Abstract
Myotonic dystrophy type 1 (DM1) is caused by expanded Cytosine-Thymine-Guanine (CTG) repeats in the 3′-untranslated region (3′ UTR) of the Dystrophia myotonica protein kinase (DMPK) gene, for which there is no effective therapy. The objective of this study is to develop genome therapy in human DM1 induced pluripotent stem (iPS) cells to eliminate mutant transcripts and reverse the phenotypes for developing autologous stem cell therapy. The general approach involves targeted insertion of polyA signals (PASs) upstream of DMPK CTG repeats, which will lead to premature termination of transcription and elimination of toxic mutant transcripts. Insertion of PASs was mediated by homologous recombination triggered by site-specific transcription activator-like effector nuclease (TALEN)-induced double-strand break. We found genome-treated DM1 iPS cells continue to maintain pluripotency. The insertion of PASs led to elimination of mutant transcripts and complete disappearance of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated neural stem cells, cardiomyocytes, and teratoma tissues. In conclusion, genome therapy by insertion of PASs upstream of the expanded DMPK CTG repeats prevented the production of toxic mutant transcripts and reversal of phenotypes in DM1 iPS cells and their progeny. These genetically-treated iPS cells will have broad clinical application in developing autologous stem cell therapy for DM1.
Introduction
Myotonic Dystrophy type 1 (Dystrophia Myotonica, DM1) is a progressive, debilitating, and multisystemic disorder with an estimated minimum prevalence of 8–10/100,000 (ref. 1). Its congenital form has high mortality (25%) before 18 months of age. Those who do survive through infancy will likely die from respiratory failure by the age of 40 (ref. 2). Adult classic DM1 (CTG repeats in the range of 100–1,000) usually presents with muscle weakness, atrophy, myotonia, frontal balding, cataracts, behavioral abnormalities, diabetes, cardiac conduct defects, and individuals who have a shortened lifespan. For the past two decades, pioneering investigators have unveiled much about the disease mechanism since the finding of the causative gene in 1992. DM1 results from an unstable CTG nucleotide repeat expansion within the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene on chromosome 19q13.3 (refs. 3,4,5,6). Normal individuals have 5–37 CTG repeats. When the repeats number exceeds 50 CTGs, it usually leads to onset of DM1, and larger repeat size correlates with increased severity of symptoms and earlier age of onset. Alleles containing 38–49 CTGs are considered premutated alleles. While patients with these alleles are asymptomatic, they have a high risk of passing a larger, pathological expansion to their offspring. Often DM1 families are first found in symptomatic children and when children are born with repeats >1,000, it generally leads to congenital DM1 (refs. 7,8,9).
Many therapeutic approaches have been contemplated and tested, including symptomatic management, supportive care, targeting mutant transcripts, and manipulating molecular abnormalities (see recent reviews).10,11 While these approaches are relatively easy to deliver, they tend to target only one aspect of the multisystemic disorder, and none of these are curative. There is no consensus as to when to intervene, even for a well-documented approach to prevent sudden cardiac death such as the placement of pacemaker. We anticipate that stem cell therapy will be promising not only in the treatment of the congenital form but also in the adult form of DM1. Throughout life, stem cells provide new cells to replace lost ones. This occurs even in the adult CNS,12,13 a system once thought to be nonreplaceable. We anticipate that providing a stem cell pool that is mutation-free as a source of new healthy cells will not only reverse the degenerative process but may also delay or prevent the onset of the disease. These disease free stem cells can be delivered upon genetic diagnosis of classic or congenital DM1 without consideration of the disease course. Human induced pluripotent stem (iPS) cells are being studied for regenerative medicine. iPS cells can be generated from somatic cells of the same patient, termed patient-specific iPS cells, which essentially overcome the hurdle of immune rejection of autologous cell transplantation. In order to prevent DM1 stem cells and their progeny from undergoing the same degenerative process after transplantation, modifying the mutant genome to generate healthy DM1 iPS cells is an important step toward autologous stem cell therapy. We will use the terms “genome therapy” and “genome-treated” in this study to refer to the modification of the genome for therapeutic purpose in lieu of “genome editing” or “genome-edited”, which was initially utilized in reverse genetics for the study of a gene or a protein. We have recently developed a strategy of genome therapy on DM1 neural stem cells (NSCs) by the incorporation of polyadenylation signals (PASs) upstream of DMPK CTG repeats, which led to premature termination of transcription, elimination of toxic mutant transcripts and reversal of disease phenotypes.14 In this study, we performed genome therapy on human DM1 iPS cells since the pluripotency of iPS cells have a broader potential for the development of stem cell therapy for DM1, a multisystemic disease.
Results
Integration of PASs into DMPK intron 9 eliminated nuclear RNA foci in DM1 iPS cells
Out of the 48 puromycin-resistant clones from one DM1 iPS cell line (DM-03), 5 had total loss of nuclear RNA foci and were subjected to subcloning. Of which 19 out of 20 subclones continued to be homogeneously foci negative. Subclone 13-3 and 33–4 were expanded and continued to be foci negative, and were used for subsequent analyses (Figure 1). Genotyping by carefully designed primer pairs for genomic polymerase chain reaction (PCR) and reverse-transcriptase PCR (RT-PCR) showed the correct insertion of the cassette in the designed TALEN cutting site in the mutant allele with intact transcription of normal allele (Figure 2a, b, c). Southern blot confirmed that the genome-treated iPS cell lines contain the PASs cassette upstream of the CTG repeats. An additional restriction enzyme EcoRI site within the PASs cassette altered the banding pattern between the DM-03 parental iPS cell line and the genome-treated iPS clones 13-3 and 33–4 (Figure 2d). Southern blot using restriction enzyme NcoI digest illustrates that the CTG expansion remains intact throughout this editing and cloning process (Figure 2e). After removal of the selective marker, the genome-treated clone continues to be foci negative (see Supplementary Figure S1) and Triplet Repeat Primed PCR (TP-PCR) confirmed that the expanded CTG repeats were left intact (see Supplementary Figure S2).
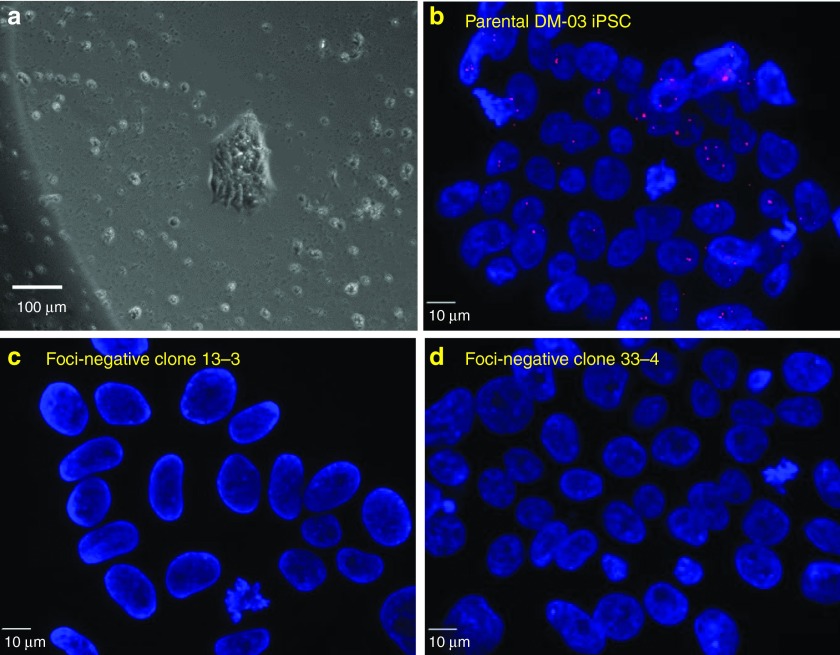
Figure 1.
Loss of nuclear RNA foci in genome-treated DM1 induced pluripotent stem (iPS) cell clones. (a) A typical Puromycin and Ganciclovir-resistant clone (phase contrast image). (b) Parental DM-03 iPS cells with nuclear RNA foci. (c, d) Nuclear foci were not detectable in the Puromycin and Ganciclovir-resistant clones of DM-03 iPS cells.
Figure 2.
Exogenous polyA signals (PASs) were integrated in the designed transcription activator-like effector nuclease (TALEN) targeting site and were transcribed contiguous with Dystrophia myotonica protein kinase (DMPK) transcription. (a) Genomic polymerase chain reaction (PCR) analysis showing the integration of the insertion cassette into the TALEN targeting site. Primer pair I9SF/TSR amplified the whole insertion cassette, detectable in Puromycin and Ganciclovir-resistant clones and donor vector but not in parental induced pluripotent stem (iPS) cells. Primer pair IVF2/IVR amplified a portion of the insertion cassette and a portion of genomic DNA downstream of the end of the 3′ homology arm. VF/VR amplified the TK resistant gene in donor backbone. CTGSF/CTGSR amplified genomic DNA as a template loading control. (b) RT-PCR showing the effect of the integration on DMPK gene transcription. Products from primer pair E8F3/PGKR1 were detected in the two foci-negative clones but not in the parental cells. Products from E8F3/E9R1 suggested upstream mRNA was intact in all of the clones. Products from E8F2/E10R2, which spans exon 8, 9, 10, and long introns, showed normal DMPK transcription in parental cells and clone 13–3 and 33–4, indicating that the normal allele was unaffected. GAPDH was amplified as a reverse transcription control. (c) Schematic overview of primer location. (d). Southern blot digested by EcoRI demonstrated insertion of the polyA signal (PAS) cassette in the mutant allele by showing the disappearance of the corresponding expanded band. This is due to the introduction of an extra EcoRI site within the PAS cassette. The normal allele remains intact. There can be two different normal allele sizes after EcoRI digestion, one ~8.6 kb and the other ~9.6 kb. Sample Negative PBL has both alleles, and sample Positive PBL has only the 9.6 kb allele and then the expansion allele. DM1-03 subject has the 9.6 kb allele and the expansion allele. (e) Southern blot digested by NcoI showed the expanded CTG repeats in DM1 parental and genome treated iPS clones (from 1793 repeats to 2087 repeats). Positive PBL is genomic DNA from peripheral blood from a DM1 expansion positive patient with ~750 CTG repeats. Negative PBL is a control peripheral blood DNA from a normal subject with two normal allele sizes.
A second DM1 patient derived iPS cell line (DM-05) has also undergone genome therapy. Out of the 7 puromycin-resistant clones from DM-05 iPS cells, one had complete loss of nuclear RNA foci and PCR genotyping also showed correct insertion of the PASs cassette in the TALEN cutting site and presence of the normal allele. The rest of the foci-positive clones reflect either an insertion of PASs in the normal allele or a random integration of the plasmid (see Supplementary Figure S3).
Genome-treated DM1 iPS cells maintain pluripotency by teratoma-forming assay
Teratomas grew from both parental and genome-treated DM1-03 and DM-05 iPS cell clones 6–8 weeks after iPS cells were injected into severe combined immunodeficiency mice. H&E staining revealed characteristic tissue structures for all three germ layers. IF staining revealed germ layer-specific markers in teratoma tissues and RNA FISH detected RNA foci in teratoma tissues derived from parental DM1 iPS cells but not in those from genome-treated iPS cells (Figure 3, Supplementary Figure S4, Supplementary Figure S5 and Supplementary Figure S6).
Figure 3.
Pluripotency assay by teratoma formation in parental Myotonic dystrophy type 1 (DM1) induced pluripotent stem (iPS) cells (DM-03) and genome-treated iPS cells. Both parental DM1 iPS cells and genome-treated iPS cells formed teratomas (upper panel) with typical three germ layer tissue structures on hematoxylin and eosin staining (middle panel). However, nuclear RNA foci were only seen in tissue derived from parental DM1 iPS cells but not in that from genome-treated iPS cells (lower panel). DM-03: parental DM 1iPS cells. 13-3: genome-treated iPS cells.
The integration of the PASs upstream of DMPK CTG repeats led to elimination of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated neural cells
In a previous study, we demonstrated complete elimination of nuclear RNA foci and reversal of alternative splicing by direct integration of the PASs upstream of the DMPK CTG repeats in DM1 NSCs.14 In this study, we conducted linear differentiation on the genome-treated DM1 iPS cells to neural cells to see whether we could recapitulate the same results. We found complete elimination of nuclear RNA foci in NSCs and neurons differentiated from genome-treated DM1-03 iPS cells (Figure 4). During this process, we noticed that early passage of NSCs has the tendency for spontaneous neuronal differentiation even when culturing the cells in a neural progenitor proliferation medium. In contrast, 6 passages after neural rosette formation, we seldom experienced spontaneously differentiated neurons. We think these early NSCs may represent primitive embryonic NSCs. To examine this further, we compared the splicing pattern of several genes in normal control, parental DM-03, and genome-treated DM-03 cell lines in both early NSCs (passage 4 after neural rosette formation) and late NSCs (passage 10 after neural rosette formation). In early NSCs, the splicing pattern of MAPT was demonstrated expression of mainly fetal isoforms (exon 2 and exon 3 exclusion). There was no difference between normal control, parental DM1, and genome-treated cells (statistical data not shown). However, the splicing patterns of MBNL 1 and MBNL 2 already displayed the reversal in genome-treated clones even though they still dominantly express the fetal isoform (exon 7 inclusion) (Figure 5, upper panel). In the late NSCs, the reversal became even more prominent in MBNL 1 and MBNL 2, and genome-treated these cells dominantly expressed the adult isoform. MAPT also showed a reversal of the mutant splicing pattern to the normal pattern in genome-treated NSCs while the parental DM-03 NSCs continued to be fetal isoform-dominant (Figure 5, lower panel and Supplementary Figure S7). Genome treatment did not affect further differentiation into neurons or astrocytes. Neurons and astrocytes expressed mature markers Neurofiliament H and GFAP, respectively, and displayed normal cell morphology (Figure 6). Similar reversal of alternative splicing was also demonstrated in linear-differentiated NSCs derived from genome-treated DM-05 iPS cells (see Supplementary Figure S8).
Figure 4.
The integration of the polyA signals upstream of DMPK CTG repeats in DM-03 induced pluripotent stem (iPS) cells led to elimination of nuclei RNA foci in linear-differentiated neural cells. (Upper panel): nuclear RNA foci were only detected in neural stem cells (NSCs) derived from parental DM-03 iPS cells while no foci were detected in normal NSCs or NSCs derived from genome-treated iPS cells. Green: nestin. (Lower panel) nuclear RNA foci were only detected in spontaneous differentiated neurons derived from parental DM-03 iPS cells while no foci were detected in normal neurons or neurons derived from genome-treated iPS cells. Green: neurofilament H. Normal: neural cells from normal iPS cells. DM-03: neural cells from parental DM1 iPS cells. 13-3 and 33-4: neural cells from genome-treated iPS cells with polyA signals (PASs) integrated in DMPK intron 9.
Figure 5.
Genome treatment reversed aberrant splicing of microtubule-associated protein tau (MAPT) and muscleblind-like splicing regulator (MBNL 1), MBNL 2 to normal patterns. (Upper panel) in early stage neural stem cells (NSCs) (passage 4 after passing from rosettes), transcripts of MAPT are mainly of the fetal isoform (exon 2, 3 exclusion) and there is no difference among NSCs from each of the induced pluripotent stem (iPS) clones. However, MBNL 1 and MBNL 2 already showed more adult isoform in normal and genome-treated NSCs even though the fetal isoform was still dominant (exon 7 exclusion ratio below 0.5), compared to parental Myotonic dystrophy type 1 (DM1) NSCs. (Lower panel) in late stage of NSCs (passage 10 after passing from rosette), most MAPT transcripts have switched to adult isoforms (exon 2 inclusion ratio over 0.5). MBNL1 and MBNL 2 have reverted to adult isoform dominant in normal and genome-treated NSCs (exon 7 exclusion ratio is >0.5) but parental NSCs continued to express mainly fetal isoform. *P < 0.05 by one-way analysis of variance.
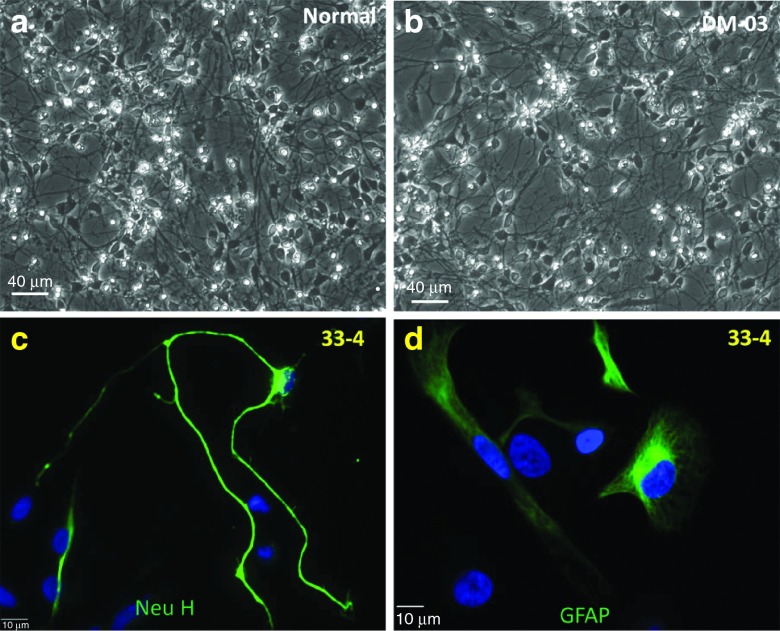
Figure 6.
Both parental and genome-treated induced pluripotent stem (iPS) cells underwent normal neuron and astrocyte differentiation. (a, b) Fourteen days after neuronal differentiation showing neural network. (c) A neuron derived from genome-treated iPS cells, showing neuronal marker (Neurofilament H) with normal neurite outgrowth. (d) Astrocytes from genome-treated iPS cells, showing astrocyte marker (GFAP).
The integration of the PASs upstream of DMPK CTG repeats led to elimination of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated cardiomyocytes
In order to further investigate whether this genome treatment has any effect in cardiomyocytes, we conducted cardiomyocyte differentiation in vitro on genome-treated DM1 iPS cells along with parental and normal iPS cells. We did not see a morphology difference during cardiac differentiation among these three lines. Functionally, they all began spontaneous contraction ~12 days after induction of differentiation and continued to contract for more than 3 weeks before they were harvested for RNA. Cardiomyocytes from genome-treated DM1 iPS cells lost nuclear RNA foci and demonstrated a reversal of aberrantly spliced genes MBNL1, MBNL2, insulin receptor (INSR) and cardiac troponin T (cTNT) (Figure 7, upper and lower panel, Supplementary Figure S9) to a normal splicing pattern.
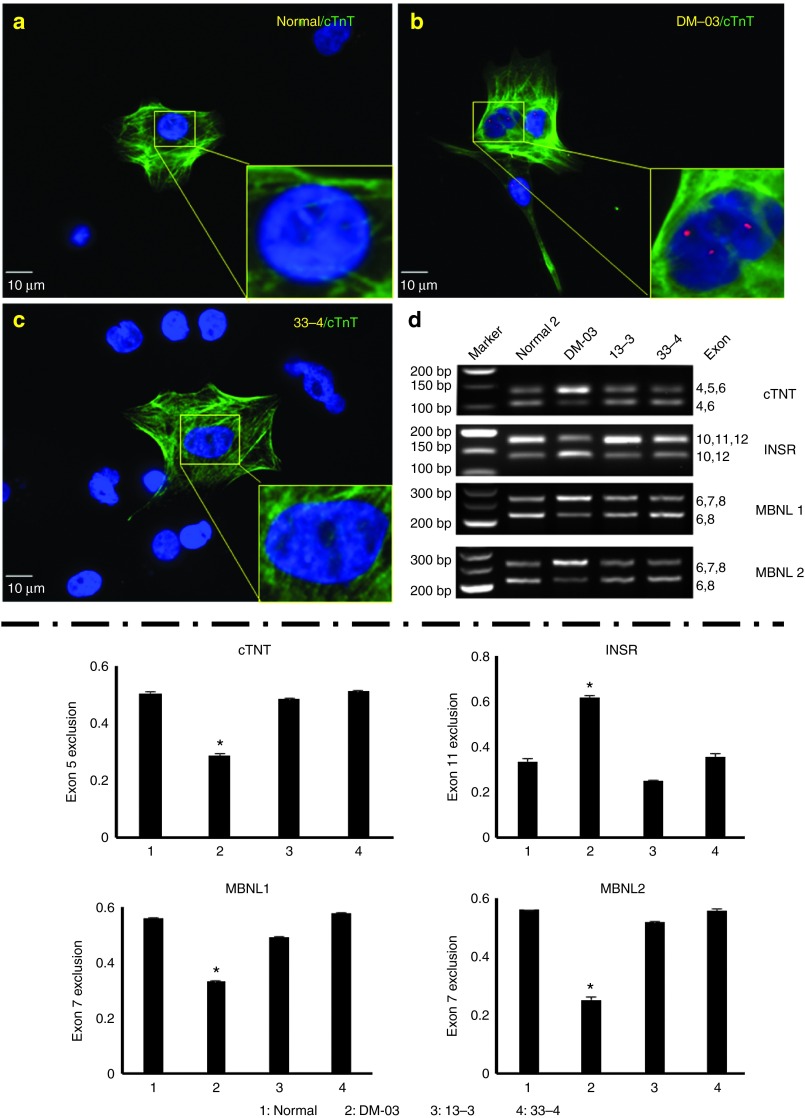
Figure 7.
Elimination of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated cardiomyocytes derived from genome-treated induced pluripotent stem (iPS) cells. (Upper panel) (a) Cardiomyocyte from normal control iPS cells showing no nuclear RNA foci. (b) Cardiomyocyte from parental Myotonic dystrophy type 1 (DM1) iPS cells showing nuclear RNA foci. (c) Cardiomyocyte from genome-treated DM1 iPS cells showing disappearance of nuclear RNA foci. (d) Agarose gel images showing the reversal of aberrant splicing pattern in cardiac troponin T (cTNT), insulin receptor (INSR), muscleblind-like splicing regulator (MBNL)1 and MBNL2. (Lower Panel) Graphs showing the statistical differences of exon exclusion, in which exon 5 or 7 exclusion is adult isoform in cTNT, MBNL1 and MBNL2 while exon 10 exclusion is fetal isoform in INSR. *P < 0.05 by one-way analysis of variance.
Discussion
Genome editing in which ectopic DNA is inserted or a part of endogenous DNA is replaced or deleted from a genome in living cells is progressing from a lab technology to clinical application owing to the advancement of gene tools, especially engineered sequence-specific endonuclease (e.g., engineered meganuclease, zinc finger nucleases, TALENs, and the CRISPR/Cas9 system).15,16,17 We have seen the first translation of this approach into a human clinical trial using genome-treated human cells.18,19,20 In the clinical trial, autologous CD4 T-cells were engineered to mimic a naturally occurring mutation in the C–C chemokine receptor type 5 (CCR5) gene that provides resistance to HIV infection using zinc finger nuclease technology. zinc finger nuclease-modified CD4 T-cells were well tolerated when reinfused and had a selective advantage over unmodified CD4-T cells after discontinuation of highly active antiretroviral therapy .20 This approach holds great promise for the cure of HIV/AIDS. It is reasonable to predict that genome therapy will soon extend to inherited monogenic disorders. In this study, we successfully generated genome-treated DM1 iPS cells by incorporating PASs upstream of the DMPK CTG expansion repeats to prevent production of mutant transcripts. These cells were pluripotent and retained their trilineage potential in vivo and were able to differentiate into functionally and morphologically normal neurons and cardiomyocytes. In comparison with genome modification of unipotent stem cells or terminally differentiated somatic cells, genome-treated iPS cells have broad potential for future clinical application for the cell-based treatment for DM1, a multisystemic disorder.
We attempted to target the 223-bp 3′UTR between the normal DMPK stop codon (TGA) and the start of the CTG repeat because this approach might maintain normal expression of the wild type DMPK protein even if both alleles were targeted. However, we found this approach selectively targeted the normal allele. None of the 84 single clones analyzed, had the mutant allele targeted (unpublished data). This has likely resulted from heterochromatin spreading and regional methylation by the expanded CTG repeats,21,22,23 which might have prevented TALEN binding or donor accessibility. In selecting the insertion site, we have to consider the effect of nonhomologous end joining. Nonhomologous end joining always competes with homology-directed repair. Double strand break in cells that are repaired by nonhomologous end joining will cause mutation by insertion or deletion (Indel) of nucleotides. We have thus avoided targeting the exons. Intron 9 was chosen based on the following protein structure and domain functions of the DMPK gene: (i) Intron 9 is 2.2 kb. We can easily select a specific TALEN pair without affecting potential functional sequences (e.g., alternative splicing branching point, etc. (ii) All the kinase domains are encoded before exon 9, including the downstream VSGGG motif which is important for DMPK autophosphorylation and phosphorylation of its targets. The C-terminus after exon 9 is involved in protein trafficking and localization.24 (iii) There is a stop codon right at the beginning of intron 9 and no new peptide of unknown function will be added. Since the transcription has been diverted toward the inserted PASs, there is no exon–exon junction complex remaining after the stop codon. The newly-formed transcripts will not be degraded by nonsense-mediated decay after the first round of translation and a truncated protein will be produced. There was concern that the truncated protein may create an unwanted dominant-negative function, but we have shown the truncated isoform naturally exists in the cells.14 Since only minor histopathological abnormalities have occurred in DMPK knockout mice, haploinsufficiency is not considered as a disease mechanism of DM1 (refs. 25,26), and imprinting is not involved in expression of the DMPK gene,27 we do not anticipate that a minor disturbance of DMPK expression in this approach will affect the cellular function. Indeed, our current data confirm that it did not affect in vitro neural and cardiac differentiation or in vivo teratoma formation, a stringent method to test pluripotency. We anticipate the unknown “side effects” will be well-tolerated.
Targeting the furthest downstream intron is more ideal because this would allow the cell to generate its own natural splicing isoforms. However, all downstream introns are <350 bp except for intron 10 (712 bp). It would be difficult to find a pair of specific TALENs or the splicing branching points will likely to be affected if nonhomologous end joining happened. If intron 10 is selected, a 34 amino acids peptide will be added to the natural protein before it reaches the next stop codon and the function of the attached peptide would be unknown. With the above considerations, intron 9 was selected for the targeted insertion of the PASs.
Allogenic transplantation of in vitro cultured myoblast from healthy donors was tested in Duchene Muscular Dystrophy but the results were not satisfactory.28,29,30,31 One of the major limitations was the reduction of regenerative and self-renewal potential of satellite cell-derived myoblasts after the long in vitro cultures required to obtain enough cells for transplantation.32 iPS cells have a great potential for cell-based therapy for skeletal muscle cell replacement (see reviews).33,34 In particular, iPS cells can provide unlimited early stage muscle precursor cells for transplantation purposes. The genome-treated iPS cells will likely survive and function better than unmodified cells carrying the mutation. As such, we are differentiating these genome-treated iPS cells into myogenic precursors for a transplantation study. These precursor cells may contribute to the satellite cell compartment, which can supply genome-treated myoblasts to join the host muscle myofibers or form new myotubes. Even though it may not replace all of the cells, the resultant mosaicism may delay the onset and progression of the disease.
There are other hurdles to generate clinically-clean iPS cells. Grafting efficiency issues, functional engraftment, purity of the delivered population, mutations acquired in vitro, oncogenic transformation, etc. Our study was designed as a first step to generate genomically-healthy iPS cells to solve one of these hurdles before such cells are applied to clinical usage. Additional experiments are needed to determine which stage of differentiation is the most optimal for cell transplantation. Data from preclinical studies using mouse models will provide further evidence regarding whether the genome-treated cells will function better than parental cells as a cellular source for transplantation. We think this approach is also applicable to other haplosufficient, autosomal dominant RNA gain-of-function, monogenetic disorders that are caused by long nucleotide repeat expansions.
In our previous study, the MAPT splicing pattern was reversed in genome-treated NSCs.14 Those NSCs were “late” NSCs. The results were reproduced in the “late” NSCs but not in the “early” NSCs derived from genome-treated DM1 iPS cells in the current study. We found that the alternative splicing patterns are quite different between “early” NSCs and “late” NSCs. This differential expression likely reflects the neural system development in the brain.35 We have been able to generate reasonably homogeneous NSCs of different stages, which will likely generate consistent, comprehensive data about the differential expression pattern between the “early” NSCs and “late” NSCs by RNAseq and bioinformatics analyses. These data might help explain this apparent dichotomy and its relationship to congenital and adult DM1 phenotypes. It will be interesting to examine the developmental process versus the degenerative process. The revelation of this dichotomy may also help with guiding cell transplantation therapy. Early embryonic primitive NSCs are different from adult NSCs. It is possible that early primitive NSCs might generate the preponderance of neurons, while late NSCs contribute mostly glia.36,37,38 Adult NSCs may still have the capacity to generate neurons, but it is regionally and temporally restricted. As yet, however, there is no direct evidence that adult-derived stem cells can create the types of projections that are normally generated in the embryo. How to identify cell sources for therapeutic use will be an important issue. The “early” NSCs in this study likely consist of embryonic-like primitive NSCs. These cells can spontaneously differentiate into neurons even while cultured under proliferative conditions. It will be important to understand whether the transplantation of these “early” NSCs can create projections that can connect into the existing neural network.
Cardiac differentiation appears normal in terms of the morphology and spontaneous contraction capabilities. However, parental DM1 cardiomyocytes do show molecular abnormalities by alternative splicing assays. Early knowledge in this area was acquired by autopsy tissues and had its limitations. We generated an isogenic cellular model system, which will reduce the intrinsic variability associated with the genetic background of each individual iPS cell line. Inclusion of exon 7 of MBNL dramatically decreases during heart development.39 Inclusion of exon 7 in tibia anterior muscle increases in human DM1 (ref. 40). Using our DM1 iPS cellular model, we have been able to show that the cardiomyocytes derived from genome-treated DM1 iPS cells can be returned to normal splicing pattern. The comparative analysis of RNAseq data among normal, parental DM1, and genome-treated DM1 cardiomyocytes at different stages of heart development will likely generate valuable knowledge into the pathogenesis of the cardiovascular system in DM1. These viable cardiomyocyte can also be utilized for electrophysiological studies and potentially provide insight for therapeutic drug development.
Conclusion
Genome treatment by incorporating exogenous PASs upstream of the DMPK CTG repeat expansion prevented the production of toxic mutant RNAs and reversal of molecular phenotypes in DM1 iPS cells and their progeny. These genetically-treated iPS cells will have broad clinical applications in developing autologous stem cell therapies for DM1.
Materials and Methods
Approach and construction of donor vectors. Genome modification was performed in human DM1 iPS cells as described previously.41 Briefly, SV40 and bovine growth hormone, PASs were inserted upstream of DMPK CTG repeats to induce premature termination of DMPK gene transcription and prevent the production of toxic mutant transcripts. In this study, a negative selectable marker, Herpes Simplex Virus-Thymidine Kinase, was engineered into the donor vector downstream of the 3′ homologous arm (see Supplementary Figure S10). Clones with random insertion containing the Herpes Simplex Virus-Thymidine Kinase cassette will be eliminated by Ganciclovir selection. The expression of mutant CUG repeats was monitored by nuclear RNA foci, the molecular hallmark of DM1, using RNA fluorescence in situ hybridization (RNA-FISH).
Cell culture, transfection, and clone selection. Normal, DM-03, and DM-05 iPS cell lines were generated previously in our lab42 and have been adapted to TeSR-E8 medium (STEMCELL Technologies, Vancouver, Canada). The “Normal 2” iPS cell line was generated from foreskin fibroblasts kindly provided by the laboratory of Naohiro Terada. For transfection, DM iPS cells were passed in small colonies using Gentle Cell Dissociation Reagent (STEMCELL Technologies) the day before transfection on a Vitronectin coated 6-well plate in TeSR-E8 medium (STEMCELL Technologies). Transfection was conducted with MACSfectin (Miltenyi Biotec, Auburn, CA). Briefly, 1 μg of each TALEN and 4 μg of donor vector pDEST-Puro-TK were mixed in 100 μl TeSR-E7 medium containing 12 μl MACSfectin. They were incubated at RT for 20 minutes and then the complex were added drop wise to 1 well of cultured cells in a 6-well plate (3 ml in each well). Medium was changed 24 hours later. Puromycin selection was started 48 hours after transfection at 0.4 μg/ml. Ganciclovir (4 μmol/l) selection was started 5 days postpuromycin selection. Selection was continued until individual clones were large enough for isolation. Fortyeight puromycin-resistant clones of DM-03 iPS cells and 7 puromycin-resistant clones of DM-05 iPS cells were picked for RNA-FISH phenotyping, subcloning, and genotyping to identify clones that had loss of nuclear RNA foci and only the mutant allele targeted.
Teratoma generation. Animal studies were approved by the Institutional Animal Care and Use Committee. Normal, DM1 (DM-03 and DM-05), and genome-treated DM1 (13-3 and 33-4 from DM-03; and T17 from DM-05) iPS cell clones were cultured in TeSR-E8 medium on 6-well plates until 90% confluent. Cells were detached with Gentle Cell Dissociation Reagent (STEMCELL Technologies) and incubated at RT for 10 minutes. Cells were resuspended in 200 μl cold phosphate-buffered saline (PBS), and mixed with 200 μl BD Matrigel (BD Bioscience, San Jose, CA, Ca# 354277) before injection (kept on ice). Injection was conducted using a 28.5 gauge syringe into the dorsal back of C57BL6 severe combined immunodeficiency mice (The Jackson Laboratory, Bar Harbor, Maine) subcutaneously. The total cell number was 2 × 106 cells in the volume of 200 μl. Mice were monitored and when the tumor reached 1–1.5 cm in diameter, mice were sacrificed and the teratoma were harvested. Tumors were fixed in 4% paraformaldhyde overnight and then processed for embedding in Tissue-Tek O. C. T compound for histological, RNA FISH and immunofluorescence (IF) staining.
RNA-FISH and IF. RNA-FISH and IF were performed as described previously.41,42 For RNA FISH on teratoma tissue, frozen sections (6 μm) were dried for 30 minutes at RT. Sections were then washed 3 times in PBS for 2 minutes and permeabilized in 0.03% TritonX 100 for 5 minutes. Hybridization with HPLC-purified, Cy3-labeled (CAG)10 DNA probe(1 ng/µl) (Integrated DNA Technologies, Coralville, IA) was performed as described in RNA FISH on fixed cells. Sections were rinsed in PBS (PH 7.4) and then processed for IF. Slides were first blocked with SuperBlock (PBS) Blocking Buffer (Life Technologies) at RT for 30 minutes, then primary antibodies Desmin (1:100), Nestin (1:100), or CDX2 (1:100) were added and incubated overnight at 4ºC. The following day, slides were washed with PBS, incubated with appropriate secondary antibodies conjugated with Alexa Fluor 488 (Invitrogen/Life Technologies) (1:500) for 30 minutes at RT. After a final wash in PBS, slides were mounted with Vectashield Mounting Medium with 4',6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Images were taken using an Olympus IX81-DSU Spinning Disk confocal microscope.
Neural differentiation. Neural differentiation was performed as described previously.41 NSCs derived from iPS cells were passed at 1:3 ratio after passing from neural rosettes. The “early” NSCs were defined as before passage 4. Whereas the “late” NSCs were defined as after passage 10. While “normal 2” and 13-3 NSCs were only passed to passage 4. Normal, parental, 33-4 and T17 NSCs were passed until passage 10. The genome-treated NSCs used in the current study are different from the genome-treated NSCs in our previously published experiment.14 In the previous paper, the NSCs were generated through direct editing of iPS cell-derived NSCs. They are “late” NSCs. In the current study, NSCs are generated through differentiation of genome-treated iPS cells. We performed alternative splicing assay in both the “early” NSCs and “late” NSCs.
For neuronal differentiation, 4 × 105 NSCs passage at 5, were plated in 6-well plates coated with Poly-L-ornithine and Laminin in triplicate in neuronal differentiation medium: 2% B-27 supplement, 2 mmol/l GlutaMAX (Life Technologies) in Neurobasal Medium (Invitrogen/Life Technologies). Medium was changed every 3–4 days. For astrocyte differentiation, 2.5 × 105 cells were plated in 6-well plates coated with Geltrex (Life Technologies) in triplicate in astrocyte differentiation medium: 1% N-2 supplement, 2 mmol/l GlutaMAX, 1% Fetal Bovine Serum in DMEM medium. Medium was changed every 3–4 days. Neuronal and astrocyte differentiation was continued for 14 days and then transferred to laminin-coated ibidiTreat μ-slides (Ibidi GmbH, Martinsried, Germany) for IF staining of neuron and astrocyte markers.
Cardiomyocyte differentiation. The 95% confluent iPS cells in a 6-well plate were treated with Gentle Cell Dissociation Reagent for 12 minutes in RT, scraped off the plate, and transferred to 15 ml tube and pipetted twice using 1 ml tips and plated at a 1:8 dilution on a Geltrex-coated 6-well plate. Cardiomyocyte differentiation was performed when cell became 70%–80% confluent using the PSC Cardiomyocyte Differentiation Kit (Thermo Fisher Scientific, # A29212-01) as described in the manufacturer's protocol. Cells became a whole sheet and focal areas started beating around day 12 and were harvested on day 20 for RNA extraction and alternative splicing assays.
For RNA-FISH and IF staining, cardiomyocytes in a 6-well plate were washed in PBS without Ca2+ or Mg2+.and then 1 ml of TrypLE (Life Technologies) was added and incubated at 37ºC for 10 minutes. Cells were detached gently by pipetting to make a single cell suspension. Next, 2 ml of Cardiomyocyte Maintenance Medium (Life Technologies) was added, and cells were filtered using a 100 μm cell strainer. Cells were centrifuged at 200×g for 5 minutes. The supernatant was aspirated and the pelleted cells were resuspended in 1 ml Cardiomyocyte Maintenance Medium. 6 × 104.cells were plated on Geltrex-coated ibidiTreat μ-slides. After 12 hours, cells were fixed in 4% paraformaldhyde and subjected to FISH and IF staining as described above. The primary antibody, anticardiac troponin T (Abcam, Cambridge, MA, abc45932), was diluted 1:400.
DNA/RNA isolation, genomic PCR, RT-PCR and TP-PCR. Genomic DNA of individual colonies was isolated with the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Total RNA was isolated using Quick-RNA MiniPrep (Zymo Research, Irvine, CA, Cat # R1055,) according to the manufacturer's protocol. Primers were designed to flank a specific genomic or cDNA region for genotyping to confirm the correct insertion of PASs as described earlier.41 Primer sequences and PCR conditions can be found in Supplementary Table S1 and in our previous publication (see Supplementary Table S1).41
Alternative splicing assays. Cells were collected and RNA was extracted for cDNA synthesis as described above. RT-PCRs of the microtubule-associated protein tau (MAPT) gene spanning exon 2, 3, muscleblind-like splicing regulator (MBNL) 1 and MBNL 2 spanning exon 7, cTNT spanning exon 5, INSR spanning exon 11 were performed as published.41,43,44,45,46 The intensity of each band on electrophoresis gel image was analyzed by Alphaview. The percentage of exon inclusion/exclusion was calculated from three independent PCR experiments. One-way analysis of variance was used for statistical analyses and P < 0.05 was accepted as statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Removal of selective marker with cre-recombinase 33–4 was infected with AAV (RGD)-Cre-GFP. Figure S2. Expanded CTG repeats remained in genome-treated clones detected by TP-PCR. Figure S3. Representative clones of DM1-05 iPS cells treated with TALENs and donor after puromycin selection. Figure S4. Disappearance of intranuclear foci in teratoma tissues foci were seen in endoderm (gland-like structure) and mesoderm (cartilage-like) tissues in teratoma derived from parental DM1 iPS cells (DM-03). Figure S5. Pluripotency assay by teratoma formation in parental DM1 iPS cells (DM-05) and genome-treated iPS cells. Figure S6. FISH and IF in teratoma tissues from parental DM1 iPS (DM-05) and genome-treated iPS cells (T17). Figure S7. Dynamic change of alternative splicing of MAPT from early to late passage of NSCs. Figure S8. The integration of the polyA signals upstream of DMPK CTG repeats in DM-05 iPS cells led to elimination of nuclei RNA foci and reversal of aberrant splicing of MAPT in linear-differentiated neural cells. Figure S9. Elimination of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated cardiomyocytes derived from genome-treated iPS cells. Figure S10. Donor construct information. Map of donor vector with puromycin resistant cassette (pDEST-puro). Table S1. Sequences of the primers.
Author Contributions
The first two authors contributed equally to the work. Study conception and design (Xia, Ashizawa, and Terada), data acquisition (Xia, Gao, Guo, Santostefano, Wang, Reid and Zeng), analysis and interpretation (Xia, Gao, Guo, Wang, Reid, Ashizawa, and Terada). Xia wrote the manuscript with input from all authors.
Acknowledgments
Research reported in this presentation was supported by NIH/NIAMS grant K08 AR064836-01 to Xia mentored by Maury S. Swanson, Tetsuo Ashizawa, Naohiro Terada, and Laura P.W. Ranum, Opportunity fund from University of Florida, ReproCELL Inc., Life Technologies, Marigold Foundation Ltd., The Evelyn L& William F. McKnight Brain Institute and Muscular Dystrophy Fund from Fraternal Order of Eagles Charity Fund. All the authors indicate no potential conflicts of interest to disclose.
Supplementary Material
References
- Romeo, V (2012). Myotonic Dystrophy Type 1 or Steinert's disease. Adv Exp Med Biol 724: 239–257. [DOI] [PubMed] [Google Scholar]
- Reardon, W, Newcombe, R, Fenton, I, Sibert, J and Harper, PS (1993). The natural history of congenital myotonic dystrophy: mortality and long term clinical aspects. Arch Dis Child 68: 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook, JD, McCurrach, ME, Harley, HG, Buckler, AJ, Church, D, Aburatani, H et al. (1992). Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell 69: 385. [DOI] [PubMed] [Google Scholar]
- Mahadevan, M, Tsilfidis, C, Sabourin, L, Shutler, G, Amemiya, C, Jansen, G et al. (1992). Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science 255: 1253–1255. [DOI] [PubMed] [Google Scholar]
- Fu, YH, Pizzuti, A, Fenwick, RG Jr, King, J, Rajnarayan, S, Dunne, PW et al. (1992). An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255: 1256–1258. [DOI] [PubMed] [Google Scholar]
- Ashizawa, T, Monckton, DG, Vaishnav, S, Patel, BJ, Voskova, A and Caskey, CT (1996). Instability of the expanded (CTG) repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics 36: 47–53. [DOI] [PubMed] [Google Scholar]
- Salehi, LB, Bonifazi, E, Stasio, ED, Gennarelli, M, Botta, A, Vallo, L, et al. (2007). Risk prediction for clinical phenotype in myotonic dystrophy type 1: data from 2,650 patients. Genet Test 11: 84–90. [DOI] [PubMed] [Google Scholar]
- The International Myotonic Dystrophy Consortium (IDMC). (2000). New nomenclature and DNA testing guidelines for myotonic dystrophy type 1 (DM1). Neurology 54: 1218–1221. [DOI] [PubMed] [Google Scholar]
- Morales, F, Couto, JM, Higham, CF, Hogg, G, Cuenca, P, Braida, C et al. (2012). Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum Mol Genet 21: 3558–3567. [DOI] [PubMed] [Google Scholar]
- Thornton, CA (2014). Myotonic dystrophy. Neurol Clin 32: 705–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, C and Hilton-Jones, D (2014). Myotonic dystrophy: diagnosis, management and new therapies. Curr Opin Neurol 27: 599–606. [DOI] [PubMed] [Google Scholar]
- Reynolds, BA and Weiss, S (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255: 1707–1710. [DOI] [PubMed] [Google Scholar]
- Lois, C and Alvarez-Buylla, A (1993). Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci USA 90: 2074–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, G, Gao, Y, Jin, S, Subramony, SH, Terada, N, Ranum, LP et al. (2015). Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 induced pluripotent stem cell-derived neural stem cells. Stem Cells 33: 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, KM and Wang, HH (2013). Genome-scale engineering for systems and synthetic biology. Mol Syst Biol 9: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, WS, Carlson, DF, Walton, MW, Fahrenkrug, SC and Hackett, PB (2012). Precision editing of large animal genomes. Adv Genet 80: 37–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio, I and Gonçalves, MA (2015). Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol 33: 280–291. [DOI] [PubMed] [Google Scholar]
- Perez, EE, Wang, J, Miller, JC, Jouvenot, Y, Kim, KA, Liu, O et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, DA, Brennan, AL, Jiang, S, Binder-Scholl, GK, Lee, G, Plesa, G et al. (2013). Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas, P, Stein, D, Tang, WW, Frank, I, Wang, SQ, Lee, G et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten, AD and Tapscott, SJ (1995). Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc Natl Acad Sci USA 92: 5465–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, DH, Thienes, CP, Mahoney, SE, Analau, E, Filippova, GN and Tapscott, SJ (2005). Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20: 483–489. [DOI] [PubMed] [Google Scholar]
- Steinbach, P, Gläser, D, Vogel, W, Wolf, M and Schwemmle, S (1998). The DMPK gene of severely affected myotonic dystrophy patients is hypermethylated proximal to the largely expanded CTG repeat. Am J Hum Genet 62: 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink, DG, van Herpen, RE, Coerwinkel-Driessen, MM, Groenen, PJ, Hemmings, BA and Wieringa, B (2003). Alternative splicing controls myotonic dystrophy protein kinase structure, enzymatic activity, and subcellular localization. Mol Cell Biol 23: 5489–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, S, Smith, DB, Rich, MM, Leferovich, JM, Reilly, P, Davis, BM et al. (1996). Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat Genet 13: 325–335. [DOI] [PubMed] [Google Scholar]
- Narang, MA, Waring, JD, Sabourin, LA and Korneluk, RG (2000). Myotonic dystrophy (DM) protein kinase levels in congenital and adult DM patients. Eur J Hum Genet 8: 507–512. [DOI] [PubMed] [Google Scholar]
- Jansen, G, Bartolomei, M, Kalscheuer, V, Merkx, G, Wormskamp, N, Mariman, E et al. (1993). No imprinting involved in the expression of DM-kinase mRNAs in mouse and human tissues. Hum Mol Genet 2: 1221–1227. [DOI] [PubMed] [Google Scholar]
- Farini, A, Razini, P, Erratico, S, Torrente, Y and Meregalli, M (2009). Cell based therapy for Duchenne muscular dystrophy. J Cell Physiol 221: 526–534. [DOI] [PubMed] [Google Scholar]
- Gussoni, E, Blau, HM and Kunkel, LM (1997). The fate of individual myoblasts after transplantation into muscles of DMD patients. Nat Med 3: 970–977. [DOI] [PubMed] [Google Scholar]
- Miller, RG, Sharma, KR, Pavlath, GK, Gussoni, E, Mynhier, M, Lanctot, AM et al. (1997). Myoblast implantation in Duchenne muscular dystrophy: the San Francisco study. Muscle Nerve 20: 469–478. [DOI] [PubMed] [Google Scholar]
- Mendell, JR, Kissel, JT, Amato, AA, King, W, Signore, L, Prior, TW et al. (1995). Myoblast transfer in the treatment of Duchenne's muscular dystrophy. N Engl J Med 333: 832–838. [DOI] [PubMed] [Google Scholar]
- Montarras, D, Morgan, J, Collins, C, Relaix, F, Zaffran, S, Cumano, A et al. (2005). Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067. [DOI] [PubMed] [Google Scholar]
- Salani, S, Donadoni, C, Rizzo, F, Bresolin, N, Comi, GP and Corti, S (2012). Generation of skeletal muscle cells from embryonic and induced pluripotent stem cells as an in vitro model and for therapy of muscular dystrophies. J Cell Mol Med 16: 1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, T, Awaya, T, Chang, H, Mizuno, Y, Niwa, A, Fukada, S et al. (2010). [Derivation of engraftable myogenic precursors from murine ES/iPS cells and generation of disease-specific iPS cells from patients with Duchenne muscular dystrophy (DMD) and other diseases]. Rinsho Shinkeigaku 50: 889. [DOI] [PubMed] [Google Scholar]
- Mazin, P, Xiong, J, Liu, X, Yan, Z, Zhang, X, Li, M et al. (2013). Widespread splicing changes in human brain development and aging. Mol Syst Biol 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, MS (1999). Multipotent and restricted precursors in the central nervous system. Anat Rec 257: 137–148. [DOI] [PubMed] [Google Scholar]
- Qian, X, Shen, Q, Goderie, SK, He, W, Capela, A, Davis, AA et al. (2000). Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28: 69–80. [DOI] [PubMed] [Google Scholar]
- He, W, Ingraham, C, Rising, L, Goderie, S and Temple, S (2001). Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci 21: 8854–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi, F and Ladd, AN (2010). Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity. RNA Biol 7: 43–55. [DOI] [PubMed] [Google Scholar]
- Nakamori, M, Sobczak, K, Puwanant, A, Welle, S, Eichinger, K, Pandya, S et al. (2013). Splicing biomarkers of disease severity in myotonic dystrophy. Ann Neurol 74: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, G, Gao, Y, Jin, S, Subramony, S, Terada, N, Ranum, LP, et al. (2015). Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 iPS-cell derived neural stem cells. Stem Cells 33: 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, G, Santostefano, KE, Goodwin, M, Liu, J, Subramony, SH, Swanson, MS et al. (2013). Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis. Cell Reprogram 15: 166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaenens, CM, Tran, H, Frandemiche, ML, Carpentier, C, Schraen-Maschke, S, Sistiaga, A et al. (2011). Mis-splicing of Tau exon 10 in myotonic dystrophy type 1 is reproduced by overexpression of CELF2 but not by MBNL1 silencing. Biochim Biophys Acta 1812: 732–742. [DOI] [PubMed] [Google Scholar]
- Hernández-Hernández, O, Sicot, G, Dinca, DM, Huguet, A, Nicole, A, Buée, L et al. (2013). Synaptic protein dysregulation in myotonic dystrophy type 1: Disease neuropathogenesis beyond missplicing. Rare Dis 1: e25553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, AV, Timchenko, LT and Cooper, TA (1998). Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280: 737–741. [DOI] [PubMed] [Google Scholar]
- Jog, SP, Paul, S, Dansithong, W, Tring, S, Comai, L and Reddy, S (2012). RNA splicing is responsive to MBNL1 dose. PLoS One 7: e48825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.