cell envelope stress [81]. Although we identified 26 of the known Cpx-regulated proteins [82] in our proteomics dataset, most were of low abundance and therefore unsuitable for reliable quantification. Nevertheless, six Cpx substrates could be quantified to high reliability (panel C in S2 Fig). Although we observed some changes in these Cpx substrates during induction of prn (panel C in S2 Fig), these changes were less severe than the complete abrogation of σE and some of its substrates. Although we cannot rule out contributions from Cpx towards lethality, the observed changes in Cpx alone are insufficient to explain the lethal phenotype in E. coli. In contrast, the complete loss of σE, which is essential, and loss of several σE-dependent essential genes is sufficient to explain lethality.
Discussion
It has previously been suggested that periplasmic chaperones participate in late steps of AT secretion, ensuring that the AT passenger remains in a high-energy, secretion competent conformation required for efficient transfer to BAM or TAM [25,31,53,83–86]. In particular, previous studies revealed a variety of effects that hint at an interplay between DegP and AT secretion, including increased degP expression upon AT expression [34,38,55], defects in cell growth and AT secretion in degP null strains [37,56,57] and binding of DegP to an AT passenger in vitro [37]. However, the results presented here represent the first identification of a lethal phenotype during AT expression in a degP deletion background. Moreover, to our knowledge this is the first observation of a link between the function of a periplasmic chaperone and the IM translocation mechanism of a secreted protein in Gram-negative bacteria
The mechanistic connection between substrate IM import mechanism and DegP dependency may arise due to differences in IM translocation kinetics. Co-translational IM translocation is reported to be slower than classical post-translational IM translocation [5,8]. Hence changing the IM translocation rate could alter the conformation of the passenger as it enters the periplasm, akin to altered co-translational folding under different translation rates [87–89] (Fig 7). We propose a model where fast IM translocation via the PhoA signal sequence results in the rapid appearance of the substrate in the periplasm and subsequent accumulation of toxic conformations. These toxic conformations can be masked or avoided by DegP. Alternatively, reducing the rate of appearance of the passenger in the periplasm can avoid toxicity, for example during the slower, co-translational IM translocation of proteins bearing the DsbA signal sequence (Fig 7). This model predicts that the IM translocation mechanism and DegP work together to maintain E. coli secreted proteins in a non-native, non-toxic unfolded conformation.
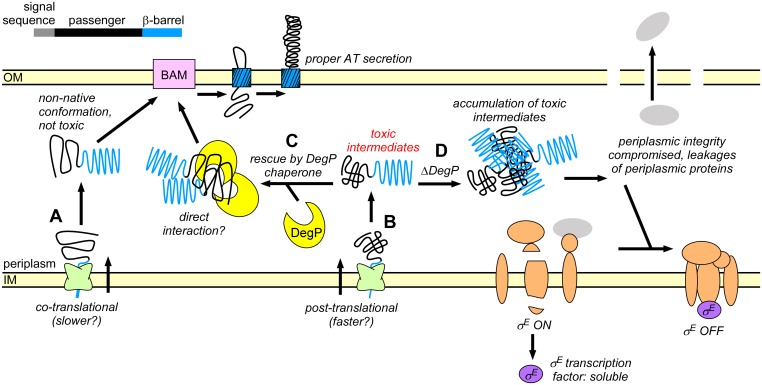
Fig 7. Model for the interplay between IM translocation, DegP chaperone activity, and the σE stress response.
(A) Co-translational IM translocation results in passenger conformations in the periplasm that are tolerated in the absence of DegP. (B) Post-translational IM translocation results in conditional lethal AT conformations in the periplasm. (C) Lethality can be prevented by interactions with the chaperone DegP. (D) In the absence of DegP, accumulation of pertactin intermediates compromises periplasmic integrity and results in leakage of periplasmic material. This leads to inactivation of the σE stress response. σE inactivation is lethal and causes cell death.
Consistent with this model, we found that swapping the pertactin N-terminal signal sequence with the co-translational DsbA signal sequence was sufficient to restore E. coli growth in the absence of DegP, whereas the mainly post-translational PhoA signal sequence recapitulated the lethal phenotype. It is important to point out that both the DsbA and PhoA signal sequences are endogenous to E. coli. Hence while we cannot exclude that expressing the heterologous B. pertussis prn AT in E. coli may exacerbate lethality, the observation that these E. coli signal sequences were sufficient to recapitulate or reverse the phenotype demonstrates that the IM translocation mechanism is sufficient to alter the fate of a secreted protein in the E. coli periplasm.
Independent of the AT signal sequence, we found that properties of the AT passenger also contribute to lethality to some extent. Fusing the Pet signal sequence to the pertactin passenger only partially rescued the lethal phenotype, suggesting that the sequence and/or conformational properties of the AT passenger contribute to the viability phenotype (Fig 5). Although the pertactin signal sequence alone was not sufficient to render the Pet AT passenger lethal in the absence of DegP, these findings are ambiguous due to the dramatically reduced level of Pet accumulation (Fig 5E). Taken together, we found that ATs can avoid DegP dependence when at least one of two criteria are met: They are either translocated across the IM co-translationally, and/or have passenger folding properties capable of avoiding conformations that lead to lethality in the absence of DegP (Fig 7).
We characterized the lethality caused by pertactin synthesis in the degP deletion strain in an unbiased manner by measuring broad changes in the proteome. Although many periplasmic proteins were depleted during lethality in the absence of DegP (Fig 6D), we did not detect significant changes in the abundance of cytosolic or OM components (panel B in S2 Fig). Prior to pertactin production, we observed measurable levels of σE and σE-induced proteins, which appears constitutive under our growth conditions. In contrast, we observed nearly complete inactivation of the σE stress response in the degP deletion strain upon prn induction. These results are consistent with lethality propagated from the periplasm, triggered by the periplasmic conformation of the pertactin passenger in the absence of DegP, which leads to collapse of the σE periplasmic stress response (Figs 6E and 7). Inactivation of σE could either be cause or consequence of leakage of soluble periplasmic material. The σE response is induced when DegS binds unfolded substrate proteins [75]. Therefore, its inactivation may start by release of DegS from its substrates, perhaps due to their dilution during periplasm leakage (Fig 7). In this scenario, unbound and inactive DegS prevents degradation of RseA, resulting in sequestration of the σE protein at the IM to RseA [75,80]. Membrane sequestration of the σE protein would also make it less likely to be detected by mass spectrometry, which could also lead to its absence in our proteomics assay. Consistent with this model, we also did not detect significant levels of other IM components of the σE machinery (DegS, RseA, RseP).
We were initially surprised that outer membrane proteins were largely unaffected by σE loss, since BamA and other OMPs are generally positively and negatively regulated by σE [74] and OMP levels overall were stable even after 5 h of prn expression. However, our results mirror those of Ades and coworkers, who found that an induced suppressor of σE function produced an analogous lethal phenotype with no apparent defects in outer membrane proteins [80]. Presumably, the unusually high stability of native OMPs [31,90] reduces their turnover rate, making them less susceptible to σE loss on our experimental time scale. Together, our unbiased characterization of changes across the proteome as cells approach death provides a unique view on time-resolved, global responses from the E. coli proteome when the periplasmic chaperone network collapses.
It is important to note that because the lethal DegP-dependent phenotype reported here may arise due to high-level production of a heterologous AT protein in E. coli, these results may have important implications for exploiting the AT secretion mechanism to produce heterologous proteins via autodisplay [91–93]. Furthermore, some ATs, including pet, are naturally encoded on plasmids. Differential reliance upon DegP could therefore prevent a Gram-negative pathogen from acquiring a plasmid via horizontal gene transfer. It will be important to investigate whether other Gram-negative species have a similar dependence upon DegP.
Supporting Information
(TIF)
(A) Quantification of DegP across all experimental conditions. (B) Quantification of representative marker proteins for the cytosol (σ70 and DnaK), the OM (LamB and BamA) and the OM-associated BamB. (C) Quantification of detectable proteins regulated by the Cpx stress response. No systematic decrease or increase of these Cpx-regulated proteins is observed during death (last condition, prn induction for 5 h in E. coli DegP deletion strain). Error bars correspond to the standard error (∑/mean) for each label-free measurement.
(TIF)
(A) E. coli deletion strains from the BW30270 series [43] were transformed with plasmids that constitutively express prn or pet, as described in Fig 2A. Error bars in bold are standard deviations from at least three biological replicates. The vertical lines for pet transformed degP::kan indicates the spread of two biological replicates. (B) prn is expressed and the passenger is secreted in fkpA::cm and skp::kan. As a control the parent strain (BW30270) was used. Secretion is routinely assayed by the production of the processed passenger in whole cell lysate samples.
(TIF)
Acknowledgments
We thank I. Drobnak, J. Renn, P. Champion, A. Buskirk and M. Ehrmann for helpful discussions. We thank the Mass Spectrometry and Proteomics Facility at Notre Dame for expert technical assistance, J. Beckwith for E. coli strains KS303 and KS476 and the anti-MBP/DegP antibody, and M. Ehrmann for plasmid pCS10 and strains E. coli BW30270, BW30270 skp::kan (JB117), BW30270 degP::kan (JB8), BW30270 fkpA::cm (JB4).
Data Availability
RAW data files, search results, search parameters and databases were deposited into MassIVE and are available for download at massive.ucsd.edu with the identifier MSV000080095 or ftp://MSV000080095@massive.ucsd.edu.
Funding Statement
This project was supported by the National Institutes of Health (US) grant R01 GM097573 to PLC. EB was supported by a Notre Dame CBBI Graduate Fellowship (National Institutes of Health grant T32 GM075762). JLC was supported by a Clare Boothe Luce Graduate Fellowship from the Henry Luce Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Braselmann E, Chaney JL, Clark PL. Folding the proteome. Trends Biochem Sci. 2013;38(7):337–44. 10.1016/j.tibs.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–67. 10.1146/annurev.biochem.77.061606.160747 [DOI] [PubMed] [Google Scholar]
- 3.Chatzi KE, Sardis MF, Karamanou S, Economou A. Breaking on through to the other side: protein export through the bacterial Sec system. Biochem J. 2013;449:25–37. 10.1042/BJ20121227 [DOI] [PubMed] [Google Scholar]
- 4.Cross BCS, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10(4):255–64. 10.1038/nrm2657 [DOI] [PubMed] [Google Scholar]
- 5.Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138(6):1164–73. 10.1016/j.cell.2009.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31(10):563–71. 10.1016/j.tibs.2006.08.004 [DOI] [PubMed] [Google Scholar]
- 7.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol. 2005;187(9):2983–91. 10.1128/JB.187.9.2983-2991.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuyama S, Fujita Y, Sagara K, Mizushima S. Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochim Biophys Acta—Protein Struct Mol Enzymol. 1992;1122:77–84. 10.1016/0167-4838(92)90130-6 [DOI] [PubMed] [Google Scholar]
- 9.Pugsley AP. The complete general secretory pathway in Gram-negative bacteria. Microbiol Rev. 1993;57(1):50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–46. 10.1111/j.1365-2958.2005.04674.x [DOI] [PubMed] [Google Scholar]
- 11.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. Protein quality control in the bacterial periplasm. Annu Rev Microbiol. 2011;65:149–68. 10.1146/annurev-micro-090110-102925 [DOI] [PubMed] [Google Scholar]
- 12.Goemans C, Denoncin K, Collet JF. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta—Mol Cell Res. 2014;1843(8):1517–28. 10.1016/j.bbamcr.2013.10.014 [DOI] [PubMed] [Google Scholar]
- 13.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. 2007;2473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa J, Heuck A, Ehrmann M, Clausen T. Molecular transformers in the cell: lessons learned from the DegP protease-chaperone. Curr Opin Struct Biol. 2010;20(2):253–8. 10.1016/j.sbi.2010.01.014 [DOI] [PubMed] [Google Scholar]
- 15.Huber D, Bukau B. DegP: a protein “death star”. Structure. 2008;16(7):989–90. 10.1016/j.str.2008.06.004 [DOI] [PubMed] [Google Scholar]
- 16.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97(3):339–47. 10.1016/s0092-8674(00)80743-6 [DOI] [PubMed] [Google Scholar]
- 17.Ge X, Wang R, Ma J, Liu Y, Ezemaduka AN, Chen PR. DegP primarily functions as a protease for the biogenesis of beta-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J. 2014;281:1226–40. 10.1111/febs.12701 [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Sauer RT. Cage assembly of DegP protease is not required for substrate-dependent regulation of proteolytic activity or high-temperature cell survival. Proc Natl Acad Sci USA. 2012;109(19):7263–8. 10.1073/pnas.1204791109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen QT, Bai XC, Chang LF, Wu Y, Wang HW, Sui SF. Bowl-shaped oligomeric structures on membranes as DegP’s new functional forms in protein quality control. Proc Natl Acad Sci USA. 2009;106(12):4858–63. 10.1073/pnas.0811780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jomaa A, Damjanovic D, Leong V, Ghirlando R, Iwanczyk J, Ortega J. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J Bacteriol. 2007;189(3):706–16. 10.1128/JB.01334-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra R, CastilloKeller M, Deng M. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182(17):4882–8. 10.1128/JB.182.17.4882-4888.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CastilloKeller M, Misra R. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. J Bacteriol. 2003;185(1):148–54. 10.1128/JB.185.1.148-154.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celik N, Webb CT, Leyton DL, Holt KE, Heinz E, Gorrell R, et al. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS ONE. 2012;7(8):e43245 10.1371/journal.pone.0043245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renn JP, Junker M, Besingi RN, Braselmann E, Clark PL. ATP-independent control of autotransporter virulence protein transport via the folding properties of the secreted protein. Chem Biol. 2012;19(2):287–96. 10.1016/j.chembiol.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drobnak I, Braselmann E, Clark PL. Multiple driving forces required for efficient secretion of autotransporter virulence proteins. J Biol Chem. 2015;290(16):10104–16. 10.1074/jbc.M114.629170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braselmann E, Clark PL. Autotransporters: The cellular environment reshapes a folding mechanism to promote protein transport. J Phys Chem Lett. 2012;3(8):1063–71. 10.1021/jz201654k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10(3):213–25. 10.1038/nrmicro2733 [DOI] [PubMed] [Google Scholar]
- 28.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc Natl Acad Sci USA. 2010;107(41):17739–44. 10.1073/pnas.1009491107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71(5):1323–32. 10.1111/j.1365-2958.2009.06607.x [DOI] [PubMed] [Google Scholar]
- 30.Leyton DL, Johnson MD, Thapa R, Huysmans GHM, Dunstan R a, Celik N, et al. A mortise-tenon joint in the transmembrane domain modulates autotransporter assembly into bacterial outer membranes. Nat Commun. 2014;5:4239 10.1038/ncomms5239 [DOI] [PubMed] [Google Scholar]
- 31.Selkrig J, Leyton DL, Webb CT, Lithgow T. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta—Mol Cell Res. 2014;1843(8):1542–50. 10.1016/j.bbamcr.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 32.Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26(7):1942–52. 10.1038/sj.emboj.7601638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leyton DL, Sevastsyanovich YR, Browning DF, Rossiter AE, Wells TJ, Fitzpatrick RE, et al. Size and conformation limits to secretion of disulfide-bonded loops in autotransporter proteins. J Biol Chem. 2011;286(49):42283–91. 10.1074/jbc.M111.306118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saurí A, ten Hagen-Jongman CM, van Ulsen P, Luirink J. Estimating the size of the active translocation pore of an autotransporter. J Mol Biol. 2012;416(3):335–45. 10.1016/j.jmb.2011.12.047 [DOI] [PubMed] [Google Scholar]
- 35.Jose J, Kramer J, Klauser T, Pohlner J, Meyer TF. Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga beta autotransporter pathway. Gene. 1996;178(1–2):107–10. [DOI] [PubMed] [Google Scholar]
- 36.Kang’ethe W, Bernstein HD. Charge-dependent secretion of an intrinsically disordered protein via the autotransporter pathway. Proc Natl Acad Sci USA. 2013;110:E4246–55. 10.1073/pnas.1310345110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol. 2009;191(21):6571–83. 10.1128/JB.00754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jong WSP, ten Hagen-Jongman CM, den Blaauwen T, Slotboom DJ, Tame JRH, Wickström D, et al. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol Microbiol. 2007;63(5):1524–36. 10.1111/j.1365-2958.2007.05605.x [DOI] [PubMed] [Google Scholar]
- 39.Junker M, Schuster CC, McDonnell A V, Sorg KA, Finn MC, Berger B, et al. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103(13):4918–23. 10.1073/pnas.0507923103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renn JP, Clark PL. A conserved stable core structure in the passenger domain beta-helix of autotransporter virulence proteins. Biopolymers. 2008;89(5):420–7. 10.1002/bip.20924 [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki K. Creating random mutagenesis libraries by megaprimer PCR of whole plasmid (MEGAWHOP) In: Arnold FH, Georgiou G, editors. Dir Evol Libr Creat. 2003. p. 23–8. [DOI] [PubMed] [Google Scholar]
- 42.Strauch KL, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA. 1988;85(5):1576–80. 10.1073/pnas.85.5.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weski J, Ehrmann M. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope. J Bacteriol. 2012;194(12):3225–33. 10.1128/JB.00221-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Champion MM, Williams EA, Pinapati RS, DiGiuseppe Champion PA. Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial Esx-1 substrates. J Proteome Res. 2014;13:5151–64. 10.1021/pr500484w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Champion MM, Campbell CS, Siegele DA, Russell DH, Hu JC. Proteome analysis of Escherichia coli K-12 by two-dimensional native-state chromatography and MALDI-MS. Mol Microbiol. 2003;47(2):383–96. 10.1046/j.1365-2958.2003.03294.x [DOI] [PubMed] [Google Scholar]
- 46.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 47.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen J V., Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 48.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–75. 10.1038/nmeth785 [DOI] [PubMed] [Google Scholar]
- 49.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–26. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnard TJ, Gumbart J, Peterson JH, Noinaj N, Easley NC, Dautin N, et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J Mol Biol. 2012;415(1):128–42. 10.1016/j.jmb.2011.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauch KL, Johnson K, Beckwith J. Characterization of DegP, a gene required for proteolysis in the cell-envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171(5):2689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dutta PR, Sui BQ, Nataro JP. Structure-function analysis of the enteroaggregative Escherichia coli plasmid-encoded toxin autotransporter using scanning linker mutagenesis. J Biol Chem. 2003;278(41):39912–20. 10.1074/jbc.M303595200 [DOI] [PubMed] [Google Scholar]
- 53.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106(45):19120–5. 10.1073/pnas.0907912106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soprova Z, Sauri A, van Ulsen P, Tame JRH, den Blaauwen T, Jong WSP, et al. A conserved aromatic residue in the autochaperone domain of the autotransporter Hbp is critical for initiation of outer membrane translocation. J Biol Chem. 2010;285(49):38224–33. 10.1074/jbc.M110.180505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saurí A, Oreshkova N, Soprova Z, Jong WSP, Sani M, Peters PJ, et al. Autotransporter β-domains have a specific function in protein secretion beyond outer-membrane targeting. J Mol Biol. 2011;412(4):553–67. 10.1016/j.jmb.2011.07.035 [DOI] [PubMed] [Google Scholar]
- 56.Purdy GE, Hong M, Payne SM. Shigella flexneri DegP facilitates IcsA surface expression and is required for efficient intercellular spread. Infect Immun. 2002;70(11):6355–64. 10.1128/iai.70.11.6355-6364.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purdy GE, Fisher CR, Payne SM. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J Bacteriol. 2007;189(15):5566–73. 10.1128/JB.00483-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325(6103):458–62. [DOI] [PubMed] [Google Scholar]
- 59.Desvaux M, Cooper LM, Filenko NA, Scott-Tucker A, Turner SM, Cole JA, et al. The unusual extended signal peptide region of the type V secretion system is phylogenetically restricted. FEMS Microbiol Lett. 2006;264(1):22–30. 10.1111/j.1574-6968.2006.00425.x [DOI] [PubMed] [Google Scholar]
- 60.Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, et al. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153(Pt 1):59–70. 10.1099/mic.0.29091-0 [DOI] [PubMed] [Google Scholar]
- 61.Chevalier N, Moser M, Koch H-G, Schimz K-L, Willery E, Locht C, et al. Membrane targeting of a bacterial virulence factor harbouring an extended signal peptide. J Mol Microbiol Biotechnol. 2004;8(1):7–18. 10.1159/000082076 [DOI] [PubMed] [Google Scholar]
- 62.Peterson JH, Szabady RL, Bernstein HD. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J Biol Chem. 2006;281(14):9038–48. 10.1074/jbc.M508681200 [DOI] [PubMed] [Google Scholar]
- 63.Jong WSP, Luirink J. The conserved extension of the Hbp autotransporter signal peptide does not determine targeting pathway specificity. Biochem Biophys Res Commun. 2008;368(3):522–7. 10.1016/j.bbrc.2008.01.122 [DOI] [PubMed] [Google Scholar]
- 64.Jong WSP, ten Hagen-Jongman CM, Ruijter E, Orru RVA, Genevaux P, Luirink J. YidC is involved in the biogenesis of the secreted autotransporter hemoglobin protease. J Biol Chem. 2010;285(51):39682–90. 10.1074/jbc.M110.167650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102(1):221–6. 10.1073/pnas.0406055102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185(19):5706–13. 10.1128/jb.185.19.5706-5713.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marrichi M, Camacho L, Russell DG, DeLisa MP. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J Biol Chem. 2008;283(50):35223–35. 10.1074/jbc.M802660200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steiner D, Forrer P, Stumpp MT, Plückthun A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat Biotechnol. 2006;24(7):823–31. 10.1038/nbt1218 [DOI] [PubMed] [Google Scholar]
- 69.Mansell TJ, Linderman SW, Fisher AC, DeLisa MP. A rapid protein folding assay for the bacterial periplasm. Protein Sci. 2010;19(5):1079–90. 10.1002/pro.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teh MY, Tran ENH, Morona R. Absence of O antigen suppresses Shigella flexneri IcsA autochaperone region mutations. Microbiology. 2012;158(11):2835–50. 10.1099/mic.0.062471-0 [DOI] [PubMed] [Google Scholar]
- 71.Baud C, Gutsche I, Willery E, de Paepe D, Drobecq H, Gilleron M, et al. Membrane-associated DegP in Bordetella chaperones a repeat-rich secretory protein. Mol Microbiol. 2011;80(6):1625–36. 10.1111/j.1365-2958.2011.07672.x [DOI] [PubMed] [Google Scholar]
- 72.Baud C, Hodak H, Willery E, Drobecq H, Locht C, Jamin M, et al. Role of DegP for two-partner secretion in Bordetella. Mol Microbiol. 2009;74(2):315–29. 10.1111/j.1365-2958.2009.06860.x [DOI] [PubMed] [Google Scholar]
- 73.Raivio TL, Silhavy TJ. The Sigma(E) and Cpx regulatory pathways: Overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2(2):159–65. 10.1016/S1369-5274(99)80028-9 [DOI] [PubMed] [Google Scholar]
- 74.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the Sigma-E stress response in related genomes. PLoS Biol. 2006;4(1):0043–59. 10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ades SE. Regulation by destruction: design of the σE envelope stress response. Curr Opin Microbiol. 2008;11(6):535–40. 10.1016/j.mib.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 76.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J Bacteriol. 2009;191(23):7279–87. 10.1128/JB.01047-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the. Genes Dev. 2006;1776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Las Peñas A, Connolly L, Gross CA. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol. 1997;24(2):373–85. . [DOI] [PubMed] [Google Scholar]
- 79.De Las Peñas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997;179(21):6862–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayden JD, Ades SE. The extracytoplasmic stress factor, sigma(E), is required to maintain cell envelope integrity in Escherichia coli. PLoS ONE. 2008;3(2):e1573 10.1371/journal.pone.0001573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vogt SL, Raivio TL. Just scratching the surface: An expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2012;326:2–11. 10.1111/j.1574-6968.2011.02406.x [DOI] [PubMed] [Google Scholar]
- 82.Price NL, Raivio TL. Characterization of the Cpx Regulon in Escherichia coli Strain MC4100. J Bacteriol. 2009;191(6):1798–815. 10.1128/JB.00798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Norell D, Heuck A, Tran-Thi T-A, Götzke H, Jacob-Dubuisson F, Clausen T, et al. Versatile in vitro system to study translocation and functional integration of bacterial outer membrane proteins. Nat Commun. 2014;5:5396 10.1038/ncomms6396 [DOI] [PubMed] [Google Scholar]
- 84.Roman-Hernandez G, Peterson JH, Bernstein HD. Reconstitution of bacterial autotransporter assembly using purified components. Elife. 2014;3:1–20. 10.7554/eLife.04234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gruss F, Zahringer F, Jakob RP, Burmann BM, Hiller S, Maier T. The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol. 2013;20(11):1318–20. 10.1038/nsmb.2689 [DOI] [PubMed] [Google Scholar]
- 86.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang HS, Easley NC, et al. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature. 2013;501(7467):385–90. 10.1038/nature12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sander IM, Chaney JL, Clark PL. Expanding Anfinsen’s Principle: Contributions of Synonymous Codon Selection to Rational Protein Design. J Am Chem Soc. 2014;136(3):858–61. 10.1021/ja411302m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, et al. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348(6233):444–8. 10.1126/science.aaa3974 [DOI] [PubMed] [Google Scholar]
- 89.Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, et al. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495(7439):111–5. 10.1038/nature11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283(39):26748–58. 10.1074/jbc.M802754200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jong WS, Soprova Z, de Punder K, ten Hagen-Jongman CM, Wagner S, Wickström D, et al. A structurally informed autotransporter platform for efficient heterologous protein secretion and display. Microb Cell Fact. 2012;11:85 10.1186/1475-2859-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jose J, Maas RM, Teese MG. Autodisplay of enzymes-Molecular basis and perspectives. J Biotechnol. 2012;161(2):92–103. 10.1016/j.jbiotec.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 93.Tozakidis IEP, Sichwart S, Jose J. Going beyond E. coli: autotransporter based surface display on alternative host organisms. N Biotechnol. 2015;32(6):644–50. 10.1016/j.nbt.2014.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(A) Quantification of DegP across all experimental conditions. (B) Quantification of representative marker proteins for the cytosol (σ70 and DnaK), the OM (LamB and BamA) and the OM-associated BamB. (C) Quantification of detectable proteins regulated by the Cpx stress response. No systematic decrease or increase of these Cpx-regulated proteins is observed during death (last condition, prn induction for 5 h in E. coli DegP deletion strain). Error bars correspond to the standard error (∑/mean) for each label-free measurement.
(TIF)
(A) E. coli deletion strains from the BW30270 series [43] were transformed with plasmids that constitutively express prn or pet, as described in Fig 2A. Error bars in bold are standard deviations from at least three biological replicates. The vertical lines for pet transformed degP::kan indicates the spread of two biological replicates. (B) prn is expressed and the passenger is secreted in fkpA::cm and skp::kan. As a control the parent strain (BW30270) was used. Secretion is routinely assayed by the production of the processed passenger in whole cell lysate samples.
(TIF)
Data Availability Statement
RAW data files, search results, search parameters and databases were deposited into MassIVE and are available for download at massive.ucsd.edu with the identifier MSV000080095 or ftp://MSV000080095@massive.ucsd.edu.