Abstract
Methyloversatilis universalis FAM5 utilizes single carbon compounds such as methanol or methylamine as a sole source of carbon and energy. Expression profiling reveals distinct sets of genes altered during growth on methylamine vs methanol. As expected, all genes for the N-methylglutamate pathway were induced during growth on methylamine. Among other functions responding to the aminated source of C1-carbon, are a heme-containing amine dehydrogenase (Qhp), a distant homologue of formaldehyde activating enzyme (Fae3), molybdenum-containing formate dehydrogenase, ferredoxin reductase, a set of homologues to urea/ammonium transporters and amino-acid permeases. Mutants lacking one of the functional subunits of the amine dehydrogenase (ΔqhpA) or Δfae3 showed no growth defect on C1-compounds. M. universalis FAM5 strains with a lesion in the H4-folate pathway were not able to use any C1-compound, methanol or methylamine. Genes essential for C1-assimilation (the serine cycle and glyoxylate shunt) and H4MTP-pathway for formaldehyde oxidation showed similar levels of expression on both C1-carbon sources. M. universalis FAM5 possesses three homologs of the formaldehyde activating enzyme, a key enzyme of the H4MTP-pathway. Strains lacking the canonical Fae (fae1) lost the ability to grow on both C1-compounds. However, upon incubation on methylamine the fae1-mutant produced revertants (Δfae1R), which regained the ability to grow on methylamine. Double and triple mutants (Δfae1RΔfae3, or Δfae1RΔfae2 or Δfae1RΔfae2Δfae3) constructed in the revertant strain background showed growth similar to the Δfae1R phenotype. The metabolic pathways for utilization of methanol and methylamine in Methyloversatilis universalis FAM5 are reconstructed based on these gene expression and phenotypic data.
Keywords: rhodocyclaceae, methyloversatilis, C1-metabolism, N-methylglutamate pathway, formaldehyde activating enzyme homologues
1. Introduction
Facultative betaproteobacterial methylotrophs, belonging to Burkholderiales and Rhodocyclales, occupy a variety of ecological niches, including soils, sediments, plant biomass, wastewater sludge, hot springs, and oil sands [1,2,3,4,5,6,7]. These bacterial lineages show exceptionally versatile metabolic capabilities, including the ability to utilize single carbon compounds, such as methanol and methylated amines [8,9,10]. Despite the relatively high abundance of these facultative methylotrophic betaproteobacteria in nature, only a few of them have been isolated in pure culture and characterized [4,5,6,9,10]. C1-utilization pathways have been investigated in Methyloversatilis universalis and Methyloversatilis thermotolerans (both are members of the family Rhodocyclaceae) and Methylibium petroleiphilum PM1 (a representative of unclassified Burkholderiales) [11,12,13]. It has been shown that a homolog of the pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenase is essential for oxidation of C1 and C2 alcohols during aerobic or anaerobic growth in M. universalis [8,13]. The same enzyme is crucial for methanol utilization even in strains possessing the canonical methanol dehydrogenase, such as Methyloversatilis sp. 18–153 or RZ94 [8,10].
In addition to methanol, all tested members of the genera Methyloversatilis are capable of utilizing methylated amines [6,10]. M. universalis FAM5 oxidizes methylamine via the N-methylglutamate pathway [12]. The pathway includes three enzymes: the N-methylglutamate synthase (NMGS, encoded by mgsABC gene cluster), which can produce N-methylglutamate from glutamate and methylamine; the N-methylglutamate dehydrogenase (NMGDH, encoded by mgdABCD gene cluster), which is predicted to produce methylene-tetrahydrofolate (H4F) and regenerated glutamate from N-methylglutamate; and the gamma-glutamylmethylamine synthetase (GMAS, gmas), a reversible enzyme which produces gamma-glutamylmethylamine (GMA) from glutamate and methylamine using ATP. The gamma-glutamylmethylamine synthetase is predicted to serve as the first enzyme of the pathway, however the exact functional role of the enzyme and its product, gamma-glutamylmethylamine, are not well defined. It has been predicted that the enzyme contributes to methylamine detoxification, by converting the potentially toxic amine into a metabolically neutral form [14]. This vision of the pathway has been supported by a number of studies, including mutagenesis, 13C-labeling and in vivo NMR investigation [12,14,15,16]. The GMA has also been proposed to be a substrate for NMGS instead of methylamine [17,18,19]. Overall, the exact topology of the pathway is still awaiting a careful enzymatic investigation. Nonetheless, several genomic studies suggest that the N-methylglutamate pathway is recruited for methylamine utilization by a number of methylotrophic and non-methylotrophic bacteria [16,17,18,19,20,21,22].
Recently the genomes of seven strains representing three Methyloversatilis species have been sequenced at the University of Washington [23] and in collaboration with the Joint Genomic Institute (JGI) [10]. The C1-pathways were found to be highly conserved in all strains sequenced. It was predicted that formaldehyde, a product of methanol or methylamine oxidation could be converted to formate by a tungsten-dependent aldehyde dehydrogenase in addition to the tetrahydrofolate (H4F) or tetrahydromethanopterin (H4MPT)-pathways [5,13,23]. Formate is further oxidized to CO2 by two NAD-dependent formate dehydrogenases [13]. Two C1-assimilation pathways, the serine cycle and Calvin-Benson-Bassham (CBB) cycle, could be predicted for sequenced Methyloversatilis spp, with the exception of M. thermotolerans 3tT, which possesses only the serine cycle [10,11,23]. Only serine cycle enzymes were induced in M. universalis cells during anaerobic growth on methanol [13]. Genome analyses also indicate that M. universalis FAM5 also possesses a heme-containing amine dehydrogenases (Qhp). It has been shown that the Qhp oxidizes a number of aliphatic and aromatic amines [24,25,26]. Qhp was highly expressed during growth of the Paracoccus denitrificans IFO 12442 on methylamine. However, the enzyme displays relatively low affinity for methylamine and is specific to n-butylamine [25]. It has been speculated that during growth of P. denitrificans on methylamine the enzyme plays a secondary role and might contribute to oxidation/detoxification of the amine [25,26]. The contribution of the enzyme to metabolism of methylamine has not been tested by mutagenesis.
Here we investigate pathways for aerobic utilization of methanol and methylamine in M. universalis FAM5. The predicted end-product of methylamine oxidation via the N-methylglutamate pathways is methylene-H4F [12]. Thus the metabolic arrangement of methylamine utilization was expected to differ from the methanol utilization network, where formaldehyde is the key end product of the methanol oxidation [8].
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
The M. universalis sp FAM5 strains used in this study are listed in Table 1. Since the wild type aggregates in liquid culture we selected a strain of the M. universalis (named here FAM5E) that does not clump. No differences in growth rate between strain FAM5 and FAM5E on plates or in liquid culture were observed (data not shown). The main advantage of the strain FAM5E is that it produces reliable reading of optical density and does not require homogenization. The strain FAM5E was used to generate and test the majority of mutants described in this study (Table 1).
Table 1.
Growth phenotypes of the Methyloversatilis universalis wild type and mutant strains.
| Strain | Description | Succinate | Methanol | Methylamine | Succinate + C1 * |
|---|---|---|---|---|---|
| FAM5 | Wild type, forms aggregates in liquid culture, KanSTetSCmR | ++++ | +++ | ++++ | ++++ |
| FAM5E | Trait of FAM5 which does not form aggregates in liquid culture, KanSTetSCmR | ++++ | +++ | ++++ | ++++ |
| ΔmgdD | FAM5-ΔmgdD:kan, KanRTetSCmR | ++++ | +++ | - | NT |
| ΔmgsC | FAM5-ΔmgsC:kan, KanRTetSCmR | ++++ | +++ | - | NT |
| Δfae1 | FAM5E-Δfae1:kan, KanRTetSCmR | ++++ | - | - | NT |
| Δfae2 | FAM5E-Δfae2:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae3 | FAM5E-Δfae3:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| ΔmtdA | FAM5E-ΔmtdA:kan, KanRTetSCmR | ++++ | - | - | +++ (1) |
| ΔqhpA | FAM5-ΔqhpA, KanSTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae1ΔqhpA | FAM5-ΔqhpAΔfae1:kan, KanRTetSCmR | ++++ | - | ++ | NT |
| Δfae2ΔqhpA | FAM5-ΔqhpAΔfae2:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae3ΔqhpA | FAM5-ΔqhpAΔfae3:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae1RΔqhpAΔfae3 | FAM5E-ΔqhpA Δfae1Δfae3:kan, KanRTetSCmR | ++++ | - | - | NT |
| Δfae1R | Revertant of FAM5E-Δfae1, KanSTetSCmR | ++++ | - | ++ | ++++ (2) |
| Δfae1Δfae3 | FAM5-Δfae1Δfae3:kan, KanRTetSCmR | ++++ | - | - | NT |
| Δfae1RΔ fae3 | FAM5-Δfae1Δfae3, KanSTetSCmR | ++++ | - | ++ | ++++ (2) |
| Δfae1Δ fae2 | FAM5-Δfae1Δfae2:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae3 | FAM5E-Δfae3, KanSTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae3Δfae2 | FAM5E-Δfae3 Δfae2:kan, KanRTetSCmR | ++++ | +++ | ++++ | NT |
| Δfae1Δfae2Δfae3 | FAM5E-Δfae1Δfae3Δfae2:kan, KanRTetSCmR | ++++ | - | - | NT |
NT, not tested; “++++” correspond to doubling time of 6 h; “+++” correspond to doubling time of 9 h; “++” correspond to 12 h doubling time; and “-” indicates no growth. Methanol (5 mM) was added to test if a mutant strain is sensitive to formaldehyde (2) or unable to produce methyl-H4folate for purine synthesis (1).
Cells were grown in previously described minimal medium [6] with succinate (20 mM), methylamine (30 mM or 5 mM), and methanol (25 mM) as growth substrates. The following antibiotic concentrations were used for M. universalis: tetracycline (Tet), 1.0 mg L−1; kanamycin (Kan), 100 mg L−1; and chloramphenicol (Cmp), 10–15 mg L−1.
The following cloning vectors were used: pCR2.1 (Invitrogen, Carlsbad, CA, USA) for cloning of PCR products, pCM184 for generation of single, double and triple mutant strains, and the pCM157 system for construction of unmarked mutations (Table 2) [27]. Escherichia coli strains were routinely cultivated at 37 °C in Luria-Bertani medium (BD Difco, Franklin Lakes, NJ, USA). When indicated, antibiotic concentrations were used: Tet, 12.5 mg L−1; Kan, 100 mg L1; and ampicillin (Amp), 100 mg L−1.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain/Plasmid | Markers/Description | Reference/Source |
|---|---|---|
| Escherichia coli One Shot TOP 10 | F-mcrA Δ (mrr-hsdRMS-mcrBC) Φ80 lacZΔM15 ΔlacX74 recA1 araD139 Δ (ara-leu)7697 galU galK rpsL StrR endA1 nupG | Invitrogen |
| Escherichia coli S17-1 | recA pro hsdR RP4-2-Tc:Mu-Km:Tn7 TmpR SpcRStrR | [28] |
| pCR2.1 | KanR, AmpR | Invitrogen |
| pDrive | KanR, AmpR | Qiagen |
| pCM184 | Broad-host-range allelic exchange vector, KanR, TetR | [27] |
| pCM157 | Cre/LoxP, TetR | [27] |
| pMgdD | pCM184 with mgdD upstream and downstream flanks | This study |
| pMgsC | pCM184 with mgsC upstream and downstream flanks | This study |
| pFAE1 | pCM184 with fae1 upstream and downstream flanks | This study |
| pFAE2 | pCM184 with fae2 upstream and downstream flanks | This study |
| pFAE3 | pCM184 with fae3 upstream and downstream flanks | This study |
| pQHNDH1 | pCM184 with qhpA upstream and downstream flanks | This study |
| pQHNDH2 | pCM184 with qhpX upstream and downstream flanks | This study |
| pMtdA | pCM184 with mtdA upstream and downstream flanks | This study |
2.2. DNA Manipulations
DNA was isolated using the QIAamp DNA Mini kit (Qiagen, Venlo, Netherlands). Plasmid DNA was purified using the Qiagen Miniprep Kit (Qiagen). E. coli transformation, restriction enzyme digestion and ligation reactions were carried out as described by Sambrook et al. [29]. Polymerase chain reaction (PCR) amplifications were performed using hot-start Taq polymerase (Qiagen) in accordance with the manufacturer’s instructions. Primers used for PCR amplification of upstream or downstream regions (approximately 450–600 bp) of each targeted gene are listed in Table S1. Amplified fragments were cloned into pCR2.1, sequenced and then sub-cloned into pCM184 using appropriate restriction sites. After verification of the nucleotide sequence, the plasmids were transformed into E. coli S17-1, and the resulting donor strains were mated with wild-type M. universalis FAM5 via biparental mating as previously described [8]. The identity of the double-crossover mutants was verified by diagnostic PCR with primers specific to the insertion sites. Mutant phenotypes were assessed on solid media and in liquid culture with succinate, pyruvate, methanol or methylamine as carbon sources.
2.3. RNA-seq Experiments
Trascriptomic experiments were carried out with M. universalis strain FAM5. Cells pre-grown on methanol (10 mM) were used for inoculation of methanol (25 mM) or methylamine (35 mM) cultures at 5:100 ratios (inoculum:fresh media). Cultures were grown to mid-exponential phase (OD600 0.4 ± 0.05). The cellular activities were terminated by addition of “stop solution” as described [30]. The cells were collected by centrifugation at 4300× g at 4 °C for 10 min. mRNA samples were isolated and enriched as previously described [30] and submitted to the University of Washington’s High-Throughput Sequencing Solutions Center on dry ice for single-read Illumina® sequencing (Department of Genome Sciences, University of Washington; http://www.htseq.org/). The RNA-Seq sequence data sets ranged from 12.19 to 25.46 million reads (36 bp) per sample. The M. universalis FAM5 genome to be used as the alignment scaffold was downloaded from MaGE (https://www.genoscope.cns.fr/agc/microscope/mage/) on 31 March 2014. The raw reads were aligned to the scaffold using BWA version 0.7.4-r385 with default parameters [31]. The alignments were post-processed and sorted into BAM files with SAMTools version 0.1.19-44428cd [32]. Reads were attributed to open reading frames (ORFs) using the htseq-count tool from the “HTSeq” framework version 0.5.4p5 in the “intersection-nonempty” mode [33]. The ratios of reads mapped per ORF in the two conditions were calculated along with p-values using DESeq2 [33]. The p-values in Table 2 were corrected for multiple testing using the q-value method of Storey [34]. Proportions of reads mapped to rRNA were 78%–90% for RNA-libraries prepared from cells grown on methanol, and 91%–92% for RNA-libraries prepared from cells grown on methylamine. For display and presentation purposes, the Reads per Kilobase per Million reads sequenced (RPKM) were calculated [35].
2.4. Enzyme Assays
All cell extracts (50 mL) for activity were prepared from methylamine-grown cultures. Cells were harvested by centrifugation at 4500× g using a Sorvall RC-5B centrifuge at 4 °C for 10 min. The supernatant was removed and the cell pellet was stored at −80 C. Cell pellets were thawed and resuspended in 1 mL of 100 mM potassium phosphate buffer, pH 7.6 or 7.2 and broken using a French press (three times, 1000 psi). The extracts were centrifuged at 28,000× g for 5 min to remove cell debris. Qhp activity was detected by measuring amine dehydrogenase activity using a spectrophotometric assay measuring the reduction rate of potassium ferricyanide (500 μM, at 420 nm) in the presence of cell free extract (100 μg), and methylamine (25 mM) as substrate at room temperature. The extinction coefficient of potassium ferricyanide is 1.02 mM−1 cm−1 at pH = 7.6. The total volume of the reaction was 200 μL. Activity was measured using a Spectramax 190 plate reader (Molecular Devices, Sunnyvale, CA, USA); a minimum of two biological replicates was assayed.
2.5. Accession Numbers
The RNA-seq data were uploaded into NCBI/GEO under accession number GSE63822.
3. Results
3.1. Gene Expression Profiles: Methanol vs. Methylamine
Whole transcriptome shotgun sequencing data were generated for M. universalis cells grown on methanol or methylamine using a high-throughput sequencing (Illumina) platform. Based on relative expression, genes (omitting rRNAs) could be grouped into six major categories: genes with very high (Reads per Kilobase Million (RPKM) ≥ 10000), high (RPKM ≥ 5000), moderate (5000 > RPKM ≥ 1000), modest (1000 > RPKM ≥ 250), and low (250 > RPKM ≥ 50) expression, and not expressed (RPKM < 50). Annotation of the M. universalis FAM5 genome predicts 4027 coding sequences [23]. Based on the expression data, 950 (24%) genes were not expressed under any condition tested. Among the silent functional modules were genes encoding denitrification reactions (nitrate and nitrite reductases, nitrate transporters), phototrophy (light-harvesting complexes) and pathways for utilization of urea, acetoin, methanesulfonate, and phenolic compounds. The majority of genes fell into low expression categories (45%). About 25% of genes displayed modest expression, 5% of genes showed moderate expression and only a small fraction (1%) of the genome showed very high/high expression.
Most of the previously recognized metabolic modules essential for oxidation of C1-compounds fell into very high to moderate categories. Expression of central metabolic pathways, such as the citric acid cycle, amino acid biosynthesis, gluconeogenesis and energy generating pathway (respiratory chain and ATP-synthase) genes remained mostly unchanged (Table 3). Genes encoding the H4MPT synthesis and the H4MPT-linked C1-transfer enzymes and the serine cycle enzymes also remained unchanged, suggesting that these pathways contribute in a similar fashion to methanol and methylamine utilization (Table 3).
Table 3.
Gene expression profile in methane or methylamine grown cells of M. universalis FAM5.
| Gene ID (Old) | Gene ID (New) | Gene Product | Methanol (RPKM) * | Methylamine (RPKM) * | Fold Change | q-Value |
|---|---|---|---|---|---|---|
| Methylamine oxidation | ||||||
| METUNv1_760110 | METUNv2_580110 | Gmas, Gamma glutamylmethylamide synthetase | 2961.80 | 19,440.25 | 6.56 | 0.65 |
| METUNv1_760111 | METUNv2_580111 | MgsA, N-methyl glutamate synthase subunit A | 2727.43 | 22,221.30 | 8.15 | 0.73 |
| METUNv1_760112 | METUNv2_580112 | MgsB, N-methyl glutamate synthase subunit B | 2680.76 | 19,852.55 | 7.41 | 0.69 |
| METUNv1_760113 | METUNv2_580113 | MgsC, N-methyl glutamate synthase subunit C | 4221.56 | 53,956.55 | 12.78 | 0.67 |
| METUNv1_760114 | METUNv2_580114 | MgdA, N-methyl glutamate dehydrogenase/oxidoreductase subunit A | 1414.90 | 13,014.45 | 9.20 | 0.74 |
| METUNv1_760115 | METUNv2_580115 | MgdB, N-methyl glutamate dehydrogenase/oxidoreductase subunit B | 388.00 | 3906.31 | 10.07 | 0.74 |
| METUNv1_760116 | METUNv2_580116 | MgdC, N-methyl glutamate dehydrogenase/oxidoreductase subunit C | 3389.10 | 26,611.05 | 7.85 | 0.75 |
| METUNv1_760117 | METUNv2_580117 | MgdD, N-methyl glutamate dehydrogenase/oxidoreductase subunit D | 701.94 | 7962.74 | 11.34 | 0.71 |
| METUNv1_760127 | METUNv2_580127 | QhpB, Quinohemoprotein amine dehydrogenase, beta subunit | 420.06 | 4164.18 | 9.91 | 0.68 |
| METUNv1_760128 | METUNv2_580128 | QhpA, Quinohemoprotein amine dehydrogenase, alpha subunit | 733.76 | 7390.84 | 10.07 | 0.65 |
| METUNv1_760130 | METUNv2_580130 | QhpC, Quinohemoprotein amine dehydrogenase, SAM-radical dependent activating subunit | 380.86 | 4283.06 | 11.25 | 0.00 |
| Methanol oxidation | ||||||
| METUNv1_770214 | METUNv2_590217 | XoxF1, PQQ-linked dehydrogenase | 2023.28 | 3380.16 | 1.67 | 0.50 |
| METUNv1_770216 | METUNv2_590218 | XoxF2, PQQ-linked dehydrogenase | 10,962.17 | 16,741.70 | 1.53 | 0.05 |
| METUNv1_590046 | METUNv2_420045 | XoxF3, PQQ-dependent dehydrogenase | 1213.63 | 659.01 | −1.84 | 0.62 |
| METUNv1_590042 | METUNv2_420041 | XoxJ3, Extracellular solute-binding protein family 3 | 4399.57 | 1403.51 | −3.13 | 0.05 |
| METUNv1_590043 | METUNv2_420042 | XoxG3, Cytochrome c class I | 3156.11 | 1089.89 | −2.90 | 0.30 |
| METUNv1_590049 | METUNv2_420048 | Mdh2, PQQ-dependent methanol/ethanol dehydrogenase | 48,100.35 | 34,793.20 | −1.38 | 0.77 |
| METUNv1_590050 | METUNv2_420049 | Mdh2J, Extracellular solute-binding protein family | 2486.88 | 903.00 | −2.75 | 0.69 |
| METUNv1_590051 | METUNv2_420050 | Mdh2G, cytochrome c-type protein | 1732.91 | 595.39 | −2.91 | 0.58 |
| Formaldehyde oxidation | ||||||
| METUNv1_580096 | METUNv2_410093 | Fae 1, Formaldehyde-activating enzyme | 18,235.00 | 11,635.85 | −1.57 | 0.82 |
| METUNv1_660037 | METUNv2_480039 | Fae 2, Formaldehyde-activating enzyme | 561.52 | 642.50 | 1.14 | 0.89 |
| METUNv1_700516 | METUNv2_520523 | Fae 3, Formaldehyde activating enzyme | 584.51 | 6861.86 | 11.74 | 0.00 |
| METUNv1_590006 | METUNv2_420006 | Orf 9, Involved in biosynthesis of tetrahydromethanopterin. Essential for formaldehyde oxidation. | 430.06 | 187.51 | −2.29 | 0.21 |
| METUNv1_580095 | METUNv2_410092 | Orf 7, Involved in tetrahydromethanopterin-linked formaldehyde oxidation. | 371.25 | 549.26 | 1.48 | 0.75 |
| METUNv1_580094 | METUNv2_410091 | Orf 5, Involved in biosynthesis of tetrahydromethanopterin | 489.69 | 493.89 | 1.01 | 0.93 |
| METUNv1_580093 | METUNv2_410090 | Mch, Methenyltetrahydromethanopterin cyclohydrolase | 620.26 | 736.89 | 1.19 | 0.87 |
| METUNv1_580092 | METUNv2_410089 | OrfY, Involved in tetrahydromethanopterin C1 transfer. | 896.40 | 819.12 | −1.09 | 0.94 |
| METUNv1_580091 | METUNv2_410088 | MtdB, NAD-dependent methylenetetrahydromethanopterin dehydrogenase | 1295.29 | 1379.77 | 1.07 | 0.88 |
| METUNv1_580088 | METUNv2_410086 | FhcB, Formyltransferase/hydrolase complex subunit B | 587.63 | 515.00 | −1.14 | 0.92 |
| METUNv1_580087 | METUNv2_410085 | FhcA, Formyltransferase/hydrolase complex subunit A | 766.77 | 744.38 | −1.03 | 0.93 |
| METUNv1_580086 | METUNv2_410084 | FhcD, Formyltransferase/hydrolase complex subunit D | 598.76 | 547.64 | −1.09 | 0.94 |
| METUNv1_580085 | METUNv2_410083 | FhcC, Formyltransferase/hydrolase complex subunit C | 634.50 | 467.32 | −1.36 | 0.88 |
| METUNv1_590040 | METUNv2_420039 | Ald, Tungsten-containing aldehyde ferredoxin oxidoreductase | 408.13 | 155.57 | −2.62 | 0.17 |
| METUNv1_490013 | METUNv2_320014 | AldB, Aldehyde dehydrogenase B | 2762.93 | 2224.63 | −1.24 | 0.94 |
| Formate oxidation | ||||||
| METUNv1_770385 | METUNv2_590384 | Fdh, Putative formate dehydrogenase subunit A | 634.02 | 668.94 | 1.06 | 0.92 |
| METUNv1_700257 | METUNv2_520260 | FdhD, NAD-linked formate dehydrogenase delta subunit | 38.00 | 76.63 | 2.02 | 0.55 |
| METUNv1_700258 | METUNv2_520261 | FdhC, Formate dehydrogenase, accessory protein | 751.50 | 1024.07 | 1.36 | 0.74 |
| METUNv1_700259 | METUNv2_520262 | FdhA, NAD-dependent formate dehydrogenase alpha subunit | 3720.44 | 5479.72 | 1.47 | 0.39 |
| METUNv1_700260 | METUNv2_520263 | FdhB, NAD-dependent formate dehydrogenase beta subunit | 2758.00 | 4434.84 | 1.61 | 0.46 |
| METUNv1_700261 | METUNv2_520264 | FdhG, NAD-dependent formate dehydrogenase gamma subunit | 1127.75 | 1882.41 | 1.67 | 0.67 |
| METUNv1_700262 | METUNv2_520265 | FdhR, Formate dehydrogenase regulator | 210.25 | 231.00 | 1.10 | 0.90 |
| METUNv1_570005 | METUNv2_400021 | FdsA, NAD-dependent, tungsten-containing formate dehydrogenase alpha subunit | 1438.46 | 1527.51 | 1.06 | 0.91 |
| METUNv1_570006 | METUNv2_400022 | FdsB, NAD-dependent, tungsten-containing formate dehydrogenase beta subunit | 951.26 | 1018.18 | 1.07 | 0.90 |
| H4F-pathway/Serine cycle/Glyoxylate shunt | ||||||
| METUNv1_460318 | METUNv2_290319 | FtfL, formate-tetrahydrofolate ligase/synthetase | 4729.66 | 1674.32 | −2.82 | 0.54 |
| METUNv1_460309 | METUNv2_290310 | Fch, Methenyltetrahydrofolate cyclohydrolase | 2091.39 | 1541.64 | −1.36 | 0.90 |
| METUNv1_460310 | METUNv2_290311 | MdtA, NADP-dependent methylenetetrahydrofolate dehydrogenase | 4363.44 | 2973.78 | −1.47 | 0.93 |
| METUNv1_460311 | METUNv2_290312 | Hpr, Hydroxypyruvate reductase, NAD(P)H-dependent. | 3620.17 | 3220.89 | −1.12 | 0.89 |
| METUNv1_460312 | METUNv2_290313 | Sga, Serine-glyoxylate aminotransferase | 10,590.93 | 8570.41 | −1.24 | 0.79 |
| METUNv1_460313 | METUNv2_290314 | GlyA, Serine hydroxymethyltransferase | 6313.10 | 5292.54 | −1.19 | 0.87 |
| METUNv1_460314 | METUNv2_290315 | MtkA, Malate thiokinase large subunit | 4955.01 | 6292.07 | 1.27 | 0.73 |
| METUNv1_460315 | METUNv2_290316 | MtkB, Malate thiokinase small subunit | 5081.45 | 5363.84 | 1.06 | 0.73 |
| METUNv1_460316 | METUNv2_290317 | Ppc1, phosphoenolpyruvate carboxylase | 2661.63 | 2768.19 | 1.04 | 0.86 |
| METUNv1_460317 | METUNv2_290318 | Mcl, malyl-CoA lyase | 3260.85 | 4275.94 | 1.31 | 0.58 |
| METUNv1_770329 | METUNv2_590331 | Gk, Glycerate kinase | 1197.01 | 1116.38 | −1.07 | 0.93 |
| METUNv1_770169 | METUNv2_590170 | Ppc2, Phosphoenolpyruvate carboxylase | 1034.15 | 1074.00 | 1.04 | 0.91 |
| METUNv1_460302 | METUNv2_290303 | Eno, Enolase | 2095.13 | 1936.51 | −1.08 | 0.92 |
| METUNv1_710053 | METUNv2_530053 | Pgm, Phosphoglyceromutase | 470.57 | 517.14 | 1.10 | 0.90 |
| METUNv1_620020 | METUNv2_450021 | Ms, Malate synthase A | 353.94 | 269.76 | −1.31 | 0.89 |
| METUNv1_620018 | METUNv2_450018 | Icl, Isocitrate lyase | 12,174.92 | 8389.98 | −1.45 | 0.85 |
| CO2 Fixation (CBB cycle) | ||||||
| METUNv1_750044 | METUNv2_570044 | CbbR, RuBisCO operon transcriptional regulator | 195.62 | 189.12 | −1.03 | 0.93 |
| METUNv1_750045 | METUNv2_570045 | CbbL, Ribulose-1,5-bisphosphate carboxylase large subunit | 127.63 | 130.88 | 1.03 | 0.94 |
| METUNv1_750046 | METUNv2_570046 | CbbS, Ribulose bisphosphate carboxylase small subunit | 41.62 | 47.50 | 1.14 | 0.92 |
| METUNv1_750047 | METUNv2_570047 | CbxX, chromosomal AAA type ATPase | 8.75 | 11.94 | 1.36 | 0.89 |
| METUNv1_750048 | METUNv2_570048 | CbbY, haloacid dehalogenase | 7.25 | 14.56 | 2.01 | 0.73 |
| METUNv1_750049 | METUNv2_570049 | CbbE, Ribulose-phosphate 3-epimerase | 13.88 | 16.13 | 1.16 | 0.91 |
| METUNv1_750050 | METUNv2_570050 | Pgp, phosphoglycolate phosphatase | 12.50 | 13.00 | 1.04 | 0.94 |
| METUNv1_750051 | METUNv2_570051 | CbbF, Fructose-1,6-bisphosphatase | 26.75 | 30.51 | 1.14 | 0.92 |
| METUNv1_750052 | METUNv2_570052 | CbbP, Phosphoribulokinase | 26.69 | 32.00 | 1.20 | 0.92 |
| METUNv1_750053 | METUNv2_570053 | CbbT, Transketolase | 24.88 | 42.56 | 1.71 | 0.73 |
| METUNv1_750054 | METUNv2_570054 | CbbG, Glyceraldehyde-3-phosphate dehydrogenase | 17.50 | 19.56 | 1.12 | 0.93 |
| METUNv1_750055 | METUNv2_570055 | CbbA, Fructose-bisphosphate aldolase | 72.75 | 65.50 | −1.11 | 0.93 |
| METUNv1_700111 | METUNv2_520114 | CbbR, RuBisCO operon transcriptional regulator | 360.50 | 255.84 | −1.41 | 0.90 |
| METUNv1_760018 | METUNv2_580018 | CbbQ, Post-translational RubisCO activator | 16.25 | 19.38 | 1.19 | 0.91 |
| Sugar Phosphate Interconversions | ||||||
| METUNv1_470279 | METUNv2_300282 | Fbp, Fructose-1,6-bisphosphatase | 403.90 | 440.81 | 1.09 | 0.91 |
| METUNv1_700104 | METUNv2_520106 | Fba, Fructose-bisphosphate aldolase | 920.68 | 1094.83 | 1.19 | 0.85 |
| METUNv1_700105 | METUNv2_520107 | Pyk, Pyruvate kinase II | 807.00 | 786.28 | −1.03 | 0.92 |
| METUNv1_700106 | METUNv2_520108 | Pgk, Phosphoglycerate kinase | 693.63 | 684.62 | −1.01 | 0.92 |
| METUNv1_700107 | METUNv2_520109 | Gapdh, Glyceraldehyde 3-phosphate dehydrogenase | 1478.94 | 1536.51 | 1.04 | 0.90 |
| METUNv1_700108 | METUNv2_520110 | Tk, Transketolase | 982.15 | 730.45 | −1.34 | 0.89 |
| METUNv1_700109 | METUNv2_520111 | Prk, Phosphoribulokinase | 380.94 | 483.87 | 1.27 | 0.82 |
| METUNv1_700110 | METUNv2_520112 | Fbp3, Fructose-1,6-bisphosphatase | 902.64 | 846.62 | −1.07 | 0.94 |
| METUNv1_470090 | METUNv2_300091 | Pps, Phosphoenolpyruvate synthase | 1610.66 | 1360.78 | −1.18 | 0.94 |
| METUNv1_460093 | METUNv2_290089 | Pck, Phosphoenolpyruvate carboxykinase (GTP) | 242.99 | 191.39 | −1.27 | 0.89 |
| METUNv1_580038 | METUNv2_410037 | Pfk, Pyrophosphate-dependent phosphofructokinase | 866.70 | 1075.27 | 1.24 | 0.85 |
| METUNv1_580036 | METUNv2_410036 | Pyrophosphate-energized inorganic pyrophosphatase | 747.52 | 887.26 | 1.19 | 0.88 |
| METUNv1_580049 | METUNv2_410048 | Pyrophosphate phosphohydrolase | 555.12 | 602.37 | 1.09 | 0.89 |
| TCA cycle | ||||||
| METUNv1_700127 | METUNv2_520130 | E1 component of pyruvate dehydrogenase | 959.88 | 1210.51 | 1.26 | 0.80 |
| METUNv1_700126 | METUNv2_520129 | E2 component of pyruvate dehydrogenase | 217.00 | 302.25 | 1.39 | 0.77 |
| METUNv1_700125 | METUNv2_520128 | E3 component of pyruvate dehydrogenase | 482.26 | 555.08 | 1.15 | 0.88 |
| METUNv1_700186 | METUNv2_520190 | Succinyl-CoA synthetase beta subunit | 705.14 | 556.31 | −1.27 | 0.91 |
| METUNv1_700187 | METUNv2_520191 | Succinyl-CoA synthetase alpha subunit | 622.44 | 414.43 | −1.50 | 0.86 |
| METUNv1_470127 | METUNv2_300128 | Fumarate hydratase class I | 659.82 | 590.00 | −1.12 | 0.93 |
| METUNv1_460167 | METUNv2_290164 | Fumarate hydratase class II (fumarase C) | 185.50 | 141.26 | −1.31 | 0.91 |
| METUNv1_460003 | METUNv2_290002 | ME1, Malic Enzyme | 952.12 | 789.15 | −1.21 | 0.93 |
| METUNv1_520009 | METUNv2_350011 | Mdh, Malate dehydrogenase | 2673.56 | 1934.79 | −1.38 | 0.91 |
| METUNv1_520011 | METUNv2_350013 | Succinate dehydrogenase cytochrome b556 subunit | 324.52 | 191.00 | −1.70 | 0.70 |
| METUNv1_520012 | METUNv2_350014 | Succinate dehydrogenase anchor subunit | 572.13 | 412.89 | −1.39 | 0.88 |
| METUNv1_520013 | METUNv2_350015 | Succinate dehydrogenase flavoprotein subunit | 4086.04 | 2492.77 | −1.64 | 0.84 |
| METUNv1_520014 | METUNv2_350016 | Succinate dehydrogenase Fe–S protein | 843.00 | 709.16 | −1.19 | 0.93 |
| METUNv1_520016 | METUNv2_350018 | Citrate synthase | 2249.29 | 2218.66 | −1.01 | 0.91 |
| METUNv1_520017 | METUNv2_350019 | E1 component of alpha-ketoglutarate dehydrogenase | 1429.24 | 1533.03 | 1.07 | 0.89 |
| METUNv1_520018 | METUNv2_350020 | E2 component of alpha-ketoglutarate dehydrogenase | 697.75 | 762.99 | 1.09 | 0.90 |
| METUNv1_520019 | METUNv2_350021 | E3 component of alpha-ketoglutarate dehydrogenase | 1364.02 | 1401.04 | 1.03 | 0.89 |
| METUNv1_620012 | METUNv2_450011 | Isocitrate dehydrogenase kinase/phosphatase | 241.63 | 308.13 | 1.28 | 0.84 |
| METUNv1_620013 | METUNv2_450012 | Isocitrate dehydrogenase (NADP+) | 2578.02 | 2899.29 | 1.12 | 0.80 |
| Amino Acid Synthesis | ||||||
| METUNv1_660052 | METUNv2_480054 | Glutamate synthase (NADPH) large chain (NADPH-GOGAT) | 3985.94 | 2866.80 | −1.39 | 0.87 |
| METUNv1_660053 | METUNv2_480055 | Glutamate synthase (NADPH) small chain (NADPH-GOGAT) | 1357.89 | 903.01 | −1.50 | 0.91 |
| METUNv1_450044 | METUNv2_280044 | Glutamine synthetase (Glutamate-ammonia ligase) | 2361.78 | 2288.13 | −1.03 | 0.56 |
| METUNv1_470103 | METUNv2_300104 | Glutamate dehydrogenase, NADP-specific (NADP-GDH) | 1040.15 | 1519.76 | 1.46 | 0.93 |
| METUNv1_470104 | METUNv2_300105 | Aspartate aminotransferase (Transaminase A) (AspAT) | 1020.04 | 941.84 | −1.08 | 0.92 |
| METUNv1_470184 | METUNv2_300183 | Putative aspartate transaminase | 617.88 | 620.11 | 1.00 | 0.93 |
| METUNv1_750043 | METUNv2_570043 | Serine-pyruvate aminotransferase | 679.01 | 606.13 | −1.12 | 0.91 |
| Oxidative Phosphorylation | ||||||
| METUNv1_750182 | METUNv2_570182 | ATP synthase F0, A chain | 886.37 | 965.57 | 1.09 | 0.83 |
| METUNv1_750183 | METUNv2_570183 | ATP synthase F0, C chain | 1218.52 | 1365.89 | 1.12 | 0.93 |
| METUNv1_750184 | METUNv2_570184 | ATP synthase F0, B chain | 2528.92 | 2775.88 | 1.10 | 0.89 |
| METUNv1_750185 | METUNv2_570185 | ATP synthase delta chain | 2885.17 | 2564.77 | −1.12 | 0.86 |
| METUNv1_750186 | METUNv2_570186 | ATP synthase subunit alpha subunit | 8915.44 | 6780.46 | −1.31 | 0.89 |
| METUNv1_750187 | METUNv2_570187 | ATP synthase gamma subunit | 5294.58 | 4705.58 | −1.13 | 0.93 |
| METUNv1_750188 | METUNv2_570188 | ATP synthase beta subunit | 9593.01 | 6768.24 | −1.42 | 0.92 |
| METUNv1_750189 | METUNv2_570189 | ATP synthase epsilon subunit | 1784.40 | 1464.79 | −1.22 | 0.90 |
| METUNv1_770336 | METUNv2_590337 | NAD(P) transhydrogenase subunit alpha | 858.91 | 858.01 | −1.00 | 0.93 |
| METUNv1_770337 | METUNv2_590338 | NAD(P) transhydrogenase subunit beta | 399.38 | 455.25 | 1.14 | 0.92 |
| METUNv1_460126 | METUNv2_290122 | NADH-quinone oxidoreductase chain A | 156.74 | 148.38 | −1.06 | 0.93 |
| METUNv1_460127 | METUNv2_290123 | NADH-quinone oxidoreductase subunit B | 451.50 | 376.26 | −1.20 | 0.94 |
| METUNv1_460128 | METUNv2_290124 | NADH (or F420H2) dehydrogenase subunit C | 388.51 | 353.76 | −1.10 | 0.94 |
| METUNv1_460129 | METUNv2_290125 | NADH-ubiquinone oxidoreductase D subunit | 754.27 | 681.62 | −1.11 | 0.92 |
| METUNv1_460130 | METUNv2_290126 | NADH-quinone oxidoreductase subunit E | 207.95 | 211.51 | 1.02 | 0.92 |
| METUNv1_460131 | METUNv2_290127 | NADH-quinone oxidoreductase subunit F | 628.76 | 528.76 | −1.19 | 0.87 |
| METUNv1_460132 | METUNv2_290128 | NADH-quinone oxidoreductase subunit G | 1248.13 | 938.27 | −1.33 | 0.93 |
| METUNv1_460133 | METUNv2_290129 | NADH-quinone oxidoreductase subunit H | 520.13 | 381.75 | −1.36 | 0.93 |
| METUNv1_460134 | METUNv2_290130 | NADH-quinone oxidoreductase subunit I | 327.57 | 337.82 | 1.03 | 0.94 |
| METUNv1_460135 | METUNv2_290131 | NADH-quinone oxidoreductase subunit J | 131.50 | 118.13 | −1.11 | 0.88 |
| METUNv1_460136 | METUNv2_290132 | NADH-quinone oxidoreductase subunit K | 58.88 | 60.63 | 1.03 | 0.91 |
| METUNv1_460137 | METUNv2_290133 | NADH-quinone oxidoreductase subunit L | 637.25 | 465.50 | −1.37 | 0.83 |
| METUNv1_460138 | METUNv2_290134 | NADH-quinone oxidoreductase subunit M | 372.75 | 302.75 | −1.23 | 0.77 |
| METUNv1_460139 | METUNv2_290135 | NADH-ubiquinone oxidoreductase, chain N | 523.88 | 339.07 | −1.55 | 0.64 |
| METUNv1_660132 | METUNv2_480138 | Ferredoxin-NADP reductase | 522.14 | 879.52 | 1.68 | 0.69 |
| METUNv1_590030 | METUNv2_420029 | Ubiquinol-cytochrome c reductase complex, cytochrome c1 | 2503.55 | 1092.90 | −2.29 | 0.58 |
| METUNv1_590031 | METUNv2_420030 | Ubiquinol-cytochrome c reductase complex, cytochrome b | 2945.38 | 1365.76 | −2.16 | 0.81 |
| METUNv1_590032 | METUNv2_420032 | Ubiquinol-cytochrome c reductase iron-sulfur subunit (Rieske iron-sulfur protein) (RISP) | 3125.54 | 1240.80 | −2.52 | 0.84 |
| METUNv1_590008 | METUNv2_420008 | Peroxidase/catalase (Catalase-peroxidase) | 3075.82 | 1883.61 | −1.63 | 0.84 |
| METUNv1_670031 | METUNv2_490032 | Cytochrome c oxidase subunit 1 | 5577.29 | 4801.32 | −1.16 | 0.93 |
| METUNv1_670030 | METUNv2_490031 | Cytochrome c oxidase subunit 2 | 4105.55 | 3835.175 | −1.07 | 0.87 |
| METUNv1_670034 | METUNv2_490035 | Cytochrome c oxidase subunit 3 | 2746.54 | 2227.3 | −1.23 | 0.89 |
| METUNv1_670033 | METUNv2_490034 | Cytochrome c oxidase assembly protein | 1365.325 | 1525.035 | 1.12 | 0.93 |
| METUNv1_580051 | METUNv2_410050 | Hemin uptake protein hemP (fragment) | 1319.89 | 1517.79 | 1.15 | 0.68 |
| METUNv1_580052 | METUNv2_410051 | Bacterioferritin-associated ferredoxin | 930.37 | 739.50 | −1.26 | 0.80 |
| METUNv1_580053 | METUNv2_410052 | Bfr, Bacterioferritin | 1845.02 | 2568.53 | 1.39 | 0.77 |
* Each sample represents the average of two biological replicates. Values represent reads per kilobase of coding sequence per million (reads) mapped (RPKM).
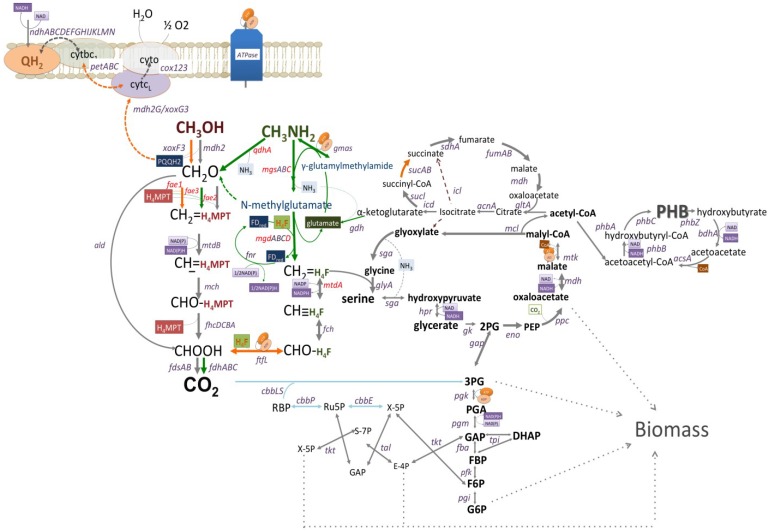
Cells growth on methanol showed a slightly higher (about 1.4 fold) abundance of transcripts for methanol dehydrogenase (mdh2) and the associated cytochrome gene (cyt) (Table 3; Figure 1). The relative abundance of formate-tetrahydrofolate ligase/formyltetrahydrofolate synthetase (ftfL) transcripts, a tungsten-containing aldehyde ferredoxin oxidoreductase, a distant homolog of the PQQ-dependent methanol dehydrogenase (xoxF3G3J3), cytochrome bc1 reductase complex, a putative NADP(H)-dependent aldo/keto reductase and a number of hypothetical proteins were higher (1.6–3 folds) in methanol grown cells (Table 3; Figure 1). Genes encoding the N-methylglutamate pathway enzymes, co-clustered conserved proteins and a putative transcriptional regulator, were up-regulated (6−10-fold) during growth on methylamine (Table 3, Figure 1). Among other genes altered by the shift to methylamine were: a seven gene cluster (qhpRADCBFE) encoding a heme-containing amine dehydrogenase (Qhp, qhpADC), and associated proteins involved in the posttranslational modification and regulation; a ferredoxin; and a distant homolog of the formaldehyde activating enzyme (fae3). The fae3 gene was almost 10 fold higher in cells shifted to methylamine.
Figure 1.
The reconstructed pathway of methanol and methylamine utilization in Methyloversaltilis universalis FAM5 based on genomic, transcriptomic and mutagenesis data. Genes mutated in this study are shown in red. Pathways upregulated (6–11 fold) by growth on methylamine are shown in green; Pathways slightly downregulated (1.5–3 fold) by growth on methylamine are shown in orange; Grey lines show pathways whose expression do not change; and Blue lines indicate steps that are not expressed on tested C1-compounds, methanol or methylamine.
3.2. Mutagenesis Studies: Methylamine Oxidation
It has previously been shown that deletion of two subunits of NMGS (encoded by mgsA and mgsB) completely abolished the ability of the strain M. universalis FAM5 to utilize methylamine; however the mutants were still able to generate small amounts of N-methylglutamate [12]. It has been speculated that the third subunit of the protein (encoded by mgsC) can contribute to the residual production. Here we mutated mgsC and showed that the lack of the gene eliminates the ability of the strain to utilize methylamine (Table 1).
We also tested involvement of the gamma subunit of the NMGDH (mgdD) in methylamine utilization. The mgdD is located at the end of the four-gene cluster encoding the NMGDH. The fourth gene has been identified in all microbes possessing the N-methylglutamate pathway; however, in contrast to the catalytic subunits (mgdA and mgdC) and mgdB, which are typically quite conserved, the gamma subunit of the enzyme is surprisingly divergent [12]. Relatively low homology (30%–45% amino acid identity) has been observed between the mgdD subunit from the Methyloversatilis species and the corresponding protein from betaproteobacterial methylotrophs, such as Methylotenera versatilis 301 or Methylophilus methylotrophus DSM 46235. The gene from Methyloversatilis species shared 25% AA identity with the mgdD gene from Agrobacterium tumefaciens. No homology was observed among mgdD genes from betaproteobacteria and gamma subunit of the NMGDH from alphaprotebacterial methylotrophs, such as Methylocellla silvestris, Methylopila spp, and Methylobacterium extorquens spp. The exact function of the mgdD gene product is still not known. We mutated the mgdD gene in M.universalis FAM5, and the mutant strain lost the ability to utilize methylamine (Table 1). These two additional mutations additionally confirmed the importance of the N-methylglutamate pathway genes for methylamine utilization by M. universalis FAM5.
The transcriptomic study presented above revealed a number of additional functions indicating that the metabolic arrangement of the methylamine utilization in M. universalis FAM5 could be more intricate (Figure 1). Two of the most noticeable differences between cells grown on methanol vs. grown on methylamine are the overexpression of the heme-containing amine dehydrogenases gene cluster and fae3 in cells grown on methylamine. We generated a strain of M. universalis FAM5 lacking the alpha subunit of the Qhp gene (ΔqhpA). The activity of the Qhp enzyme was reduced to background level in the mutant strain (Table 4). However, the strain lacking Qhp gene (ΔqhpA) was able to grow on methylamine similarly to wild type (Table 1). The phenotype of the fae3 mutant is described below.
Table 4.
Activity of the heme-containing amine dehydrogenase in wild type and ΔqhpA-mutant strains upon growth on methylamine.
| Strain | Enzyme Activity (μmol min−1 mg−1 Protein) |
|---|---|
| FAM5 (WT) | 30 ± 9 |
| ΔqhpA | 6 ± 3 |
3.3. Mutagenesis Studies: Metabolic Arrangement Downstream from the Methylamine Oxidation
During growth on methanol, formaldehyde, the end product of alcohol oxidation, is oxidized to formate via the H4MPT pathway. Formate is either oxidized to CO2 to generate energy, or converted to methylene-H4F via an FtfL-Fch-MtdA variant of the H4F pathway. It has been predicted that the end product of the N-methylglutamate pathway is methylene-H4F, which could be incorporated directly into the serine cycle. However it is not known how it is oxidized. The FtfL-Fch-MtdA variant of the H4F pathway is typically linked to assimilation rather that oxidation [36]. In order to investigate the fate of the methylene-H4F in M. universalis FAM5 upon growth on methylamine, two possible scenarios were investigated: (1) The H4F pathway contributes to methylene-H4F oxidation by operating in the reverse direction; (2) The H4F pathway in M. universalis FAM5 contributes only to formate assimilation, and thus it should not be essential for growth on methylamine. However, in order to generate energy and reducing equivalent from C1-oxidation, the M. universalis FAM5 cells should possess an additional system for converting methylene-H4F to formaldehyde, or transfer C1-units from H4F to H4MPT. One candidate for C1-unit decoupling/shuttling is fae3, a homolog of formaldehyde activating enzyme, which has a very strong expression during growth on methylamine, but not methanol.
To verify these scenarios, mutations in H4F (ΔmtdA) and H4MPT pathways (Δfae1) were made. In addition we mutated homologs of the formaldehyde dehydrogenase (Δfae2 and Δfae 3), and generated multiple mutants (Δfae1Δfae2, Δfae1Δfae3, Δfae2Δfae3 and Δfae1Δfae2Δfae3). In addition, a strain lacking fae1 and qhpA was constructed. The mutant phenotypes are shown in Table 1.
Strains lacking mtdA or fae1 genes were not able to grow on methanol or methylamine. Interestingly, revertants were observed for the fae1-mutant. The number of revertants arising on methylamine plates was 10 ± 3 per 109 cells plated. The revertant strains grew on methylamine, albeit with a growth defect compared to the wild type strain (the growth rate was about 30% of the wild type). The revertant strains did not restore the ability to utilize methanol for growth. Single Δfae 2 or Δfae3 mutations, or the double Δfae2Δfae3 mutation displayed a growth rate similar to the wild-type strain on both methanol and methylamine. Δfae1Δfae2 and Δfae1Δfae3 double mutants were not able to grow on C1-compounds, similarly to Δfae1. No revertants were observed for Δfae1Δfae3 mutant upon transfer to methylamine plates. However, the incorporation of the Δfae3 mutation into the revertant Δfae1R strain did not abolish the ability of the strain to utilize methylamine. A similar phenotype was observed for a triple Δfae1RΔfae2Δfae3 mutant. The double Δfae1ΔqhpA mutant phenotype was similar to the single Δfae1 mutant. Like the latter, there were revertants of the double mutant (Δfae1ΔqhpAR) that regained the ability to utilize methylamine for growth. The frequency of reversion was similar to the single fae1-mutant. The triple Δfae3Δfae1ΔqhpAR mutant retained the ability to use methylamine.
4. Discussion
Methylotrophic capabilities of the members of families Rhodocyclaceae/Burholderiaceae have relatively recently been discovered [1,3,4,5,7], and thus not much is known about the pathways for the single carbon utilization in this betaproteobacterial lineage. The M. universalis is the first representative of the family isolated in a pure culture and formally described [6]. The genome of the strain has been sequenced opening up new approaches for investigation of pathways for single carbon or methylated multicarbon compounds investigation [23]. Over the past few years the strain became the model system for understanding molecular mechanisms of C1-carbon utilization and denitrification [8,12,13].
The mutagenesis performed in this study suggests that the N-methylglutamate pathway is the main route for methylamine oxidation. Based on the available genomic and enzymatic data the end product of the N-methylglutamate pathway is methylene-H4F [12,16,20]. Thus it could be predicted that, first, the H4folate pathway contributes to C1-oxidation; and, second, the H4MPT-linked transfer should not be required for growth on methylamine. M. universalis FAM5 possesses the Mtd-Fch-FtfL variant of the H4MPT pathway, which is typically associated with formate assimilation, rather than formaldehyde/methylene-H4F oxidation [25,26]. It has been shown that this portion of the pathway is not essential for growth on methylamine in Methylobacterium spp. [16,20]. We found here that the mtdA mutant is not able to grow on both C1-compounds. The phenotypic data indicate that the enzyme of the pathway can play some additional role in C1-utilization in M. universalis.
We also found that the H4MPT pathway is essential for growth on methylamine. That observation indicates that M. universalis somehow produces methylene-H4MPT during growth on methylamine. Since the heme-dependent amine dehydrogenase (which is predicted to produce formaldehyde) is not essential for growth on methylamine, the C1-units entering H4MPT pathway should come from methylamine oxidation via the N-methylglutamate pathway. Based on the available evidences it could be proposed that the NMGDH releases both methylene-H4F and formaldehyde and that the ratio of the produced compounds depends on the intracellular pool of H4F. It has been shown that the pool of H4F is only a quarter of the pool of H4MPT in facultative methylotrophs [37]. In M.universalis, the relative expression of the dihydrofolate reductase (folA), the only enzyme essential for biosynthesis of H4F (folA) from a supplied source of folate, showed similar levels of expression in methanol or methylamine grown cells. That suggests that the overall capacity of the cells to produce the folate-cofactor did not change. If methylene-H4folate is the only product of methylamine oxidation, the cellular needs for H4F should be much higher upon growth on methylamine. Because formaldehyde is a toxic compound, it is reasonable to speculate that formaldehyde is produced only upon the depletion of the H4F pool. The Fae1 and/or Fae3 enzymes handle the cellular formaldehyde pool, and deliver C1-units into the H4MPT pathway for oxidation. However, while this scenario explains the growth deficiencies of the fae1 mutant, it fails to explain the revertants. It could be speculated that in vivo the N-methylglutamate dehydrogenase uses both H4F and H4MPT cofactors. The preferable compound is H4F, which could be substituted with H4MPT upon depletion of H4F and buildup of H4MPT (due to the lack of Fae enzymes). The latter scenario is also supported by the previous study of methylamine utilization via the NMG-pathway [16], which showed that the H4MPT-biosynthesis, rather than the Fae-driven condensation of formaldehyde, is essential for growth of Methylobacterium extorquens PA1 on methylamine.
The initial reconstruction of the methylotrophy in methylotrophic Rhodocyclaceae/Burholderiaceae has been made based on enzymatic and genetic investigations [13,38,39]. In this study we further evaluated pathways for aerobic utilization of methanol and methylamine via transcriptomics and mutagenesis. The updated reconstruction of the core metabolic pathways in M. universalis FAM5 is shown in Figure 1. It has been demonstrated that methanol oxidation via PQQ-dependent enzymes is operating as a redox arm and is coupled with ATP generation with 0.6–1 mol of ATP produced per 1 mol of methanol oxidized [40]. Thus, the growth on the C1-alcohol is predicted to be reducing-power limited. Interestingly, the transcriptomic studies show that cells of M. universalis FAM5 expressed complex III (cytochrome bc1). As the majority of the cellular energy needs are fulfilled by the first step of methane oxidation, it could be speculated that M.universalis FAM5 uses complex III to supply reducing power for carbon assimilation upon growth on methanol. Methylamine oxidation in M.universalis FAM5 might also be connected to NADPH production, via a ferredoxin-NADP reductase. An uphill electron transfer has been proposed for a number of chemolitotrophs [41,42,43], but it has never been investigated in non-phototrophic methylotrophs. Here we present the first evidence that methylotrophic bacteria might develop a mechanism for restoring reducing power upon growth on C1-compounds. The exact role of the complex III and FNR in cellular energetics will be evaluated in further investigations.
5. Conclusions
Overall, the metabolic arrangement of C1-utilization in M. universalis appears to be quite complex and to some degree redundant. Methylamine utilization is supported by two different enzymatic systems: the N-methylglutamate pathways and heme-dependent amine dehydrogenase. Genes encoding both enzymatic systems are chromosomally co-located and show a very high level of expression upon switch from growth on methanol to growth on methylamine. Like other heme-containing amine dehydrogenases, the enzyme from M. universalis FAM5 is capable of direct conversion of methylamine to formaldehyde. However, from the ΔqhpA phenotypic data it could be suggested that the enzyme is not essential, at least under the growth condition tested, for methylamine utilization.
The functional implication of the enzymatic redundancy of the C1-utilization is not apparent in the experiments described here. It could be speculated, that in the presence of heterogeneous substrates, the simultaneous activation of different enzymatic pathways might provide some benefit. However, it remains to be determined via additional simulation experiments or in-situ investigations.
Acknowledgments
This research was supported by a grant (MCB-0604269) from the US National Science Foundation.
Supplementary Files
Author Contributions
M.G.K. designed the experiments, performed the RNA-seq experiments, compiled and analyzed the data and wrote the paper; A.L. and N.C.M.G. designed and performed mutagenesis experiments; C.M.G performed the enzymatic testing, and D.A.C.B. analyzed the RNA-seq data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Baytshtok V., Kim S., Yu R., Park H., Chandran K. Molecular and biokinetic characterization of methylotrophic denitrification using nitrate and nitrite as terminal electron acceptors. Water Sci. Technol. 2008;58:359–365. doi: 10.2166/wst.2008.391. [DOI] [PubMed] [Google Scholar]
- 2.Cai T., Qian L., Cai S., Chen L. Biodegradation of benazolin-ethyl by strain Methyloversatilis sp. cd-1 isolated from activated sludge. Curr. Microbiol. 2011;62:570–577. doi: 10.1007/s00284-010-9746-7. [DOI] [PubMed] [Google Scholar]
- 3.Gangwar P., Alam S.I., Bansod S., Singh L. Bacterial diversity of soil samples from the western Himalayas, India. Can. J. Microbiol. 2009;55:564–577. doi: 10.1139/W09-011. [DOI] [PubMed] [Google Scholar]
- 4.De Marco P., Pacheco C.C., Figueiredo A.R., Moradas-Ferreira P. Novel pollutant-resistant methylotrophic bacteria for use in bioremediation. FEMS Microbiol. Lett. 2004;234:75–80. doi: 10.1016/j.femsle.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Doronina N.V., Kaparullina E.N., Trotsenko Y.A. Methyloversatilis thermotolerans sp. nov., a novel thermotolerant facultative methylotroph isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2014;64:158–164. doi: 10.1099/ijs.0.055046-0. [DOI] [PubMed] [Google Scholar]
- 6.Kalyuzhnaya M.G., de Marco P., Bowerman S., Pacheco C.C., Lara J.C., Lidstrom M.E., Chistoserdova L. Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int. J. Syst. Evol. Microbiol. 2006;56:2517–2522. doi: 10.1099/ijs.0.64422-0. [DOI] [PubMed] [Google Scholar]
- 7.Ramos-Padron E., Bordenave S., Lin S., Bhaskar I.M., Dong X., Sensen C.W., Fournier J., Voordouw G., Gieg L.M. Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environ. Sci. Technol. 2011;45:439–446. doi: 10.1021/es1028487. [DOI] [PubMed] [Google Scholar]
- 8.Kalyuzhnaya M.G., Hristova K., Lidstrom M.E., Chistoserdova L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: Implications for environmental detection of methylotrophy and evidence for convergent evolution. J. Bacteriol. 2008;190:3817–3823. doi: 10.1128/JB.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakatsu C.H., Hristova K., Hanada S., Meng X.Y., Hanson J.R., Scow K.M., Kamagata Y. Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int. J. Syst. Evol. Microbiol. 2006;56:983–989. doi: 10.1099/ijs.0.63524-0. [DOI] [PubMed] [Google Scholar]
- 10.Smalley N.E., Taipale S., de Marco P., Doronina N.V., Kyrpides N., Shapiro N., Woyke T., Kalyuzhnaya M.G. Functional and genomic diversity of methylotrophic Rhodocyclaceae: description of the new species Methyloversatilis discipulorum sp. nov. Int. J. Syst. Evol. Microbiol. 2015 doi: 10.1099/ijs.0.000190. in press. [DOI] [PubMed] [Google Scholar]
- 11.Hristova K.R., Schmidt R., Chakicherla A.Y., Legler T.C., Wu J., Chain P.S., Scow K.M., Kane S.R. Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol. Appl. Environ. Microbiol. 2007;73:7347–7357. doi: 10.1128/AEM.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latypova E., Yang S., Wang Y.S., Wang T., Chavkin T.A., Hackett M., Schäfer H., Kalyuzhnaya M.G. Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 2010;75:426–439. doi: 10.1111/j.1365-2958.2009.06989.x. [DOI] [PubMed] [Google Scholar]
- 13.Lu H., Kalyuzhnaya M.G., Chandran K. Comparative proteomic analysis reveals insights into anoxic growth of Methyloversatilis universalis FAM5 on methanol and ethanol. Environ. Microbiol. 2012;14:2935–2945. doi: 10.1111/j.1462-2920.2012.02857.x. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y. Metabolism of methylamine in the tea plant (Thea sinensis L.) Biochem. J. 1973;132:753–763. doi: 10.1042/bj1320753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones J.G., Bellion E. In vivo 13C and 15N NMR studies of methylamine metabolism in Pseudomonas species. J. Biol. Chem. 1991;266:11705–11713. [PubMed] [Google Scholar]
- 16.Nayak D.D., Marx C.J. Methylamine utilization via the N-methylglutamate pathway in Methylobacterium extorquens PA1 involves a novel flow of carbon through C1 assimilation and dissimilation pathways. J. Bacteriol. 2014;196:4130–4139. doi: 10.1128/JB.02026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony C. The Biochemistry of Methylotrophs. Academic Press INC; London, UK: 1982. p. 431. [Google Scholar]
- 18.Chen Y., McAleer K.L., Murrell J.C. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl. Environ. Microbiol. 2010;76:4102–4104. doi: 10.1128/AEM.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Scanlan J., Song L., Crombie A., Rahman M.T., Schäfer H., Murrell J.C. γ-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 2010;76:4530–4537. doi: 10.1128/AEM.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruffaz C., Muller E.E., Louhichi-Jelail Y., Nelli Y.R., Guichard G., Bringel F. Genes of the N-methylglutamate pathway are essential for growth of Methylobacterium extorquens DM4 with monomethylamine. Appl. Environ. Microbiol. 2014;80:3541–3550. doi: 10.1128/AEM.04160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wischer D., Kumaresan D., Johnston A., Khawand M.E.L., Stephenson J., Hillebrand-Voiculescu A.M., Chen Y., Murrell C.J. Bacterial metabolism of methylated amines and identification of novel methylotrophs in Movile Cave. ISME J. 2015;9:195–206. doi: 10.1038/ismej.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck D.A.C., McTaggart T.L., Setboonsarng U., Vorobev A., Goodwin L., Shapiro N., Woyke T., Kalyuzhnaya M.G., Lidstrom M.E., Chistoserdovaand L. Multiphyletic origins of methylotrophy in Alphaproteobacteria, exemplified by comparative genomics of Lake Washington isolates. Environ. Microbiol. 2015;17:547–554. doi: 10.1111/1462-2920.12736. [DOI] [PubMed] [Google Scholar]
- 23.Kittichotirat W., Good N.M., Hall R., Bringel F., Lajus A., Médigue C., Smalley N.E., Beck D., Bumgarner R., Vuilleumier S., et al. Genome sequence of Methyloversatilis universalis FAM5T, a methylotrophic representative of the order Rhodocyclales. J. Bacteriol. 2011;193:4541–4542. doi: 10.1128/JB.05331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durham D.R., Perry J.J. Purification and characterization of a heme-containing amine dehydrogenase from Pseudomonas putida. J. Bacteriol. 1978;134:837–843. doi: 10.1128/jb.134.3.837-843.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai T., Deguchi T., Frébort I., Tanizawa K., Okajima T. Identification of genes essential for the biogenesis of quinohemoprotein amine dehydrogenase. Biochemistry. 2014;53:895–907. doi: 10.1021/bi401625m. [DOI] [PubMed] [Google Scholar]
- 26.Takagi K., Torimura M., Kawaguchi K., Kano K., Ikeda T. Biochemical and electrochemical characterization of quinohemoprotein amine dehydrogenase from Paracoccus denitrificans. Biochemistry. 1999;38:6935–6942. doi: 10.1021/bi9828268. [DOI] [PubMed] [Google Scholar]
- 27.Marx C.J., Lidstrom M.E. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 28.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol. 1984;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 29.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1989. p. 1626. [Google Scholar]
- 30.Matsen J.B., Yang S., Stein L.Y., Beck D., Kalyuzhnaya M.G. Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part I. Transcriptomic study. Front. Microbiol. 2013;7:1–13. doi: 10.3389/fmicb.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Pyl T.P., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. PNAS. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 36.Crowther G.J., Kosály G., Lidstrom M.E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 2008;190:5057–5062. doi: 10.1128/JB.00228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagemeier C.H., Chistoserdova L., Lidstrom M.E., Thauer R.K., Vorholt J.A. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 2000;267:3762–3769. doi: 10.1046/j.1432-1327.2000.01413.x. [DOI] [PubMed] [Google Scholar]
- 38.Chistoserdova L. Modularity of methylotrophy, revisited. Environ. Microbiol. 2011;13:2601–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 39.Chistoserdova L., Kalyuzhnaya M.G., Lidstrom M.E. The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 2009;63:477–499. doi: 10.1146/annurev.micro.091208.073600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthony C. Methanol oxidation and growth yields in methylotrophic bacteria: A review. Acta Biotechnol. 1983;3:261–268. doi: 10.1002/abio.370030310. [DOI] [Google Scholar]
- 41.Elbehti A., Brasseur G., Lemesle-Meunier D. First evidence for existence of an uphill electron transfer through the bc1 and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J. Bacteriol. 2000;182:3602–3606. doi: 10.1128/JB.182.12.3602-3606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osyczka A., Moser C.C., Daldal F., Dutton P.L. Reversible redox energy coupling in electron transfer chains. Nature. 2004;427:607–612. doi: 10.1038/nature02242. [DOI] [PubMed] [Google Scholar]
- 43.Sievert S.M., Scott K.M., Klotz M.G., Chain P.S., Hauser L.J., Hemp J., Hügler M., Land M., Lapidus A., Larimer F.W., et al. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.