Abstract
Cytomegalovirus is an attractive cancer vaccine platform because it induces strong, functional CD8+ T-cell responses that accumulate over time and migrate into most tissues. To explore this, we used murine cytomegalovirus expressing a modified gp100 melanoma antigen. Therapeutic vaccination by the intraperitoneal and intradermal routes induced tumor infiltrating gp100-specific CD8+ T-cells, but provided minimal benefit for subcutaneous lesions. In contrast, intratumoral infection of established tumor nodules greatly inhibited tumor growth and improved overall survival in a CD8+ T-cell-dependent manner, even in mice previously infected with murine cytomegalovirus. Although murine cytomegalovirus could infect and kill B16F0s in vitro, infection was restricted to tumor-associated macrophages in vivo. Surprisingly, the presence of a tumor antigen in the virus only slightly increased the efficacy of intratumoral infection and tumor-specific CD8+ T-cells in the tumor remained dysfunctional. Importantly, combining intratumoral murine cytomegalovirus infection with anti-PD-L1 therapy was synergistic, resulting in tumor clearance from over half of the mice and subsequent protection against tumor challenge. Thus, while a murine cytomegalovirus-based vaccine was poorly effective against established subcutaneous tumors, direct infection of tumor nodules unexpectedly delayed tumor growth and synergized with immune checkpoint blockade to promote tumor clearance and long-term protection.
Introduction
Despite substantial efforts over many years, vaccines that elicit effective antitumor immunity are rare.1 Much of the failure of these treatments comes from tumor immune evasion due to features of the tumor microenvironment2 and tumor-specific T-cells becoming dysfunctional or even failing to migrate into the tumor.3,4 Nevertheless, the presence of spontaneously generated tumor-reactive T-cells correlates with improved prognosis in cancer patients5 suggesting that tumor-specific T-cells can effectively delay tumor growth given the right conditions. Thus, recent work has explored therapies that modulate the tumor environment directly.6,7
Many viruses have been explored for their ability to cause tumor cell destruction and provide “danger” signals within the tumor environment, leading to some preclinical and recent clinical successes. The most developed of these so-called oncolytic viruses are based on herpes simplex virus, adenovirus, vaccinia virus, measles virus, and reovirus,8 with the herpes simplex platform (T-VEC) recently completing a phase 3 clinical trial, in which a 26% objective response rate and 16% durable response rate were reported in stage IIIb, IIIc, and IV melanoma patients.9 T-VEC has recently been approved by the FDA for treatment in cutaneous melanoma.10 Each of these oncolytic viruses was designed to replicate rapidly in tumor cells and directly induce tumor cell lysis with the hope that this would liberate tumor antigens in an inflammatory (i.e., immune stimulatory) environment. Pre-existing antiviral immunity directed against the oncolytic virus may be able to terminate the therapy by clearing the virus,11 requiring this variable to be evaluated for each therapeutic agent. Notably however, several clinical studies have found no correlation between pre-existing immunity and clinical results after treatment.9,12,13,14
Cytomegalovirus (CMV) is a β-herpesvirus that establishes an asymptomatic but life-long infection, leading to exceptionally large cellular and humoral immune responses. Recent interest in developing a CMV-based vaccine has arisen from its ability to induce enormous populations of CD8+ T-cells specific for virally-encoded epitopes, better known as memory inflation.15,16,17,18,19 CMV can be manipulated to express genes of interest for vaccination18,20 and such CMV-based vectors have been profoundly protective in a nonhuman primate model of HIV infection.21,22,23 Most people in the world are infected with CMV.24 However, previous CMV infection does not preclude reinfection, and as a result, CMV-infected monkeys (and presumably people) can be vaccinated and boosted several times with CMV.21,25 Moreover, CMV-specific CD8+ T-cells do not show evidence of exhaustion in immune competent people26 and are able to migrate into almost any tissue in the body.27,28,29 Thus, CMV-based vaccines are in development for clinical trials.
Relatively little is known about CMV-based vaccines for cancer. Vaccination with murine-CMV (MCMV) expressing prostate-specific antigen was able to delay tumor growth and increase survival in a Tramp-prostate-specific antigen model.30 In addition, MCMV expressing the tyrosinase-related protein 2, a common melanoma antigen, induced antibodies that provided prophylactic protection and therapeutic delay in the subcutaneous B16F10 melanoma model.31 Lastly, systemic infection with MCMV expressing an altered gp100 peptide induced the accumulation of gp100-specific CD8+ T-cells in the periphery and reduced the growth of B16F10 cells in the lungs of mice in both prophylactic and therapeutic settings, likely in a T-cell dependent manner.32
We also generated an MCMV viral vector encoding an altered version of the melanoma peptide gp100 (gp100S27P). This altered peptide has been shown to induce a potent cytotoxic T lymphocyte response that can cross-react with the native-gp100 antigen.33 This MCMV-gp100S27P vaccine (hereafter referred to as MCMV-gp100) induced robust expansion of gp100-specific CD8+ T-cells, which migrated into subcutaneously implanted B16F0 tumors, but had little therapeutic efficacy. Remarkably, we found that direct intratumoral (IT) infection of well-established tumor nodules with either wild-type MCMV or MCMV-gp100 was markedly more effective than MCMV-gp100 intraperitoneal (IP) and intradermal (ID) vaccination. Prolonged survival was dependent on CD8+ T-cells and the therapeutic efficacy was not abrogated by previous MCMV infection. Although infection of B16F0s with MCMV in vitro reduced tumor cell growth and led to cell death, IT infection with MCMV resulted primarily in infection of tumor associated macrophages (TAMs). Importantly, MCMV IT infection synergized with the blockade of the PD-1/PD-L1 inhibitory pathway to induce primary tumor clearance and long-term protection independent of the presence of gp100 in the vaccine. These data show that while systemic vaccination with MCMV-gp100 alone is ineffective for subcutaneous lesions, IT MCMV infection promotes T-cell dependent tumor inhibition that can synergize with immune checkpoint blockades.
Results
Construction and characterization of MCMV-gp100S27P
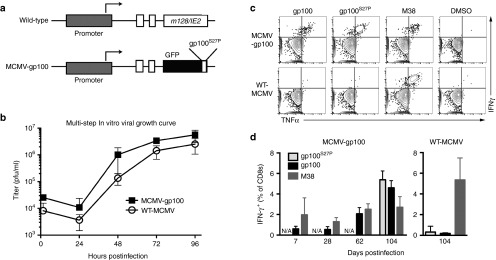
A recombinant strain of MCMV was created that expresses GFP fused to an altered version of the gp10025-33 peptide (gp100S27P). This fusion construct was inserted into the IE2 locus and under the control of the endogenous MCMV IE2 promoter (MCMV-gp100, Figure 1a), a strategy that has been used to stimulate robust T-cell responses to recombinant antigens in the MCMV backbone.34,35,36 The growth of MCMV-gp100 was similar to that of its wild-type counterpart as seen by multistep in vitro growth curves (Figure 1b). Infection of C57BL/6 mice with MCMV-gp100 induced the accumulation of CD8+ T-cells in the blood that responded to the altered and native gp100 peptides (Figure 1c,d). In contrast, WT-MCMV infection did not elicit gp100-specific CD8+ T-cells (Figure 1c,d). The representative gating strategies for these data and subsequent flow cytometry experiments are shown in Supplementary Figure S1.
Figure 1.
Construction and characterization MCMV-GFP-gp100S27P. (a) Schematic of recombinant strain MCMV-GFP-gp100S27P (MCMV-gp100) in which the altered gp100 peptide was fused to EGFP and cloned into the IE2 region of the MCMV genome. (b) The growth of MCMV-gp100 versus WT-MCMV in M2-10B4s. Data represent pooled results from two independent experiments and show the mean ± SD. (c) Representative FACS plots of CD8+ T-cell cytokine production after stimulation with the indicated peptides ex vivo. T-cells were obtained from the peripheral blood on day 104, postinfection with either MCMV-gp100 or WT-MCMV. (d) CD8+ T-cell responses to the indicated peptides over time, assessed as in c. Data is represented as the mean value ± SD from a total of five animals per group. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV; EGFP, enhanced green fluorescent protein.
Therapeutic IP and ID vaccination with MCMV-gp100 induces minimal growth delay of B16F0 tumors
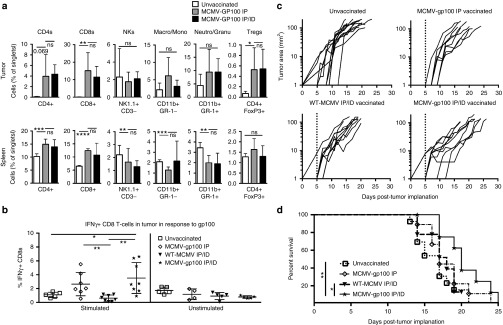
To determine the therapeutic efficacy of MCMV-gp100 vaccination, B16F0 tumors were subcutaneously implanted in the flank and mice were vaccinated 5 days later with MCMV-gp100. Recent work has shown that the site of infection or vaccination can influence the migration of CD8+ T-cells and subsequent protection.37,38 Therefore, we vaccinated mice by the IP route alone or in combination with an ID vaccination in the skin adjacent to the tumor. In both cases, vaccination caused increased infiltration of CD4+ T-cells, CD8+ T-cells, and FoxP3+ regulatory T-cells, but no increase of NK Cells, Neutrophils, Granulocytes, Macrophages, or Monocytes (Figure 2a). Moreover, vaccination with MCMV-gp100 by either route induced an increased frequency of gp100-reactive T-cells within the tumor, as measured by intracellular cytokine stimulation (Figure 2b) or by using gp100-specific Pmel-I TCR transgenic T-cells (data not shown). However, there was only a small effect on tumor growth in comparison with unvaccinated animals (Figure 2c) and only the combined IP/ID routes of vaccination improved survival compared to unvaccinated mice or mice infected via the IP/ID routes with WT-MCMV (Figure 2d). Moreover, median survival was only increased by 3 days and this was not significantly greater than mice vaccinated with MCMV-gp100 by the IP route alone (Figure 2d). These data suggest that systemic and dermal-localized MCMV-gp100 vaccinations were able to cause expansion and tumor infiltration of gp100-specific CD8+ T-cells, but were ineffective as therapeutic treatment of subcutaneous B16F0 melanoma lesions.
Figure 2.
Intraperitoneal (IP) and intradermal (ID) infection with MCMV-gp100 induced poor antitumor responses. Animals received 1 × 105 B16F0s subcutaneously on day 0 followed by IP or IP/ID vaccination with MCMV-gp100 or WT-MCMV on day 5, post implantation. The data shown is combined from three separate experiments. (a) Lymphocytes in the tumor (top panel) and spleen (bottom panel) after MCMV-gp100 vaccination. Data are represented as the mean ± SD. Significance was assessed by unpaired t-tests, ns, P > 0.05; *P < 0.05; **P < 0.01;, ***P < 0.001; ****P < 0.0001; NKs, NK cells; Neutro, Neutrophil; Granu, Granulocyte; Macro, Macrophage; Mono, Monocyte; Treg, regulatory T cell. (b) IFN-γ production of CD8+ T-cells recovered from tumors at sacrifice and stimulated or not ex vivo with the native gp100 peptide (n = 5–9 mice). Represented as the mean ± SD. (c) Tumor growth curves showing the growth, by area, of individual tumors from unvaccinated (n = 13) MCMV-gp100 IP vaccinated (n = 9), WT-MCMV IP/ID vaccinated (n = 9), and MCMV-gp100 IP/ID vaccinated mice (n = 10). The dotted line indicates the day of vaccination. (d) Kaplan–Meier survival curve of treated animals. Significance was assessed by the logrank test, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV.
IT infection with MCMV significantly delays tumor growth and improves overall survival
As systemic vaccination was unremarkable, we turned to alternative infection routes. Recent work has shown that the introduction of therapies directly into tumors can lead to therapeutic responses.6,7 We found that MCMV-gp100 could infect B16F0s in vitro at low and high multiplicities of infection (MOI) and spread through the culture, as seen by GFP-expression of infected cells (Supplementary Figure S2c and data not shown), although the recovery of infectious virus from B16F0s was poor compared with the well-characterized M2-10B4 cell line (Supplementary Figure S2a,b). At a high MOI, most B16F0s in the culture were infected (Supplementary Figure S2c) and this correlated with poor growth of the B16F0s and cell death (Supplementary Figure S2d,e). In addition, infected B16F0 cells expressed more MHC-I, MHC-II and the costimulatory molecule CD86 compared with uninfected cells in the same wells (data not shown). Thus, MCMV infection of B16F0s inhibits tumor growth, kills infected cells, and makes these cells better targets for the immune system. Together, these data suggest that MCMV may be oncolytic after direct infection of established tumors.
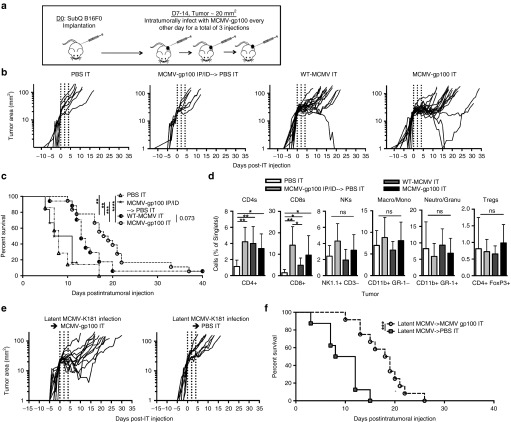
To determine whether IT infection with MCMV would improve the therapeutic impact of vaccination, mice were implanted with B16F0s subcutaneously, as above. When tumors were ~20 mm2 (~7–14 days after tumor implantation), they were injected directly with WT-MCMV, MCMV-gp100, or phosphate-buffered saline (PBS), every other day for three treatments (Figure 3a). For comparison, another group was vaccinated by IP and ID routes as above (Figure 2), and then given PBS by the IT route. As shown in Figure 3b, direct IT infection with MCMV had a marked effect on the growth of established tumors. Mice treated with PBS IT or MCMV-gp100 IP/ID + PBS IT had an average daily tumor growth rate after the IT injection of 21 and 19% respectively, and the tumor size doubled every 3.6 and 3.7 days respectively (Figure 3b). Strikingly, when mice were infected with either WT-MCMV or MCMV-gp100 by the IT route, the average daily growth rate post IT injection was reduced to 10 and 8% respectively, and the doubling time was increased to 7.3 and 9.4 days respectively, all of which were significantly slower than the controls (Figure 3b). This correlated with substantially increased survival of the host (Figure 3c). Interestingly, the presence of the gp100 epitope in the vaccine did relatively little to improve the outcome. One mouse in each group cleared its tumor (Figure 3b) and the average daily tumor growth rate and tumor doubling time between WT-MCMV IT and MCMV-gp100 IT treated mice were not significantly different. Mice given MCMV-gp100 IT survived slightly longer than those treated with WT-MCMV IT (P = 0.073, Figure 3c), but the difference was not dramatic. In addition, MCMV-gp100 IT infection slowed the growth of MC38 tumors, a transplantable colon adenocarcinoma that does not express gp100 (average daily growth rate of 4% for MCMV-injected tumors compared with 8% for PBS-injected tumors, 7 days after IT injection, P = 0.042, Supplementary Figure S3a,b). These data further suggest that MCMV IT infection delays tumor growth in a manner that is largely independent of the gp100 antigen in the vaccine.
Figure 3.
Intratumoral (IT) infection with MCMV induced tumor growth delay, regression, and improved survival. (a) The treatment schedule of MCMV IT infection. All tumors were initially injected at a tumor area of ~ 20 mm2. Each IT infection consisted of 5 × 105 plaque forming units. (b–d) The data shown is combined from four separate experiments. (b) Tumor growth, represented as change in tumor area (mm2) over time, is shown from the day of the first IT injection. MCMV-gp100 IP/ID vaccination was given on day 5, post tumor implantation followed by PBS IT on the schedule shown in a. PBS IT (n = 6); MCMV-gp100 IP/ID → PBS IT (n = 6); WT-MCMV IT (n = 18); MCMV-gp100 IT (n = 18). Vertical dotted lines represent days of IT injection. (c) Kaplan–Meier survival curve of the different treatment groups from day of tumor implantation until tumors were above 100 mm2. Significance was assessed by a logrank test, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (d) Tumor lymphocyte infiltration at time of sacrifice. Data represented as the mean ± SD. Significance was assessed by an unpaired t-test, ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001, P < 0.0001. NKs, NK cells; Neutro, Neutrophil; Granu, Granulocyte; Macro, Macrophage; Mono, Monocyte; Treg, regulatory T cell. (e,f) Mice latently infected with MCMV-K181 for 8 or 52 weeks received B16F0s and were infected following the schedule described in a with MCMV-gp100 (n = 8 mice infected 8 weeks previously and n = 4 mice infected 52 weeks previously) or PBS (n = 8 mice). (e) Tumor growth from the day of IT infection. (f) Kaplan–Meier survival curve of different treatment groups. Significance was assessed by a logrank test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV; PBS, phosphate-buffered saline.
Infection of mice bearing B16F0s by any route, including IT, induced similar increases in the accumulation of CD4+ and CD8+ cells in the tumor, without any other obvious changes in the frequencies of cell populations (Figure 3d). Notably, there were more activated (KLRG-1+) NK cells in tumors 7 days after IT infection when compared with systemically vaccinated animals (Supplementary Figure S4). Interestingly however, there were fewer activated NK cells that expressed Ly49H, an NK cell activating receptor that is ligated by the viral m157 protein39 (Supplementary Figure S4). Collectively, these data suggest that MCMV IT infection promotes CD4+ and CD8+ T-cell entry into subcutaneous tumors and an increase in activated NK cells.
Prior MCMV infection does not prevent the therapeutic effect of IT infection
Pre-existing antiviral immunity may be able to restrict the efficacy of oncolytic viruses by clearing the virus.11 More than half of people in the USA and most people in the world are already infected with CMV.24 Therefore, we tested whether IT infection would delay tumor growth in mice that had been infected with a wild-type strain of MCMV (MCMV-K181) 8 or 52 weeks prior to tumor implantation. Importantly, prior MCMV infection had no significant effect on the survival induced by MCMV-gp100 IT infection (Figures 3e,f) or the daily tumor growth rate measured after MCMV-gp100 IT infection (11 versus 8% for MCMV immune versus naive animals). Thus, pre-existing MCMV-specific immunity did not limit the therapeutic benefit of MCMV IT infection.
MCMV infects tumor-associated macrophages after MCMV IT infection
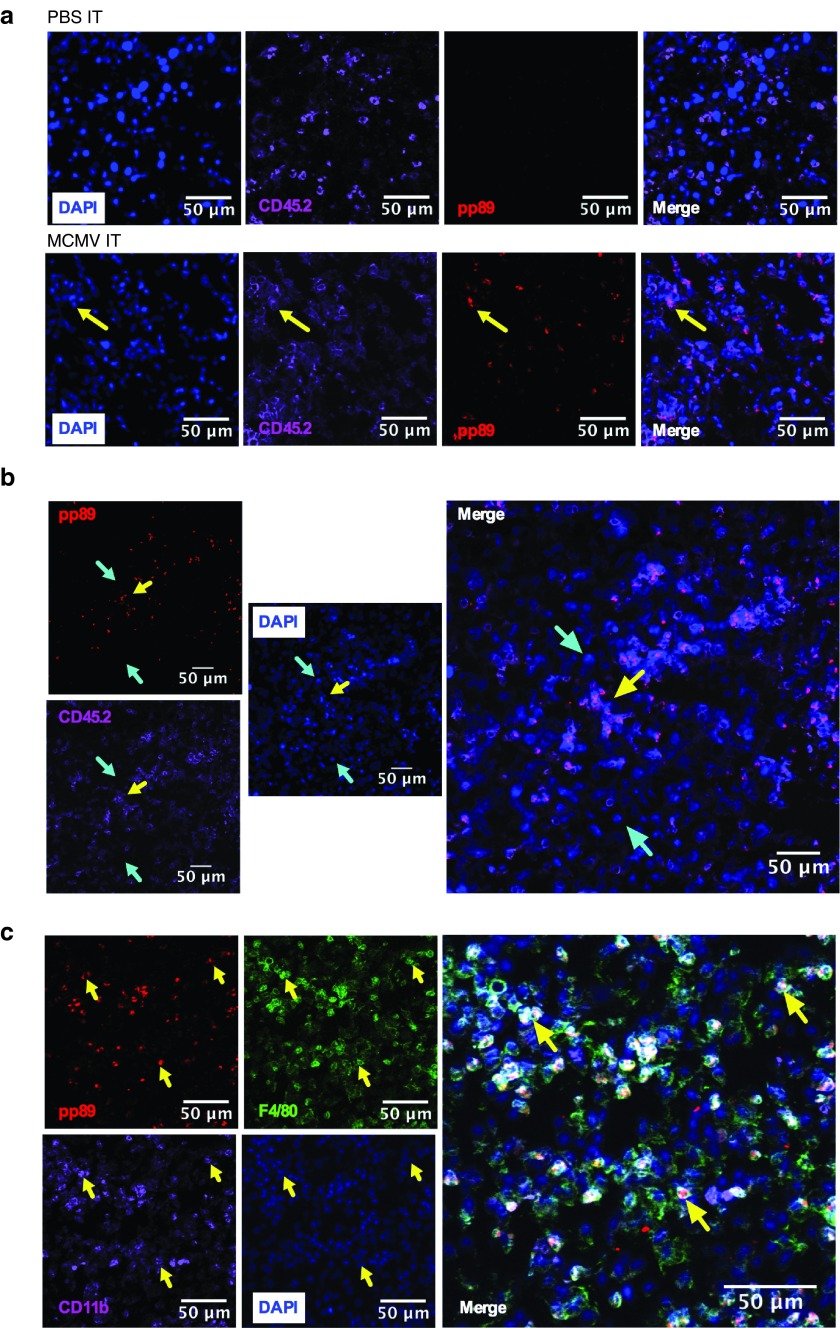
MCMV could infect and kill B16F0s (Supplementary Figure S2) and MC38s (not shown) in vitro, suggesting that it could be acting like an oncolytic virus. However, MCMV can also infect many other cells in the tumor environment including endothelial cells, fibroblasts and macrophages. To determine which cells were infected by MCMV after IT inoculation, B16F0 tumors were recovered one day after the last MCMV IT injection. Infected cells were identified histologically by the presence of nuclear-localized viral pp89, an immediate early protein expressed by MCMV infected cells shortly after infection.40 Viral pp89 (red) was only detected in tumors IT injected with MCMV (Figure 4a) and colocalized with 4',6-diamidino-2-phenylindole (DAPI) staining of the nucleus (Figure 4a and data not shown). Remarkably, pp89 staining was almost exclusively associated with CD45+ hematopoietic cells in the tumor (Figure 4a,b; yellow arrows). Further analyses revealed that infected cells also expressed CD11b and F4/80 (Figure 4c, yellow arrows). These data show that MCMV primarily infected tumor associated macrophages (TAMs) and not tumor cells, suggesting that MCMV was not acting as an oncolytic virus.
Figure 4.
MCMV infects TAMs after IT therapy. Mice received IT injections with WT-MCMV or MCMV-gp100 as in Figure 3. Tumors were harvested 1 day after the last IT injection and processed for histology. Yellow arrows indicate pp89 positive cells. Cyan arrows indicate pp89 negative cells. (a,b) Immunofluorescence staining of pp89 (red) in tumors IT injected with PBS or MCMV. Tumors were also stained for hematopoietic cells (CD45.2, purple) and costained with DAPI (blue). (c) pp89 (red) expression colocalizes with macrophages expressing CD11b (purple) and F4/80 (green) cells, after MCMV IT infection. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV; PBS, phosphate-buffered saline; DAPI, ; TAMs, tumor associated macrophages; IT, intratumoral.
Therapeutic efficacy of MCMV IT infection depends on CD8+ T-cells
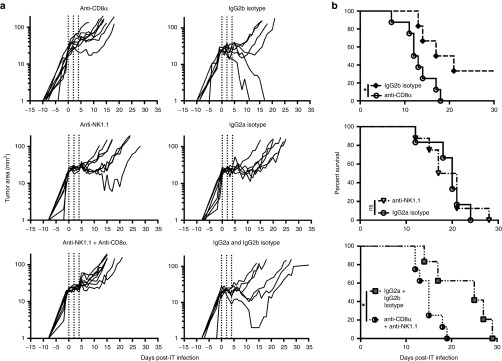
Since MCMV was likely not acting as an oncolytic virus, we wished to determine the roles of CD8+ T-cells and NK cells in the therapy. To this end, CD8+ T-cells and/or NK cells were depleted before the implantation of B16F0 tumors and throughout the MCMV IT therapy. Depletion of CD8+ T-cells significantly reduced survival after MCMV-gp100 IT infection, while depletion of NK1.1 alone had no effect (Figure 5a,b). Moreover, combined depletion of CD8s and NK cells was not different from depletion of CD8+ T-cells alone. Thus MCMV-IT therapy depended on CD8+ T-cells to prolong survival.
Figure 5.
Survival benefit after MCMV IT therapy depends on CD8+ T-cells. Mice were depleted of CD8+ and/or NK1.1+ cells as described in the materials and methods (n = 8 mice per group) or the relevant isotype control antibodies (n = 6 mice per group). (a) Tumor growth, represented as change in tumor area (mm2) over time, is shown from the day of the first intratumoral (IT) injection. (b) Kaplan Meier survival curves of the different antibody depletion groups compared to the relevant isotype controls from day of tumor implantation until tumors were above 100 mm2. Significance was assessed by a logrank test, *P < 0.05. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV.
Tumor-specific T-cells are markedly dysfunctional within the tumor and PD-L1 blockade greatly enhances tumor growth delay and regression induced by MCMV IT treatment
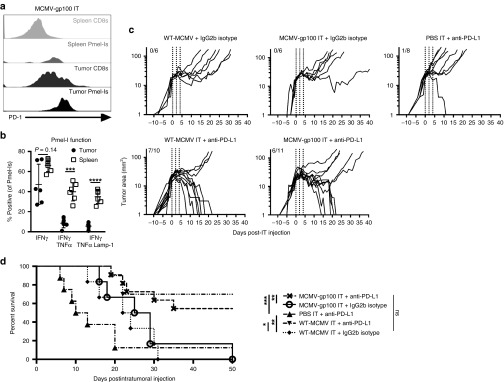
Since the MCMV IT therapy was dependent on CD8+ T-cells, gp100-specific Pmel-I transgenic T-cells were used to explore tumor-specific T-cells after IT therapy. Naive mice were given 104 Pmel-I T-cells expressing the Thy1.1. congenic marker, and B16F0 cells were implanted 1 day later. As above, recipients were IT infected when the tumors reached ~20 mm2. Animals were sacrificed 7 days after the initial IT infection and tumor-infiltrating T-cells were assessed. With only 104 Pmel-I T-cells transferred, the donor cells were undetectable in recipients infected with WT-MCMV, with the exception of one animal (Supplementary Figure S5a and data not shown). In contrast, IT infection with MCMV-gp100 induced expansion and migration of Pmel-I T-cells to the tumor in all mice (Supplementary Figure S5a). Notably, these cells expressed high levels of the inhibitory molecule PD-1 (Figure 6a and Supplementary Figure S5b) and were dysfunctional for cytokine production and degranulation compared with Pmel-I cells in the spleens of the same animals (Figure 6b). PD-L1 was also detectable on cells within tumors in slightly higher levels after MCMV IT injections than PBS IT injections (Supplementary Figure S5c), although there were no differences in PD-1 expression (data not shown). To test whether blocking PD-1/PD-L1 interactions in the tumor could improve MCMV IT therapy, WT-MCMV IT or MCMV-gp100 IT infection was combined with anti-PD-L1 antibody blockade. Remarkably, combining IT infection with PD-L1 blockade resulted in clearance of the established tumors from more than half of the mice and markedly improved overall survival regardless of which virus was used, effects that were not seen with any of these therapies alone (Figure 6c,d). Importantly, there was no significant survival difference between groups in which PD-L1 blockade was combined with MCMV-gp100 IT or WT-MCMV IT therapy (Figure 6d). Therefore, IT infection with MCMV synergized with anti-PD-L1 checkpoint blockade, regardless of the presence of gp100 in the vaccine.
Figure 6.
IT MCMV treatment combined with anti-PD-L1 therapy profoundly improves B16F0 tumor growth delay and survival. For a and b, Mice received 1 × 104 Pmel-Is one day prior to tumor implantation and were IT infected with MCMV as in Figure 3. (a) Representative histograms of the PD-1 expression of CD8+ T-cells or Pmel-Is 7 days post infection. (b) Ex vivo cytokine production and degranulation in response to native gp100 stimulation of Pmel-Is 7 days post infection and represented as the mean ± SD. Significance was assessed by a paired t-test, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (c) Mice bearing B16F0 tumors were treated with anti-PD-L1 or an isotype control antibody beginning on the day of MCMV IT infection. Shown is the tumor growth as in Figure 3 for the indicated groups of mice. Vertical dotted lines represent days of MCMV IT infection. Fractions in each graph represent the number of animals that cleared the tumor out of the number of animals tested. (d) Kaplan–Meier survival curve of the mice in each treatment group. Significance was assessed by a logrank test, P > 0.05 is nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. CMV, cytomegalovirus; MCMV, vaccination with murine-CMV; PBS, phosphate-buffered saline; IT, intratumoral.
Complete regression of primary tumors results in resistance or rejection of secondary B16F0 tumor challenges
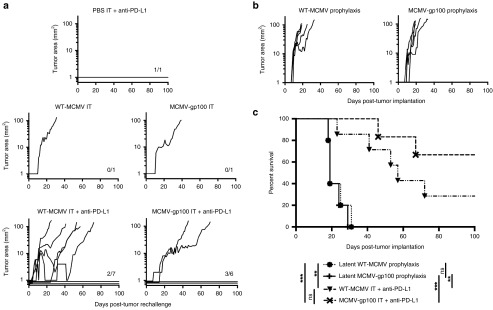
To determine whether clearance of tumors would result in protection against tumor challenge, the animals that cleared primary tumors after the various treatments described above were rechallenged with 2 × 105 B16F0s in the opposite flank 50–60 days after initial tumor implantation and at least 2 weeks after primary tumor clearance. Secondary tumors completely failed to grow in 5 of 15 mice that received any type of IT MCMV infection (Figure 7a, observed for >100 days). Moreover, in two of the cases that we found tumor growth, the challenge tumor did not appear until 27 (WT-MCMV IT + anti-PD-L1) or 140 days (MCMV-gp100 IT + anti-PD-L1, data not shown) post challenge. The one mouse that cleared its primary tumor after anti-PD-L1 treatment alone also rejected the secondary tumor (Figure 7a). Regardless of whether the primary tumor had been cleared after treatment with WT-MCMV IT + anti-PD-L1 or MCMV-gp100 IT + anti-PD-L1, there was no significant difference in the survival of these mice upon rechallenge (Figure 7c). Nevertheless, to determine whether some of this enhanced tumor resistance could be attributed to the fact that the challenged mice had been previously infected with MCMV-gp100, we implanted B16F0s into animals that had been infected with WT-MCMV or MCMV-gp100 by the IP route >100 days previously, but were not previously given tumors. Even though large gp100-specific T-cell populations were evident after vaccination with MCMV-gp100 (Figure 1c), there was little effect on tumor growth or survival in these mice compared with mice infected with WT-MCMV (Figure 7b,c) and no marked growth delay or tumor resistance, unlike mice that had previously cleared a tumor (Figure 7a). Importantly, depletion of CD8+ T-cells > 90 days after rechallenge did not enable late tumor growth in mice that rejected the challenge tumor (data not shown), suggesting that either long-term protection was CD8+ T-cell independent or the animals were cured of their tumors.
Figure 7.
Primary tumor clearance after MCMV IT treatment induces resistance or rejection of secondary tumor challenges. Any animal that cleared a primary tumor was rechallenged with 2 × 105 B16F0s in their opposite flank 2–3 weeks after initial tumor clearance. (a) Shown is the tumor growth starting from the day of tumor rechallenge. For the sake of clarity and fitting the data to a log scale, individual tumor area lines are spaced out below 1 mm2 when no nodule was evident. Fractions in each graph represent the number of animals that rejected tumor challenge out of the number of animals tested. (b) Mice were infected by the IP route with 2 × 105 plaque forming units WT-MCMV or MCMV-gp100 and 2 × 105 B16F0s were implanted subcutaneously 106 days later. Shown is the tumor growth as displayed in Figure 2. T-cell responses in the blood of these mice, prior to tumor implantation, are shown in Figure 1c,d. (c) Kaplan-Meier survival curve of rechallenged mice from WT-MCMV IT + anti-PD-L1 treated, MCMV-gp100 IT + anti-PD-L1 treated and prophylactically WT-MCMV or MCMV-gp100 vaccinated mice. Significance was assessed by a logrank test, P > 0.05 is nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
These data strongly imply that IT MCMV infection combined with PD-L1 blockade induced broad immunity to the B16F0 melanoma, subsequently preventing tumor growth at a distal site, independent of the gp100 antigen encoded in the viral genome or large numbers of circulating gp100-specific T-cells. Collectively, these data suggest that MCMV infects TAMs after IT infection, resulting in an unexpectedly potent, CD8+ T-cell-dependent, antitumor effect that can act synergistically with blockade of the PD-1 pathway to clear established tumors and promote systemic antitumor immunity.
Discussion
Direct modulation of the tumor microenvironment can markedly improve both local and systemic antitumor effects. Recent evidence suggests that IT administration of several different therapies induces better antitumor responses in animals, many times correlating with effects on distant tumors.6,7 Thus, IT therapies are currently being explored for both cutaneous and noncutaneous cancers.7 Our data show that systemic vaccination with MCMV-gp100 by the IP and ID routes induced migration of antigen-specific CD8+ T-cells into tumor tissue, but relatively poor antitumor effects (Figure 2). However, IT infection with MCMV induced marked tumor growth delay and prolonged survival in both B16F0 (melanoma) and MC38 (colon adenocarcinoma) models (Figure 3 and Supplementary Figure S3). Remarkably, this was true even for MCMV lacking expression of tumor-associated antigens or in mice that were already infected with MCMV (Figure 3 and Supplementary Figure S3).
MCMV does not fit the typical definition of an oncolytic virus. Oncolytic viruses are typically defined by their ability to replicate rapidly and somewhat selectively in tumor cells, inducing tumor cell death and subsequent antitumor and antiviral immune responses.41,42 While MCMV could infect and kill B16F0s in culture (Supplementary Figure S2), it only seemed to infect TAMs in vivo (Figure 4), suggesting that IT therapy is not working through direct tumor lysis. TAMs are associated with tumor progression by inducing a proangiogenic environment and suppressing antitumor immune responses.43,44 CMV infects monocytes and macrophages, inducing monocyte migration, tissue entry, and differentiation into macrophages.45,46 CMV infection of macrophages shifts them to an immune stimulatory phenotype by inducing upregulation of Toll-like receptors and increasing Th1 cytokine production,46,47,48 subsequently leading to increased T-cell proliferation.47 Moreover, it is possible that MCMV infection of TAMs could decrease the macrophage production of proangiogenic factors, such as vascular endothelial growth factor decreasing blood flow to tumors and slowing growth.43,44 All these possibilities must be addressed in future studies as possible mechanisms for MCMV IT therapy. It is worth noting that the MCMV immediate early protein pp89, which is expressed early after infection, may have different expression levels in different cell types. Thus it is possible that we only detected a subset of infected cells in the tumor that expressed pp89 at high levels, and that MCMV is still infecting the tumor cells themselves, or the tumor vasculature.49 This caveat aside, our data suggest that MCMV IT therapy works by altering TAMs and their interaction with tumors.
It is also worth noting that CMV almost certainly affects the tumor microenvironment beyond the cells that are directly infected. Contact with viral particles or gene-products likely triggers cell signaling and gene expression by cells in the tumor environment, regardless of infection. For example, CMV glycoproteins have been described to activate Toll-like receptor 2 (refs. 50,51) and CMV particles can activate the epidermal growth factor receptor,52,53,54 leading to an array of cellular responses. In addition, MCMV is a potent stimulator of NK cells and γδ T-cells,39,55 both of which might have antitumor effects. In B6 mice, NK cells expressing Ly49H are specifically expanded in response to the viral m157 protein.39 However, this population was not expanded in the tumors of mice vaccinated with MCMV by the IT route (Supplementary Figure S4). Rather, the tumor-infiltrating NK cells were largely Ly49H-, KLRG-1+, possibly suggesting that tumor-localized NK cells were activated in response to the tumor. Despite this, NK cell depletion had no effect on the MCMV IT therapy (Figure 5), suggesting NK cells were not important for the therapeutic outcome. Additional experiments will be needed to explore the impact of MCMV on other cells in the tumor and the tumor environment as a whole, after injection of live or inactivated viral particles.
Ultimately, improved survival after MCMV IT therapy depended on CD8+ T-cells but not NK cells (Figure 5). Interestingly however, IT injection of MCMV did not increase CD8+ T-cell frequencies above that induced by IP or ID infections. Although our data show that gp100-specific Pmel-I T-cells were markedly dysfunctional in the tumor after MCMV-gp100 IT vaccination (Figure 6 and Supplementary Figure S5), our preliminary data suggest that Pmel-I function was actually improved by IT therapy compared with MCMV-gp100 IP vaccination (data not shown). Moreover, blockade of the PD-1/PD-L1 pathway was synergistic with IT MCMV infection, leading to clearance of tumors from over half of the mice (Figure 6). This tumor clearance correlated with systemic antitumor immunity that could resist a secondary tumor challenge in the opposite flank, and was seemingly independent of the gp100 epitope encoded in the viral genome as well as gp100-specific T-cells induced by prophylactic vaccination (Figure 7). Thus, we propose that MCMV IT infection of TAMs within B16F0 tumors, in combination with blockade of the PD-1 pathway, improved the endogenous antitumor immunity.
Our data indicated that IP vaccination alone was ineffective for subcutaneous B16 tumors, which contrasts with recent work by Qiu and colleagues, who found that IP vaccination with a similar MCMV-gp100 vector was sufficient to delay the growth of lung nodules after IV injection of B16 melanoma cells.32 We favor the possibility that the different outcomes reflect the differences in tumor location. For example, gp100-specific CD8+ T-cells may more easily traffic to lung tumors as these nodules will be well exposed to the blood supply56 and we have shown that many circulating MCMV-specific T-cells are localized to the lung vasculature after IP infection.28 Moreover, after IP infection, MCMV may more readily infect macrophages in lung nodules as compared with subcutaneous nodules. Alternatively, it is possible that tumors growing in each location depend on different immune inhibitory mechanisms that are more or less easily overcome by MCMV-driven T-cells. Finally, it is notable that the MCMV-gp100 vaccine used by Qiu and colleagues expressed a variant of the gp100 antigen that differed by two amino acids from the native sequence (gp100E25K, S27P), whereas the epitope used in our study differed by only one amino acid (gp100S27P), a difference that could, in theory, have a substantial impact on the efficacy or function of gp100-specific T-cells. Future work will be required to test these ideas.
Overall, our study is the first to show that MCMV may have superior therapeutic efficacy for cutaneous melanomas after direct intratumoral injections, and that this route of vaccination can synergize with immune checkpoint blockades to clear tumors and induce protection against distal tumors, without virally encoded tumor antigens. This study builds on recent data suggesting that CMV may be an effective antitumor therapy and suggests that the route of infection and tumor location may be critical factors in defining the efficacy of this platform.
Materials and Methods
Mice and tumor models. C57BL/6J mice and gp100-specific Pmel-I T-cell transgenic mice expressing the Thy1.1 congenic marker (B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J) were purchased from Jackson Laboratory, (Bar Harbor, ME) and bred in house for use in all experiments. Donor and recipient mice were sex-matched for all adoptive transfers. For most experiments, mice were between 6 and 12 weeks old at the time of tumor implantation. For the data shown in Figure 3e,f, mice were 6–12 weeks old at the time of primary MCMV infection and tumors were implanted 8 or 52 weeks later. For primary tumors, mice were subcutaneously challenged in the shaved right flank with 1 × 105 B16F0s (kindly provided by Dr Vitali Alexeev) or with 5 × 105 MC38s (kindly provided by Dr Adam Snook) suspended in Hank's balanced salt solution (HBSS) (CellGro, Manassas, VA). For tumor rechallenge experiments (Figure 7), animals that had cleared a primary tumor or prophylactically vaccinated animals were rechallenged with 2 × 105 B16F0s in the shaved left flank. In all cases, tumor area was calculated by multiplying the length and width (in millimeters) of the tumor as measured with a 6-inch digital caliper (Neiko). Animals were sacrificed when the tumor was growing exponentially and had exceeded ~100 mm2 in area, or when the tumors had ulcerated, or the animals had become moribund. The Thomas Jefferson University Institutional Animal Care and Use Committee reviewed and approved all protocols.
Virus strains, cell lines, and in vitro infections. To produce the recombinant strain MCMV-gp100S27P, the sequence encoding the altered gp100S27P peptide (EGPRNQDWL) was fused to the 3′ end of the sequence encoding GFP, upstream of the stop codon, as done previously with other antigens.36 The fusion construct was recombined with MCMV encoded with a bacterial artificial chromosome (BAC, strain MW97.01, hereafter called WT-MCMV57) and targeted to replace the m128 exon (IE2 gene) using established techniques.20 Viral stocks were prepared on M2-10B4 stromal cells as previously described.58 In brief, 2–4 × 106 cells were infected at an MOI of 0.01. Cells were collected 5–6 days later, homogenized by douncing, and the supernatant was ultracentrifuged to concentrate the virus. Viral stocks were suspended in media containing serum and frozen at −80°C until use. Viral titers were determined by plaque assay without centrifugal enhancement using M2-10B4s as previously described.58 In brief, subconfluent layers of M2-10B4s were infected with lysates at several different titrations, covered with viscous media, incubated for 5 days, and stained with crystal violet for plaque counting. The single- and multistep growth analyses shown in Figure 1 and Supplementary Figure S2 were performed by infecting M2-10B4 cells or B16F0 cells with an MOI = 0.1 (multistep), or an MOI = 10 (single step), harvesting lysates at the indicated times and measuring viral growth by plaque assay as described above. In all cases, M2-10B4s and MC38s were grown in Roswell Park Memorial Institute media (RPMI) (CellGro) + 1% PenStrep (Gemini Bio-Products, Sacramento, CA) + 10% fetal bovine serum (Benchmark serum, Gemini). B16F0s were grown in Dulbecco's modified essential medium (CellGro) + 1% PenStrep + 10% fetal bovine serum.
Infections and vaccinations of mice. For infection of mice without tumors (Figure 1, Figure 3e,f, and Figure 7b,c), animals received 2 × 105 plaque forming units of MCMV-gp100, WT-MCMV, or MCMV-K181 by the IP route in a single injection of 100 μl. For IP and ID infections of tumor-bearing mice, animals received 5 × 105 plaque forming units of the indicated virus in a single injection of 100 μl for IP infection and 25 μl for ID infection. In all cases, ID infection was performed in the skin next to the tumor implantation site. For IT injections, animals received 5 × 105 plaque forming units of the indicated virus, diluted in PBS, in 30 μl volume or 30 μl of PBS alone every other day for three total injections.
Adoptive transfer of Pmel-I T-cells. Spleens were harvested from naive Pmel-I transgenic mice, passed through a 70 μm cell strainer to form single cell suspensions and washed twice with T-cell media (RPMI 1640 (Corning, NY) with L-glutamine + 10% fetal bovine serum + 1% PenStrep and 5 × 10–5 mol/l β-mercaptoethanol (MilliporeSigma, Darmstadt, Germany)). Total splenocytes were counted on a Z2 Coulter Particle Count and Size Analyzer (Beckman Coulter) and the sample was assessed for frequency of CD8+ T-cells by flow cytometry. Based on these data, total splenocytes were suspended in PBS so that the desired number of CD8+ T-cells was present in 100 μl, which is the volume that was retro-orbitally injected into recipient C57BL/6 mice.
Lymphocyte isolation, analyses, and intracellular cytokine staining. Spleens were suspended in T-cell media and mechanically processed through a 70 μm nylon cell strainer to achieve a single cell suspension. For recovery of lymphocytes from tumors, tumor masses were placed in tumor digestion media (1× HBSS (Cellgro), 0.1 mg/ml Collagenase A (Worthington, Lakewood, NJ), 60 U/ml DNase I (Roche, Indianapolis, IN))59 and minced using the gentleMACS Octo Dissociator using C Tubes (Miltenyi Biotec, Bergisch Gladbach, Germany). Minced tumors in digestion media were incubated at 37°C for 30 minutes with continuous rotation. Digested tumors were minced again using the gentleMACS Octo Dissociator, then washed twice with T-cell media and mechanically filtered through a 70 μm nylon filter to make a single cell suspension. Lymphocytes were then either directly assessed by flow cytometry or tested for their ability to produce cytokines upon stimulation. For analyses of cytokine production by cells from spleens and tumors, 1–2 × 106 cells were incubated in T-cell media in a round bottom 96-well plate for 5 hours at 37° C in 5% CO2. The final incubation volume was 100 μl and contained 1 μg/ml of the indicated peptide (synthesized by Genemed Synthesis, San Antonio, TX) and 1 μg/ml brefeldin A (GolgiPlug, BD Biosciences, San Jose, CA), as well as fluorescently labeled antibody specific for CD107a. At the end of the incubation, cells were washed twice with ice-cold Fluorescence-activated cell sorting buffer (PBS, 0.05% Sodium Azide, 1% FBS) and stained with antibodies specific for surface proteins followed by analyses of intracellular IFN-γ and TNF-α using the BD Cytofix/Cytoperm kit (BD Biosciences) and following the manufacturers instructions. In Figure 1, ~150 μl of peripheral blood was collected from the retro-orbital sinus. Red blood cells were lysed for 5 minutes in red blood cell lysis buffer (150 mmol/l NH4Cl, 10 mmol/l NaHCO3), the remaining white blood cells were washed twice, and resuspended in T-cell media. Approximately 1/5 of the recovered cells were added to individual wells and incubated as described above for 3 hours and without the antibodies specific for CD107a. For regulatory T-cell staining, cells were fixed with FOXP3 Fix/Perm buffer (Biolegend, San Diego, CA) for 10 minutes on ice and then permeabilized for 15 minutes with FOXP3 Perm buffer (Biolegend) before staining.
Antibodies and FACS analysis. Analyses of lymphocytes were performed using antibodies specific for the following molecules: CD3 (clone 500A2), CD4 (clone GK1.5), CD8α (clone 53.6.7), CD8β (clone YTS156.7.7), Thy1.1 (clone OX-7), PD-1 (clone 29F.1A12), PD-L1 (clone 10F.9G2), H-2Db (clone KH95), H-2Kb (clone AF6-88.5), CD80 (16-10A1), CD86 (GL-1), NK1.1 (clone PK136), CD11b (clone ICRF44), GR-1 (clone RB6-8C5), FoxP3 (clone 150D), I-A/I-E (clone M5/114.15.2), IFN-γ (clone XMG1.2), TNF-α (clone MP6-XT22), KLRG-1 (clone MAFA), Ly49H (clone 3D10), and CD107a (clone 1D4B). All antibodies were purchased from Biolegend or BD Biosciences. Stained cells were analyzed using the LSR II flow cytometer (BD Biosciences) and FlowJo Software version 10 (TreeStar, Ashland, OR).
In vivo antibody blockades. To deplete CD8+ T-cells or NK cells, mice were treated with 300 μg of anti-CD8α (clone 53–6.72) and/or anti-NK1.1 (clone PK136) every 3 days for a total of eight treatments, starting 2 days before tumor implantation. Treatment resulted in > 90% depletion of target cells (data not shown). As controls, additional animals were treated with an irrelevant IgG2a antibody (isotype control for anti-NK1.1, clone C1.18.4), or IgG2b antibody (isotype control for anti-CD8α, clone LTF-2) following the same schedule. To study the effect of PD-L1 blockade on IT infection, mice were treated with 400 μg of anti-PD-L1 (clone 10F.9G2) by the IP route on the first day of IT treatment, followed by an additional 200 μg anti-PD-L1 given every third day by the IP route for a total of six treatments. As a control, additional animals were treated with the IgG2b isotype control clone LTF-2, following the same schedule. All antibodies were purchased from Bio-X-Cell (West Lebanon, NH).
Fluorescence microscopy. Isolated tumors were frozen in Fisher HealthcareTM Tissue-Plus OCT (Fisher Scientific, Waltham, MA) and cut into 6–8 μm sections using a cryostat. Samples were fixed in cold acetone for 10 minutes and rehydrated with Tris-buffered saline (TBS) for 20 minutes, blocked with blocking buffer (TBS + 3% BSA and 0.1% Tween-20) for 20 minutes and stained with antibodies specific for CD31 (clone 390), CD45.1/2 (clone A20/104), CD11b (clone M1/70), F4/80 (clone BM8) and/or MCMV pp89 (clone 6/58/1 (ref. 60)) in blocking buffer for 1 hour and later costained with DAPI (Prolong Gold antifade, Life Technologies). The anti-pp89 antibody was purified from hybridoma supernatant using Pierce Protein A/G Magnetic Beads (Fisher Scientific), concentrated using Amicon Ultra-0.5 or 15 Centrifugal Filter Unit with Ultracel-100 membrane (Millipore), and labeled using Mix-N-Stain CF555 Antibody Labeling Kit (Sigma-Aldrich, St. Louis, MO). Anti-pp89 flourophore conjugation was confirmed by staining infected and uninfected M2-10B4s with the labeled antibody (data not shown). Images were generated with an LSM 510 Meta confocal laser scanning microscope (Carl Zeiss), the Zeiss AIM 4.2 SP1 software (Carl Zeiss, Oberkochen, Germany), and ImageJ (Fiji).
Statistical analysis. Prism Version 6.0 d and SAS 9.4 were used for graph creation and statistical analyses. For statistical significance, *P < 0.05; **P < 0.01; *** P < 0.001; ****P < 0.0001. Tumor growth rates and doubling times were analyzed with a mixed-effects linear regression, an extension of ordinary linear regression for repeated measures over time. Heuristically, the model estimates a tumor growth curve for each animal and then appropriately averages these curves to estimate the group's average trajectory. This approach accounts for the within-animal correlation of tumor sizes over time and the potential uneven timing of readings. Tumor size was log-transformed before the analyses and was modeled as a function of time, experimental group, and their interaction. The main aim was to compare growth rates over time across the experimental groups. Results were expressed in terms of the average daily increase of tumor size and the tumor doubling time. For the MC38 tumors, Supplementary Figure S3, tumor growth was analyzed by fitting a quadratic curve to each data set and analyzing the subsequent daily tumor growth rate. We also used Kaplan–Meier survival curves and the logrank test to analyze the time tumors needed to reach 100 mm2 (overall survival, the approximate tumor size when animals are typically sacrificed).
SUPPLEMENTARY MATERIAL Figure S1. Representative gating strategies for CD8+ T-cells recovered from blood, tumors and spleens. Figure S2. MCMV-gp100 infection of B16F0s in vitro induced cell death. Figure S3. MCMV IT therapy prolongs survival of MC38-tumor bearing animals. Figure S4. MCMV IT therapy induces more activated NK cells in tumors that do not recognize m157 on CMV infected cells than systemic infection on D7 post infection. Figure S5. Tumor antigen-specific CD8+ T-cells in the tumor were PD-1hi and dysfunctional after MCMV IT infection.
Acknowledgments
This work was supported by grants from the American Cancer Society (ACS-IRG-08-060-04 and ACS-RSG-15-184-01) and the NIH (RO3 CA174979), all awarded to C.M.S. and by the NIH (R21 CA127181), awarded to A.B.H. Both A.B.H. and C.M.S. have a financial interest in UbiVac CMV for the development of spread-defective CMV-based therapeutics. Neither the funding bodies nor UbiVac CMV had any role in the design of the experiments or the interpretation of the data. Histological samples were analyzed in the SKCC bioimaging shared resource, supported by Cancer Center Support Grant 5P30CA056036-17. The authors would also like to thank Yolanda Covarrubias for her help with the immune fluorescence imaging and the optimization of our staining protocol.
Supplementary Material
References
- Klebanoff, CA, Acquavella, N, Yu, Z and Restifo, NP (2011). Therapeutic cancer vaccines: are we there yet? Immunol Rev 239: 27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, RD, Old, LJ and Smyth, MJ (2011). Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331: 1565–1570. [DOI] [PubMed] [Google Scholar]
- Wherry, E. J. (2011). T cell exhaustion. Nature Immunol 131: 492–499. [DOI] [PubMed] [Google Scholar]
- Hailemichael, Y and Overwijk, WW (2014). Cancer vaccines: Trafficking of tumor-specific T-cells to tumor after therapeutic vaccination. Int J Biochem Cell Biol 53: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi, F, Scolyer, RA, Rumcheva, P, Moncrieff, M, Murali, R, McCarthy, SW et al. (2012). Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 30: 2678–2683. [DOI] [PubMed] [Google Scholar]
- Singh, M and Overwijk, WW (2015). Intratumoral immunotherapy for melanoma. Cancer Immunol Immunother 64: 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabelle, A, Kohrt, H, Caux, C and Levy, R (2014). Intratumoral immunization: a new paradigm for cancer therapy. Clin Cancer Res 20: 1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miest, TS and Cattaneo, R (2014). New viruses for cancer therapy: meeting clinical needs. Nat Rev Microbiol 12: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andtbacka, RH, Kaufman, HL, Collichio, F, Amatruda, T, Senzer, N, Chesney, J et al. (2015). Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol 33: 2780–2788. [DOI] [PubMed] [Google Scholar]
- FDA approves first-of-its-kind product for the treatment of melanoma (2015). <http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm469571.htm>.
- Barlett, D. L. et al. (2013). Oncolytic viruses as therapeutic cancer vaccines. Molecular Cancer 13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, J, Reid, T, Ruo, L, Breitbach, CJ, Rose, S, Bloomston, M et al. (2013). Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat Med 19: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki, O, Parviainen, S, Juhila, J, Turkki, R, Linder, N, Lundin, J et al. (2015). Immunological data from cancer patients treated with Ad5/3-E2F-Δ24-GMCSF suggests utility for tumor immunotherapy. Oncotarget 6: 4467–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woller, N, Gürlevik, E, Ureche, CI, Schumacher, A and Kühnel, F (2014). Oncolytic viruses as anticancer vaccines. Front Oncol 4: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtappels, R, Pahl-Seibert, MF, Thomas, D and Reddehase, MJ (2000). Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T-cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J Virol 74: 11495–11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtappels, R, Grzimek, NK, Simon, CO, Thomas, D, Dreis, D and Reddehase, MJ (2002). Processing and presentation of murine cytomegalovirus pORFm164-derived peptide in fibroblasts in the face of all viral immunosubversive early gene functions. J Virol 76: 6044–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, H, Sierro, S, V Cuero, A and Klenerman, P (2003). Population analysis of antiviral T cell responses using MHC class I-peptide tetramers. Clin Exp Immunol 134: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, U, Wagner, M, Sierro, S, Oxenius, A, Hengel, H, Dumrese, T et al. (2004). Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J Virol 78: 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munks, MW, Gold, MC, Zajac, AL, Doom, CM, Morello, CS, Spector, DH et al. (2006). Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J Immunol 176: 3760–3766. [DOI] [PubMed] [Google Scholar]
- Borst, EM, Benkartek, C and Messerle, M (2007). Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr Protoc Immunol Chapter 10: Unit 10.32. [DOI] [PubMed] [Google Scholar]
- Hansen, SG, Vieville, C, Whizin, N, Coyne-Johnson, L, Siess, DC, Drummond, DD et al. (2009). Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, SG, Ford, JC, Lewis, MS, Ventura, AB, Hughes, CM, Coyne-Johnson, L et al. (2011). Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, SG, Piatak, M Jr, Ventura, AB, Hughes, CM, Gilbride, RM, Ford, JC et al. (2013). Immune clearance of highly pathogenic SIV infection. Nature 502: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate, SL, Dollard, SC and Cannon, MJ (2010). Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988-2004. Clin Infect Dis 50: 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, SG, Powers, CJ, Richards, R, Ventura, AB, Ford, JC, Siess, D et al. (2010). Evasion of CD8+ T-cells is critical for superinfection by cytomegalovirus. Science 328: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertoghs, KM, Moerland, PD, van Stijn, A, Remmerswaal, EB, Yong, SL, van de Berg, PJ et al. (2010). Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J Clin Invest 120: 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro, S, Rothkopf, R and Klenerman, P (2005). Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur J Immunol 35: 1113–1123. [DOI] [PubMed] [Google Scholar]
- Smith, CJ, Turula, H and Snyder, CM (2014). Systemic hematogenous maintenance of memory inflation by MCMV infection. PLoS Pathog 10: e1004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, CJ, Caldeira-Dantas, S, Turula, H and Snyder, CM (2015). Murine CMV infection induces the continuous production of mucosal resident T-cells. Cell Rep 13: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyushnenkova, EN, Kouiavskaia, DV, Parkins, CJ, Caposio, P, Botto, S, Alexander, RB et al. (2012). A cytomegalovirus-based vaccine expressing a single tumor-specific CD8+ T-cell epitope delays tumor growth in a murine model of prostate cancer. J Immunother 35: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G, Smith, T, Grey, F and Hill, AB (2013). Cytomegalovirus-based cancer vaccines expressing TRP2 induce rejection of melanoma in mice. Biochem Biophys Res Commun 437: 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Z, Huang, H, Grenier, JM, Perez, OA, Smilowitz, HM, Adler, B et al. (2015). Cytomegalovirus-based vaccine expressing a modified tumor antigen induces potent tumor-specific CD8(+) T-cell response and protects mice from Melanoma. Cancer Immunol Res 3: 536–546. [DOI] [PubMed] [Google Scholar]
- van Stipdonk, MJ, Badia-Martinez, D, Sluijter, M, Offringa, R, van Hall, T and Achour, A (2009). Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100. Cancer Res 69: 7784–7792. [DOI] [PubMed] [Google Scholar]
- Dekhtiarenko, I, Jarvis, MA, Ruzsics, Z and Čičin-Šain, L (2013). The context of gene expression defines the immunodominance hierarchy of cytomegalovirus antigens. J Immunol 190: 3399–3409. [DOI] [PubMed] [Google Scholar]
- Farrington, LA, Smith, TA, Grey, F, Hill, AB and Snyder, CM (2013). Competition for antigen at the level of the APC is a major determinant of immunodominance during memory inflation in murine cytomegalovirus infection. J Immunol 190: 3410–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turula, H, Smith, CJ, Grey, F, Zurbach, KA and Snyder, CM (2013). Competition between T-cells maintains clonal dominance during memory inflation induced by MCMV. Eur J Immunol 43: 1252–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim, LM, Jones, CM, Gebhardt, T, Preston, CM and Carbone, FR (2008). CD8(+) T-cell attenuation of cutaneous herpes simplex virus infection reduces the average viral copy number of the ensuing latent infection. Immunol Cell Biol 86: 666–675. [DOI] [PubMed] [Google Scholar]
- Liu, L, Zhong, Q, Tian, T, Dubin, K, Athale, SK and Kupper, TS (2010). Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat Med 16: 224–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, LL (2008). Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 8: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, GM, Ebeling-Keil, A and Koszinowski, UH (1987). Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol 61: 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca, C and Schlom, J (2011). Viral vector-based therapeutic cancer vaccines. Cancer J 17: 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty, BD, Breitbach, CJ, Stojdl, DF and Bell, JC (2014). Going viral with cancer immunotherapy. Nat Rev Cancer 14: 559–567. [DOI] [PubMed] [Google Scholar]
- Gabrilovich, DI, Ostrand-Rosenberg, S and Bronte, V (2012). Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 12: 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanmee, T, Ontong, P, Konno, K and Itano, N (2014). Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 6: 1670–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley-Bauer, LP, Roback, LJ, Wynn, GM and Mocarski, ES (2014). Cytomegalovirus hijacks CX3CR1(hi) patrolling monocytes as immune-privileged vehicles for dissemination in mice. Cell Host Microbe 15: 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, MS, Bentz, GL, Alexander, JS and Yurochko, AD (2004). Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J Virol 78: 4444–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, C, Varani, S, Wang, L, Walther, P, Zhou, S, Straschewski, S et al. (2013). Human cytomegalovirus infection of M1 and M2 macrophages triggers inflammation and autologous T-cell proliferation. J Virol 87: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G, Smith, MS, Smith, PM and Yurochko, AD. (2008). Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation towards a M1 macrophage. J Immunol 181: 698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Berg, PJ, Yong, SL, Remmerswaal, EB., van Lier, RA and ten Berge, IJ. (2012). Cytomegalovirus-induced effector T-cells cause endothelial cell damage. Clin Vaccine Immunol 19: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szomolanyi-Tsuda, E, Liang, X, Welsh, RM, Kurt-Jones, EA and Finberg, RW (2006). Role for TLR2 in NK cell-mediated control of murine cytomegalovirus in vivo. J Virol 80: 4286–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme, KW, Guerrero, M and Compton, T (2006). Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol 177: 7094–7102. [DOI] [PubMed] [Google Scholar]
- Wang, X, Huong, S-M Chiu, M L, Raab-Traub, N,Huang, E-S. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424: 452–456. [DOI] [PubMed] [Google Scholar]
- Chan, G, Nogalski, MT and Yurochko, AD (2009). Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci USA 106: 22369–22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz, GL and Yurochko, AD (2008). Human CMV infection of endothelial cells induces an angiogenic response through viral binding to EGF receptor and beta1 and beta3 integrins. Proc Natl Acad Sci USA 105: 5531–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell, S, Dietz, M, Schneider, A, Holtappels, R, Mach, M and Winkler, TH (2015). Control of murine cytomegalovirus infection by γδ T-cells. PLoS Pathog 11: e1004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannmark, U, Johansson, BR, Bryant, JL, Unger, ML, Hokland, ME, Goldfarb, RH et al. (1995). Microvessel origin and distribution in pulmonary metastases of B16 melanoma: implication for adoptive immunotherapy. Cancer Res 55: 4627–4632. [PubMed] [Google Scholar]
- Wagner, M, Jonjic, S, Koszinowski, UH and Messerle, M (1999). Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol 73: 7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbach, KA, Moghbeli, T and Snyder, CM (2014). Resolving the titer of murine cytomegalovirus by plaque assay using the M2-10B4 cell line and a low viscosity overlay. Virol J 11: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, ED, Enriquez, HL, Fu, YX and Engelhard, VH (2010). Tumor masses support naive T cell infiltration, activation, and differentiation into effectors. J Exp Med 207: 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase, MJ, Fibi, MR, Keil, GM and Koszinowski, UH (1986). Late-phase expression of a murine cytomegalovirus immediate-early antigen recognized by cytolytic T lymphocytes. J Virol 60: 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.