Abstract
A series of poly(esteramine)s (PEAs) constructed from low molecular weight polyethyleneimine (LPEI) and Pluronic were evaluated for the delivery of antisense oligonuclotides (AOs), 2′-O-methyl phosphorothioate RNA (2′-OMePS) and phosphorodiamidate morpholino oligomer (PMO) in cell culture and dystrophic mdx mice. Improved exon-skipping efficiency of both 2′-OMePS and PMO was observed in the C2C12E50 cell line with all PEA polymers compared with PEI 25k or LF-2k. The degree of efficiency was found in the order of PEA 01, PEA 04 > PEA 05 > others. The in vivo study in mdx mice demonstrated enhanced exon-skipping of 2′-OMePS with the order of PEA 06 > PEA 04, PEA 07 > PEA 03 > PEA 01 > others, and much higher than PEI 25k formulated 2′-OMePS. Exon-skipping efficiency of PMO in formulation with the PEAs were significantly enhanced in the order of PEA 02 > PEA 10 > PEA 01, PEA 03 > PEA 05, PEA 07, PEA 08 > others, with PEA 02 reaching fourfold of Endo-porter formulated PMO. PEAs improve PMO delivery more effectively than 2′-OMePS delivery in vivo, and the systemic delivery evaluation further highlight the efficiency of PEA for PMO delivery in all skeletal muscle. The results suggest that the flexibility of PEA polymers could be explored for delivery of different AO chemistries, especially for antisense therapy.
Keywords: antisense delivery, exon-skipping, muscular dystrophy, nanoparticle, polyethylenimine modification
Introduction
Antisense therapy is a powerful strategy for inducing post-transcriptional modifications and thereby regulating target genes associated with disease. Particularly, the antisense oligonucleotide (AO)-mediated exon-skipping has been demonstrated as one of the most promising experimental therapies for the treatment of Duchenne muscular dystrophy (DMD), applicable for as many as 90% of DMD patients.1,2,3,4,5,6,7,8,9,10,11 “DMD”, an X-linked inherited muscle degenerative disorder caused by mutations in the dystrophin gene, is the most common and serious form of childhood muscle wasting disease, with an incidence of about 1 in 3,500 live male births and premature lethality from respiratory or cardiac failure at around 30 years of age.1,2,3,4 Recent trials in vitro and in vivo in mdx mice, canine models of DMD as well as clinic demonstrate that AO-mediated exon-skipping is able to induce dystrophin expression and improve muscle function. However, preliminary reports also indicate that the levels of dystrophin expression and clinical benefits of the current regime were limited, and statistical significance in preventing disease progression has not yet been convincingly obtained. This is most likely the consequence of inadequate dystrophin induction with suboptimal dosages.11,12,13,14,15,16,17,18,19 It has been well established in a number of animal models that dystrophin levels are correlated with the functional outcome of muscle, and significant therapeutic value can be achieved only with high-dose AO regimes, considerably higher than the doses being used in ongoing clinical trials. However, long-term treatments with high dose could be cost-inhibitive and potential toxic risk.8,16 Among the several AO chemical structures, only 2′ O-methyl-phosphorothioate RNA (2′-OMePS) and phosphorodiamidate morpholino oligomer (PMO) have been tested in clinical trials reaching phase III. The 2′-OMePS backbone are resistant to nuclease degradation and relatively stable in biological systems compared with natural DNA and RNA, but the negative charges may restrict their overall uptake.9,10 Furthermore, toxicity has limited the ceiling of dosage for clinical trials. In contrast, PMO, is a neutral molecule under physiological condition, having the deoxyribose rings replaced with morpholino linked through phosphorodiamidate intersubunits. PMO has exhibited excellent stability and lower toxicity compared with other counterparts.20,21 However, the relatively uncharged nature of PMOs associated with poor cell uptake and fast clearance in bloodstream has been identified as a major obstacle for effective delivery. These factors taken together suggest that an important challenge remaining in AO exon-skipping is to improve delivery without increasing toxicity.19,22,23
To improve delivery efficiency, arginine-rich cell penetrating peptides or guanidine-dendran modified PMO have been reported to enhance the uptake of PMOs.11,12,13,14,15 The results showed increased exon-skipping and dystrophin levels in skeletal and cardiac muscle by systemic delivery in mdx mice. Peptide-modified 2′-OMePS enhanced cardiac uptake and exon-skipping in mdx mice.24 However, the cationic peptide chemical modification to AOs is associated with higher toxicity, with LD50 near 100 mg/kg, making it unsuitable for use in clinical applications.11,12,15 Furthermore, the complicated synthesis and purification in covalent modification increase cost significantly, and potential immune responses to the targeting peptide could prevent repeated administration.
Polymeric-mediated delivery strategy for gene delivery remains attractive, because of the polymer's structural flexibility, capacity for delivery of larger therapeutic cargo, ease in handling with greater safety, and less expensive than viral vector. Among them, the low molecular weight polyethylenimine (LPEI) constructed modifications have validated promise as antisense oligomer delivery vehicles related to their molecular size, optional hydrophilic-lipophilic balance (HLB) and low cytotoxicity, as well as their buffering capacity.25,26,27,28,29,30,31,32 2′-OMePS mediated with PEG550-PEI2000 copolymer enhanced exon-skipping in skeletal muscle, but the effect is limited. This is presumably due to the hydrophilic nature of the polymer hampering the interactions between the polymer with 2′-OMePS and cell or tissue membranes.25,26 To approach this dynamic system, we have recently developed a series of amphiphilic cationic polymers, including the polycarbamates (PCMs) composed of Pluronic and LPEI, Tween 85 modified LPEIs, the branched poly(ester amine)s composed of tris[2-(acryloyloxy)ethyl]isocyanurate (TAEI) and LPEI.28,29,30,31,32 These polymers are effective in improving AO delivery in vitro and in vivo in mdx mice. The results indicate that delivery efficiency and cytotoxicity depend heavily on the molecular size, HLB, and inclusion of cationic components. It also suggests that hydrophobic components of a polymer are beneficial to forming stable complexes with oligonucleotides, and to pass through the hydrophobic membrane. Given the encouraging results of our earlier studies, a series of poly(ester amine)s (PEAs) based on different Pluronics and LPEIs (Mw: 0.8k, 1.2kDa) modification by Michael addition reaction were recently prepared. Their structures in correlation to DNA condensation capability, cytotoxicity, and the delivery efficiency in vitro and in vivo has been studied and the best formulation for plasmid DNA (pDNA) delivery increased transgene expression efficiency 5- and 19-fold of PEI 25k in vitro and in vivo, respectively.33 In the current study, we examined this series of PEAs as vectors for AO delivery both PMO and 2′-OMePS in vitro and in vivo in mdx mice.
Results and discussion
Synthesis and characterization of poly(esteramine)s (PEAs)
A series of poly(esteramine)s (PEAs) constructed from LPEIs and Pluronics were synthesized as previously reported.33 Pluronic were first end-capped with acryloyl chloride and then reacted with PEI by Michael addition reaction. The synthesis, nomenclature and characteristics of the cationic amphiphilic polymers are described in Supplementary Materials (Supplementary Scheme S1 and Table S1).
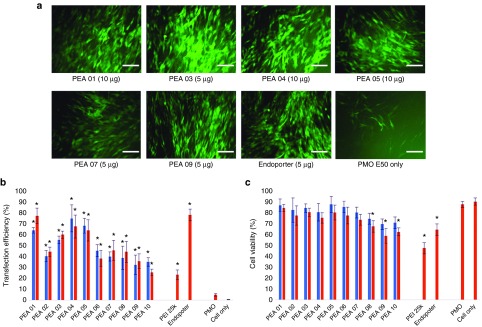
Uncharged PMO delivery. We next examined the same PEAs for neutral PMO delivery. PMOE50 (5′-AACTTCCTCTTTAACAGAAAAGCATAC-3′) with previously confirmed efficacy of targeted removal of human dystrophin exon 50 at a fixed amount (10 µg/ml) was formulated with each polymer.28,29,30 The TE of PMOE50 alone was below 5%. However, TE of PMOE50 with two doses (10 µg/ml, 20 µg/ml) of PEAs was over 60% for PEA 01/03/04/05 at the dose 10 µg/ml. The highest TE was achieved with PEA 01/04, with over 75%, which was close to Endo-porter mediated PMO delivery (TE = 78%). Endo-porter at this concentration caused cell death of up to 35%, whereas over 75% cells survived with PEA 01/04 (Figure 4), only 22% TE with much higher toxicity was observed with PEI 25k. The TE achieved with individual PEA for PMO delivery was similar to that for 2′-OMePS delivery. However, exceptions were observed with PEA 03/08 showing higher efficiency for PMO delivery than for 2′-OMePS delivery, which indicated the hydrophobic interaction is more predominant than charge interaction between the PEAs and uncharged PMO. The results that amphiphilic PEAs also provide higher PMO delivery efficiency than PEI, and reaches similar levels to that of commercially available cationic peptide (Endo-porter) demonstrates their potential to be applied in vivo for achieving therapeutic values.
Figure 4.
Delivery efficiency and toxicity of PEAs mediated PMO in C2C12 cell line determined by fluorescence microscopy and flow cytometer. (a) Representative fluorescent microscopy images of PMOE50 -induced exon-skipping in C2C12E50 cell line. The images were taken 48 hours after treatment (scale bar = 500 µm). (b) Transfection efficiency of PMOE50 formulated with polymers (Student's t-test, *P ≤ 0.05 compared with PMO only). (c) Cell viability (Student's t-test, *P ≤ 0.05 compared with untreated cells as a control). Five micrograms of PMOE50 were formulated with PEAs (5,10 µg), and PEI-25k (2 µg), Endo-porter (5 µg) formulated as controls in 500 µl 10% FBS-DMEM medium, respectively. The results are presented as the mean ± SD in triplicate.
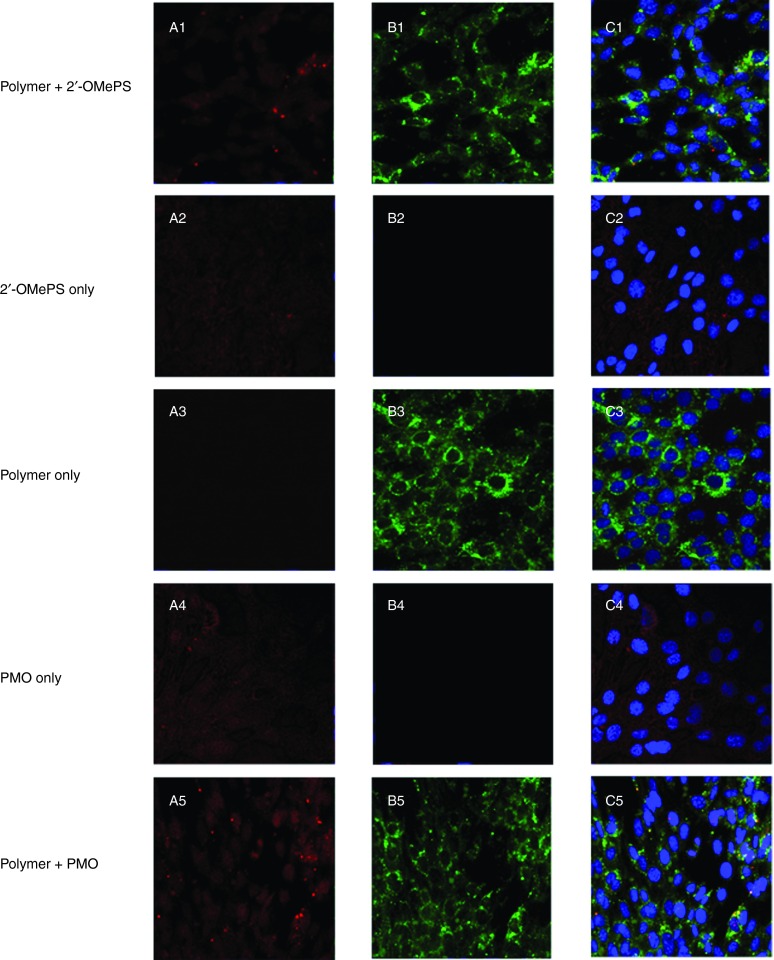
Cell uptake study. To better understand how the PEA polymers improve the delivery or transfection efficiencies of AOs, we directly visualized the cellular uptake of polymer/AO polyplexes, wherein the FITC-labeled PEA 01 was complexed with Cy3-labeled oligonucleotide at a weight ratio of 5:1 (Rw = 5) and examined under confocal microscope (Figure 5). In the absence of the PEA 01 polymer, the oligonucleotide alone showed weak diffuse signal within the treated cells. In contrast, signals for polymer formulated AO were predominantly located in cytoplasmic and perinuclear areas. Intensive signals of the labeled polymer were also observed at the plasma membrane. While the two signals for PEA 01 and oligonucleotide were generally codistributed as indicated by yellow or light blue (merging green, red, and blue) on the merged image presented, punctate signal in cytoplasm was primarily green for polymers. Similar patterns of signal distribution were observed for both polymer/PMO and polymer/2′-OMePS complexes except that PMO signal was more diffuse than 2′-OMePS in the absence of polymers. This is probably due to the nonspecific interaction with the cell membrane relevant to its uncharged nature.
Figure 5.
Intracellular interaction of FITC-labeled PEA formulated with Cy3-labeled-oligonucleotide (Cy3-Oligo) in C2C12 cell line (5 µg polymer in 500 µl 10% FBS-DMEM with Cy3-Oligo 1 µg). (A1-A5) Red fluorescence from Cy3-Oligo; (B1-B5) Green fluorescence arising from FITC-polymer; (C1-C5) Merged image. The yellow fluorescence indicates that FITC-polymer colocalized with Cy3-Oligo. The images were obtained using a Zeiss LSM-710 inverted confocal microscope with 63× magnification.
Evaluation of 2′-OMePS and PMO delivery in C2C12E50 myoblast cell lines expressing GFP/hDysE50
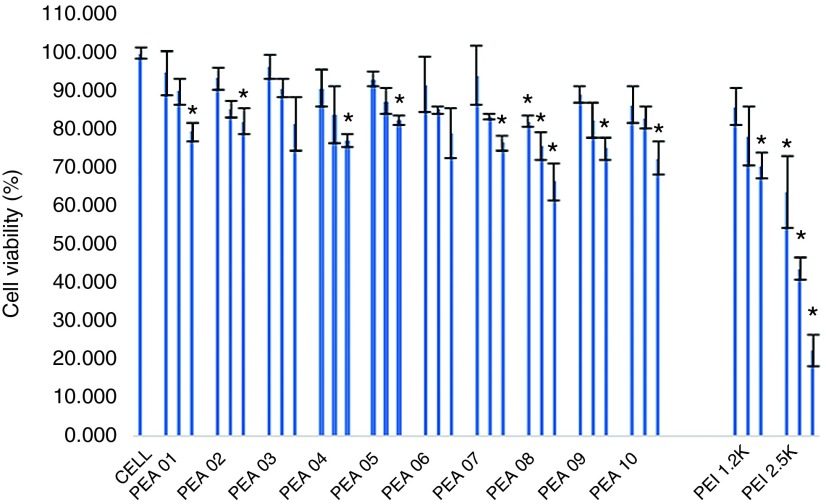
Cytotoxicity study. Cytotoxicity of the polymers in C2C12E50 cells was evaluated by (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS)-based assay (Figure 1). Cells treated with PEI 2k maintained over 70% cell viability at a dose of 20 µg/ml. In contrast, cell viability was only 22% with PEI 25k at the same dose and remained low with 10 and 4 µg/ml treatment (42 and 63%, respectively). At the dose of 20 µg/ml, cell survival rate was over 75% with all PEAs, except for PEA 08, which displayed relatively higher toxicity (67% live cells) due to its more hydrophobic compared with others.28,30 The cytotoxicity data suggests that the toxicity of the Pluronic-modified LPEI does not increase with the growth in polymer size. We reason this is due to the substitution of the primary and/or some secondary amines of PEI with the amphiphilic Pluronics.34 Toxicity of Pluronic is limited indicated by their wide use as pharmaceutical adjuvant resulted from their excellent biocompatibility and environmental sensitivity; some of which have been approved by US Food and Drug Administration (FDA).35
Figure 1.
The cytotoxicity of the PEAs polymers in C2C12E50 cell lines via MTS-based cell viability assay. The polymer dosages are 4, 10, 20 µg/ml from left to right for each sample. Cells were seeded in 96-well plates at an initial density of 1 × 104 cells/well in 200 µl growth media. The results are presented as the mean ± SD in triplicate (Student's t-test, *P ≤ 0.05 compared with untreated cell as a control).
Uncharged PMO delivery. We next examined the same PEAs for neutral PMO delivery. PMOE50 (5′-AACTTCCTCTTTAACAGAAAAGCATAC-3′) with previously confirmed efficacy of targeted removal of human dystrophin exon 50 at a fixed amount (10 µg/ml) was formulated with each polymer.28,29,30 The TE of PMOE50 alone was below 5%. However, TE of PMOE50 with two doses (10 µg/ml, 20 µg/ml) of PEAs was over 60% for PEA 01/03/04/05 at the dose 10 µg/ml. The highest TE was achieved with PEA 01/04, with over 75%, which was close to Endo-porter mediated PMO delivery (TE = 78%). Endo-porter at this concentration caused cell death of up to 35%, whereas over 75% cells survived with PEA 01/04 (Figure 4), only 22% TE with much higher toxicity was observed with PEI 25k. The TE achieved with individual PEA for PMO delivery was similar to that for 2′-OMePS delivery. However, exceptions were observed with PEA 03/08 showing higher efficiency for PMO delivery than for 2′-OMePS delivery, which indicated the hydrophobic interaction is more predominant than charge interaction between the PEAs and uncharged PMO. The results that amphiphilic PEAs also provide higher PMO delivery efficiency than PEI, and reaches similar levels to that of commercially available cationic peptide (Endo-porter) demonstrates their potential to be applied in vivo for achieving therapeutic values.
Cell uptake study. To better understand how the PEA polymers improve the delivery or transfection efficiencies of AOs, we directly visualized the cellular uptake of polymer/AO polyplexes, wherein the FITC-labeled PEA 01 was complexed with Cy3-labeled oligonucleotide at a weight ratio of 5:1 (Rw = 5) and examined under confocal microscope (Figure 5). In the absence of the PEA 01 polymer, the oligonucleotide alone showed weak diffuse signal within the treated cells. In contrast, signals for polymer formulated AO were predominantly located in cytoplasmic and perinuclear areas. Intensive signals of the labeled polymer were also observed at the plasma membrane. While the two signals for PEA 01 and oligonucleotide were generally codistributed as indicated by yellow or light blue (merging green, red, and blue) on the merged image presented, punctate signal in cytoplasm was primarily green for polymers. Similar patterns of signal distribution were observed for both polymer/PMO and polymer/2′-OMePS complexes except that PMO signal was more diffuse than 2′-OMePS in the absence of polymers. This is probably due to the nonspecific interaction with the cell membrane relevant to its uncharged nature.
Transfection study for AOs
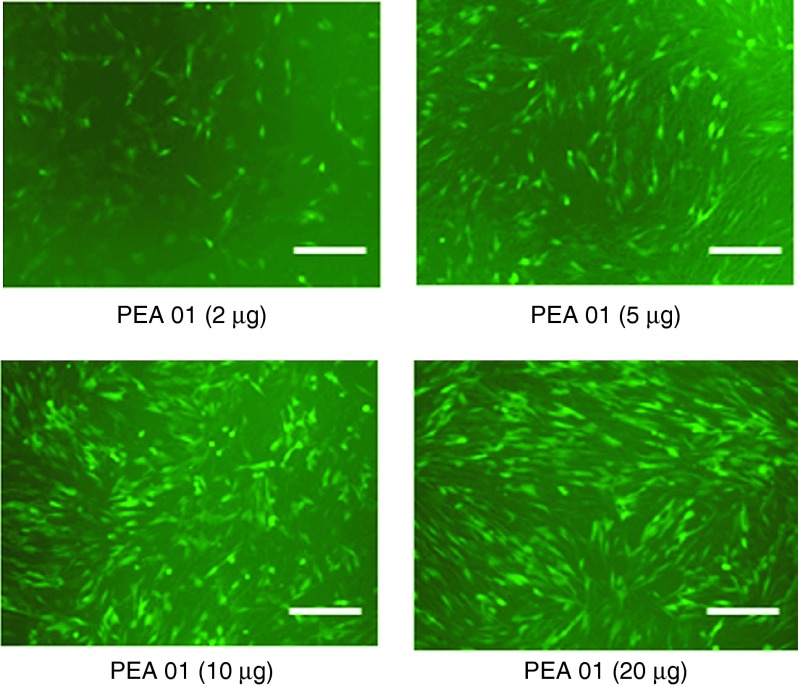
Negative charged 2′-OMePS delivery. We further examined the effect of PEAs at four different dosages (4, 10, 20, and 40 µg/ml) on exon-skipping of 2′-OMePS in the C2C12E50 cell line. The AO sequence (5′-GGGAUCCAGUAUACUUACAGGCUCC-3′) targeting the inserted human dystrophin exon 50 within the GFP coding region was used. The cells were treated with the 2′-OMePSE50 at a fixed amount (4 µg/ml) formulated with each PEA. Transfection efficiency (TE) of formulated 2′-OMePSE50 was visualized under fluorescence microscopy 2 days after delivery. The results exhibited that all PEAs at the dose of 10 µg/ml produced enhanced GFP expression as compared to the 2′-OMePSE50 alone. The highest levels of GFP expression were achieved at the dose of 20 µg/ml with most PEAs. No further GFP enhancement was observed when the dose of PEA increased up to 40 µg/ml, probably due to the aggregation or some toxicity of polymer-AO complex under the high-dose ratio (Rw = 10). As an example illustrated in Figure 2, PEA 01-mediated 2′-OMePS delivery dose-dependently increased GFP expression, resulting from the oligonucleotide-induced skipping of the human dystrophin exon 50 and the restoration of the GFP reading frame. Further, the delivery efficiency of PEAs (at the dose of 10–20 µg/ml) formulated 2′-OMePS (4 µg/ml) were measured quantitatively by flow cytometer (Figure 3), reaching over 50% with PEA 01/02/04/05, and over 75% with PEA 01/04 at the dose of 10 µg/ml. In contrast, 2′-OMePS alone, and 2′-OMePS formulated with PEI 25k or Lipofectamine-2000 (LF-2k) exhibited around 1.7, 11.5, and 36.4% GFP-positive cells, respectively at the optimal concentration. The 2′-OMePS with PEA 01 formulation achieved the highest efficiency with over 46-fold of that obtained with 2′-OMePS alone. Clearly, the results suggest that a combination of middle size amphiphilic part and cationic ingredients together is preferred as carrier to enhance delivery efficiency of 2′-OMePS. In agreement with the viability results from polymer alone, cytotoxicity of the PEA/2′-OMePS complexes was dramatically lower than that of PEI 25k or LF-2k formulated 2′-OMePS complexes.
Figure 2.
Dose–response GFP expression of polymer PEA 01 formulated 2′-OMePSE50. C2C12E50 cells were treated with 2 µg 2′-OMePS and the polymer in 500 µl 10% FBS-DMEM and incubated under 37 °C, 10% CO2 environment. The images were taken 48 hours after transfection (scale bar = 500 µm).
Figure 3.
Delivery efficiency and toxicity of PEAs mediated 2′-OMePS in C2C12 cell line determined by fluorescence microscopy and flow cytometer. (a) Representative fluorescence images of 2′-OMePSE50-induced exon-skipping in C2C12E50 cell line. The images were taken 48 hours after treatment (scale bar = 500 µm). (b) Transfection efficiency of 2′-OMePSE50 formulated with the polymers (Student's t-test, *P ≤ 0.05 compared with 2′-OMePS only). (c) Cell viability (Student's t-test, *P ≤ 0.05 compared with untreated cells as a control). Two micrograms of 2′-OMePSE50 were formulated with PEAs (5,10 µg), PEI-25k (2 µg) and LF-2k (4 µg) in 500 µl 10% FBS-DMEM medium. The results are presented as the mean ± SD in triplicate.
Uncharged PMO delivery. We next examined the same PEAs for neutral PMO delivery. PMOE50 (5′-AACTTCCTCTTTAACAGAAAAGCATAC-3′) with previously confirmed efficacy of targeted removal of human dystrophin exon 50 at a fixed amount (10 µg/ml) was formulated with each polymer.28,29,30 The TE of PMOE50 alone was below 5%. However, TE of PMOE50 with two doses (10 µg/ml, 20 µg/ml) of PEAs was over 60% for PEA 01/03/04/05 at the dose 10 µg/ml. The highest TE was achieved with PEA 01/04, with over 75%, which was close to Endo-porter mediated PMO delivery (TE = 78%). Endo-porter at this concentration caused cell death of up to 35%, whereas over 75% cells survived with PEA 01/04 (Figure 4), only 22% TE with much higher toxicity was observed with PEI 25k. The TE achieved with individual PEA for PMO delivery was similar to that for 2′-OMePS delivery. However, exceptions were observed with PEA 03/08 showing higher efficiency for PMO delivery than for 2′-OMePS delivery, which indicated the hydrophobic interaction is more predominant than charge interaction between the PEAs and uncharged PMO. The results that amphiphilic PEAs also provide higher PMO delivery efficiency than PEI, and reaches similar levels to that of commercially available cationic peptide (Endo-porter) demonstrates their potential to be applied in vivo for achieving therapeutic values.
Cell uptake study. To better understand how the PEA polymers improve the delivery or transfection efficiencies of AOs, we directly visualized the cellular uptake of polymer/AO polyplexes, wherein the FITC-labeled PEA 01 was complexed with Cy3-labeled oligonucleotide at a weight ratio of 5:1 (Rw = 5) and examined under confocal microscope (Figure 5). In the absence of the PEA 01 polymer, the oligonucleotide alone showed weak diffuse signal within the treated cells. In contrast, signals for polymer formulated AO were predominantly located in cytoplasmic and perinuclear areas. Intensive signals of the labeled polymer were also observed at the plasma membrane. While the two signals for PEA 01 and oligonucleotide were generally codistributed as indicated by yellow or light blue (merging green, red, and blue) on the merged image presented, punctate signal in cytoplasm was primarily green for polymers. Similar patterns of signal distribution were observed for both polymer/PMO and polymer/2′-OMePS complexes except that PMO signal was more diffuse than 2′-OMePS in the absence of polymers. This is probably due to the nonspecific interaction with the cell membrane relevant to its uncharged nature.
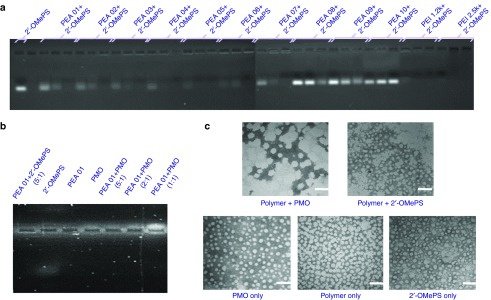
Affinity study between polymer and oligonucleotide
The affinity between polymer and oligonucleotide is an important parameter for their efficient delivery into cells. We studied the interaction between the polymer and oligonucleotide by electrophoretic mobility and particle morphology investigations.
Electrophoretic mobility. We first evaluated the mixture of the PEAs and negative 2′-OMePS at various weight ratio to determine the interaction between the two components. As illustrated in Figure 6a, most PEA polymers, except for PEA 08/09/10, showed a high binding capacity with 2′-OMePS at Rw = 5, resulting in condensed and/or charge-neutralized complexes indicated by the shifting of the bands stained with ethidium bromide (EtBr) toward significantly higher molecular size. However, their binding ability was not as strong as PEI 25K at the same doses. As we know, super binding ability may hinder the release of delivery cargo in the cells, thus the balance between the AO condensation and release is essential or rational as effective non-viral vectors. These results are consistent with the expectation that negatively charged oligonucleotides are progressively neutralized by the increasing amount of positive charged PEA polymers. The weak binding of PEA 08/09/10 with 2′-OMePS is likely due to the smaller molecular weight or lower PEI content compared to the other PEAs. On the other hand, PMO is considered charge-neutral, and its affinity to cationic amphiphile is likely limited, but expected through the hydrophobic interaction and possibly hydrogen-bind. As illustrated in Figure 6b for a comparison of these two AOs formulated with PEA 01 at different weight ratios, electrophoresis results confirmed a weak positive charge of PMO as the EtBr stained PMO only slightly shifted to the negative electrode. In contrast, the 2′-OMePS alone shifted markedly to the positive electrode. Both PEA 01/2′-OMePS polyplex and PEA 01/PMO polyplex at a weight ratio (5:1) remained almost entirely within the well. The intensity of EtBr stained band of the PMO/PEA 01 complex became weaker as weight ratio decreased from 5:1 to 1:1, which supports the notion that PMO itself is largely charge neutral and relies on PEA for binding to EtBr (more intense signal with increasing weight ratio). The results demonstrate that lipophilic components in a polymer are critical for the effective condensation between the polymer carrier and uncharged AOs, and further affect delivery behaviors.
Figure 6.
Affinity between polymer and oligonucleotide. (a) Electrophoretic mobility of Polymer/2′-OMePS complexes at three weight ratio of Rw = 1, 2, and 5 (from left to right for each polymer, 1 µg oligonucleotide in a total of 24 µl medium was used. The first lane on the left is loaded with 1 µg oligonucleotide only). (b) PEA 01 with 2′-OMePS/PMO at different weight ratios (5 µg PEA 01 used in this experiment in total of 24 µl). (c) Negative staining TEM images of PEA 01/oligonucleotide condensates: PEI 01/PMO (10 µg, 5 µg) and PEI 01/2′-OMePS (10 µg, 2 µg) in 500 µl 0.9% Saline (scale bar = 100 nm).
Morphology study. The particle size of cargo-polymer complex is important for effective gene transfection, and less than 200 nm particles are generally considered preferable for effective delivery. In our previous study, we found the particle of polymer/PMO complex is hard to determine by dynamic light scattering (DLS) in diluted solution. We chose to examine the most effective PEA 01/AO polyplex by transmission electron microscopy (TEM) to evaluate the particle size. As illustrated in Figure 6c, the polymer PEA 01 itself formed particles of different-sizes likely because of aggregation, whereas PMO oligonucleotides alone formed particles with the size below 50 nm, probable resulted from hydrophobic interactions and hydrogen-bond among PMO microstructure. The polyplex of PEA 01/PMO at a weight ratio of 10/5 formed particles with an average diameter around 60–100 nm. The PEA 01/2′-OMePS polyplex formed smaller particles around 20–50 nm, probably the result of strong interactions including charge interaction and hydrophobic interactions among them. The mechanisms of interaction between the PMO and the PEA molecules are unclear, but the chemical nature of PMO likely creates a hydrophobic interaction with the PEA and possible hydrogen-bond interaction between them. The surface charges of the polyplex may stabilize it in a biological environment for a longer period than PMO alone, although the positively charged groups within the PEA hardly play a key role for the interaction with uncharged PMO.
Protein adsorption test
Adsorption of polymers to surface of serum proteins and other components are known to be able to trigger thrombosis and blood coagulation, which can lead to life-threatening situations. In system circulation, serum proteins could bind to polymer/DNA complexes in a nonspecific manner, leading to aggregation and severely decreased TE.36 The protein adsorption assay of the polymers was performed to confirm their serum tolerance. As shown in Supplementary Figure S1, after the addition of bovine serum albumin (BSA), the PEI 25k solution turned turbid, while the solution of PEAs remained clear. The BSA adsorptions to PEAs were 33.0, 28.5, 38.9, 35.1, 28.0, 45.8, 32.0, 34.5, 37.4, and 38.9% for PEA 01, PEA 02, PEA 03, PEA 04, PEA 05, PEA 06, PEA 07, PEA 08, PEA 09, and PEA 10, respectively, while adsorption to PEI 25k was 98.5% by quantitative measurement. Such phenomenon might be ascribed to the amphiphilic property of PEAs compared with PEI 25k, and the relatively dispersed positive charges within PEA polymers, thus preventing their aggregation with negatively charged serum proteins. The relatively higher adsorption rate with PEA 06 is probably related to its higher molecular size and/or more hydrophilic property. Thus as expected, BSA adsorption was primarily caused by electrostatic interactions between negatively charged BSA and the positively charged polymers.
Delivery of AOs with PEAs in vivo
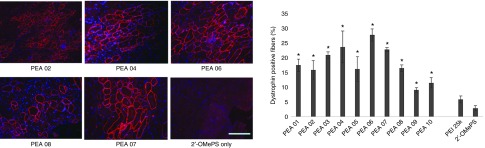
Delivery of 2′-OMePS locally. First, we evaluated the effects of the PEAs for 2′-OMePS delivery in vivo by intramuscular (i.m.) injection to the TA muscles of mdx mice aged 4–5 weeks with the 2′-OMePSE23 targeting mouse dystrophin exon 23. The mdx mouse contains a nonsense mutation in exon 23 and lacks the functional dystrophin protein. Targeted removal of the mutated exon 23 is able to restore the reading-frame of dystrophin transcripts and thus the expression of dystrophin protein. All PEAs were examined at the dose of 10 µg mixed with 5 µg of 2′-OMePSE23 in 40 µl saline. Treated TA muscles were harvested 2 weeks after injection. Immunohistochemistry showed that the numbers of dystrophin-positive fibers increased up to 3–10-folds in the muscles treated with same amount of 2′-OMePS formulated with PEA polymers. In particular, dystrophin-positive fibers were induced to 19, 16, 23, 25, 17, 30, 24, 17, 10, and 12% with PEA 01 to PEA 10 formulation, respectively. In contrast, PEI 25k-mediated 2′-OMePS produced around 5% positive fibers (Figure 7). PEA 03/04/06/07, and especially the PEA 06 increased dystrophin induction more effectively than other PEAs, probably due to its larger molecular size with more favorable positive surface charge of PEA 06/2′-OMePS complex.
Figure 7.
Dystrophin exon-skipping and protein expression following i.m. administration of 2′-OMePSE23 without and with polymers in TA muscle of mdx mice (aged 4–5 weeks) 2 weeks after treatment. Muscles were treated with 5 µg 2′-OMePSE23 and 10 µg PEAs (5 µg PEI 25k as comparison) in 40 µl saline. 2′-OMePSE23 only was used as control: (Left) Restoration of dystrophin in TA muscle was detected by immunohistochemistry with rabbit polyclonal antibody P7 against dystrophin. Blue nuclear staining with DAPI (4,6-diamidino-2-phenylindole). Scale bar = 200 µm. (Right) The numbers of dystrophin-positive fiber in a single cross-section induced by 2′-OMePSE23 with/without polymer formulation (mean ± SD, n = 5, Student's t-test, *P ≤ 0.05 compared with 2′-OMePSE23).
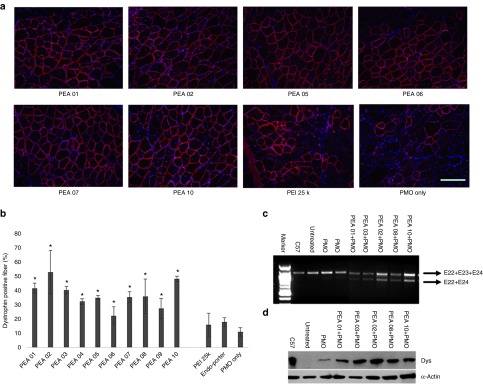
Delivery of PMO locally. Next, we examined the PEAs for PMO delivery in vivo by i.m. injection of PMOE23. All PEAs were also examined at the dose of 10 µg and mixed with 2 µg PMOE23 before injection. The treated TA muscles were harvested 2 weeks later. The results were illustrated in Figure 8. Immunohistochemistry showed that PMOE23 alone induced a maximum of 11% dystrophin positive fibers in one cross-section of the TA muscle. Dystrophin-positive fibers increased dramatically in the muscles treated with PEA formulated PMOE23, achieving over 35% positive fibers with all PEAs except for PEA 06/09. PMO with PEI 25k and Endo-porter produced 16%, 18% dystrophin-positive fibers, respectively (Figure 8b). In particular, PEA 02 and PEA 10 improved the efficiency up to 54 and 48%. Interestingly, PEA 06, which is most effective for 2′-OMePS delivery was least effective for PMO delivery with only 22% dystrophin-positive fibers. This low efficiency of PEA 06 in PMO delivery was probably due to poor affinity between PMO and the polymer being the largest molecule and more hydrophilic as compared with other PEAs. The levels of exon-skipping and dystrophin expression were further confirmed by reverse transcription polymerase chain reaction (RT-PCR, Figure 8c) and western-blot (Figure 8d). Hematoxylin and Eosin (H&E) staining of the muscle sections from the treated mice with of PEA revealed no signs of elevated inflammation, fiber degeneration and regeneration when compared to the saline-treated control TA muscles in both AOs delivery, confirming their low tissue toxicity as demonstrated for pDNA delivery in mdx mice.33 The results from the current study indicate that AO delivery efficiency and toxicity of PEA polymers are closely related to the molecular size, composition and HLB of the polymers as well as the nature of AOs.
Figure 8.
Dystrophin exon-skipping and protein expression following i.m. administration of PMOE23 with polymers in TA muscle of mdx mice (aged 4–5 weeks) 2 weeks after treatment. Muscles were treated with PMOE23 (2 µg) and PEAs (10 µg) or PEI 25k (2 µg)/Endo-porter (5 µg) in 40 µl saline. PMOE23 (2 µg) only was used as control: (a) Restoration of dystrophin in TA muscle was detected by immunohistochemistry with rabbit polyclonal antibody P7. Blue nuclear staining with DAPI (4,6-diamidino-2-phenylindole). (scale bar = 200 µm). (b) The percentage of dystrophin-positive fibers in a single cross-section induced by PMOE23 with/without polymer formulation (mean ± SD, n = 5, Student's t-test, *P ≤ 0.05 compared with PMOE23). (c) Detection of exon 23 skipping by RT-PCR. Total RNA (100 ng) of from each sample was used for amplification of dystrophin mRNA from exon 20 to exon 26. The upper bands (indicated by E22+E23+E24) correspond to the normal mRNA and the lower bands (indicated by E22+E24) correspond to the mRNA with exon E23 skipped. (d) Western blot demonstrates the expression of dystrophin protein from treated mdx mice compared with C57BL/6 and untreated mdx mice. Dys: dystrophin detected with a monoclonal antibody ManDys 1. α-Actin was used as the loading control.
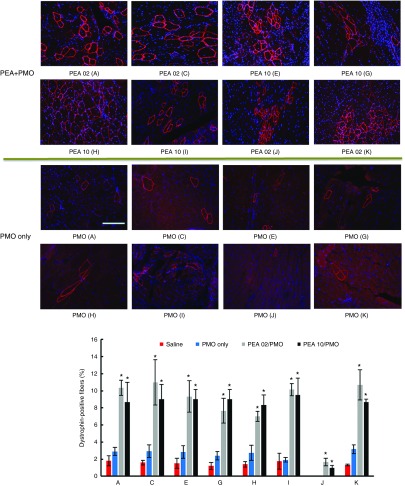
Delivery of PMO systemically. DMD is a systemic disease, affecting body-wide muscles including cardiac muscle, systemic treatment therefore is indispensable. Based on the results in vitro and in vivo locally, we selected the two most effective polymers (PEA 02 and PEA 10) to evaluate their effects on PMO systemic delivery by intravenous (i.v.) injection at the dose of 0.5 mg formulated with 1 mg PMOE23. The control PMOE23 alone induced dystrophin expression in less than 3% of muscle fibers in all skeletal muscles and no detectable dystrophin in cardiac muscle 2 weeks after injection. PMOE23 formulated with both PEAs produced more dystrophin positive fibers over 7% in all skeletal muscles (Figure 9). Similar to the results with local delivery, PEA 02 formulated PMO was slightly more effective with over 10% positive fibers in TA, Quadriceps, Diaphragm and Bicep muscles. Variation in number of dystrophin-positive fibers among different muscles is consistent with our early report that induction of dystrophin is not homogenously distribute and mechanism(s) of the uptake remains to be determined.11,12,13 Also important is that both PEA-medicated PMO delivery produced groups of dystrophin positive fibers although limited to some areas in the cardiac muscle, whereas only occasional 1 or 2 dystrophin-positive fibers were observed in the heart treated with PMO alone. This result together with the lack of detectable toxicity in muscles, liver, and kidney after systemic treatment further solidify the potential of PEA copolymers as AO delivery carriers for the treatment of muscular dystrophies.
Figure 9.
Dystrophin expression in different muscles of mdx mice (aged 4–5 weeks) 2 weeks after systemic administration of PMO with polymers. Each mouse was injected with 1 mg PMOE23 with and without polymer (0.5 mg). Upper panel, immunohistochemistry with antibody P7 for the detection of dystrophin (scale bar = 200 µm). Down panel, percentage of dysrophin-positive fibers in different muscle tissues (mean ± SD, n = 3, Student's t-test, *P ≤ 0.05 compared with 1 mg PMO. A: Tibialis anterior; C: Quadriceps; E: Gastronemieus; G: Abdomen; H: Intercostals; I: Diaphragm; J: Heart; K: Bicep).
Conclusions
The study of PEA polymers for two antisense oligonucleotides (2′-OMePS and PMO) delivery in vitro and in vivo of dystrophic mdx mice revealed that all PEAs showed improved exon-skipping efficiency with 2′-OMePS in vitro over PEI 25k or LF-2k, and the degree of efficiency was found in the order of PEA 01, PEA 04 > PEA 05 > others. Enhanced exon-skipping of 2′-OMePS with low toxicity were also demonstrated in vivo in the order of PEA 06 > PEA 04, PEA 07 > PEA 03 > PEA 01 > others. PEAs improved exon-skipping efficiency of PMO in vitro in the order: PEA 01, PEA 04, PEA 05 > PEA 03 > others, especially with PEA 01 being the most effective with efficiency close to Endo-porter formulated PMO. Significant enhancement in exon-skipping in vivo was achieved with the PEAs in the order of PEA 02 > PEA 10 > PEA 01, PEA 03 > PEA 05, PEA 07, PEA 08 > others. The most effective PEA 02 achieved fourfold or higher increase in dystrophin induction over Endo-porter formulated PMO by local delivery, and improved the systemic delivery in body-wide muscles including cardiac muscle. Delivery performance depends on both the hydrophobic and electrostatic interaction between the polymer and short, single-stranded AOs. The results indicate the importance of amphiphilic nature of the cationic carriers for effective oligonucleotide delivery.37 The hydrophobic interaction between the PEA polymers and AOs likely results in a more stable complex in primarily hydrophilic microenvironments and could also enhance interactions between polymer/AO complex-plasma membrane both in vitro and in vivo. The variability of individual PEA polymers for delivery of negatively charged 2′-OMePS and uncharged PMO highlights the complexity of the interaction between polymer and their delivery cargos, and the difference in the delivery mechanism. The flexibility of the PEA polymers as nano-vehicles with excellent biocompatibility for both large size pDNA and small oligonucleotides in vitro and in vivo, suggests their potential for broad therapeutic spectrum.
Materials and methods
Materials. A series of cationic amphiphlic PEAs constructed from Pluronics and low molecular weight (Mw) polyethyleneimene (LPEI) were synthesized as previous reported.33 Cell culture media Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, L-glutamine and HEPES buffer solution (1M), penicillin-streptomycin, and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) were purchased from Invitrogen (Carlsbad, CA). An arbitrary single-stranded 20-mer deoxyoligonucleotide was used as a model antisense oligonucleotide (AO), with the sequence 5′-GGCCAAACCTCGGCTTACCT-3′ (phosphodiester), for the physicochemical study of polymer-oligonucleotide polyplex. AOs modified by 2′-O-methylation and phosphorthioation 2′-OMePSE50 (5′-GGGAUCCAGUAUACUUACAGGCUCC-3′) targeting human dystrophin gene exon 50, 2′-OMePSE23 (5′-GGCCAAACCUCGGCUUACCU-3′) targeting mouse dystrophin gene exon 23, FL-Oligonucleotide (5′-GGCCAAACCUCGGCUUACCU-3′) used for delivery in vitro and in vivo were purchased from GenScript (Piscataway, NJ). Phosphorodiamidate morpholino oligomer PMOE50 (5′-AACTTCCTCTTTAACAGAAAAGCATAC-3′), PMOE23 (5′-GGCCAAACCTCGGCTTACCTGAAAT-3′) and FL-PMO as well as Endo-porter were purchased from Gene Tools (Philomath, OR). Pluronics were supplied by BASF (Florham Park, NJ). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
In vitro study Cell viability. Cytotoxicity was evaluated in C2C12E50 cell line using the MTS-based assay. Cells were seeded in a 96-well tissue culture plate at 1 × 104 cells per well in 200 µl 10% FBS-DMEM. Cells achieving 70–80% confluence were exposed to polymer at different doses for 24 hours, followed by the addition of 20 µl of Cell Titer 96Aqueous One Solution Proliferation Kit (Promega Corporation, Madison, MI). After further incubation for 4 hours, the absorbance was measured at 570 nm using Tecan Infinite F500 Plate reader (Tecan US, Morrisville, NC) to obtain the metabolic activity of the cell. Untreated cells were taken as controls with 100% viability and wells without cells as blanks, the relative cell viability was calculated by: (Atreated-Abackground) × 100/(Acontrol-Abackground). All viability assays were carried out in triplicate.
In vitro transfection. The C2C12 E50 cell has a human dystrophin exon 50 sequence (hDysE50) placed inside the coding sequence of GFP gene under control of an actin promoter. Upon the effect of antisense oligonucleotide targeting exon hDysE50, the hDysE50 sequence is spliced out and the GFP coding sequence rejoined back in-frame.
C2C12 or C2C12E50 cells were cultured in 10% FBS-DMEM and kept in a 10% CO2 humidified incubator at 37 °C. Approximately 5 × 104 cells per well were seeded in a 24-well plate in 500 µl medium and allowed to reach 70–80% confluence prior to transfection experiments. Cell culture medium was replaced before the addition of polymer/2′-OMePSE50 formulation with varying weight ratios. PEI 25k was used as positive control for delivery. Transfection efficiency was visualized using an Olympus IX71 fluorescent microscope (Olympus America, Milville, NY). Digital images were taken using the Olympus DP Controller and DP Manager software (Olympus America, Milville, NY). Transfection efficiency was also examined using flow cytometry to quantitatively gauge the GFP expression level. Cells were washed with PBS (1×) and released from the culture vessel with 0.05% Trypsin-EDTA, neutralized by FBS, pelleted by centrifugation and then resuspended in 1 ml PBS. Samples were run on a FACS Calibur flow cytometer (BD, Franklin Lakes, NJ). Minimum 1 × 104 cells were counted and analyzed with CellQuest Pro (BD, Franklin Lakes, NJ) software package.
Fluorescence labeling of polymers. The polymer was labeled with Fluorescein isothiocyanate (FITC) as described in a previous study with some modification.26 The over excess FITC dissolved in DMSO, was added drop-wise to polymer in PBS (pH 7.4) solution. The solution was then incubated at room temperature with continuous stirring in the dark overnight. The labeled polymer was dialyzed with MWCO 2000 membrane against PBS (pH 7.4) for 24 hours and then distilled water for 12 hours. The product was finally lyophilized and kept at −20 °C.
Cellular uptake and intracellular localization. For cellular uptake and intracellular localization study, Cy3-labeled oligonucleotide was combined with FITC-labeled polymer at a predetermined ratio, followed by imaging under confocal microscopy. C2C12 cells were seeded onto eight-well glass Lab-Tek II chamber slides (Scientific, Ocala, FL) at 5 × 103 cells/well, and cultured to 70% confluence before the addition of polymer-oligonucleotide formulation for testing. Eight hours after addition of the samples, the cells were washed with warm PBS (0.1M, pH 7.4) to remove any residual polymer-oligonucleotide complex not taken up by cells. Cells were then counterstained with Hoechst 33258 (Life Technologies, Carlsbad, CA) for 20 minutes at 37 °C. Subsequently, the cells were washed with PBS three times and then incubated with 200 µl PBS. The cells were examined using a Zeiss LSM-710 inverted confocal microscope (Carl Zeiss Microscopy, LLC; Thornwood, NY), and the resulting images were analyzed for uptake and localization by single channel images. Colocalization of polymer/oligonucleotide to the lysosome was visualized by merging channel images.
Affinity study between polymer and oligonucleotides Agarose gel electrophoresis. All polymer-oligonucleotide complexes were freshly prepared before use with different polymer/oligonucleotide weight ratios by vortex of oligonucleotide and polymer stock solutions. The complexes were incubated at room temperature for 30 minutes in 24 µl volume and loaded on 1% agarose gel with ethidium bromide (EtBr, 0.1 µg/ml) included into the gel to visualize the localization of the oligonucleotide. Electrophoresis was carried out with tris-acetate (TAE) buffer at 100 V for 40 minutes and analyzed on UV illuminator.
Morphology analysis. The polymer-oligonucleotide polyplex was prepared in 200 µl 0.9% NaCl, pH 7.4 medium, and corresponding polymer and oligonucleotide only as a comparison. The solutions were analyzed using transmission electron microscopy (TEM, Phillips CM-10, Philips Electronic North America, Andover, MA). Samples were prepared using negative staining with 1% phosphotungstic acid. Briefly, one drop of sample solution was placed on a formvar and carbon coated grid (Electron Microscopy Sciences, Hatfield, PA) for 1 hour, and the grid was blotted dry, followed by staining for 3 minutes. The grids were, once again, blotted dry. Samples were analyzed at 60 kV. Digital images were captured with a digital camera system from the 4 pi Analysis (Durham, NC).
Protein adsorption assay. Briefly, 1 ml of polymer solution (1.0 mg/ml) was mixed with 1 ml of bovine serum albumin (BSA) solution (1.0 mg/ml). After shaking for 0.5 hours at 37 °C followed by centrifugation, 1.0 ml of the supernatant was carefully collected, and then the concentration of BSA in the supernatant was monitored with a Nanodrop 2000 UV-Visible assay measured absorption at 280 nm. The protein adsorbed on the polyplexes was calculated (Ai-A) ×100/Ai. Ai is absorption of BSA only at the concentration of 0.5 mg/ml, A is the absorption corresponding Polymer formulated BSA.
In vivo delivery and antibodies, immunohistochemistry. This study was carried out in strict accordance with the recommendations in Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee (IACUC), Carolinas Medical Center (Breeding protocol: 10-13-07A; Experimental protocol: 10-13-08A). All injections were performed under isoflurane anesthesia, and all efforts were made to minimize suffering.
Animals and injections. Dystrophic mdx mice aged 4–5 weeks were used for in vivo testing (five mice each in the test and control groups) unless otherwise stated. Four muscles were examined for each experimental group.
The 2′-OMePSE23 (+02-18) (5′-GGCCAAACCUCGGCUUACCU-3′) and PMOE23 (5′-GGCCAAACCTCGGCTTACCTGAAAT-3′) targeting the boundary sequences of exon and intron 23 of the mouse dystrophin gene were used. For intramuscular (i.m.) injections, 5 µg 2′-OMePSE23 or 2 µg PMOE23 with or without polymer was used in 40 µl saline for each tibialis anterior (TA) muscle. For intravenous (i.v.) injection, 1 mg PMOE23 with or without polymer was used in 100 µl saline. Mice were euthanized by CO2 inhalation following 2 weeks treatment and muscles were snap-frozen in liquid nitrogen-cooled isopentane and stored at −80 °C.
Reverse transcription polymerase chain reaction (RT-PCR). Total RNA was extracted from tissue using TRIzol and 100 ng of RNA template was used for a 50 µl RT-PCR with the Stratascript One-Tube RT-PCR System (Stratagene, Santa Clara, CA). The primer sequences for the RT-PCR were Ex20Fo 5′-CAGAATTCTGCCAATTGCTGAG-3′ and Ex26Ro 5′-TTCTTCAGCTTGTGTCATCC-3′ for amplification of mRNA from exons 20 to 26. The conditions were 43 °C for 15 minutes, 94 °C for 2 minutes, then cycled 30 times at 94 °C for 30 seconds, 65 °C for 30 seconds, and 68 °C for 1 minute. The products were examined by electrophoresis on a 2% agarose gel. Bands with the expected size for the transcript with exon 23 deleted were extracted and sequenced.
Antibodies, immunohistochemistry, and western blots. Sections of 6 µm were cut from the muscles and stained with a rabbit polyclonal antibody P7 for the dystrophin protein and detected by goat anti-rabbit Igs Alexa 594 (Invitrogen, Waltham MA). The number of dystrophin-positive fibers in one section was counted using the Olympus BX51 fluorescent microscope (Olympus America). Digital images were taken with the Olympus DP Controller and DP Manager software (Olympus America). Protein extraction and Western blot were performed as described previously.11,12,28,29,30 Briefly, the membrane was probed with NCL-DYS1 monoclonal antibody against dystrophin rod domain (1:200 dilution, Vector Laboratories, Burlingame, CA) followed by HRP-conjugated goat anti-mouse IgG (1:3000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) and the ECL Western Blotting Analysis System (Perkin-Elmer, Waltham, MA). The intensity of the bands with appropriate size was measured and compared with that from normal muscles of C57BL mice using NIH ImageJ software. Loading control of α-Actin was detected by rabbit anti-actin antibody (Sigma, St. Louis, MO).
Statistical analysis. The data were analyzed for statistical significance via Student's t-test with a value of P ≤ 0.05 being considered statistically significant. Data were expressed as mean ± SD.
SUPPLEMENTARY MATERIAL Figure S1. Protein adsorption of synthesized PEA polymers and PEI 1.2k, PEI 25k: (Upper) Adsorption quantity of BSA. Table S1. Characteristics of PEA polymers. Scheme S1. Synthesis of the PEI-Pluronic copolymers (PEAs). Supplementary Materials
Acknowledgments
The authors thank Daisy M. Ridings and Ben J. Wagner with the Electron Microscopy Core Laboratory for the negative staining and TEM electron micrographs; also thank David M Foureau and Fei Guo for their technical assistance in Flow Cytometry and analysis. Furthermore, the authors gratefully acknowledge the financial support by the Carolinas Muscular Dystrophy Research Endowment at the Carolinas HealthCare Foundation and Carolinas Medical Center, Charlotte, NC. The authors declare no conflicts of interest in relation to this paper.
Supplementary Material
References
- Hoffman, EP, Brown, RH Jr and Kunkel, LM (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51: 919–928. [DOI] [PubMed] [Google Scholar]
- Koenig, M, Beggs, AH, Moyer, M, Scherpf, S, Heindrich, K, Bettecken, T et al. (1989). The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 45: 498–506. [PMC free article] [PubMed] [Google Scholar]
- Wagner, KR, Lechtzin, N and Judge, DP (2007). Current treatment of adult Duchenne muscular dystrophy. Biochim Biophys Acta 1772: 229–237. [DOI] [PubMed] [Google Scholar]
- McNeil, DE, Davis, C, Jillapalli, D, Targum, S, Durmowicz, A and Coté, TR (2010). Duchenne muscular dystrophy: Drug development and regulatory considerations. Muscle Nerve 41: 740–745. [DOI] [PubMed] [Google Scholar]
- Kole, R and Krieg, AM (2015). Exon skipping therapy for Duchenne muscular dystrophy. Advanced Drug Delivery Reviews 87: 104–107. [DOI] [PubMed] [Google Scholar]
- Lu, QL, Cirak, S and Partridge, T (2014). What can we learn from clinical trials of exon skipping for DMD? Mol Ther Nucleic Acids 3: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, K, Foster, H and Dickson, JG (2006). Gene therapy progress and prospects: Duchenne muscular dystrophy. Gene Ther 13: 1677–1685. [DOI] [PubMed] [Google Scholar]
- Järver, P, O'Donovan, L and Gait, MJ (2014). A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther 24: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, T and Wood, MJ (2013). Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum Gene Ther 24: 479–488. [DOI] [PubMed] [Google Scholar]
- Eckstein, F (2000). Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev 10: 117–121. [DOI] [PubMed] [Google Scholar]
- Wu, B, Moulton, HM, Iversen, PL, Jiang, J, Li, J, Li, J et al. (2008). Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proc Natl Acad Sci USA 105: 14814–14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B, Lu, P, Cloer, C, Shaban, M, Grewal, S, Milazi, S et al. (2012). Long-term rescue of dystrophin expression and improvement in muscle pathology and function in dystrophic mdx mice by peptide-conjugated morpholino. Am J Pathol 181: 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B, Lu, P, Benrashid, E, Malik, S, Ashar, J, Doran, TJ et al. (2010). Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino. Gene Ther 17: 132–140. [DOI] [PubMed] [Google Scholar]
- Moulton, HM, Wu, B, Jearawiriyapaisarn, N, Sazani, P, Lu, QL and Kole, R (2009). Peptide-morpholino conjugate: a promising therapeutic for Duchenne muscular dystrophy. Ann N Y Acad Sci 1175: 55–60. [DOI] [PubMed] [Google Scholar]
- Yin, H, Moulton, HM, Seow, Y, Boyd, C, Boutilier, J, Iverson, P and Wood, MJ (2008). A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum Mol Genet 17: 3909–3918. [DOI] [PubMed] [Google Scholar]
- Goemans, NM, Tulinius, M, van den Akker, JT, Burm, BE, Ekhart, PF, Heuvelmans, N et al. (2011). Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med 364: 1513–1522. [DOI] [PubMed] [Google Scholar]
- Cirak, S, Arechavala-Gomeza, V, Guglieri, M, Feng, L, Torelli, S, Anthony, K et al. (2011). Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet 378: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, JR, Rodino-Klapac, LR, Sahenk, Z, Roush, K, Bird, L, Lowes, LP et al.; Eteplirsen Study Group. (2013). Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol 74: 637–647. [DOI] [PubMed] [Google Scholar]
- van Deutekom, JC, Janson, AA, Ginjaar, IB, Frankhuizen, WS, Aartsma-Rus, A, Bremmer-Bout, M et al. (2007). Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357: 2677–2686. [DOI] [PubMed] [Google Scholar]
- Summerton, J and Weller, D (1997). Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev 7: 187–195. [DOI] [PubMed] [Google Scholar]
- Yano, J and Smyth, GE (2012). New antisense strategies: chemical synthesis of RNA oligomers. Adv Polym Sci 249: 1–48. [Google Scholar]
- ‘t Hoen, PA, van der Wees, CG, Aartsma-Rus, A, Turk, R, Goyenvalle, A, Danos, O et al. (2006). Gene expression profiling to monitor therapeutic and adverse effects of antisense therapies for Duchenne muscular dystrophy. Pharmacogenomics 7: 281–297. [DOI] [PubMed] [Google Scholar]
- Hoffman, EP (2007). Skipping toward personalized molecular medicine. N Engl J Med 357: 2719–2722. [DOI] [PubMed] [Google Scholar]
- Jirka, SM, Heemskerk, H, Tanganyika-de Winter, CL, Muilwijk, D, Pang, KH, de Visser, PC et al. (2014). Peptide conjugation of 2′-O-methyl phosphorothioate antisense oligonucleotides enhances cardiac uptake and exon skipping in mdx mice. Nucleic Acid Ther 24: 25–36. [DOI] [PubMed] [Google Scholar]
- Sirsi, SR, Schray, RC, Guan, X, Lykens, NM, Williams, JH, Erney, ML et al. (2008). Functionalized PEG-PEI copolymers complexed to exon-skipping oligonucleotides improve dystrophin expression in mdx mice. Hum Gene Ther 19: 795–806. [DOI] [PubMed] [Google Scholar]
- Williams, JH, Sirsi, SR, Latta, DR and Lutz, GJ (2006). Induction of dystrophin expression by exon skipping in mdx mice following intramuscular injection of antisense oligonucleotides complexed with PEG-PEI copolymers. Mol Ther 14: 88–96. [DOI] [PubMed] [Google Scholar]
- Gebhart, CL, Sriadibhatla, S, Vinogradov, S, Lemieux, P, Alakhov, V and Kabanov, AV (2002). Design and formulation of polyplexes based on pluronic-polyethyleneimine conjugates for gene transfer. Bioconjug Chem 13: 937–944. [DOI] [PubMed] [Google Scholar]
- Wang, M, Wu, B, Lu, P, Cloer, C, Tucker, JD and Lu, Q (2013). Polyethylenimine-modified pluronics (PCMs) improve morpholino oligomer delivery in cell culture and dystrophic mdx mice. Mol Ther 21: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M, Wu, B, Lu, P, Tucker, JD, Milazi, S, Shah, SN et al. (2014). Pluronic–luronic b) livery in cell culture and dystrophic ystphosphorothioate oligonucleotide in cell culture and dystrophic mdx mice. Gene Ther 21: 52–59. [DOI] [PubMed] [Google Scholar]
- Wang, M, Wu, B, Tucker, JD, Lu, P, Bollinger, LE and Lu, Q (2015). Tween 85 grafted PEIs enhanced delivery performance of antisense 2′-o-methyl phosphorothioate oligonucleotides in vitro and in dystrophic mdx mice. J Mater Chem B 3: 5330–5340. [DOI] [PubMed] [Google Scholar]
- Wang, M, Wu, B, Tucker, JD, Lu, P, Cloer, C and Lu, Q (2014). Evaluation of Tris[2-(acryloyloxy)ethyl]isocyanurate cross-linked polyethylenimine as antisense morpholino oligomer delivery vehicle in cell culture and dystrophic mdx mice. Hum Gene Ther 25: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M, Wu, B, Tucker, JD, Lu, P and Lu, Q (2015). Tris[2-(acryloyloxy)ethyl]isocyanurate cross-linked polyethylenimine enhanced exon-skipping of antisense 2ntOmethyl phosphorothioate oligonucleotide in vitro and in vivo. J Nanomed Nanotech 6: 261. [Google Scholar]
- Wang, M, Wu, B, Tucker JD, Lu, P and Lu, Q (2016). Poly(ester amine) constructed from polyethylenimine and pluronic for gene delivery in vitro and in vivo. Drug Delivery. DOI: 10.3109/10717544.2016.1162877. [DOI] [PubMed]
- Huang, Y, Rao, Y, Chen, J, Yang, VC and Liang, W (2011). Polysorbate cationic synthetic vesicle for gene delivery. J Biomed Mater Res A 96: 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta, DA and Sosnik, A (2007). Poly(ethylene oxide)-poly(propylene oxide) block copolymer micelles as drug delivery agents: improved hydrosolubility, stability and bioavailability of drugs. Eur J Pharm Biopharm 66: 303–317. [DOI] [PubMed] [Google Scholar]
- Sun, J, Zeng, F, Jian, H and Wu, S (2013). Conjugation with betaine: a facile and effective approach to significant improvement of gene delivery properties of PEI. Biomacromolecules 14: 728–736. [DOI] [PubMed] [Google Scholar]
- Alshamsan, A, Haddadi, A, Incani, V, Samuel, J, Lavasanifar, A and Uludağ, H (2009). Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol Pharm 6: 121–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.