Abstract
The sulfur oxygenase reductase (Sor) catalyzes the oxygen dependent disproportionation of elemental sulfur, producing sulfite, thiosulfate and sulfide. Being considered an “archaeal like” enzyme, it is also encoded in the genomes of some acidophilic leaching bacteria such as Acidithiobacillus caldus, Acidithiobacillus thiooxidans, Acidithiobacillus ferrivorans and Sulfobacillus thermosulfidooxidans, among others. We measured Sor activity in crude extracts from Sb. thermosulfidooxidans DSM 9293T. The optimum temperature for its oxygenase activity was achieved at 75 °C, confirming the “thermophilic” nature of this enzyme. Additionally, a search for genes probably involved in sulfur metabolism in the genome sequence of Sb. thermosulfidooxidans DSM 9293T was done. Interestingly, no sox genes were found. Two sor genes, a complete heterodisulfidereductase (hdr) gene cluster, three tetrathionate hydrolase (tth) genes, three sulfide quinonereductase (sqr), as well as the doxD component of a thiosulfate quinonereductase (tqo) were found. Seven At. caldus strains were tested for Sor activity, which was not detected in any of them. We provide evidence that an earlier reported Sor activity from At. caldus S1 and S2 strains most likely was due to the presence of a Sulfobacillus contaminant.
Keywords: Sulfobacillus thermosulfidooxidans, Acidithiobacillus caldus, sulfur metabolism, sulfur oxygenase reductase, genome
1. Introduction
Control of biological sulfur oxidation is important in bioleaching operations for the industrial bioleaching of metal sulfides or heavy metal recovery from industrial wastes [1]. Moderately thermoacidophilic leaching bacteria such as At. caldus and Sb. thermosulfidooxidans, are frequently found in leaching operations [2,3]. Sulfobacillus are Gram-positive, rod shaped, spore forming bacteria able to use S°, reduced inorganic sulfur compounds (RISC), ferrous iron and/or metal sulfide minerals as energy sources under chemolithoautotrophic or mixotrophic conditions [4]. Heterotrophic growth is also possible under low concentrations of organic substrates [5]. Complete genome sequences for Sb. thermosulfidooxidans DSM 9293T (NCBI taxon: 929705) and “Cutipay” strain [6], as well as for Sb. acidophilus strains DSM 10332T [7] and TPY [8] are available. At. caldus is a moderately thermophilic acidophilic Gram-negative bacterium able to chemolithoautotrophically oxidize S° and RISC such as tetrathionate or thiosulfate [9]. Although it cannot oxidize ferrous iron or pyrite, it can grow on RISCs resulting from pyrite oxidation in combination with iron oxidizers like Leptospirillum spp. [10]. It is also able to oxidize arsenopyrite [11]. Complete genome sequences exist for At. caldus DSM 8584T [12] and At. caldus SM-1[13].
In thermophilic archaea, such as Acidianus ambivalens, a sulfur oxygenase reductase (Sor) catalyzes an oxygen-dependent S° disproportionation reaction to thiosulfate, sulfite and hydrogen sulfide. In this one 3 moles of S° are converted into 1 mole sulfite and 2 moles of hydrogen sulfide [14,15]. It does not require addition of external cofactors for its activity. Although no energy conservation occurs during Sor catalysis, its reaction products can be further oxidized by other enzymes to sulfate [16]. Recombinant Sor enzymes from A. ambivalens [17], Acidianus tengchongensis [18], Aquifex aeolicus [19] and Halothiobacillus neapolitanus [20] have been expressed in E. coli and their activity has been reported.
Several other enzymes for sulfur oxidation are conserved among thermophilic and mesophilic acidophiles. Sulfite is substrate for sulfite acceptor oxidoreductases (Saor), which catalyze its oxidation to sulfate [21]. In addition sulfite can abiotically react with an excess of S° to form thiosulfate, which is substrate for a thiosulfate:quinone oxidoreductase (Tqo), catalyzing the generation of tetrathionate and feeding the electrons into the quinone pool in the cytoplasmic membrane. A. ambivalens Tqo is composed of two subunits, named DoxD and DoxA [22]. Interestingly, in At. ferrooxidans both dox genes are fused and duplicated, named doxDA1 and doxDA2 [23]. At. caldus Dox proteins have similar sizes as At. ferrooxidans DoxDA, but just the DoxD domain is found to be present [24]. In addition to its generation by the abiotic reaction of S° with sulfite, thiosulfate is generated by the reaction of tetrathionate hydrolase (Tth), which in A. ambivalens catalyzes its decomposition to sulfate, thiosulfate and S° [16]. The Acidianus Tth is biochemically and phylogenetically similar to the Acidithiobacillus Tth [25]. Both enzymes are located outside the cell and have optimal activities at acidic pH [26].
Apart from the Saor activity, the enzymes adenylylsulfate (Aps) reductase and adenylyl transferase (Apat) are involved in the generation of ATP from sulfite by substrate level phosphorylation [27]. The third product, hydrogen sulfide, is oxidized back to S° by the membrane bound sulfide:quinone oxidoreductase (Sqr) [28,29,30]. All electrons made available from sulfur oxidation in the course of Sqr, Sar and Tqo activities reduce Caldariella quinones (CQ) but not cytochromes [16,22]. The bacterial “Sox” (sulfur-oxidizing) system consists of a set of dehydrogenases and other periplasmic proteins which catalyze the oxidation of sulfide, S°, thiosulfate and sulfite to sulfate, accompanied by subsequent electron transfers through di- and mono-heme cytochromes [31]. The sox gene cluster of Paracoccus pantotrophus comprises 15 genes, encoding among others for the periplasmic proteins SoxXA, SoxYZ, SoxB, Sox(CD)2, which interact with each other [32,33]. SoxXA is composed by the diheme citochrome SoxA and the monoheme cytochrome SoxX. The SoxYZ complex does not contain cofactors and is probably involved in the reaction cycle. Sox(CD)2 is composed of the molybdoprotein SoxC and the diheme cytochrome C protein SoxD. An incomplete sox system, where soxCD orthologous genes are missing, has been found encoded in the genomes of At. caldus [12] and Acidithiobacillus ferrivorans [34]. Enzyme reconstitution assays with P. pantotrophus Sox, have shown that the absence of the tetrameric protein Sox(CD)2 reduced the activity of the Sox pathway from 8 mol of electrons/mol of thiosulfate to two mols of electrons/mol of thiosulfate [35]. The Sox multienzyme-complex is absent in the mesophilic acidophilic leaching bacterium At. ferrooxidans [36], in which a sulfur dioxygenase (Sdo) has been proposed to be responsible for the S° oxidation step [37]. Recently a deletion mutant strain of a putative At. ferrooxidans ATCC 23270T Sdo was constructed. The mutant strain still possessed Sdo activity, suggesting a dissimilatory function of this enzyme and the presence of other enzyme(s) responsible for Sdo activity [38]. By bioinformatics and transcriptomic analyses it has also been suggested that the gene cluster hdrABC (heterodisulfide reductase) and some of its accessory proteins, which are conserved in several acidithiobacilli as well as in sulfur oxidizing archaea, could catalyze a similar sulfur oxidation reaction as Sdo [36]. However, biochemical evidence to support this proposal is missing. Proteins containing Rhodanese domain(s) are ubiquitous sulfur transferase enzymes that catalyze the transfer of a sulfane sulfur atom from a donor to an appropriate sulfur acceptor in vitro. These can belong to the thiosulfate:cyanide sulfurtransferase (TST) or the 3-mercaptopyruvate sulfurtransferases (MSTs) family [39].
At. caldus ATCC 53993T possesses a sor gene encoded on its genome sequence [12]. Previously we reported the presence of Sor enzyme activity in At. caldus strains S1 and S2 [40]. However, our attempts to measure Sor activity in At. caldusT and some other strains were unsuccessful (see Materials and Methods). Four sor sequences were obtained from metagenomic DNA samples from a bioreactor treating gold concentrates. This reactor contained species of Leptospirillum, Sulfobacillus, Acidithiobacillus and Sphingomonas. One of these sor genes (DQ480734) was cloned and expressed in E. coli. The recombinant Sor showed an optimal oxygenase activity of 3.76 U/mg at 75–80 °C and pH 7.5. This protein was attributed to belong to At. caldus SM-1 [41]. However, further analysis of the complete genome sequence of At. caldus SM-1 showed that the sor gene was missing in this strain. Its deletion was explained by an event of transposition of the element ISAtc1 [13].
BLAST searches revealed the presence of sor genes encoded in the genomes of Sb. thermosulfidooxidans DSM 9293T and in S. acidophilus strains TPY and DSM 10332T. In this article we report that Sb. thermosulfidooxidans DSM 9293T crude extracts possess Sor activity. We also provide evidence that the previously reported Sor activity in At. caldus strains S1 and S2 was most likely due to the presence of a Sulfobacillus contaminant in strains S1 and S2.
2. Materials and Methods
2.1. Strains Used in This Study
Sb. thermosulfidooxidans DSM 9232T, the At. caldus strains: DSM 8584T, DSM 9466 (former C-SH12), S1, S2, MNG, f, and #6 were used. Strains S1 and S2 were provided by Zhou H. (Central South University of Changsha, China). Strains MNG, f, & 6# were described by Rawlings et al. [42].
All A. caldus strains as well as Sulfolobus metallicus DSM 6482T were grown in Mackintosh (Mac) basal salt medium [43], at pH 2.5, supplemented with 5 g/L S° and traces of ferric sulfate (~1 mg/L). Media were autoclaved at 110 °C for 90 min. S. metallicus was used as a positive control for Sor enzyme activity tests. For Sb. thermosulfidooxidans and S. metallicus, 0.2 g/L yeast extract was added after autoclaving. Batch cultures (10 L) of Sb. thermosulfidooxidans or At. caldus for Sor enzyme assays were grown at 45 °C with aeration and stirring at 300 rpm. S. metallicus cultures (5L) were incubated at 65 °C without shaking. When necessary, S° was removed by low speed centrifugation for 5 min at 120× g before cell harvesting. Additionally, Sb. thermosulfidooxidans was also grown in Mac basal salt solution with 2 g/L ferrous iron ions (supplied as FeSO4·7 H2O) and 0.2 g/L yeast extract). During our experiments, after detecting Sulfobacillus contamination, At. caldus strains S1 and S2 were repurified by three consecutive rounds of maximal serial 10-fold dilutions in Mac medium amended with S°.
2.2. Molecular Biology Techniques
DNA was extracted as described [44]. PCR reactions were done in a final reaction volume of 25 μL using 20–50 ng of genomic DNA template, 1× Green Flexi buffer, 2.5 mM MgCl2, 1 mM dNTPs, 10 pmol of each single primer and 0.5 U GoTaq® DNA polymerase (Promega®, MI, Wisconsin, USA). Reactions were incubated in an Eppendorf Mastercyler 5332, (Hamburg, Germany). The following temperature program was used: five minutes initial denaturation at 95 °C followed by 30 to 40 cycles of denaturation for 30 s at 95 °C, primer annealing for 30 to 45 s at 50 to 58 °C, depending on each primer pair used (Table 1), and 0.5 to 1.5 min of extension at 72 °C, depending on the size of the expected amplicon. A final extension step was done for 3 min at 72 °C.
Table 1.
Polymerase chain reaction (PCR) primers used in this study.
| Primer | Sequence 5ʹ→3ʹ | Target Gene | Amplicon Size | References |
|---|---|---|---|---|
| 16s_27fw | agagtttgatcctggctcag | 16S rDNA | ~1.5 kb | Lane et al. 1991 [46] |
| 16s_1492rv | gcctaccttgttacgactt | Bacteria | ||
| Arch25F | cyggttgatcctgccrg | 18S rDNA | ~1.5 kb | Achenbach and Woese 1995 [47] |
| Arch1492R | tacggytaccttgttacgactt | Archaea | ||
| sorC1-F | Gtiggiccnaargtntgy * | Sor | ~230 bp | Chen et al. 2007 [41] |
| sorH1-R | rtgcatntcytcrtgrtc | |||
| bsor_1F | gtccttcgagaccatgatgmargtnggncc | bacterialsor (CODEHOP) | ~800 bp | This study |
| bsor_2R | ccgccactgggcctsytccatcatng | |||
| PCJ2_for | caggcctcccagcaggtnggnccnaa | sor(CODEHOP) | 840 bp | This study |
| PCJ3_rev | ctcccgccatgaggtgtcctccatnayngg | |||
| SULFO170F | caatcccgcatacgttcc | 16S rDNA | 436 bp | De Wulf-Durand et al. 1997 [45] |
| SULFO606R | aaaccgctacgtatcgcac | Sulfobacillus spp. | ||
| CALD460F | atccgaatacggtctgcta | 16S rDNA | ~1 kb | De Wulf-Durand et al. 1997 [45] |
| CALD1475R | tataccgtggtcgtcgcc | At. caldus | ||
| THIO458F | gggtgctaatawcgcctgctg | 16S rDNA | ~1 kb | De Wulf-Durand et al. 1997 [45] |
| THIO1473R | taccgtggtcatcgccct | At. thiooxidans | ||
| LEPTO176F | cgaatagtatccggttccg | 16S rDNA | 503 bp | De Wulf-Durand et al. 1997 [45] |
| LEPTO679R | aaattccgcttccctctcc | Leptospirillumspp. | ||
| FERRO458F | gggttctaatacaatctgct | 16S rDNA | ~1 kb | De Wulf-Durand et al. 1997 [45] |
| FERRO1473 | taccgtggtaaccgccct | At. ferrooxidans | ||
| T7 | taatacgactcactataggg | Promoterregions | 158 bp | Promega® pGEM-T vector manual |
| SP6 | atttaggtgacactatagaa | in pGEM®-T vector |
* i, inosine.
Purity tests of At. caldus S1 and S2 strains were done by a two-stage nested polymerase chain reaction (PCR)-mediated detection method [45]. Additionally, At. caldus strains S1 and S2 were tested for archaeal contamination [46,47]. At. caldus sor genes were amplified with consensus-degenerate hybrid oligonucleotide primer (CODEHOP)-PCR primers [48]. For this, the primer pairs PCJ2_for-PCJ3_rev and bsor_1F-bsor_2R (Table 1) were designed based on the alignments of amino acid sequences of all Sor proteins, or just the bacterial ones available at the time of this study, respectively. The latter ones included At. caldusT (EET26704.1), uncultured bacterium BSB (gi:94470458), Halothiobacillus neapolitanus C2 (ACX96058.1) and Desulfomicrobium baculatum DSM 4028 (ACU89275.1). Positive sor gene amplicons were cloned using the pGEM®-T vector system (Promega®). Ligation reactions were transformed in competent E. coli DH5α cells. Plasmids were isolated using Roti®-Prep Plasmid MINI Kit (Carl Roth, Karlsruhe, Germany). The presence of a cloned insert was confirmed by PCR using T7 and SP6 primers, adjacent to its cloning site. DNA sequencing was done in “Zentraler DNA-Sequenzierservice”, Universitätsklinikum Essen.
2.3. Bioinformatics and Phylogeny Analyses
Gene sequences were analyzed in the databases of the National Center for Biotechnology Information NCBI (www.ncbi.nlm.nih.gov), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.genome.jp/kegg/) and the DOE Joint Genome Institute (JGI) (https://signon.jgi.doe.gov/), in which genome sequences are available upon registration. To compare gene or protein sequences, multiple sequence alignments with Clustal W (www.ebi.ac.uk/Tools/msa/clustalw2/) were done [49]. All Sor sequences found after BLAST searches in NCBI & JGI databases at the time of this study were used. Additionally, the At. caldus Sor sequences obtained in this study as well as the four clones (DQ480731-DQ480734) containing Sor sequences previously attributed to At. caldus SM-1 [41] were included. For phylogenetic analysis, sequences were aligned using the multiple sequence comparison by log-expectation (MUSCLE) tool [50] and a maximum likelihood analysis with the substitution model (WAG) was conducted. Support was evaluated using 100 bootstrap replications. The phylogenetic tree was edited using MEGA5 [51].
2.4. Cell Harvest and Preparation of Cell-Free Extracts
Ten liters of batch cultures were harvested by centrifugation at 8700× g for 10 min. After removal of S°, cells were pelleted at 8700× g for 10 min and washed twice with a solution containing 2 mM NH4Cl, 0.1 mM MgCl2, 1 mM CaCl2, pH 3 [37]. Cell pellets were resuspended at 1/10 (w/v) in 100 mM Tris-HCl, pH 7.5. Afterwards, cells were broken using a French® Press (Thermo Electron Corporation; French Pressure Cell Press, Milford, MA, USA) in four passages of 10–15 mL. Crude extracts were dispatched in 2 mL aliquots and centrifuged at 20,800× g for 20 min at 4 °C. Supernatants were combined and protein concentrations were measured as described [52].
2.5. Sor Enzyme Assays
Sor enzyme assays for Sb. thermosulfidooxidans were performed aerobically at 45 °C and from 65 °C to 80 °C (in 5 °C intervals). Reaction mixtures (25 mL) contained 20 mL of “dispersed elemental S°” [37] and 5 mL of crude extracts (0.2 mg/mL protein) in 100 mM Tris-HCl, pH 7.5. Supelco glass serum bottles of 43 mm by 73 mm (Sigma-Aldrich, Darmstadt, Germany) were used. Immediately after mixing, bottles were closed with rubber lids (Butyl septum; Ochs GmbH, Bovenden, Germany) in order to avoid hydrogen sulfide loss. Bottles were stirred at 180 rpm during enzyme measurements. Under our assay conditions, Sor activity was tested with ~0.04 mg/mL total protein and ~17 mM dispersed S°. Samples (1.5 mL) were taken off with a syringe after 1 min, from 5 to 30 min (in 5 min intervals) and at 40 min. These samples were immediately filtered through nylon filters (Rotilabo®-Spritzenfilter 0.45 µm, Carl Roth, Karlsruhe, Germany). Additionally, for the determination of the reductase activity (sulfide production) 200 µL of these samples were fixed with addition of 200 µL of 2% w/v Zn-acetate. The sum of sulfite, sulfate and thiosulfate, as equivalent for oxygenase activity, was quantified by ion-exchange chromatography as further described. Specific activities were calculated from the linear increase of the reaction products. One Unit (U) of enzyme activity was defined as 1 μmol of formed sulfite, sulfate and thiosulfate (oxygenase) or hydrogen sulfide (reductase) per min per mg of protein. Optimum pH values for Sb. thermosulfidooxidans Sor activity were determined at 75 °C between pH 6.5–8.5 (in 0.5 pH steps). The optimum temperature of Sor activity was determined in the range of 65 °C–80 °C (in 5 °C steps) at pH 7.5. To determine non-enzymatic reactions, control assays with addition of 40 mg/L Bovine Serum Albumin (Sigma®) were done. These values were subtracted from the assays with crude extracts. Sor enzyme assays for At. caldus strains were done as mentioned at 45 °C and 65 °C at pH 7.5. Additionally, Sor activity of S. metallicus was measured at pH 8 and 65 °C as positive control.
2.6. Determination of Thiosulfate, Sulfite, Sulfate and Sulfide
Thiosulfate, sulfite and sulfate were quantified by ion-exchange chromatography and conductivity detection as described ([53] Schippers, 2002 #789). The DIONEX system DX-500 (Thermo Scientific, USA) G with an AS 3500 autosampler, ASRD ULTRA II 2 mm suppressor, conductivity detector CD20,gradient generator EG 50 in combination with the EluGen cartridge EGC II KOH (Thermo Scientific, USA), guard column AG17C 2 × 50 mm and separation column AS17C 2 × 250 mm (Thermo Scientific, USA) were used. A KOH gradient was applied starting with 10 mM for 1 min followed by a linear increase to 50 mM over 4.5 min. Afterwards, the concentration declined over 1 min to 10 mM and it was retained for an additional min before the next measurement. Chromatograms were processed with Chromeleon 6.70 software (Dionex, Thermo Scientific, USA) . Sulfide was determined using themethylene-blue-method with dimethylene-p-phenylendiamine and ferric iron solutions [54]. Samples were measured at 670 nm (Biochrom Novaspec 4049 Spectrophotometer, Cambridge, England).
3. Results
3.1. Sor Activity in Sb. Thermosulfidooxidans
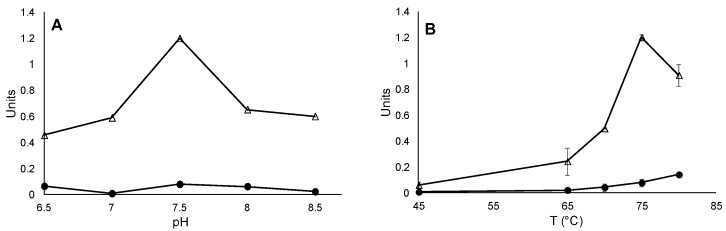
After observing the presence of sor genes encoded in genomes of sulfobacilli, we measured Sor activity in crude extracts of Sb. thermosulfidooxidans. Optimum pH and temperature values for Sor activity were determined. It showed the highest specific oxygenase activity (1.2 U/mg) at 75 °C and pH 7.5. The reductase activity at this condition was 77 mU/mg (Figure 1). Interestingly, a higher reductase activity (140 mU/mg) was measured at 80 °C. The optimum conditions for reductase activity were not determined since enzyme activities at higher temperatures were not analyzed. Neither oxygenase nor reductase activities were found when Sb. thermosulfidooxidans cells grown on ferrous iron were analyzed, suggesting the presence of possible regulatory mechanisms controlling Sor expression (not shown). To validate our assays, we measured S. metallicus Sor as positive control. Although Sor enzyme activity has not been characterized earlier in this archaeon, the presence of sor gene transcripts has been reported. Higher levels of expression were found in S° grown cells, compared to iron and pyrite grown ones [55]. At its optimal growth temperature (65 °C), a specific oxygenase activity of 0.22 U/mg and a specific reductase activity of 50 nU/mg were measured in crude extracts. No detailed parameters for S. metallicus optimum temperature or pH were determined since it was not the main goal of our study.
Figure 1.
Determination of Sb. thermosulfidooxidans Sor properties in crude extracts. Optimal pH values (A); and temperature (B) were determined for the oxygenase (triangles) and reductase (circles) enzyme activities. In (A) experiments were done at 75 °C; and in (B) at pH 8. Standard deviation values from triplicates are shown.
3.2. Genes Probably Involved in RISC Metabolism of Sb. Thermosulfidooxidans
Sb. thermosulfidooxidans is able to oxidize S°, thiosulfate and tetrathionate [56]. The sequences of proteins involved in RISC oxidation in At. ferrooxidans [36] and At. caldus [24] were used to search for homologous genes encoded in the Sb. thermosulfidooxidans genome database. Several genes likely to be involved in RISC oxidation were found. Among them, two sor genes (Sulth_1627 and Sulth_1798) and one complete cluster of hdr genes (Sulth_1021-Sulth_1026) were found. The doxD component of the Tqo (Sulth_1689), three putative sqr (Sulth_0548; Sulth_0580 and Sulth_0946) and three putative tth genes (Sulth_0921; Sulth_1188; Sulth_3251) were identified as well (Table 2). Contrary to At. caldus, doxD and tth were not found clustered in Sb. thermosulfidooxidans. Several genes encoding proteins with a rhodanese domain were also found. Interestingly, no sox genes were found.
Table 2.
Proteins related to sulfur metabolism encoded in Sb. thermosulfidooxidans genome.
| Locus_Tag | Protein Annotation | Homologous in At. caldus | BlastP Identity |
|---|---|---|---|
| Sulth_0548 | FAD-dependent pyridine nucleotide-disulfideoxidoreductase | Sqr_1 (WP_004871912) | 65% |
| Sulth_0580 | FAD-dependentpyridinenucleotide-disulfideoxidoreductase | Sqr_1 (WP_004871912) | 58% |
| Sulth_0921 | Pyrrolo-quinolinequinone repeat-containing protein | Tetrathionate hydrolase WP_004873216.1 | 40% |
| Sulth_0946 | FAD-dependentpyridinenucleotide-disulfideoxidoreductase | Sulfidequinoneoxidorreductase Sqr_1 (WP_004871912) | 62% |
| Sulth_1021 | Heterodisulfidereductase, subunit C | Heterodisulfidereductase, subunit C HdrC (WP_038472248.1) | 52% |
| Sulth_1022 | Heterodisulfidereductase, subunit B | Heterodisulfidereductase, subunit B HdrB (WP_051620817.1) | 59% |
| Sulth_1023 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase | pyridinenucleotide-disulfideoxidoreductase (WP_004868630.1) | 41% |
| Sulth_1024 | Hypotheticalprotein | Hypotheticalprotein (WP_004868631.1) | 30% |
| Sulth_1025 | Iron-sulfur cluster-binding protein | Heterodisulfidereductase, subunit C HdrC(WP_004868632.1) | 32% |
| Sulth_1026 | unknown function DUF224 cysteine-rich region domain protein | Heterodisulfidereductase, subunitB HdrB (WP_004868633.1) | 38% |
| Sulth_1046 | DsrEfamilyprotein | Disulfidereductase(WP_004868633.1) | 31% |
| Sulth_1188 | Pyrrolo-quinolinequinone repeat-containing protein | Tetrathionate hydrolase (WP_004873216.1) | 31% |
| Sulth_1355 | Adenylyl-sulfate kinase | Adenylyl sulfate kinase (WP_004868315.1) | 40% |
| Sulth_1366 | Sulfate adenylyltransferrase | Adenylyl sulfate kinase (WP_004868315.1) | 39% |
| Sulth_1433 | Sulfate adenylyltransferrase | Adenylyl sulfate kinase (WP_004868315.1) | 38% |
| Sulth_1435 | Sulfate adenylyltransferrase | Adenylyl sulfate kinase (WP_004868315.1) | 44% |
| Sulth_1627 | Sulfuroxygenasereductase | Sulfuroxygenasereductase (WP_004871908.1) | 48% |
| Sulth_1680 | Rhodanese like protein | Sulfur transferase(WP_004872361.1) | 32% |
| Sulth_1689 | Tqo small subunit DoxD domain-containing | Quinol oxidase (WP_004873215.1) | 34% |
| Sulth_1798 | Sulfuroxygenasereductase | Sulfur transferase(WP_004872361.1) | 47% |
| Sulth_1878 | Rhodanese-likeprotein | Sulfurtransferase(WP_004872361.1) | 29% |
| Sulth_2335 | Rhodanese-likeprotein | Sulfurtransferase(WP_004868554.1) | 35% |
| Sulth_2366 | Nitratereductase | Formate dehydrogenase (WP_004868564.1) | 50% |
| Sulth_2367 | Sulfur reductase beta subunit | Ferredoxin (WP_004868562.1) | 55% |
| Sulth_2368 | DMSO reductase anchor subunit | dimethyl sulfoxidereductase subunit C (WP_004872154.1) | 27% |
| Sulth_2770 | Heterodisulfidereductase, subunit C | Heterodisulfidereductasesubunit C (WP_038472248.1) | 47% |
| Sulth_2771 | Heterodisulfidereductase, subunit B | Heterodisulfidereductasesubunit B (WP_051620815.1) | 50% |
| Sulth_2772 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase | Pyridinenucleotide-disulfideoxidoreductase(WP_004868887.1) | 42% |
| Sulth_3040 | Rhodanese-likeprotein | Sulfurtransferase(WP_004872361.1) | 30% |
| Sulth_3251 | Pyrrolo-quinolinequinone repeat-containing protein | Tetrathionate hydrolase (WP_004873216.1) | 54% |
| Sulth_3294 | Rhodanese-likeprotein | Sulfurtransferase(WP_004872361.1) | 31% |
3.3. Does At. caldus Possess an Active Sor Enzyme?
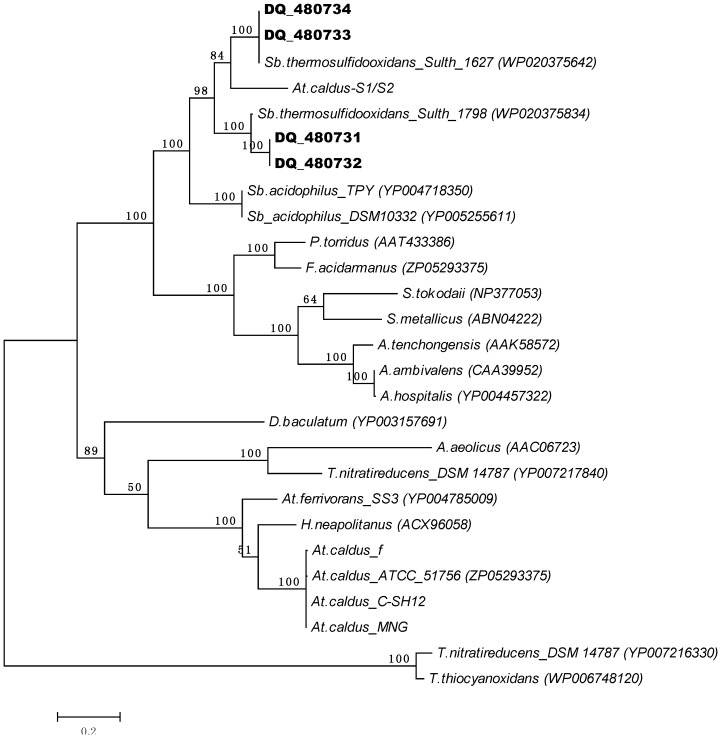
Previously, we had reported Sor activity in At. caldus S1 and S2 strains [40]. However, after several attempts we could not detect Sor activity in any of the seven At. caldus strains studied. Additionally, no sor gene is encoded in the genome sequence of strain SM-1 [13]. To answer the question of how conserved the sor gene is in At. caldus, we screened seven strains by PCR with CODEHOP primers designed based on alignments of bacterial Sor sequences. Positive sor gene amplicons were detected in six of them. These amplicons (~800 bp), representing 80% of the complete Sor protein, were cloned and sequenced. A phylogenetic tree showed that these At. caldus Sor sequences clustered within the acidithiobacilli branch (Figure 2), which also includes the Sor from H. neapolitanus and At. ferrivorans SS3 [34]. Surprisingly, the Sor aminoacid sequences obtained from At. caldus S1 and S2 strains, which are 100% identical (shown as S1/S2), clustered within the Sulfobacillus Sor branch (Figure 2). In this context, it is highly probable that our previously reported Sor activity in these strains was due to the presence of a Sulfobacillus contaminant. The purity of these cultures was checked by nested PCR using 16S rDNA primers for Sulfobacillus and Acidithiobacillus, confirming the presence of Sulfobacillus (Figure S1). Sequence analysis of the obtained sor amplicon from the Sulfobacillus contaminant strain revealed a high similarity with Sulfobacillus L15 (data not shown). Further PCR tests were done to discard the presence of At. thiooxidans, At. ferrooxidans, Leptospirillum sp. and archaea in S1 and S2 strains. After re-purification of At. caldus S1 and S2 strains in our laboratory, their sor gene sequences were 100% identical to the At. caldusT (not shown). Nevertheless, no Sor activity was detected in S° grown cells under our assay conditions.
Figure 2.
Maximum likelihood phylogenetic tree showing relationship amongst bacterial and archaeal Sor proteins. Sor aminoacidic sequences of Sb. thermosulfidooxidans DSM 9293T (WP_020375642 and WP-_020375834), A. aeolicus VF5 (NP_21332), H. neapolitanus C2 (YP_003263105), At. ferrivorans SS3 (YP_004785009), At. caldus ATCC 51756; DSM 8589 (ZP_05293375), A. tengchongensis (AAK58572), A. ambivalens (CAA39952), Acidianus hospitalis(YP_004457322), Sulfolobus tokodaii (NP_377053), Picrophilus torridus (AAT43386), Ferroplasma acidarmanus fer1 (ZP_01708456), Desulfomicrobium baculatum DSM 4028 (YP_003157691), Sb. acidophilus DSM 10332T (YP_005255611), Sb. acidophilus TPY (YP_004718350), S. metallicus (ABN04222), Thioalkalivibrio nitratireducens DSM14787_1 (YP_007217840), and DSM14787_2 (YP_007216330), Thioalkalivibrio thiocyanoxidans (WP_006748120) were used. Additionally, Sor sequences from four metagenomic clones (DQ480731, DQ480732, DQ480733/ ABF20540, DQ48074/ABF20541) [41] and the SOR sequences obtained from At. caldus strains MNG, C-SH12, f and S1 and S2, obtained in this study (see text), were included.
4. Discussion
Sb. thermosulfidooxidans crude extracts possess an active Sor enzyme. Our results are in agreement with the “thermophilic” nature of Sor. The recombinant Sor from H. neapolitanus was shown to be active in a temperature range of 10–99 °C with an optimum at 80 °C [20]. In Sb. thermosulfidooxidans Sor reaction products such as sulfite, thiosulfate and sulfide can be further metabolized and coupled with energy conservation by enzymes such as Saor, Tqo, Tth and Sqr, which have been found to be encoded in its genome sequence. The low reductase activities measured may be related to (i) the utilization of crude extracts, in which the presence of enzymes such as Sqr may contribute to their degradation; and (ii) some hydrogen sulfide loss prior to its fixation. Since two sor genes were found, further research is needed to elucidate their regulation and their connection with other proteins likely involved in RISC oxidation in Sb. thermosulfidooxidans.
Several proteins with a Rhodanese domain were found to be encoded in the Sb. thermosulfidooxidans genome sequence. These may contribute to the oxidation of persulfides or polysulfides by acting as sulfur transferases [39]. This bacterium also possesses the hdr gene cluster, which could also be responsible for S° oxidation in At. ferrooxidans as well as some other acidophiles [36]. Considering this, in Sb. thermosulfidooxidans, S° produced from hydrolysis of tetrathionate by Tth or oxidation of H2S by Sqr could be accumulated in the form of polysulfides, which after being transferred into the cytoplasm, can be further oxidized via Sor or Hdr. Although no biochemical evidence for involvement of the Hdr complex in S° oxidation in acidophiles has been demonstrated yet, we recently found several Hdr proteins expressed by shotgun proteomics of At. ferrooxidans ATCC 23270T biofilm formation process on pyrite, [57]. Recently, a comparison was done among isolates and environmental Sulfobacillus genomes. For this, five new draft genomes of Sulfobacillus spp. were assembled from metagenomic data obtained from the Iron Mountain, California. These sequences were compared with Sb. acidophilus TPY [8] and Sb. thermosulfidooxidans Cutipay [6]. The analysis showed the presence of sor genes in two of the five genomes assembled, while one Hdr cluster was found in all of them [58].
Chen et al. reported four sor sequences obtained from metagenomic DNA samples from a bioreactor containing Leptospirillum, Sulfobacillus, Acidithiobacillus and Sphingomonas spp. One sor gene sequence was cloned and expressed in E. coli and the recombinant Sor was active [41]. Due to increased amounts of genomic information we reanalyzed these four sor sequences (Genbank accession numbers DQ480731–DQ480734) by Blast in the JGI database. These Sor proteins clustered within Sulfobacillus Sor proteins, showing 99%–100% identities with Sulfobacillus sequences (Sulth_1627 and Sulth_1798) and 44%–48% with At. caldus sequences (Figure 2). These results, plus the absence of a sor gene in the At. caldus SM-1 genome sequence [13], strongly suggest that a part of the Sulfobacillus sor gene was cloned and attributed to belong to At. caldus SM-1.
Whether Sor contributes to the overall sulfur oxidation in At. caldus is, in our opinion, still an open question. Sor enzymes contain a mononuclear non-Heme iron site as the putative redox-active cofactor [17]. By site directed mutagenesis it has been shown that the three Fe coordinating residues H86, H90 and E114 as well as the C31 (in A. ambivalens numbering), are essential for catalysis [59]. Analysis of At. caldus Sor sequence shows conservation of all of the residues relevant for the coordination of iron as well as the C31 (Supplementary Figure S2). Several strains possess a sor gene but to the best of our knowledge, its enzyme activity has not been successfully measured in any At. caldus strain, neither in crude extracts nor cell fractions. No significant differences on the levels of sor transcripts were reported between At. caldusT cells grown with tetrathionate or S° as electron donors [24]. In the same study, no protein spot could be identified as Sor in a two dimensional polyacrilamyde gel electrophoresis (2D-PAGE). We have also measured very low levels of the sor gene transcript by Real time reverse transcription (RT)-PCR in At. caldusT, and no significant differences were found when cells grown with S° or thiosulfate as energy sources were analyzed (not shown). Recently, by high throughput proteomics we detected >1300 proteins from sulfur and thiosulfate At. caldus grown cells. The Sor protein, encoded by the gene ACA_0302, was not detected in any sample from both growth conditions [60]. A proteomic study of the response of At. caldus towards suboptimal pH conditions showed that several proteins involved in sulfur oxidation such as HdrABC and Sqr were induced when cells were incubated at pH 1.1 [61]. Although Sor was not detected in this study, a test of Sor activity in At. caldus at acid pH range might be helpful to completely elucidate the presence of Sor activity in this bacterium. Recently, a sor mutant of At. caldus MTH-04 strain was produced and a differential gene expression study was done by microarrays. No obvious differences were observed in the growth of the sor mutant and the wild type strain in media with S° as energy source [62]. However, since enzyme activities were not measured in this study, the question whether Sor was active or not in wild type At. caldus cells remains open.
5. Conclusions
In this study we provide evidence that Sb. thermosulfidooxidans possess Sor activity and that the previously Sor activity reported in At. caldus strains S1 and S2 most likely was due to the presence of a Sulfobacillus contaminant.
Acknowledgments
The authors would like to thank Shelly M. Deane and Douglas E. Rawlings (University of Cape Town, Cape Town, South Africa) and H.B. Zhou (Central South University Changsha, Changsha, China) for providing At. caldus strains MNG, f ,#6, and S1, S2 strains, respectively. Pablo Aguilar (Universidad Católica del Norte) is also acknowledged for his assistance in the phylogenetic analyses.
Supplementary Files
Author Contributions
Claudia Janosch planned and carried out most of the experimental work. Francisco Remonsellez carried out phylogenetic analyses. Mario Vera planned experimental work and wrote the manuscript, with contributions of Claudia Janosch, Wolfgang Sand and Francisco Remonsellez.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rawlings D.E., Johnson D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology. 2007;153:315–324. doi: 10.1099/mic.0.2006/001206-0. [DOI] [PubMed] [Google Scholar]
- 2.Vera M., Schippers A., Sand W. Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulfide oxidation-part A. Appl. Microbiol. Biotechnol. 2013;97:7529–7541. doi: 10.1007/s00253-013-4954-2. [DOI] [PubMed] [Google Scholar]
- 3.Brierley C.L., Brierley J.A. Progress in bioleaching: Part B: Applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 2013;97:7543–7552. doi: 10.1007/s00253-013-5095-3. [DOI] [PubMed] [Google Scholar]
- 4.Norris P.R., Clark D.A., Owen J.P., Waterhouse S. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology. 1996;142:775–783. doi: 10.1099/00221287-142-4-775. [DOI] [PubMed] [Google Scholar]
- 5.Karavaiko G.I., Krasil’nikova E.N., Tsaplina I.A., Bogdanova T.I., Zakharchuk L.M. Growth and carbohydrate metabolism of Sulfobacilli. Mikrobiologiia. 2001;70:293–299. [PubMed] [Google Scholar]
- 6.Travisany D., di Genova A., Sepulveda A., Bobadilla-Fazzini R.A., Parada P., Maass A. Draft genome sequence of the Sulfobacillus thermosulfidooxidans cutipay strain, an indigenous bacterium isolated from a naturally extreme mining environment in northern chile. J. Bacteriol. 2012;194:6327–6328. doi: 10.1128/JB.01622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson I., Chertkov O., Chen A., Saunders E., Lapidus A., Nolan M., Lucas S., Hammon N., Deshpande S., Cheng J.F., et al. Complete genome sequence of the moderately thermophilic mineral-sulfide-oxidizing firmicute Sulfobacillus acidophilus type strain Nal(T) Stand. Genomic Sci. 2013;6:1–13. doi: 10.4056/sigs.2736042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., Chen Y., Liu Q., Hu S., Chen X. Complete genome analysis of Sulfobacillus acidophilus strain Tpy, isolated from a hydrothermal vent in the pacific ocean. J. Bacteriol. 2011;193:5555–5556. doi: 10.1128/JB.05684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallberg K.B., Lindstrom E.B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology. 1994;140:3451–3456. doi: 10.1099/13500872-140-12-3451. [DOI] [PubMed] [Google Scholar]
- 10.Okibe N., Johnson D.B. Biooxidation of pyrite by defined mixed cultures of moderately thermophilic acidophiles in pH-controlled bioreactors: Significance of microbial interactions. Biotechnol. Bioeng. 2004;87:574–583. doi: 10.1002/bit.20138. [DOI] [PubMed] [Google Scholar]
- 11.Dopson M., Lindstrom E.B. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Microb. Ecol. 2004;48:19–28. doi: 10.1007/s00248-003-2028-1. [DOI] [PubMed] [Google Scholar]
- 12.Valdes J., Quatrini R., Hallberg K., Dopson M., Valenzuela P.D., Holmes D.S. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 2009;191:5877–5878. doi: 10.1128/JB.00843-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You X.Y., Guo X., Zheng H.J., Zhang M.J., Liu L.J., Zhu Y.Q., Zhu B., Wang S.Y., Zhao G.P., Poetsch A., et al. Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J. Genet. Genomics. 2011;38:243–252. doi: 10.1016/j.jgg.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Kletzin A. Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: Purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 1989;171:1638–1643. doi: 10.1128/jb.171.3.1638-1643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urich T., Gomes C.M., Kletzin A., Frazao C. X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science. 2006;311:996–1000. doi: 10.1126/science.1120306. [DOI] [PubMed] [Google Scholar]
- 16.Kletzin A., Urich T., Müller F., Bandeiras T.M., Gomes C.M. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 2004;36:77–91. doi: 10.1023/B:JOBB.0000019600.36757.8c. [DOI] [PubMed] [Google Scholar]
- 17.Urich T., Bandeiras T.M., Leal S.S., Rachel R., Albrecht T., Zimmermann P., Scholz C., Teixeira M., Gomes C.M., Kletzin A. The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre. Biochem. J. 2004;381:137–146. doi: 10.1042/BJ20040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C.W., Chen Z.W., He Z.G., Zhou P.J., Liu S.J. Purification and properties of the sulfur oxygenase/reductase from the acidothermophilic archaeon, Acidianus strain s5. Extremophiles. 2003;7:131–134. doi: 10.1007/s00792-002-0304-5. [DOI] [PubMed] [Google Scholar]
- 19.Pelletier N., Leroy G., Guiral M., Giudici-Orticoni M.T., Aubert C. First characterisation of the active oligomer form of sulfur oxygenase reductase from the bacterium Aquifex aeolicus. Extremophiles. 2008;12:205–215. doi: 10.1007/s00792-007-0119-5. [DOI] [PubMed] [Google Scholar]
- 20.Veith A., Botelho H.M., Kindinger F., Gomes C.M., Kletzin A. The sulfur oxygenase reductase from the mesophilic bacterium Halothiobacillus neapolitanus is a highly active thermozyme. J. Bacteriol. 2012;194:677–685. doi: 10.1128/JB.06531-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kappler U., Dahl C. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 2001;203:1–9. doi: 10.1111/j.1574-6968.2001.tb10813.x. [DOI] [PubMed] [Google Scholar]
- 22.Muller F.H., Bandeiras T.M., Urich T., Teixeira M., Gomes C.M., Kletzin A. Coupling of the pathway of sulphur oxidation to dioxygen reduction: Characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 2004;53:1147–1160. doi: 10.1111/j.1365-2958.2004.04193.x. [DOI] [PubMed] [Google Scholar]
- 23.Valenzuela L., Chi A., Beard S., Orell A., Guiliani N., Shabanowitz J., Hunt D.F., Jerez C.A. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol. Adv. 2006;24:197–211. doi: 10.1016/j.biotechadv.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Mangold S., Valdes J., Holmes D.S., Dopson M. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front Microbiol. 2011;2:17. doi: 10.3389/fmicb.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugaytsova Z., Lindstrom E.B. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 2004;271:272–280. doi: 10.1046/j.1432-1033.2003.03926.x. [DOI] [PubMed] [Google Scholar]
- 26.Protze J., Muller F., Lauber K., Nass B., Mentele R., Lottspeich F., Kletzin A. An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front. Microbiol. 2011;2:68. doi: 10.3389/fmicb.2011.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann P., Laska S., Kletzin A. Two modes of sulfite oxidation in the extremely thermophilic and acidophilic archaeon Acidianus ambivalens. Arch. Microbiol. 1999;172:76–82. doi: 10.1007/s002030050743. [DOI] [PubMed] [Google Scholar]
- 28.Wakai S., Kikumoto M., Kanao T., Kamimura K. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 2004;68:2519–2528. doi: 10.1271/bbb.68.2519. [DOI] [PubMed] [Google Scholar]
- 29.Brasseur G., Levican G., Bonnefoy V., Holmes D., Jedlicki E., Lemesle-Meunier D. Apparent redundancy of electron transfer pathways via bc(1) complexes and terminal oxidases in the extremophilic chemolithoautotrophic Acidithiobacillus ferrooxidans. Biochim. Biophys. Acta. 2004;1656:114–126. doi: 10.1016/j.bbabio.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Brito J.A., Sousa F.L., Stelter M., Bandeiras T.M., Vonrhein C., Teixeira M., Pereira M.M., Archer M. Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry. 2009;48:5613–5622. doi: 10.1021/bi9003827. [DOI] [PubMed] [Google Scholar]
- 31.Friedrich C.G., Quentmeier A., Bardischewsky F., Rother D., Orawski G., Hellwig P., Fischer J. Redox control of chemotrophic sulfur oxidation of Paracoccus pantotrophus. In: Dahl C., Friedrich C.G., editors. Microbial Sulfur Metabolism. Springer; Berlin, Germany: 2008. pp. 139–150. [Google Scholar]
- 32.Friedrich C.G., Rother D., Bardischewsky F., Quentmeier A., Fischer J. Oxidation of reduced inorganic sulfur compounds by bacteria: Emergence of a common mechanism? Appl. Environ. Microbiol. 2001;67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welte C., Hafner S., Kratzer C., Quentmeier A., Friedrich C.G., Dahl C. Interaction between sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 2009;583:1281–1286. doi: 10.1016/j.febslet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 34.Liljeqvist M., Valdes J., Holmes D.S., Dopson M. Draft genome of the psychrotolerant acidophile Acidithiobacillus ferrivoransSS3. J. Bacteriol. 2011;193:4304–4305. doi: 10.1128/JB.05373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bardischewsky F., Quentmeier A., Rother D., Hellwig P., Kostka S., Friedrich C.G. Sulfur dehydrogenase of Paracoccus pantotrophus: The Heme-2 domain of the molybdoprotein cytochrome C complex is dispensable for catalytic activity. Biochemistry. 2005;44:7024–7034. doi: 10.1021/bi047334b. [DOI] [PubMed] [Google Scholar]
- 36.Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D.S., Bonnefoy V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics. 2009;10:394. doi: 10.1186/1471-2164-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohwerder T., Sand W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology. 2003;149:1699–1710. doi: 10.1099/mic.0.26212-0. [DOI] [PubMed] [Google Scholar]
- 38.Wang H., Liu S., Liu X., Li X., Wen Q., Lin J. Identification and characterization of an ethe1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl. Microbiol. Biotechnol. 2014;98:7511–7522. doi: 10.1007/s00253-014-5830-4. [DOI] [PubMed] [Google Scholar]
- 39.Acosta M., Beard S., Ponce J., Vera M., Mobarec J.C., Jerez C.A. Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: Structural and functional characterization of the proteins. Omics. 2005;9:13–29. doi: 10.1089/omi.2005.9.13. [DOI] [PubMed] [Google Scholar]
- 40.Janosch C., Thyssen C., Vera M., Bonnefoy V., Rohwerder T., Sand W. Sulfur oxygenase reductase in different Acidithiobacillus caldus-like strains. Adv. Mater. Res. 2009;71–73:239–242. doi: 10.4028/www.scientific.net/AMR.71-73.239. [DOI] [Google Scholar]
- 41.Chen Z.W., Liu Y.Y., Wu J.F., She Q., Jiang C.Y., Liu S.J. Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl. Microbiol. Biotechnol. 2007;74:688–698. doi: 10.1007/s00253-006-0691-0. [DOI] [PubMed] [Google Scholar]
- 42.Rawlings D.E., Coram N.J., Gardner M.N., Deane S.M. Thiobacillus caldus and Leptospirillum ferrooxidans are widely distributed in continuous—Flow biooxidation tanks used to treat a variety of metal-containing ores and concentrates. In: Part A., Amils R., Ballester A., editors. Biohydrometallurgy and the Environment toward the Mining of the 21st Century. Elsevier Press; Amsterdam, The Netherland: 1999. pp. 777–786. [Google Scholar]
- 43.Mackintosh M. Nitrogen fixation by Thiobacillus ferrooxidans. J. Gen. Microbiol. 1978;105:215–218. doi: 10.1099/00221287-105-2-215. [DOI] [Google Scholar]
- 44.Aljanabi S.M., Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Wulf-Durand P., Bryant L.J., Sly L.I. PCR-mediated detection of acidophilic, bioleaching-associated bacteria. Appl. Environ. Microbiol. 1997;63:2944–2948. doi: 10.1128/aem.63.7.2944-2948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane D.J. 16s/23s rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; Chichester, UK: 1991. pp. 115–175. [Google Scholar]
- 47.Achenbach L., Woese C. 16s and 23s rRNA-like primers. In: Sowers R., Schreier H.J., editors. A Laboratory Manual Archaea. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1995. pp. 521–523. [Google Scholar]
- 48.Rose T.M., Henikoff J.G., Henikoff S. CODEHOP (Consensus-Degenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins D.G. ClustalW: Multiple alignment of DNA and protein sequences. Methods Mol. Biol. 1994;25:307–318. doi: 10.1385/0-89603-276-0:307. [DOI] [PubMed] [Google Scholar]
- 50.Edgar R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 53.Weiß J. Ionenchromatographie. VCH, Verlagsgesellschaft mbH; Weinheim, Germany: 1991. Anionenaustauch Cromatographie, Kapitel 3; pp. 32–174. [Google Scholar]
- 54.Wasserchemische Gesellschaft, Fachgruppe in der GDCh. Gemeinschaft mit dem Normenausschuss Wasserwesen (NAW) im DIN e.V. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlammuntersuchung: 95 Lieferung. Wiley-VCH Verlag GmbH; Weinheim, Germany: 2015. Teil 26, Photometrische Bestimmung des gelösten Sulfids, Normenausschluß Wasserwesen. [Google Scholar]
- 55.Bathe S., Norris P.R. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 2007;73:2491–2497. doi: 10.1128/AEM.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krasil’nikova E.N., Tsaplina I.A., Zakharchuk L.M., Bogdanova T.I. Effects of exogenous factors on the activity of enzymes involved in carbon metabolism in thermoacidophilic bacteria of the genus Sulfobacillus. Prikl. Biokhim. Mikrobiol. 2001;37:418–423. [PubMed] [Google Scholar]
- 57.Vera M., Krok B., Bellenberg S., Sand W., Poetsch A. Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics. 2013;13:1133–1144. doi: 10.1002/pmic.201200386. [DOI] [PubMed] [Google Scholar]
- 58.Justice N.B., Norman A., Brown C.T., Singh A., Thomas B.C., Banfield J.F. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genomics. 2014;15:1107. doi: 10.1186/1471-2164-15-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urich T., Kroke A., Bauer C., Seyfarth K., Reuff M., Kletzin A. Identification of core active site residues of the sulfur oxygenase reductase from Acidianus ambivalens by site-directed mutagenesis. FEMS Microbiol. Lett. 2005;248:171–176. doi: 10.1016/j.femsle.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 60.Janosch C., Vera M. Biofilm Centre, Universität Duisburg-Essen; Essen, Germany: 2014. Unpublished work. [Google Scholar]
- 61.Mangold S., Rao Jonna V., Dopson M. Response of Acidithiobacillus caldus toward suboptimal pH conditions. Extremophiles. 2013;17:689–696. doi: 10.1007/s00792-013-0553-5. [DOI] [PubMed] [Google Scholar]
- 62.Chen L., Ren Y., Lin J., Liu X., Pang X. Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS ONE. 2012;7:e39470. doi: 10.1371/journal.pone.0039470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.