Abstract
Methylotrophs, which can utilize methane and/or methanol as sole carbon and energy sources, are key players in the carbon cycle between methane and CO2, the two most important greenhouse gases. This review describes the relationships between methylotrophs and plants, and between methanotrophs (methane-utilizers, a subset of methylotrophs) and heterotrophic bacteria. Some plants emit methane and methanol from their leaves, and provide methylotrophs with habitats. Methanol-utilizing methylotrophs in the genus Methylobacterium are abundant in the phyllosphere and have the ability to promote the growth of some plants. Methanotrophs also inhabit the phyllosphere, and methanotrophs with high methane oxidation activities have been found on aquatic plants. Both plant and environmental factors are involved in shaping the methylotroph community on plants. Methanotrophic activity can be enhanced by heterotrophic bacteria that provide growth factors (e.g., cobalamin). Information regarding the biological interaction of methylotrophs with other organisms will facilitate a better understanding of the carbon cycle that is driven by methylotrophs.
Keywords: plant colonization, symbiosis
1. Introduction
Methane and methanol, which are called C1 compounds, are ubiquitous in nature and important intermediates in the global carbon cycle. Methane is produced in anaerobic environments by methanogenic archaea, and is also generated abiotically by biomass burning, coal mining, and the oil industry. Methane is the second most important greenhouse gas after CO2, and its annual emission is estimated to be 580 Tg [1], an amount that has been increasing since the industrial era, and as such, the reduction of atmospheric methane has become a major social demand. It was discovered in 2006 that plant cells produce methane (12–370 ng per g dry weight h−1) [2], which is distinct from the microbially produced methane that is transported by hydrophytes from underground. Subsequent studies showed that various terrestrial plant species produce methane from pectin at significant rates corresponding to 2%–12% of the total global methane release [3,4]. Methanol is a volatile atmospheric carbon compound and its annual emission is estimated to be 150 Tg [5]. Methanol is principally generated from plant pectin and lignin during plant growth and decay (1.5–45.7 μg per g dry weight h−1) [6,7].
Methylotrophs are defined as microbes that can utilize C1 compounds as the sole source of carbon and energy. They include bacteria, yeasts, fungi, and archaea. Methanotrophs, which are the methane-utilizing methylotrophs, are limited to aerobic members of the γ-Proteobacteria, α-Proteobacteria and Verrucomicrobia, and anaerobic archaea and “Candidatus Methylomirabilis oxyfera” [8,9]. Almost all methanotrophs are obligate C1 utilizers, i.e., they are only capable of growth on methane or methanol [10,11]. In the last decade, some methanotroph species in the genera Methylocella, Methylocapsa and Methylocystis were found to be facultative methanotrophs that can utilize organic acids (e.g., acetate) and ethanol [12,13,14]. On the other hand, the other group of methylotrophs, the methanol-utilizing methylotrophs, belong to diverse phyla, and most of them are facultative methylotrophs capable of growth on both single-carbon and multi-carbon compounds [15].
Most of the methane that is released into the atmosphere is oxidized by hydroxyl radicals in the troposphere [16]. Methanotrophs are the only biological sink for methane: they oxidize methane that is released from terrestrial and aquatic environments before it reaches the atmosphere, as well as atmospheric methane [17]. Methanotrophs convert methane into organic compounds, which are further utilized by other organisms. Stable isotope probe (SIP) experiments showed that the methane-derived carbon compounds were found not only in methanol-utilizing methylotrophs and other heterotrophic bacteria, but also in fungi, protists, mussels, and plants [18,19,20,21,22,23]. Therefore, methanotrophs play a critical role in incorporating the carbon atom of methane into the global carbon cycle.
Since some plants emit methane and methanol, the phyllosphere, defined as the aerial part of plants, has been recognized as a habitat for methylotrophs. Phyllospheric methanol-utilizing methylotrophs were discovered in the 1980s [24], and since then, our understanding of their ecology and physiology has increased. On the other hand, studies on phyllospheric methanotrophs progressed after a report in 2006 on methane emission from plants [2].
Here we review recent research advances demonstrating positive relationships between methylotrophs and plants, and between methanotrophs and heterotrophic bacteria.
2. Methanol-Utilizing Methylotrophs on Plants
2.1. Positive Effects of Methylobacterium Species on Plant Growth
The phyllosphere is a well-known habitat of methanol-utilizing methylotrophs, and leaf surfaces are colonized by a large population of these bacteria, which include the genera Methylobacterium, Methylophilus, Methylibium and Hyphomicrobium [25,26,27]. Methylobacterium species, which represent the main genus among the leaf microbial community, are known to have mutualistic relationships with some plants [28,29,30]. In summary, they are known to promote plant growth through plant hormone production and nutrient uptake support. In addition, they may affect plant health by suppressing pathogen growth and inducing systemic resistance [31]. These abilities of Methylobacterium species can be used in agricultural plant cultivation [32,33,34,35]. Further information regarding the positive effects of Methylobacterium species on plant growth can be found in several excellent articles [36,37].
2.2. Methylobacterium Community in the Phyllosphere
Many factors are involved in shaping the microbial community in the phyllosphere. It is known that phyllosphere microorganisms come from seeds, neighboring plants, soil, air, and aerosols [28,37]. In terms of bacterial traits, growth, nutrient availability, stress resistance, attachment, and motility contribute to colonization and survival on plants [37]. Among the limited carbon sources present on plant leaves [38,39], methanol is assumed to be abundant [40], which provides an advantage for colonization of methanol-utilizing methylotrophs on plants [41]. Also, the microbial community composition on leaves is affected by the plant genotype, plant age, soil type, climate and geography, the major driving forces being the plant genotype and geography [28,42,43,44].
The Methylobacterium community from leaves of Arabidopsis thaliana, Medicago truncatula, and surrounding plant species at five sampling sites located at distances ranging from 19 to 190 km was analyzed by culture-independent metagenome sequencing [45]. The results showed that the site of cultivation and plant species had strong effects on Methylobacterium community composition in the phyllosphere. The Methylobacterium communities of Arabidopsis plants from a given site were more similar to those of other plant species from the same site than to communities of Arabidospsis plants from a different site. The Methylobacterium population represented a constant fraction of the phyllosphere bacterial population.
Plant species affect the population size of Methylobacterium in the phyllosphere [45,46]. Leaves of vegetable species that were cultivated proximately in a home garden (ca. 100 m2) were found to each harbor a different Methylobacterium population [46]. Green perilla and eggplant harbored a large population of 107 CFU/g fresh weight, while tomato and okra leaves showed populations of 105 CFU/g fresh weight. Green perilla leaves purchased at some supermarkets also had populations comparable to those on leaves from the garden. These findings indicate that the plant species affects the size of Methylobacterium populations on leaves.
Efficient plant colonization by Methylobacterium was shown to be closely linked to bacterial phylogeny, by competitive tests for colonization of Arabidopsis plants by various Methylobacterium strains [47]. The phylogenetic group consisting of the species M. radiotolerans, M. mesophilicum, and M. fujisawaense represent strong colonizers. Specific interactions at the species level, with the association of specific Methylobacterium species with specific plant species, were observed [48]. The seeds and leaves of red perilla (Perilla frutescens crispa (Thunb.) Makino) harbored a large population of Methylobacterium, dominated by M. fujisawaense-related strains, whereas green perilla (Perilla frutescens viridis (Makino) Makino) harbored some Methylobacterium species (M. radiotolerans, M. fujisawaense and M. komagatae). When red perilla was cultivated at four distantly located sites (four prefectures in Japan) using locally purchased soils, almost all Methylobacterium strains isolated from the harvested leaves and seeds had 16S rRNA gene sequences (1.5 kb) identical to those of Methylobacterium sp. OR01, which was isolated from the parental seeds. This specific association was replicated in a second year trial using seeds harvested in the first year. These results suggested the following life cycle: (1) M. fujisawaense-related cells attached to red perilla seeds competitively proliferate in soils as plants grow, followed by colonization of leaves, ultimately becoming major members of the leaf microbial community; (2) the Methylobacterium cells are closely attached to seeds and are inherited by the next generation of plants. In this case, plant species influences the dominant Methylobacterium species regardless of geographical and environmental factors.
3. Plant-Associated Methanotrophs
3.1. Characteristics of Methanotrophs Inhabiting the Phyllosphere
In contrast to methanol-utilizing methylotrophs, knowledge of methanotrophs in the phyllosphere is limited. The finding of methane emission from plants [2] provided the idea that in addition to methanol, methanotrophs can utilize methane on leaf surfaces. The phyllosphere represents a unique environment [49] in comparison with major methanotroph habitats such as soil, lakes, and wetlands.
The first question was whether methanotrophs inhabit the phyllosphere. Attempts were made to cultivate methanotrophs from natural plant samples with mineral medium and methane. Cultivable methanotrophs were present on leaves and flowers of woody and herbaceous plants at relative high frequency (12%, 41/336 sample) [50]. Doronina et al. cultivated methanotrophs from linden buds and spruce needles [51]. Studies of bacterial communities using a culture-independent metagenomic technique detected methanotrophs on leaves of soybeans, rice, and Tamarix [52,53,54]. The appearance frequencies of sequences assigned to methanotrophs estimated that methanotrophs are minor members of the population (below 1%) in the phyllosphere community. Unfortunately, many metagenomic studies have not described the minor members, and thus little information has been obtained about phyllospheric methanotrophs, despite increasing amounts of metagenome data from plant samples.
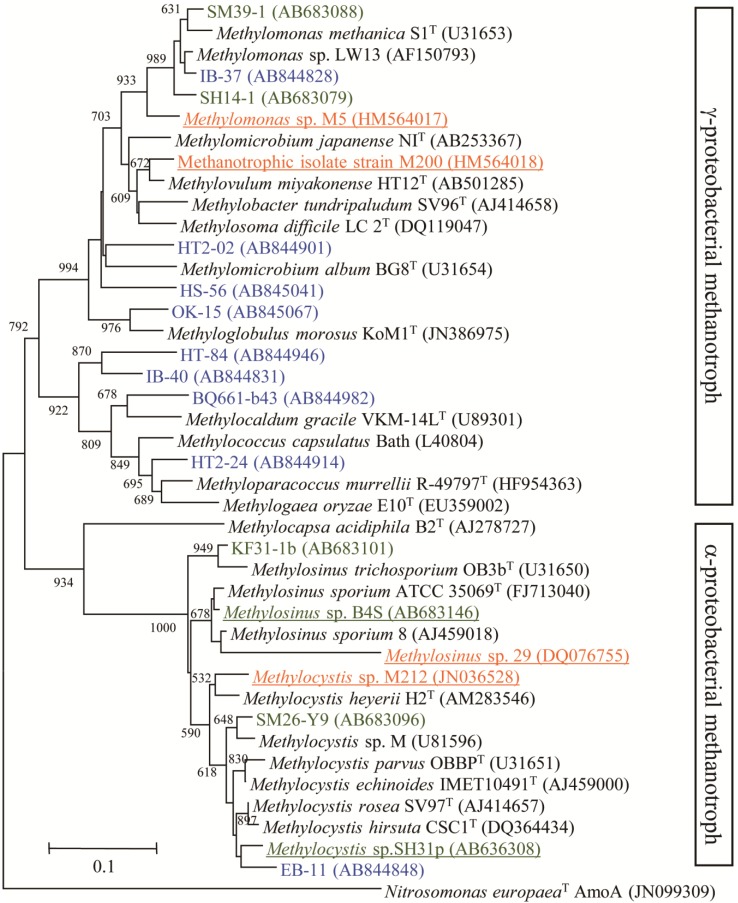
Cultured methanotrophs from phyllosphere samples included γ- and α-Proteobacteria in the genera Methylomonas, Methylobacter, Methylosinus, and Methylocystis [50,51] (Figure 1). Up to three methanotroph species were present in each plant leaf sample. Metagenome sequencing revealed similar methanotrophs in the genera Methylobacter, Methylococcus, and Methylosinus [52,53,54]. These phyllospheric methanotrophs were neither affiliated with novel species nor belonged to phyllosphere-specific phylogenetic lineages. Instead, these genera have been found widespread in a variety of environments [8,55] and are assumed to be major players in the carbon cycle.
Figure 1.
Phylogenetic tree of PmoA sequences from plant-derived methanotrophic isolates (bold letters) and methanotrophic clones retrieved from cultures. Methanotrophs that originated from plant samples are shown in colored letters: Plant tissues in the phyllosphere (green, [50]), macrophytic algae (blue, [56]) and Sphagnum mosses (orange, [57]). Bar, 0.1 substitutions per amino acid position.
Growth of methanotrophs in the phyllosphere were analyzed by inoculating the leaf isolate Methylosinus sp. B4S on Arabidopsis leaves [58]. The inoculated methanotroph did not proliferate on leaves, but survived at least for 15 days, based on GFP expression and enumeration of methanotroph cells. This property can partially explain the high prevalence of methanotrophs with small population densities on leaves.
Methanotrophs use two types of methane monooxygenase (MMO) enzymes, particulate methane monooxygenase (pMMO) and soluble methane monooxygenase (sMMO), depending on copper availability in the environment [11,59]. Transcription analysis of MMO genes in Methylosinus sp. B4S showed higher expression of mmoX (encoding a subunit of sMMO) than that of pmoA (encoding a subunit of pMMO) on leaves [58]. This indicates that the plant leaf is an environment where the bioavailability of copper is low, allowing expression of sMMO genes.
PhyR, a regulator of the general stress response, was first identified as an abundantly produced protein in Methylobacterium extorquens AM1 and was shown to be necessary for plant colonization [60]. PhyR induces resistance to stresses such as heat, UV, reactive oxygen species, osmotic pressure, and drought [61,62], which are experienced in the phyllosphere. The phyR gene is distributed in α-Proteobacteria [63]. In the methanotroph Methylosinus sp. B4S, its function was also verified to regulate UV and heat resistances [58].
3.2. High Methane Oxidation by Methanotrophs on Aquatic Plants
Aqueous environments such as wetlands, paddy fields and lakes represent the largest source of methane (40%–50%) [1], in which much of the methane produced by methanogens is oxidized by methanotrophs before it can be released into the atmosphere. It is unclear whether macrophytes generate methane or methanol. Although methanotrophs have access to high concentrations of methane in these environments, allowing a large population, O2 availability is a limiting factor for methane oxidation. Since methane and O2 concentrations are oppositely correlated relative to water depth, methane oxidation activity is high at the oxic/anoxic interface [64].
Methanotrophs are known to associate with some aquatic plants in sediments and the water phase, and attention has been paid to the methane oxidation activity in the rhizosphere due to O2 release from roots [65,66]. It was recently revealed that shoots (including leaves) of aquatic plants are colonized by methanotrophs with high methane oxidation potentials. Among the tested shoot organs, fully-submerged macrophytic algae collected from a freshwater lake (i.e., Egeria densa, Cabomba caroliniana, and Chara braunii) had much higher oxidation rates (3.7–37 μmol CH4 g−1·h−1) than emergent parts of terrestrial and aquatic plants [56]. This value is higher than that of rice roots (0.2–0.4 μmol CH4 g−1·h−1) [67]. Other studies reported methane oxidation of submerged macrophytic algae [68,69,70], and showed that shoots oxidize more methane than roots (i.e., Myriophyllum exalbescens) [71].
The high methane oxidation potential of shoots is also found in the methanotroph-moss symbiotic system. Methane oxidation associated with Sphagnum mosses in peat bogs occurred at a significant rate (0.3–1.2 μmol CH4 g−1·h−1), and CO2 generated via methane oxidation provided 10%–30% of the carbon source for mosses [23]. The methanotrophs also provide accessible nitrogen (NH4+) for Sphagnum mosses by carrying out N2 fixation [72]. The vegetated environment of the mosses is an important factor affecting methane oxidation capacity [23,73,74]; rates were much higher in submerged moss than in non-submerged moss, although the methanotrophic communities were similar. It has been speculated that this is because of lower methane availability and higher stress levels in the aerial phase than the water phase.
In both the macrophytic algae and moss relationships, such high methane oxidation was attributed to a large population of associated methanotrophs [56,74,75]. No methane oxidation, indicating the absence of a significant methanotroph population, was observed in water from the vegetated environments. The associated methanotrophic community was different between the two plant ecosystems (Figure 1). Submerged macrophytic algae in the lake were dominated by γ-proteobacterial methanotrophs belonging to Methylosarcina and Methylocaldum [56], while Sphagnum mosses in peat bogs were dominantly associated with α-proteobacterial methanotrophs belonging to Methylocystis [57,73]. The methanotroph members were shown to move between mosses through water [75], indicating that mosses are a reservoir of methanotrophs. Together, vegetated environments for these aquatic plants can be a source of plant-associated methanotrophs, since γ- and α-proteobacterial methanotrophs are abundant in freshwater lake sediments and peat soils, respectively [22,76,77,78].
In summary, macrophytic algae and mosses in the aqueous phase are important habitats for methanotrophs. Plant-associated methanotrophs represent a significant population, and thus are expected to actively oxidize methane in natural environments. Such a large plant-associated population may be due in part to the availability of O2 provided by plants as well as the environment, which allows access to nutrients and protection from environmental stress.
4. Stimulation of Methanotrophic Activity by Heterotrophic Bacteria
Syntrophic relationships between methanotrophs and heterotrophic bacteria and between methanotrophs and methanol-utilizing bacteria have been long known. When environmental samples were incubated using methane as a sole carbon source in liquid culture, methanotrophs grew together with methanol-utilizing bacteria and other heterotrophic bacteria [55,79,80]. This relationship is represented as a methane-driven food web, in which methanotrophs provide other microbes with carbon compounds such as methanol, proteins (amino acids), polysaccharides, and nucleic acids [81,82]. However, information about the benefits that methanotrophs obtain from other bacteria is limited. One example is the removal of toxic methanol in batch cultures. Methanotrophs that are sensitive to methanol were rescued by removal of methanol from the culture by a methanol-utilizing Hyphomicrobium sp. [81]. Similarly, we observed that methane oxidation by the methanotroph Methylocystis sp. SS2C was initiated only after methanol in the culture was consumed by Methylobacterium sp. [83]. Explanations for the inhibition of methanotrophic growth by methanol include the low tolerance of methanotrophs to formaldehyde generated by the oxidation of methanol [80], and the competitive inhibition of MMO between methane and methanol [84].
Stimulation of methanotrophic growth by bacteria was investigated using a co-culture system with one methanotrophic strain and each of nine heterotrophic strains [79]. The bacterial strains that were used were from a methane-enrichment culture originating from a forest soil sample. Methane oxidation and growth of the methanotrophs were strongly stimulated by three rhizobia (Rhizobium sp., Mesorhizobium sp., and Sinorhizobium sp.). The growth stimulating factor produced by the partner strains was identified as cobalamin (vitamin B12). This cobalamin-dependent stimulation occurred for diverse γ-proteobacterial methanotrophic genera (Methylovulum, Methyloparacoccus, Methylomonas). These methanotroph strains did not produce cobalamin, and showed very weak or no growth in the absence of cobalamin, indicating that cobalamin is an essential growth factor. In some cases cobalamin auxotrophy may be a strain-specific effect because of the observed difference in the cobalamin-requirement for growth between Methyloparacoccus murrellii strains OS501 and R-49797 [85]. Accordingly, some γ-proteobacterial methanotrophs depend on cobalamin produced by bacteria and archaea in natural environments [86].
A search of the genome of Mvul. miyakonense HT12 (Accession: NZ_AQZU00000000) identified three candidate genes encoding cobalamin-dependent enzymes [86,87], ethanolamine ammonia-lyase, methionine synthase, and ribonucleotide reductase. The genes encoding cobalamin-independent enzymes for the latter two enzymes were also present in the genome. The physiological function of cobalamin in γ-proteobacterial methanotrophs needs further investigation.
Further co-culture experiments were carried out between nine methanotrophic strains and 25 heterotroph strains [88]. The tested methanotrophs could grow on methane without external cobalamin. Heterotrophic strains exerted positive or negative effects on the growth of co-culture depending on the methanotroph tested. No single heterotroph had a positive relationship with all of the tested methanotrophs. The strongest growth stimulation was observed in the co-culture of Methylomonas sp. M5 and Cupriavidus taiwanensis LMG 19424. Co-cultures consisting of γ-proteobacterial methanotrophs (Methylomonas sp. R-45363 and R-45383, and Methylosarcina fibrate DSM 13736) showed highly stimulated growth. Based on genomic analysis, this stimulation was correlated with the ability of the partner heterotroph to synthesize quinone, pyridoxine, and cobalamin.
The two studies described above [79,88] showed that growth of methanotrophs was supported by the specific heterotrophs. On the other hand, Ho et al. revealed that heterotroph diversity affected the methane oxidation rate [89]. The heterotroph diversity in the consortia (that is heterotroph richness) was generated by selecting up to 10 heterotrophic species in two phyla (Proteobacteria and Firmicutes). Increased numbers of heterotroph species in the consortia, regardless of the heterotroph combination, significantly stimulated methane oxidation by Methylomonas methanica. Methane oxidation was not affected by any single heterotrophs, and was decreased by addition of heterotroph spent medium. Therefore, heterotroph richness may provide versatility in metabolic capacity to relieve accumulated inhibitory compounds, thus enhancing methane oxidation. These findings imply that community-level interactions between methanotrophs and heterotrophs contribute to methane oxidation in natural ecological systems.
5. Conclusions and Perspective
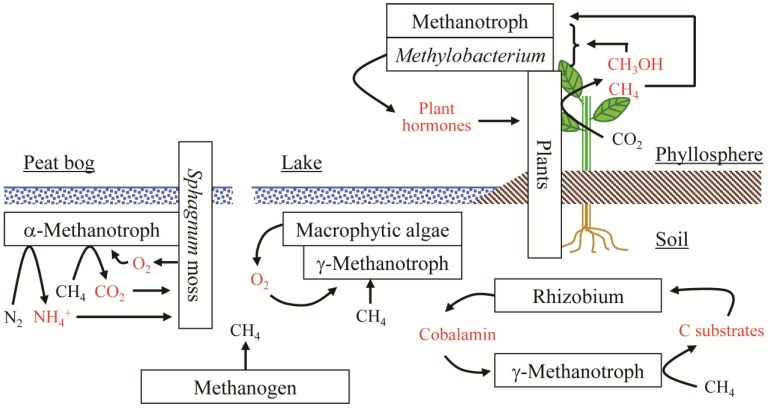
Methylotrophs inhabit soil, water, and plants, and drive the carbon cycle through interaction with neighboring organisms such as plants and bacteria (Figure 2). Plants provide methylotrophs with habitats as well as compounds including growth substrates. In the phyllosphere, Methylobacterium species utilize methanol emitted by plants and affect plant growth. Methanotrophs, despite their small populations, also proliferate or survive in the phyllosphere. Recent studies revealed that the population and community composition of Methylobacterium are affected by plant genotype and vegetated environments. However, information available regarding the traits of plants and bacteria that are involved in shaping the methylotroph community on plants is limited. The strategies and mechanisms used by methylotrophs to select and to adapt to host plants remain to be determined.
Figure 2.
Positive relationships of methylotrophs. In the phyllosphere, C1 compounds generated by plants allow methanol-utilizing methylotrophs (e.g., Methylobacterium species) and methanotrophs to populate. In aquatic environments, methanotrophs belonging to γ- and α-Proteobacteria dominantly colonize on macrophytic algae and Sphagnum mosses, respectively, which supply O2 for methane oxidation by the methanotrophs. In several environments including soils, growth of some γ-proteobacterial methanotrophs require cobalamin supplied from heterotrophic bacteria such as rhizobial species.
Methanotrophs have interactions with plants under water through the exchange of excreted compounds. A much larger population of methanotrophs is assembled on macrophytes than in the water column. Macrophytes provide O2 for methane oxidation, while methanotrophs provide macrophytes with CO2 and NH4+. This mutualism helps to increase the biomass of both methanotrophs and macrophytes, thus stimulating methane oxidation in this ecosystem.
Trophic interactions between methanotrophs and heterotrophs have been observed in terrestrial and aquatic environments. Heterotrophic bacteria function as stimulators of methane oxidation by methanotrophs, e.g., through cobalamin production. This has important implications, which suggest that not only the methanotrophs but also microbial community members besides methanotrophs are involved in methane oxidation activity in the environment. Further studies of methylotrophs and their surrounding organisms and environments could result in the discovery of new biological interactions.
Acknowledgments
This paper was supported in part by grants-in-aid for scientific research (B) (25281063 to H.Y.) from the Japan Society for the Promotion of Science. It was also supported in part by the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.IPCC . Climate Change 2007: The Physical Science Basis. Summary for Policymakers. Cambridge University Press; Cambridge, UK: 2007. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. [Google Scholar]
- 2.Keppler F., Hamilton J.T.G., Brass M., Rockmann T. Methane emissions from terrestrial plants under aerobic conditions. Nature. 2006;439:187–191. doi: 10.1038/nature04420. [DOI] [PubMed] [Google Scholar]
- 3.Keppler F., Hamilton J.T.G., McRoberts W.C., Vigano I., Brass M., Rockmann T. Methoxyl groups of plant pectin as a precursor of atmospheric methane: Evidence from deuterium labelling studies. New Phytol. 2008;178:808–814. doi: 10.1111/j.1469-8137.2008.02411.x. [DOI] [PubMed] [Google Scholar]
- 4.Bruhn D., Moller I.M., Mikkelsen T.N., Ambus P. Terrestrial plant methane production and emission. Physiol. Plant. 2012;144:201–209. doi: 10.1111/j.1399-3054.2011.01551.x. [DOI] [PubMed] [Google Scholar]
- 5.Galbally I.E., Kirstine W. The production of methanol by flowering plants and the global cycle of methanol. J. Atmos. Chem. 2002;43:195–229. [Google Scholar]
- 6.Nemecekmarshall M., Macdonald R.C., Franzen F.J., Wojciechowski C.L., Fall R. Methanol emission from leaves: Enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development. Plant Physiol. 1995;108:1359–1368. doi: 10.1104/pp.108.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macdonald R.C., Fall R. Detection of substantial emissions of methanol from plants to the atmosphere. Atmos. Environ. 1993;27:1709–1713. [Google Scholar]
- 8.Nazaries L., Murrell J.C., Millard P., Baggs L., Singh B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013;15:2395–2417. doi: 10.1111/1462-2920.12149. [DOI] [PubMed] [Google Scholar]
- 9.Ettwig K.F., Butler M.K., Le Paslier D., Pelletier E., Mangenot S., Kuypers M.M., Schreiber F., Dutilh B.E., Zedelius J., de Beer D., et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 10.Semrau J.D., DiSpirito A.A., Vuilleumier S. Facultative methanotrophy: False leads, true results, and suggestions for future research. FEMS Microbiol. Lett. 2011;323:1–12. doi: 10.1111/j.1574-6968.2011.02315.x. [DOI] [PubMed] [Google Scholar]
- 11.Semrau J.D., DiSpirito A.A., Yoon S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 12.Dedysh S.N., Knief C., Dunfield P.F. Methylocella species are facultatively methanotrophic. J. Bacteriol. 2005;187:4665–4670. doi: 10.1128/JB.187.13.4665-4670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunfield P.F., Belova S.E., Vorob’ev A.V., Cornish S.L., Dedysh S.N. Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 2010;60:2659–2664. doi: 10.1099/ijs.0.020149-0. [DOI] [PubMed] [Google Scholar]
- 14.Belova S.E., Baani M., Suzina N.E., Bodelier P.L.E., Liesack W., Dedysh S.N. Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 2010;3:36–46. doi: 10.1111/j.1758-2229.2010.00180.x. [DOI] [PubMed] [Google Scholar]
- 15.Kolb S. Aerobic methanol-oxidizing bacteria in soil. FEMS Microbiol. Lett. 2009;300:1–10. doi: 10.1111/j.1574-6968.2009.01681.x. [DOI] [PubMed] [Google Scholar]
- 16.Guenther A. The contribution of reactive carbon emissions from vegetation to the carbon balance of terrestrial ecosystems. Chemosphere. 2002;49:837–844. doi: 10.1016/s0045-6535(02)00384-3. [DOI] [PubMed] [Google Scholar]
- 17.Aronson E.L., Allison S.D., Helliker B.R. Environmental impacts on the diversity of methane-cycling microbes and their resultant function. Front. Microbiol. 2013;4:225. doi: 10.3389/fmicb.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murase J., Frenzel P. A methane-driven microbial food web in a wetland rice soil. Environ. Microbiol. 2007;9:3025–3034. doi: 10.1111/j.1462-2920.2007.01414.x. [DOI] [PubMed] [Google Scholar]
- 19.Petersen J.M., Dubilier N. Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 2009;1:319–335. doi: 10.1111/j.1758-2229.2009.00081.x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Q., Conrad R., Lu Y. Cross-feeding of methane carbon among bacteria on rice roots revealed by DNA-stable isotope probing. Environ. Microbiol. Rep. 2009;1:355–361. doi: 10.1111/j.1758-2229.2009.00045.x. [DOI] [PubMed] [Google Scholar]
- 21.Radajewski S., Webster G., Reay D.S., Morris S.A., Ineson P., Nedwell D.B., Prosser J.I., Murrell J.C. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology. 2002;148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- 22.He R., Wooller M.J., Pohlman J.W., Quensen J., Tiedje J.M., Leigh M.B. Diversity of active aerobic methanotrophs along depth profiles of arctic and subarctic lake water column and sediments. ISME J. 2012;6:1937–1948. doi: 10.1038/ismej.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghoebarsing A.A., Smolders A.J., Schmid M.C., Rijpstra W.I., Wolters-Arts M., Derksen J., Jetten M.S., Schouten S., Sinninghe Damste J.S., Lamers L.P., et al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153–1156. doi: 10.1038/nature03802. [DOI] [PubMed] [Google Scholar]
- 24.Corpe W.A., Basile D.V. Methanol-utilizing bacteria associated with green plants. Dev. Ind. Microbiol. 1982;23:483–493. [Google Scholar]
- 25.Lopez-Velasco G., Welbaum G.E., Boyer R.R., Mane S.P., Ponder M.A. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J. Appl. Microbiol. 2011;110:1203–1214. doi: 10.1111/j.1365-2672.2011.04969.x. [DOI] [PubMed] [Google Scholar]
- 26.Reisberg E.E., Hildebrandt U., Riederer M., Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS ONE. 2013;8:e78613. doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisberg E.E., Hildebrandt U., Riederer M., Hentschel U. Phyllosphere bacterial communities of trichome-bearing and trichomeless Arabidopsis thaliana leaves. Anton. Leeuw. 2012;101:551–560. doi: 10.1007/s10482-011-9669-8. [DOI] [PubMed] [Google Scholar]
- 28.Vorholt J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 29.Fedorov D.N., Doronina N.V., Trotsenko Y.A. Phytosymbiosis of aerobic methylobacteria: New facts and views. Microbiology. 2011;80:443–454. [PubMed] [Google Scholar]
- 30.Trotsenko Y.A., Ivanova E.G., Doronina N.V. Aerobic methylotrophic bacteria as phytosymbionts. Microbiology. 2001;70:623–632. [Google Scholar]
- 31.Madhaiyan M., Poonguzhali S., Senthilkumar M., Seshadri S., Chung H.Y., Yang J.C., Sundaram S., Sa T.M. Growth promotion and induction of systemic resistance in rice cultivar Co-47 (Oryza sativa L.) by Methylobacterium spp. Bot. Bull. Acad. Sinica. 2004;45:315–324. [Google Scholar]
- 32.Tani A., Takai Y., Suzukawa I., Akita M., Murase H., Kimbara K. Practical application of methanol-mediated mutualistic symbiosis between Methylobacterium species and a roof greening moss, Racomitrium japonicum. PLoS ONE. 2012;7:e33800. doi: 10.1371/journal.pone.0033800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meena K.K., Kumar M., Kalyuzhnaya M.G., Yandigeri M.S., Singh D.P., Saxena A.K., Arora D.K. Epiphytic pink-pigmented methylotrophic bacteria enhance germination and seedling growth of wheat (Triticum aestivum) by producing phytohormone. Anton. Leeuw. 2012;101:777–786. doi: 10.1007/s10482-011-9692-9. [DOI] [PubMed] [Google Scholar]
- 34.Madhaiyan M., Poonguzhali S., Kang B.G., Lee Y.J., Chung J.B., Sa T.M. Effect of co-inoculation of methylotrophic Methylobacterium oryzae with Azospirillum brasilense and Burkholderia pyrrocinia on the growth and nutrient uptake of tomato, red pepper and rice. Plant Soil. 2010;328:71–82. [Google Scholar]
- 35.Verginer M., Siegmund B., Cardinale M., Muller H., Choi Y., Miguez C.B., Leitner E., Berg G. Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol. Ecol. 2010;74:136–145. doi: 10.1111/j.1574-6941.2010.00942.x. [DOI] [PubMed] [Google Scholar]
- 36.Kwak M.J., Jeong H., Madhaiyan M., Lee Y., Sa T.M., Oh T.K., Kim J.F. Genome information of Methylobacterium oryzae, a plant-probiotic methylotroph in the phyllosphere. PLoS ONE. 2014;9:e106704. doi: 10.1371/journal.pone.0106704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 38.Morgan J.V., Tukey H.B. Characterization of leachate from plant foliage. Plant Physiol. 1964;39:590–593. doi: 10.1104/pp.39.4.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiala V., Glad C., Martin M., Jolivet E., Derridj S. Occurrence of soluble carbohydrates on the phylloplane of maize (Zea Mays L.): Variations in relation to leaf heterogeneity and position on the plant. New Phytol. 1990;115:609–615. [Google Scholar]
- 40.Kawaguchi K., Yurimoto H., Oku M., Sakai Y. Yeast methylotrophy and autophagy in a methanol-oscillating environment on growing Arabidopsis thaliana leaves. PLoS ONE. 2011;6:e25257. doi: 10.1371/journal.pone.0025257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sy A., Timmers A.C., Knief C., Vorholt J.A. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 2005;71:7245–7252. doi: 10.1128/AEM.71.11.7245-7252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkel O.M., Burch A.Y., Elad T., Huse S.M., Lindow S.E., Post A.F., Belkin S. Distance-decay relationships partially determine diversity patterns of phyllosphere bacteria on Tamarix trees across the Sonoran Desert. Appl. Environ. Microbiol. 2012;78:6187–6193. doi: 10.1128/AEM.00888-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siefert A., Fridley J.D., Ritchie M.E. Community functional responses to soil and climate at multiple spatial scales: When does intraspecific variation matter? PLoS ONE. 2014;9:e111189. doi: 10.1371/journal.pone.0111189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redford A.J., Bowers R.M., Knight R., Linhart Y., Fierer N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010;12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knief C., Ramette A., Frances L., Alonso-Blanco C., Vorholt J.A. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J. 2010;4:719–728. doi: 10.1038/ismej.2010.9. [DOI] [PubMed] [Google Scholar]
- 46.Mizuno M., Yurimoto H., Yoshida N., Iguchi H., Sakai Y. Distribution of pink-pigmented facultative methylotrophs on leaves of vegetables. Biosci. Biotechnol. Biochem. 2012;76:578–580. doi: 10.1271/bbb.110737. [DOI] [PubMed] [Google Scholar]
- 47.Knief C., Frances L., Vorholt J.A. Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microb. Ecol. 2010;60:440–452. doi: 10.1007/s00248-010-9725-3. [DOI] [PubMed] [Google Scholar]
- 48.Mizuno M., Yurimoto H., Iguchi H., Tani A., Sakai Y. Dominant colonization and inheritance of Methylobacterium sp. strain OR01 on perilla plants. Biosci. Biotechnol. Biochem. 2013;77:1533–1538. doi: 10.1271/bbb.130207. [DOI] [PubMed] [Google Scholar]
- 49.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iguchi H., Sato I., Sakakibara M., Yurimoto H., Sakai Y. Distribution of methanotrophs in the phyllosphere. Biosci. Biotechnol. Biochem. 2012;76:1580–1583. doi: 10.1271/bbb.120281. [DOI] [PubMed] [Google Scholar]
- 51.Doronina N.V., Ivanova E.G., Suzina N.E., Trotsenko Y.A. Methanotrophs and methylobacteria are found in woody plant tissues within the winter period. Microbiology. 2004;73:702–709. [PubMed] [Google Scholar]
- 52.Ikeda S., Anda M., Inaba S., Eda S., Sato S., Sasaki K., Tabata S., Mitsui H., Sato T., Shinano T., et al. Autoregulation of nodulation interferes with impacts of nitrogen fertilization levels on the leaf-associated bacterial community in soybeans. Appl. Environ. Microbiol. 2011;77:1973–1980. doi: 10.1128/AEM.02567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkel O.M., Burch A.Y., Lindow S.E., Post A.F., Belkin S. Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Environ. Microbiol. 2011;77:7647–7655. doi: 10.1128/AEM.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., von Mering C., Vorholt J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanson R.S., Hanson T.E. Methanotrophic bacteria. Microbiol. Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida N., Iguchi H., Yurimoto H., Murakami A., Sakai Y. Aquatic plant surface as a niche for methanotrophs. Front. Microbiol. 2014;5:30. doi: 10.3389/fmicb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kip N., Ouyang W., van Winden J., Raghoebarsing A., van Niftrik L., Pol A., Pan Y., Bodrossy L., van Donselaar E.G., Reichart G.J., et al. Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl. Environ. Microbiol. 2011;77:5643–5654. doi: 10.1128/AEM.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iguchi H., Sato I., Yurimoto H., Sakai Y. Stress resistance and C1 metabolism involved in plant colonization of a methanotroph Methylosinus sp. B4S. Arch. Microbiol. 2013;195:717–726. doi: 10.1007/s00203-013-0922-6. [DOI] [PubMed] [Google Scholar]
- 59.Lieberman R.L., Rosenzweig A.C. Biological methane oxidation: Regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit. Rev. Biochem. Mol. Biol. 2004;39:147–164. doi: 10.1080/10409230490475507. [DOI] [PubMed] [Google Scholar]
- 60.Gourion B., Rossignol M., Vorholt J.A. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. USA. 2006;103:13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gourion B., Francez-Charlot A., Vorholt J.A. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J. Bacteriol. 2008;190:1027–1035. doi: 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaczmarczyk A., Campagne S., Danza F., Metzger L.C., Vorholt J.A., Francez-Charlot A. Role of Sphingomonas sp. strain Fr1 PhyR-NepR-sEcfG cascade in general stress response and identification of a negative regulator of PhyR. J. Bacteriol. 2011;193:6629–6638. doi: 10.1128/JB.06006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staron A., Mascher T. General stress response in a-proteobacteria: PhyR and beyond. Mol. Microbiol. 2010;78:271–277. doi: 10.1111/j.1365-2958.2010.07336.x. [DOI] [PubMed] [Google Scholar]
- 64.Borrel G., Jezequel D., Biderre-Petit C., Morel-Desrosiers N., Morel J.P., Peyret P., Fonty G., Lehours A.C. Production and consumption of methane in freshwater lake ecosystems. Res. Microbiol. 2011;162:832–847. doi: 10.1016/j.resmic.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Van Bodegom P., Stams F., Mollema L., Boeke S., Leffelaar P. Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl. Environ. Microbiol. 2001;67:3586–3597. doi: 10.1128/AEM.67.8.3586-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu L., Ma K., Lu Y. Rice roots select for type I methanotrophs in rice field soil. Syst. Appl. Microbiol. 2009;32:421–428. doi: 10.1016/j.syapm.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 67.Eller G., Frenzel P. Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl. Environ. Microbiol. 2001;67:2395–2403. doi: 10.1128/AEM.67.6.2395-2403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heilman M.A., Carlton R.G. Methane oxidation associated with submersed vascular macrophytes and its impact on plant diffusive methane flux. Biogeochemistry. 2001;52:207–224. [Google Scholar]
- 69.King G.M. Associations of methanotrophs with the roots and rhizomes of aquatic vegetation. Appl. Environ. Microbiol. 1994;60:3220–3227. doi: 10.1128/aem.60.9.3220-3227.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calhoun A., King G.M. Regulation of root-associated methanotrophy by oxygen availability in the rhizosphere of two aquatic macrophytes. Appl. Environ. Microbiol. 1997;63:3051–3058. doi: 10.1128/aem.63.8.3051-3058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorrell B.K., Downes M.T., Stanger C.L. Methanotrophic bacteria and their activity on submerged aquatic macrophytes. Aquat. Bot. 2002;72:107–119. [Google Scholar]
- 72.Larmola T., Leppanen S.M., Tuittila E.S., Aarva M., Merila P., Fritze H., Tiirola M. Methanotrophy induces nitrogen fixation during peatland development. Proc. Natl. Acad. Sci. USA. 2014;111:734–739. doi: 10.1073/pnas.1314284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kip N., van Winden J.F., Pan Y., Bodrossy L., Reichart G.J., Smolders A.J.P., Jetten M.S.M., Damste J.S.S., Op den Camp H.J.M. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 2010;3:617–621. [Google Scholar]
- 74.Liebner S., Zeyer J., Wagner D., Schubert C., Pfeiffer E.M., Knoblauch C. Methane oxidation associated with submerged brown mosses reduces methane emissions from Siberian polygonal tundra. J. Ecol. 2011;99:914–922. [Google Scholar]
- 75.Putkinen A., Larmola T., Tuomivirta T., Siljanen H.M.P., Bodrossy L., Tuittila E.S., Fritze H. Water dispersal of methanotrophic bacteria maintains functional methane oxidation in Sphagnum mosses. Front. Microbiol. 2012;3:15. doi: 10.3389/fmicb.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rahalkar M., Deutzmann J., Schink B., Bussmann I. Abundance and activity of methanotrophic bacteria in littoral and profundal sediments of lake constance (Germany) Appl. Environ. Microbiol. 2009;75:119–126. doi: 10.1128/AEM.01350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsutsumi M., Kojima H., Fukui M. Vertical profiles of abundance and potential activity of methane-oxidizing bacteria in sediment of Lake Biwa, Japan. Microb. Environ. 2012;27:67–71. doi: 10.1264/jsme2.ME11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y., Dumont M.G., McNamara N.P., Chamberlain P.M., Bodrossy L., Stralis-Pavese N., Murrell J.C. Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 2008;10:446–459. doi: 10.1111/j.1462-2920.2007.01466.x. [DOI] [PubMed] [Google Scholar]
- 79.Iguchi H., Yurimoto H., Sakai Y. Stimulation of methanotrophic growth in co-cultures by cobalamin excreted by rhizobia. Appl. Environ. Microbiol. 2011;77:8509–8515. doi: 10.1128/AEM.05834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bowman J. The methanotrophs—The families Methylococcaceae and Mzethylocystaceae. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., editors. The Prokaryotes. Springer; New York, NY, USA: 2006. pp. 266–289. [Google Scholar]
- 81.Wilkinson T.G., Topiwala H.H., Hamer G. Interactions in a mixed bacterial population growing on methane in continuous culture. Biotechnol. Bioeng. 1974;16:41–59. doi: 10.1002/bit.260160105. [DOI] [PubMed] [Google Scholar]
- 82.Lamb S.C., Garver J.C. Batch- and continuous-culture studies of a methane-utilizing mixed culture. Biotechnol. Bioeng. 1980;22:2097–2118. doi: 10.1002/bit.260221009. [DOI] [PubMed] [Google Scholar]
- 83.Iguchi H., Yurimoto H., Sakai Y. (Kyoto University, Kyoto, Japan). 2012. Unpublished work.
- 84.Colby J., Stirling D.I., Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoefman S., van der Ha D., Iguchi H., Yurimoto H., Sakai Y., Boon N., Vandamme P., Heylen K., de Vos P. Methyloparacoccus murrellii gen. nov., sp. nov., a methanotroph isolated from pond water. Int. J. Syst. Evol. Microbiol. 2014;64:2100–2107. doi: 10.1099/ijs.0.057760-0. [DOI] [PubMed] [Google Scholar]
- 86.Roth J.R., Lawrence J.G., Bobik T.A. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 87.Sanudo-Wilhelmy S.A., Gomez-Consarnau L., Suffridge C., Webb E.A. The role of B vitamins in marine biogeochemistry. Annu. Rev. Mar. Sci. 2014;6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 88.Stock M., Hoefman S., Kerckhof F.M., Boon N., de Vos P., de Baets B., Heylen K., Waegeman W. Exploration and prediction of interactions between methanotrophs and heterotrophs. Res. Microbiol. 2013;164:1045–1054. doi: 10.1016/j.resmic.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 89.Ho A., de Roy K., Thas O., de Neve J., Hoefman S., Vandamme P., Heylen K., Boon N. The more, the merrier: Heterotroph richness stimulates methanotrophic activity. ISME J. 2014;8:1945–1948. doi: 10.1038/ismej.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]