Abstract
A naturally occurring 32-base pair deletion of the HIV-1 co-receptor CCR5 has demonstrated protection against HIV infection of human CD4+ T cells. Recent genetic engineering approaches using engineered nucleases to disrupt the gene and mimic this mutation show promise for HIV therapy. We developed a megaTAL nuclease targeting the third extracellular loop of CCR5 that we delivered to primary human T cells by mRNA transfection. The CCR5 megaTAL nuclease established resistance to HIV in cell lines and disrupted the expression of CCR5 on primary human CD4+ T cells with a high efficiency, achieving up to 80% modification of the locus in primary cells as measured by molecular analysis. Gene-modified cells engrafted at levels equivalent to unmodified cells when transplanted into immunodeficient mice. Furthermore, genetically modified CD4+ cells were preferentially expanded during HIV-1 infection in vivo in an immunodeficient mouse model. Our results demonstrate the feasibility of targeting CCR5 in primary T cells using an engineered megaTAL nuclease, and the potential to use gene-modified cells to reconstitute a patient's immune system and provide protection from HIV infection.
Keywords: Gene therapy, gene editing, HIV, megaTAL, CCR5
Introduction
Acquired Immunodeficiency Syndrome (AIDS) is caused by infection with the retrovirus Human Immunodeficiency Virus (HIV) and the subsequent collapse of the patient's immune system. In the last two decades, anti-retroviral therapies (ARTs) have been dramatically successful in controlling viral titers in infected individuals, and thus extending their life expectancy.1,2 These regimens also reduce rates of viral transmission between individuals.3 However, ART has significant disadvantages, including substantial drug-associated side effects, a lifetime commitment to a strict and expensive drug regimen, and the inability to eliminate the HIV reservoir.4,5
HIV requires two of its host's cell-surface receptors, CD4 and a G-protein coupled receptor, most commonly CCR5, to fuse to and infect human T cells. A naturally-occurring CCR5 allele, containing a 32-base pair deletion (CCR5Δ32), produces a premature stop codon that prevents surface expression of CCR5 and confers HIV resistance.6,7,8 Although studies have shown that this allele may be associated with an increased risk for symptomatic West Nile virus infection9, it has not been associated with other significant health impacts. In European populations, heterozygote and homozygote carriers occur at a frequency of ~10% and 1%, respectively.10 Over the past decade, nuclease-mediated gene editing has been pursued as a potential therapeutic strategy designed to mimic the CCR5Δ32 phenotype. The Berlin patient, an HIV-positive male with leukemia who underwent two bone marrow transplants using a CCR5Δ32 homozygous donor, has demonstrated sustained viral control in the absence of ART,11,12 thereby highlighting the importance of this mutation in a transplant setting. Shortly after, several additional subjects were treated by receiving bone marrow from donors lacking the protective CCR5Δ32 allele, based upon the rationale that myeloablative conditioning prior to transplantation combined with graft-versus-host disease may be enough to eradicate the HIV reservoir.13,14 These subjects appeared to control the virus in the absence of ART in the early post-transplant period, but in contrast to the Berlin patient's outcome, the virus eventually rebounded,13 supporting the importance of the homozygous CCR5Δ32 donor cells in controlling HIV infectivity.
The cases described above demonstrate the importance of the CCR5Δ32 mutation in a transplant setting; mimicking the CCR5Δ32 phenotype using nuclease-mediated gene disruption is thus being pursued as a therapeutic strategy for HIV. Rare-cleaving nucleases are engineered to bind and cleave at a DNA sequence of interest, introducing double-strand breaks which the cell may repair using the non-homologous end-joining (NHEJ) pathway. This repair pathway is error-prone and frequently results in mutation-causing insertions and deletions (indels) at the break site. Several groups are developing methodologies to use a zinc finger nuclease to disrupt CCR5 in T cells or CD34+ hematopoietic stem cells ex vivo for autologous transplantation.15,16,17,18,19,20,21,22,23 A recent phase 1 clinical trial transferring autologous CCR5 zinc finger nuclease-treated T cells to HIV-positive patients showed improvements in peripheral CD4 T cell numbers and decreased viral load during ART interruption.21 The patient with the longest delay in HIV resurgence was retrospectively identified as being heterozygous for the CCR5Δ32 allele, reinforcing the importance of efficient and bi-allelic gene disruption for producing CCR5− cells that would be resistant to HIV infection and allow patients to control viremia in the absence of ART.
By fusing a reprogrammed homing endonuclease (HE), also known as a meganuclease, to a transcription activator-like effector (TALE) DNA binding domain, we have developed a hybrid nuclease platform, called a megaTAL, targeting the CCR5 gene.24 We previously showed that this nuclease exhibits a high level of NHEJ and could be used to achieve targeted gene delivery at CCR5 via homologous recombination in primary human T cells.25 In this study, we evaluated the efficiency of this nuclease to disrupt CCR5 and subsequently protect cells from HIV infection in vitro. We also investigated the ability of this nuclease to protect CD4+ T cells during active HIV-1 infection in vivo using immunodeficient mice. Our study is an important step toward the ultimate goal of providing a population of immune cells that are resistant to HIV-1 infection, that could be used to reconstitute the patient's immune system.
Results
Successful reprogramming of the I-OnuI HE to target CCR5
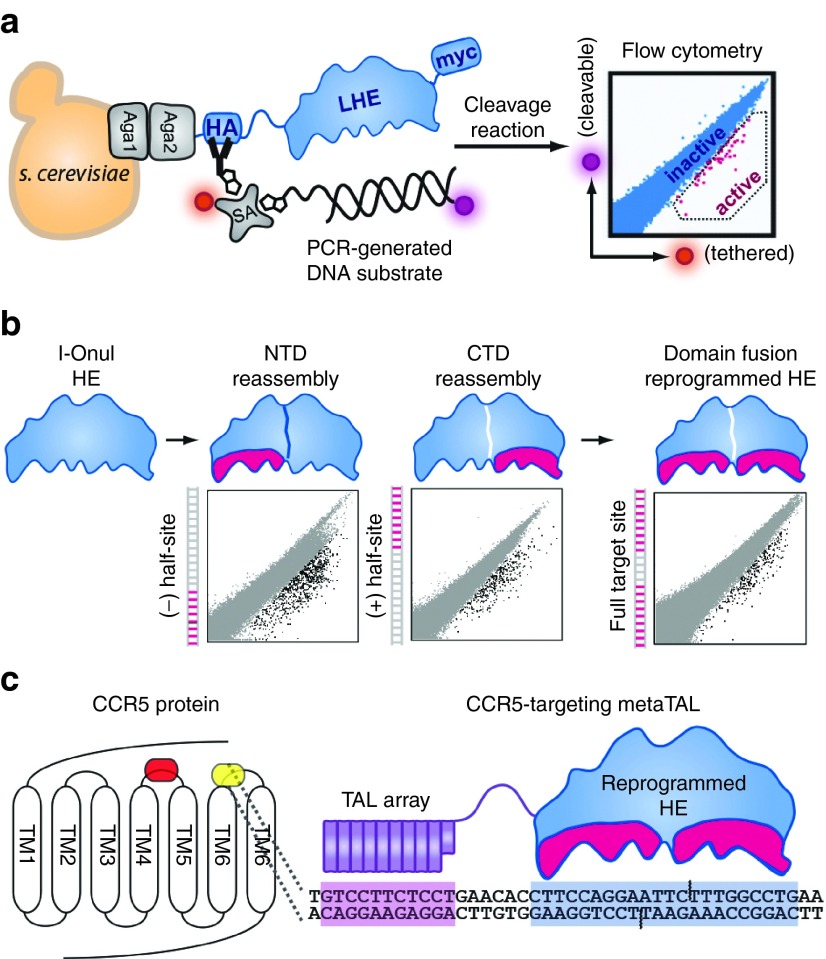
Yeast surface display technology can be used to reprogram the specificity of LAGLIDADG-motif homing endonucleases (LHEs), also known as meganucleases (Figure 1a).26 A target sequence in CCR5 was identified which comprised the central-4 binding motif of the LHE, I-OnuI, a sequence required for efficient DNA hydrolysis and double-stranded break formation. The enzyme's C-terminal domain and N-terminal domains were reprogrammed separately by screening degenerate libraries harboring mutations in the DNA recognition interface of each domain (Figure 1b). Following domain reprograming, pools of successfully reprogrammed domains were fused and screened to arrive at a fully reprogrammed HE that could recognize the target CCR5 sequence (Figure 1c). The reprogrammed LHE was subsequently assembled to a TALE DNA binding domain via a flexible linker; this megaTAL architecture was utilized to increase the binding constant (Kon) of the meganuclease. This approach has been successful in generating similar reagents which exhibit desirable characteristics for translatable applications.24,26,27
Figure 1.
Assembly of a reprogrammed megaTAL targeting the CCR5 gene. (a) Schematic depiction of yeast surfaced display technology used to reprogram the specificity of LAGLIDADG-motif homing endonuclease (LHE) I-OnuI. (b) The N-terminal (NTD) and C-terminal domains (CTD) of the LHE component of the megaTAL were reprogrammed separately to achieve activity against 9-base pair “half-sites” that were subsequently assembled to produce the fully reprogrammed enzyme recognizing the complete CCR5 target site. (c) CCR5 is a G-protein coupled receptor with seven transmembrane (TM) domains whose extracellular facing loops comprise, in part, the binding interface used by HIV-1 to gain entry to CD4+ T cells. We targeted the third extracellular loop (ECL3) located between the sixth (TM6) and seventh (TM7) transmembrane domains (yellow shading), downstream of the CCR5Δ32 deletion (orange shading). This region was selected for disruption, as this loop contains a cysteine residue that contributes to the structural integrity of CCR5 and its ability to function as an HIV-1 coreceptor.52 The megaTAL targeting the CCR5 ECL3 loop is shown schematically, with a modular transcription activator-like effector array (purple) linked to the reprogrammed I-OnuI LHE (HE; blue) and their corresponding target sequences separated by 6 base pairs of noncontacted DNA. The amino acid sequence for the CCR5 megaTAL has been previously published.25 Aga1 and Aga2, subunits of a-agglutinin heterodimer; HA, hemagglutinin; SA, streptavidin; Myc, c-myc tag.
megaTAL disruption of CCR5 in a reporter cell line prevents CCR5 expression and HIV entry
We previously reported the use of the CCR5 megaTAL nuclease to disrupt the CCR5 locus in primary human T cells.25 To determine whether CCR5 megaTAL disruption of CCR5 confers HIV resistance, we simultaneously tested its abilities to disrupt cell surface expression of the protein and to provide protection from live HIV infection using the GHOST(3) CCR5+ (GHOST Hi-5) cell line, a human osteosarcoma cell line stably transduced with human CD4 and CCR5 expression cassettes to allow HIV infection, as well as an HIV-2 long terminal repeat green fluorescent protein (LTR-GFP) reporter (activated by TAT expression) to indicate HIV infection.28 We used electroporation to introduce capped and poly-A tailed mRNA encoding the CCR5 megaTAL or blue fluorescent protein (BFP; negative control) in these cells. Untransfected control cells and cells receiving BFP mRNA retained equivalently high levels of surface CCR5 expression by flow cytometry 1 week following electroporation (98.8% and 98.3%, respectively). In contrast, cells electroporated with CCR5 megaTAL mRNA had a mean CCR5 expression of 13.5% (Figure 2a), consistent with disrupted expression in 87% of engineered cells.
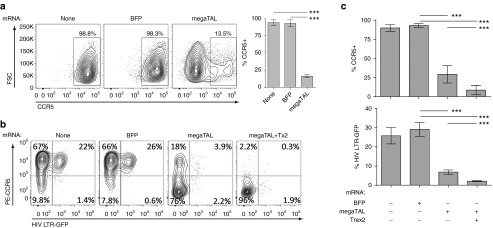
Figure 2.
megaTAL efficiently disrupts CCR5 in Ghost Hi-5 cells and protects against HIV in vitro. (a) Representative flow cytometry data (left) and statistical representations (bar graphs; right) of CCR5 surface expression in an untransfected sample or samples electroporated with either blue fluorescent protein or CCR5 megaTAL mRNA (n = 3). (b) Representative flow cytometry data showing surface CCR5 expression versus intracellular GFP expression. Phycoerythrin-labeled anti-CCR5 antibody was used to assess CCR5 surface expression (vertical axis), and HIV infection was tracked by HIV-2 LTR-driven GFP expression (horizontal axis) in Ghost Hi-5 cells infected 1 week following mRNA transfection using the CCR5-tropic virus HIV BaL. HIV infection preferentially occurs in CCR5+ cell populations (TR). (c) Statistical representations (n = 6) of Ghost Hi-5 cells shown in b, showing CCR5 surface staining (upper panel) and GFP expression (lower panel). All data represent mean ± SD. P values calculated using Student's t-test. Results of statistical analyses in this and subsequent figures as follows: *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant.
To verify that this CCR5 disruption was protective against HIV infection, we transfected GHOST Hi-5 cells with BFP, CCR5 megaTAL, or CCR5 megaTAL and Trex2 mRNA. Trex2 is a DNA end-processing enzyme that increases mutagenic NHEJ versus precise end-joining following nuclease cleavage.29 Transfection of Ghost Hi-5 cells with mRNA resulted in ~90% efficiency in BFP marker gene expression with >80% viability (Supplementary Figure S1a). One week following mRNA transfection, we added an R5 tropic virus, HIV-1Ba-L, to the cell culture media; R5 viruses were previously shown to induce peak LTR-driven GFP expression in GHOST Hi-5 cells 48-hours postprimary infection.28 A high proportion of untreated and BFP mRNA-treated control cells retained surface expression of CCR5, totaling 89% and 92%, respectively (Figure 2b,c, summing top right and top left quadrants), and 23–27% of these cells expressed the GFP reporter indicative of HIV infection at the 48-hour time point (Figure 2b,c, summing top right and bottom right quadrants). Alternatively, cells transfected with the CCR5 megaTAL alone or with Trex2 mRNA were only 22% and 2.5% CCR5+, respectively (top right and top left quadrants), with fewer HIV-infected (GFP+) cells (6.1% and 2.2%, respectively in top right and bottom right quadrants). Overall, the CCR5 megaTAL disrupted CCR5 surface expression in ~85% of GHOST Hi-5 cells, reducing HIV infection by ~80%; a process that was further enhanced by the addition of Trex2. HIV infection was restricted to the unmodified, CCR5hi populations. While the presence of synthetic DNA or RNA in the cytosol could activate antiviral effects that might interfere with HIV infection, no statistically significant difference was seen in infection rates in control mRNA-treated cells. This suggests that the reduction of GFP expression (and therefore HIV infection) seen in the megaTAL and megaTAL + Trex2 samples is not a result of mRNA transfection. These results are consistent with the hypothesis that the CCR5 megaTAL disrupts CCR5 in a manner that will mimic the CCR5Δ32 allele, providing protection from HIV infection by preventing CCR5 expression on the cell surface.
CCR5 megaTAL efficiently disrupts CCR5 in primary human CD4+ T cells
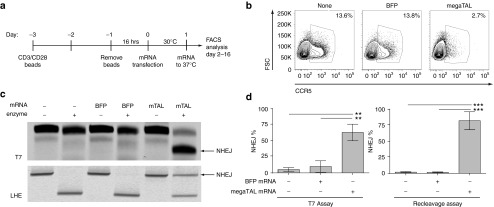
Having demonstrated that megaTAL-induced CCR5 disruption can prevent in vitro HIV infection in the Ghost Hi-5 cell line, we wanted to demonstrate the efficacy of this approach in primary human T cells. We previously reported gene modification (8–60%) at CCR5 in primary human T cells using this megaTAL alone or in association with adeno-associated virus-delivered donor DNA templates.25 Using the procedure shown (Figure 3a), we found that we could transfect primary human CD4+ T cells with mRNA with high efficiency without compromising viability (Supplementary Figure S1b). Transfection of primary human T cells with CCR5 megaTAL mRNA resulted in ~80% fewer CCR5+ cells by surface staining 4 days after transfection in comparison with either untransfected or BFP mRNA-transfected cells (Figure 3b). Note that CCR5 expression is highly variable among T cell developmental subsets and in resting versus stimulated cells.30 Therefore, genomic DNA (gDNA) was collected from transfected cells for determination of cutting rates (measured as indels) using two alterative molecular assays. First, we used the T7 endonuclease I (T7E1) assay, and identified 63% of alleles with indels in megaTAL mRNA-treated cells compared with a background level of 5–10% in untreated and BFP mRNA-treated cells. Of note, the uniform spectrum of indels generated by megaTAL cleavage (consisting primarily of 1–4 base pair deletions)25 is predicted to lower the frequency of DNA-mismatch bubble formation, the substrate for T7EI. Thus this assay is likely to underrepresent the indel readout. Deletions at this site are predicted to abolish HE cleavage due to the critical importance of the central four (C4) nucleotide positions.31 Hence, we also utilized a re-cleavage assay (RCA) to assess NHEJ frequency (Supplementary Figure S2). As opposed to measuring heteroduplex formation (as in the T7E1 assay), the RCA quantifies in vitro nuclease digestion of a genomic polymerase chain reaction (PCR) amplicon that contains the megaTAL target site, using a recombinant LHE protein identical to that used to create the CCR5 megaTAL. Because LHEs have binding-dependent activity, a mutation at the CCR5 site arising from an NHEJ event in cellulo would abolish LHE activity in vitro. By RCA, we observed background NHEJ rates of <2% in untreated and BFP mRNA-transfected cells and 60–80% NHEJ at CCR5 in CCR5 megaTAL mRNA-treated cells (Figure 3c,d). Thus, based upon both assays, the CCR5 megaTAL lead to 60–80% CCR5 gene disruption in primary human T cells.
Figure 3.
megaTAL efficiently disrupts CCR5 in primary CD4+ cells. (a) Timeline representing workflow in primary CD4+ cells relative to time of transfection (t = 0), beginning with the addition of anti-CD3/CD28 beads on cryo-preserved CD4+ T cells. (b) Surface staining of CCR5 in primary CD4+ cells comparing expression in untreated cells or cells transfected with BFP or CCR5 megaTAL mRNA. (c) Representative agarose gels quantifying CCR5 modification by T7 endonuclease assay (surveyor assay; top panel) or by re-cleavage assay (RCA) using a fluorescently labeled forward primer (lower panel). (d) Molecular quantification (n = 3) by T7 (left) or RCA (right) of CCR5 disruption in primary CD4+ cells. Values are calculated from fluorescent densitometry (%NHEJ = NHEJ band/sum of NHEJ + undisrupted bands).
Stable engraftment of gene modified primary T cells in vivo
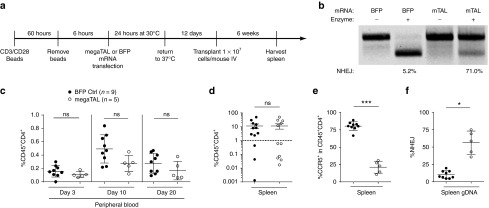
We next tested the engraftment and subsequent stability of megaTAL-treated cells in immunodeficient, NOD-SCID-IL-2Rγnull (NSG) mice (Figure 4a). Mice were transplanted with primary human CD4+ T cells that were treated with either CCR5 megaTAL or control BFP mRNA. CCR5 megaTAL-treated input cells were modified at 71% of CCR5 alleles by RCA, with an assay background of 5% in BFP mRNA-treated cells (Figure 4b). Six weeks post-transplant, we found that some of the mice did not meet our criteria of engraftment (>1% of splenic cells hCD45+CD4+) and results from these mice were excluded from additional analyses (including six animals in the megaTAL-treated and three in BFP control cohorts, respectively). We identified hCD45+CD4+ cells in peripheral blood samples throughout the experiment in both treatment groups and found no significant differences in the frequency of circulating hCD45+CD4+ (Figure 4c). Similarly, we found equivalent levels of splenic engraftment as measured by hCD45+CD4+ expression in BFP and CCR5 megaTAL mRNA-treated cells (11.3% and 11.4%, respectively; Figure 4d). As predicted, engrafted hCD45+CD4+ showed significantly lower levels of CCR5 surface expression in megaTAL-treated cells relative to control BFP mRNA-treated cells (21.2% and 80.4%, respectively; Figure 4e). To determine if reduced CCR5 expression was a result of megaTAL-mediated gene modification at CCR5, we interrogated splenic gDNA using the RCA assay. Control animals exhibited a mean CCR5 disruption rate of 10.2%, while animals transplanted with megaTAL mRNA-treated cells exhibited a mean disruption rate of 56.3% (Figure 4f), confirming the stability of cells with megaTAL-mediated CCR5 disruption in vivo.
Figure 4.
Primary CD4+ cells modified by CCR5 megaTAL engraft in the immunodeficient NSG murine model and show stable modification of CCR5. (a) Timeline representing workflow for NSG mouse experiment. (b) Re-cleavage assay (RCA) of input cells shows 71% modification at CCR5 in cells treated with CCR5 megaTAL mRNA and 5.2% background modification in cells transfected with blue fluorescent protein. (c) Mice were bled on days 3, 10, and 20 post-transplant. Surface staining of hCD45 and hCD4 was used to track engraftment in peripheral blood over time. (d) Six weeks post-transplant, mice were killed and spleens were harvested. Splenic engraftment of human T cells was tracked through surface staining of hCD45 and hCD4. (e,f) Samples with >1% splenic T engraftment were subsequently analyzed for: (e) surface expression of CCR5 by flow cytometry and (f) molecular quantification of CCR5 disruption by RCA analysis of genomic DNA (gDNA).
Gene modification protects CD4+ T cells from in vivo HIV infection
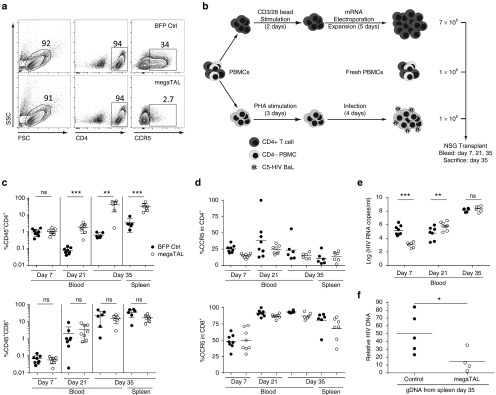
Having established the stability of CCR5 modified cells in vivo in the absence of HIV, we wanted to challenge these engineered T cell populations using the same in vivo system in the context of an HIV infection. As above, we modified primary human CD4+ T cells by transfection with either BFP control mRNA or CCR5 megaTAL mRNA. By surface staining, BFP and CCR5 megaTAL mRNA-treated CD4+ cells were 34% and 2.7% CCR5+, corresponding to a relative disruption level >90% in megaTAL-treated T cells (Figure 5a). We transplanted each mouse with 7 × 106 gene edited or control primary CD4+ T cells, 1 × 106 infected CD4-depleted peripheral blood mononuclear cells (PBMCs), and 1 × 106 freshly thawed PBMCs (see workflow, Figure 5b) from the same donor to prevent toxicity resulting from mixed lymphocyte reactions.32 Throughout the transplant, human CD4+ T cells and HIV levels in peripheral blood were monitored over time as shown in Figure 5b.
Figure 5.
megaTAL editing of CCR5 in primary CD4+ T cells protects against HIV in vivo in an NSG murine model. (a) Flow cytometry data of primary CD4 cells electroporated with either blue fluorescent protein or CCR5 megaTAL mRNA (day 5 post-electroporation). Data show surface staining for hCD4 and CCR5 prior to transplant. (b–f) NSG mice were transplanted with 7 × 106 modified or control CD4+ cells, 1 × 106 HIV-BaL infected CD4-depleted peripheral blood mononuclear cells (PBMCs), and 1 × 106 freshly thawed PBMCs from the same donor. (b) Experimental workflow detailing manipulation of transplanted cells represented. Mice were bled on days 7, 21, and 35 and subsequently killed on day 35 for spleen harvesting. (c) Engraftment of CD4+CD45+ (top) and CD8+CD45+ (bottom) T cells tracked in blood and spleens. (d) Surface expression of CCR5 in CD4+ cells (top) and CD8+ cells (bottom) tracked in blood and spleens. (e) HIV plasma viremia was tracked with quantitative PCR (qPCR) throughout transplant. (f) HIV qPCR in genomic DNA from human CD4+ cells isolated from the spleen 35 days post-engraftment, showing the viral load in both experimental groups. P < 0.05.
The frequency of hCD45+CD3+CD4+ cells in circulating blood was significantly higher in mice receiving megaTAL-treated cells than those receiving control cells by days 21 and 35 post-transplant/infection. This difference was also observed in splenic cells, where human CD4+ cells represented 31.8% and 3.4% of total cells in megaTAL or control recipient animals, respectively. By comparison, no statistical difference in hCD45+CD3+CD8+ cells was observed at any time in blood or spleens in either group (17.1% and 31.4%; megaTAL and BFP mRNA, respectively; Figure 5c). This suggests that in the presence of HIV-BaL, megaTAL-treated cells have a selective advantage relative to BFP mRNA control cells in the presence of HIV-1Ba-L. Within hCD45+ T cells, surface expression of CCR5 was indistinguishable between megaTAL- and BFP-treated cells in either CD4+ or CD8+ cells (Figure 5d). In this experiment, all CD4+ T cells are expected to have low levels of CCR5 expression due to either HIV infection of CCR5+ cells or due to megaTAL disruption of the locus. Consistent with this idea, we noted that the low levels of CCR5 observed in CD4+ cells in HIV infection experiments were not observed in the absence of HIV (Figure 4e). By comparison, untransfected CD8+ cells maintained a CCR5 expression level similar to those seen by untreated cells in the absence of HIV. On day 35, we observed a 100-fold increase in the proportion of CD4 cells present in recipients of megaTAL-treated compared with control-treated T cells in the presence of HIV. Together, these data show that HIV-1Ba-L exhibits cytotoxic properties on wild-type CCR5+CD4+ cells in this model; this negative selection is prevented by megaTAL disruption of CCR5.
In addition to analyzing cell populations in circulating blood, we collected serum at each time point for analysis by quantitative PCR. On day 7, when there were equivalent CD4+ proportions in recipients of both megaTAL and BFP control mRNA-treated cells, recipients of megaTAL-treated cells exhibited lower plasma viremia (Figure 5e). Interestingly, at day 35, the group receiving the megaTAL-treated cells had a 10-fold greater proportion of circulating human CD4+ T cells (Figure 5c), yet with statistically equivalent plasma viremia. Analysis of CD4+ T cells purified from the spleens of control or megaTAL-treated mice showed relatively reduced levels of HIV provirus in megaTAL-treated CD4+ T cells. (Figure 5f).
Discussion
Here we demonstrate the development of a megaTAL nuclease targeting CCR5. We show that the CCR5 megaTAL nuclease disrupts the expression of CCR5 and can prevent HIV infection in cell lines. The CCR5 megaTAL also disrupts CCR5 in primary human CD4+ T cells with a high efficiency, and we show that these gene-modified T cells stably engraft in immunodeficient mice. Furthermore, we demonstrate that CCR5 megaTAL-modified T cells are preferentially expanded during HIV-1 infection in vivo.
CCR5Δ32 homozygous individuals are nearly completely protected against R5-tropic HIV-1 infection, while those who are heterozygous show only modest improvements in protection and AIDS progression.33 As a T cell therapy, a phase 1 clinical trial evaluating the safety of zinc finger nucleases (delivered by adenovirus) to disrupt CCR5 suggested that bi-allelic disruption may be necessary for successful clinical applications of this technology.21 In generating a nuclease for disrupting the CCR5 locus, we strove to develop an enzyme and delivery platform with a high targeting efficiency, to improve the frequency of cells with bi-allelic modifications.
LHEs have several characteristics that make them desirable nucleases in the development of therapeutics involving gene-editing. Prior to the development of the megaTAL architecture,24 the lower binding constant (Kon) of LHE's relative to the DNA-binding features of other gene editing nucleases has limited their editing efficiency. The addition of the TALE DNA binding domain, effectively adding an anchor to the nuclease domain, resolved this limitation, and indeed megaTALs can achieve nearly complete allelic editing efficiencies.34 Furthermore, unlike the TALEN and zinc finger nucleases that require dimerization, and CRISPR/Cas9 nucleases that require codelivery of a guide-RNA, megaTALs are monomeric and can achieve gene modification using a single cistron, a feature that may promote their application to clinical settings requiring scalability and ease of delivery.
We chose to deliver the megaTAL coding sequence by transient transfection of mRNA for several reasons. mRNA can be efficiently introduced into primary T cells using electroporation and achieves high levels of reporter mRNA expression for short periods of time with little loss of cell viability (Supplementary Figure S1b).25 Unlike many viral delivery methods, mRNA transfection avoids safety risks due to genomic integration, as well as long-term expression of the nuclease, which could increase the risk of off-target cutting and cell toxicity. Other methods of transferring a coding sequence that avoid genomic integration include integration-deficient lentivirus and recombinant adenovirus, which would be more expensive to produce for clinical scale applications35 and may elicit immune responses in patients.36,37 Finally, mRNA production and cell delivery technology has already been established under large-scale good manufacturing practice conditions.38
Here we report CCR5 disruption rates in primary T cells of ~60–80%, judging by molecular (RCA) analyses of gDNA and flow cytometry of CCR5 surface expression. These results are slightly higher than rates of HDR-mediated gene knock-in of a GFP expression cassette into CCR5 using this megaTAL.25 Importantly, in the previous study, up to 80% of the modified cells had bi-allelic homology-driven modifications at CCR5. In order to confirm that this megaTAL-mediated disruption of CCR5 is protective against HIV infection, we challenged CCR5 megaTAL nuclease-treated cells with replication-competent virus using several models. We first used the GHOST Hi-5 cell line in vitro model. Although the expressed CCR5 in these cells is produced from randomly integrated expression cassettes, rather than the endogenous loci, in both cases CCR5 disruption should result in similar downstream effects: the introduction of premature missense or nonsense mutations in CCR5 coding sequence and the loss of surface expression. Note that the megaTAL target site is within the last exon of CCR5, so nonsense-mediated RNA decay is not considered a possible mechanism for mutation-induced loss of protein at the cell surface whether at the endogenous locus or in the cDNA transgene. Despite having more integrated CCR5 alleles than a primary cell, we found that at least 85% of GHOST Hi-5 cells lost CCR5 expression at the cell surface by flow cytometry, and the percentage of cells infected by HIV diminished by over 70%, confirming the effectiveness of our targeting strategy.
Our next HIV infection model was performed in vivo using humanized mice. In uninfected NSG mice, CCR5 megaTAL-edited T cells had equivalent engraftment and stability relative to control cells. While a slightly higher number of mice transplanted with CCR5 megaTAL-treated cells failed to engraft (six mice in the CCR5 megaTAL treated versus three in the control cohort), the overall level of engraftment in both groups was equivalent. In previously published data, our group has shown that CCR5 megaTAL-edited T cells engraft equivalently to unmodified T cells.25 In the absence of HIV, the fraction of engrafted human T cells that were CCR5hi in peripheral blood was significantly decreased in the CCR5 megaTAL cohort relative to mice receiving control cells, evidence that the gene editing strategy produces a viable cell product with the expected phenotype. In the presence of HIV, mice that received megaTAL-treated cells had 100-fold more circulating human CD4+ T cells and a 10-fold increase in splenic human CD4+ cells at the 35-day endpoint than mice in the control cohort. Our approach successfully selected for CD4+CCR5− cells and reduced the amount of integrated HIV DNA in edited splenic cells, reproducing HIV challenge data from other groups that were using a zinc finger nuclease targeted to ECL1 to disrupt CCR5 in human T cells or in CD34+ hematopoietic stem cells.15,16,17,18,19,20,21,22,23,32 However, while the CCR5 megaTAL cohort repressed blood viremia early on in the in vivo experiment, viremia equalized between experimental groups by the end of the study. We speculate this is likely due to the cotransfer of monocytes in PBMCs with the monocyte-tropic R5 virus as well as the delayed kinetics of HIV infection in megaTAL-treated cohort, resulting in the expansion and subsequent infection of unsuccessfully edited cells, and potentially leading to an increase in total viremia. Although we were able to demonstrate that megaTAL-modified CD4+ T cells preferentially expand in the context of HIV infection in vivo, the progression of HIV infection in humans is not replicated in these mice. Notably, the CCR5-megaTAL binding site is conserved and could be used to disrupt CCR5 in pigtail macaques, a nonhuman primate species where the SHIV (a simian immunodeficiency virus envelope with HIV genetic core) has been used to model human HIV infection. Hence, future studies in our group will investigate CCR5 disruption and subsequent protection from SHIV in a macaque model.
The disruption of CCR5 in the context of HIV raises concerns of HIV tropism. In an attempt to mimic the “Berlin Patient” cure strategy, the “Essen Patient”39 received a single bone marrow transplant and was removed from ART, resulting in the emergence of CXCR4 (X4)-tropic viral variants. Recent studies using recombinant adenoviral vector delivered zinc finger nucleases have demonstrated the feasibility of stably disrupting both CXCR4 and CCR5 in CD4 T cells to prevent dual-tropic HIV infection.18 A similar dual-receptor targeting approach could be implemented with the simple addition of a second mRNA, encoding a CXCR4 engineered megaTAL, to the electroporation reaction.
The megaTAL nuclease platform has other potential applications for HIV therapies beyond CCR5 disruption in T cells (and mobilized CD34+ cells).25 We have previously introduced several clinically relevant gene products into T cells at the CCR5 locus, including expression cassettes for the C46 peptide,40 which acts as a fusion inhibitor for HIV, and an anti-HIV envelope chimeric antigen receptor (CAR). It has been shown that partial suppression of CCR5 and expression of the C46 peptide can have synergistic benefits to protect cells from R5-tropic HIV41,42,43 or X4 and dual-tropic HIV infections.44 Thus, surface expression of C46 from the CCR5 locus as a result of homology-driven integration is likely to result in multi-tropic HIV protection.
Finally, a major challenge in HIV cure efforts is the development of a therapy that eliminates the reservoir of HIV-1 that persists in latently infected cells. Our group has successfully delivered several CARs, including an anti-HIV envelope CAR25 that selectively activates modified T cells when cocultured with HIV-infected cells in vitro.25 Concerns have been raised that the addition of an HIV-recognizing epitope on CD8+ T cells will mediate HIV infection of effector cells.45,46,47,48 By delivering the HIV-CAR into the CCR5 locus, effector cells might be protected from subsequent infection, increasing the persistence and potency of cellular therapies in the context of HIV. The ability to deliver these gene cassettes could contribute to a combinatorial gene therapy approach that could contribute to virus eradication.
In conclusion, we have used a novel nuclease platform to achieve up to 80% disruption of CCR5 in primary human CD4+ T cells. These cells engraft and traffic normally after transplantation into humanized mice and provide protection from HIV-1 infection. While the clinical safety, efficacy, and implementation of therapies using gene-modified T cells need further investigation, our data highlight the potential of this therapy for the treatment of HIV.
Materials and methods
megaTAL nuclease design. The CCR5 megaTAL enzyme was designed by bluebird bio, Inc. using a proprietary method relying on directed evolution and its amino acid sequence has been previously described.25 The enzyme recognizes a 38-base pair stretch of sequence located in the sixth transmembrane domain of the CCR5 gene.
Primary CD4+ T cell isolation and culture. Blood was drawn from consented adult healthy donors at Seattle Children's Research Institute in accordance to IBC protocols. Primary CD4 cells were isolated from either whole blood using RosetteSep Human CD4+ T Cell Enrichment Kit (StemCell Technologies, Vancouver, Canada) or from PBMCs using the Human CD4+ T Cell Isolation Kit (Miltenyi Biotec, Auburn, CA). Cells were grown in RPMI (Hyclone, Logan, Utah) supplemented with 20% fetal bovine serum, and human cytokines IL-2 (50 ng/ml), IL-7 (5 ng/ml), and IL-15 (5 ng/ml; all from PeproTech) and cultured at 0.5 × 106–3 × 106 cells/ml. Cells were activated using Dynabeads Human T-Activator CD3/CD28 (LifeTechnologies) for 48 hours at a 1:1 cell to bead ratio.
mRNA production and electroporation. CCR5 megaTAL and BFP mRNA coding sequences were manufactured as previously described by our group and electroporated in a similar fashion.25
Isolation of gDNA and PCR amplification. gDNA was extracted from 0.5 × 106 to 1.5 × 106 cells using a QiaCube (Qiagen) and following manufacturer's recommended settings and provided reagents. gDNA was eluted and stored in Qiagen Buffer AE. To amplify the CCR5 locus for NHEJ assessment, 2 µl of a 10 ng/µl gDNA dilution was mixed with 48 µl of Platinum HiFi master mix (1× Platinum HiFi buffer, 10 mmol/l dNTPs, 50 mmol/l MgSO4, 10 mmol/l each forward (TCATTACACCTGCAGCTCTC) and A.647 conjugated reverse (CAGTGGATCGGGTGTAAACTG) primers, 0.4 µl Platinum HiFi Taq (2.5 U/µl, LifeTechnologies, Carlsbad, CA). PCR was performed with denaturation at 94 °C for 2 minutes followed by 35 cycles of 94 °C for 20 seconds, 60 °C for 20 seconds, 68 °C for 1 minute, and a final extension at 68 °C for 10 minutes. PCR products were purified using QiaQuick PCR Purification Kit (Qiagen, Hilden, Germany).
Endonuclease assays. Indel frequency at the megaTAL cleavage site was measured using a T7E1 assay as described25 and using a RCA. For the T7E1 assay, 100 ng of PCR product were denatured and re-annealed in 1× New England Biolabs Buffer 2 at a total volume of 10 µl. Samples were then incubated at 37 °C for 1 hour in T7 endonuclease I (10,000 U/ml, New England Biolabs, Ipswich, MA). The reactions were terminated by adding 500 mmol/l ethylenediaminetetraacetate. For the RCA, 100 µl of PCR product was incubated at 37 °C for 16 hours in CCR5 HE and Cleavage buffer (400 mmol/l Tris Acetate, 200 mmol/l potassium acetate, 2 mmol/l dithiothreitol, 20 mmol/l MgCl2). Reactions were terminated by addition of buffer with 50 mmol/l Tris pH 8.0, 5 mmol/l ethylenediaminetetraacetate, 0.5% sodium dodecyl sulfate, 0.5 mg/ml Proteinase K, and 25% glycerol. Reaction products from both assays were run on an 0.8% agarose gel, imaged using Odyssey (LI-COR Biosciences, Lincoln, NE), and bands containing the A.647 labeled reverse primer were quantified using the ImageStudioLite (LI-COR Biosciences).
Flow cytometry. Cells were analyzed on an LSR II or Canto 2 (BD Biosciences, San Jose, CA) and analyzed using FlowJo X (Treestar, Ashland, OR). Cells were stained at 4 °C for 10 minutes in FACS buffer (phosphate-buffered saline, 2% fetal bovine serum). The following antibodies from BD Bioscience were used: CCR5 clone 3A9, CD4 clone OKT4, and CD3 clone OKT3. Dead cells were excluded from analysis using Near-IR Fixable Live/Dead stain (LifeTechnologies).
Virus production. Replication competent HIV-1Ba-L was from Suzanne Gartner, Mikulas Popovic, and Robert Gallo49,50 and obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. HIV-1Ba-L was propagated in PHA-activated human PBMCs, the supernatant was collected and sterile filtered (0.22 μm Steriflip, EMD Millipore, Darmstadt, Germany) on days 3 and 5 postinoculation. The virus was titered in GHOST Hi-5 cells against a virus standard.
HIV-1 assay in GHOST Hi-5 cells. GHOST Hi-5 cells expressing multiple copies of CCR5 were from Dr Vineet N. KewalRamani and Dr Dan R. Littman28 and obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. They were maintained in complete RPMI with 20% fetal bovine serum and supplemented with 500 μg/ml G418, 100 μg/ml hygromycin, and 1 μg/ml puromycin. Cells were infected with HIV-1Ba-L at a multiplicity of infection of 15 and harvested using Accutase buffer (LifeTechnologies) at 48 hours. HIV-infected samples were fixed in 1% paraformaldehyde in neutral-buffered saline prior to flow cytometry.
T cell transfer and HIV infection mouse models. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice obtained from Jackson laboratories, Bar Harbor, ME, were housed in specific pathogen free facilities at either the Seattle Children's Research Institute (SCRI) or the Fred Hutch Cancer Research Center (FHCRC; for experiments using the HIV infection model). Both facilities are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care in accordance with National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. All procedures using mice were approved by the Institutional Animal Care and Use Committee of either SCRI or FHCRC. To study engineered T cells in the absence of live HIV, 1 × 107 megaTAL-treated or control T cells in phosphate-buffered saline were injected intravenously into unconditioned mice. Peripheral blood was obtained 3, 10, and 20 days after T cell injection to monitor engraftment, and mice were killed 6 weeks post-transplant to harvest spleens and assess engraftment.
For the in vivo HIV challenge, we infected PHA-activated PBMCs infected with HIV-1Ba-L (MOI ~ 0.4) for 72 hours. 1 × 106 autologous PBMCs were mixed with 7 × 106 CD4 T cells (either megaTAL or control BFP mRNA treated) and 1 × 106 infected PBMCs; this cell mixture was suspended in 100 µl of phosphate-buffered saline and injected intraperitoneally into unirradiated mice (n = 8 each group, megaTAL or control). Peripheral blood samples were collected on days 7, 21, and 35 post-cell transfer for flow cytometric analyses of cells and for collection of serum for viral RNA quantitation. Mice were killed at 35 days upon developing symptoms of graft-versus-host disease, and spleens were collected. After erythrocyte lysis, spleen cells were passed through a 70-micron strainer then cells stained with antibodies for cell surface markers for flow analysis.
Detection of proviral DNA. Molecular analysis of HIV infection in CD4+ cells was performed as previously described.51 CD4+ cells were purified from HIV-infected mice using selection with CD4 magnetic beads (Miltenyi). Genomic DNA was purified with PureLink DNA extraction kit (Thermo Fisher) and the integrated HIV provirus was detected using HIV-specific primers. The iTAQ universal SYBR kit was used for PCR amplication and the CFX96 real-time PCR instrument was used for sample analysis.
Plasma viremia. Total RNA was extracted from plasma samples using QiAmp viral RNA kit (Qiagen). Quantification of viral RNA was performed using TaqMan One-Step RT-PCR Master Mix Reagents Kit (LifeTechnologies 4309169). The following oligonucleotides were designed to amplify our target: Forward Primer 5ʹ-GCC TCA ATA AAG CTT GCC TTG A-3ʹ, Reverse Primer 5ʹ-GGC GCC ACT GCT AGA GAT TTT-3ʹ, Probe 5ʹ-6FAM-AAG TAG TGT GTG CCC GTC TGT TRT KTG ACT-TAMRA-3ʹ. Samples values were calculated relative to a standard curve created by making serial dilutions of a stock virus titered using Acrometrix HIV-1 panel (Thermo Fisher Scientific, Waltham, MA). Each sample was run in duplicate on two separate plates (mean of four reactions plotted). Each plate had a no-template negative control and a phosphate-buffered saline control to ensure the absence of contamination. The uninfected controls were run on a separate plate from the infected reactions to minimize aerosols or contamination. With the exception of day 35, all samples were run on the same day using the same standard curve to minimize run-to-run variability.
Statistical analysis. All statistical analyses were performed using GraphPad Prism software (GraphPad, San Diego, CA). A minimum of three data points was used for each analysis. P values were calculated using Student's t-test.
SUPPLEMENTARY MATERIAL Figure S1. Viability and transfection efficiencies of Ghost Hi-5 cells and primary human T cells. Figure S2. Workflow of the homing endonuclease re-cleavage assay.
Acknowledgments
This work was supported by grants from the National Institutes of Health, Bethesda, MD: R42 GM085876 (to A.A.); U19 AI096111, R01 HL116217, R01 AI080326 (to H.P.K.); RL1 HL092553, PL1 HL092557 (to D.J.R.); and R01 AI118500 (to Thor Wagner and D.J.R.). Additional support was provided by the Seattle Children's Program for Cell and Gene Therapy. H.P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research. D.J.R. received support as recipient of the Children's Guild Association Endowed Chair in Pediatric Immunology. This content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. J.J. and A.A. are full-time employees of bluebird bio, Inc. which has filed for a patent application covering the CCR5 megaTAL nuclease. B.D.S. is currently a full-time employee at Juno Therapeutics. The remaining authors declare no competing financial interests. A.A., H.P.K., and D.J.R. are the principal investigators of the study. J.J. and A.A designed and tested the megaTAL and cellular delivery parameters. B.S. and Brett Kaiser produced and provided the recombinant CCR5-LHE used in the re-cleavage assay (RCA). G.S.R.I., B.D.S., and A.A. developed T cell optimization protocols and generated cells for T cell editing experiments. B.D.S. and G.S.R.I. designed and coordinated the mouse engraftment studies. G.S.R.I. and A.A. designed and conducted in vitro HIV experiments. M.H., B.P., K.S., and G.S.R.I. designed and conducted molecular analysis. P.M.Y., A.A. and J.P.K. designed and conducted the HIV infection experiments in mice. G.S.R.I. and B.P. conducted all follow-up analysis and experiments. G.S.R.I. assembled the figures. G.S.R.I. and B.P. wrote the manuscript. All authors reviewed and approved the manuscript.
Supplementary Material
References
- Robbins, RN, Spector, AY, Mellins, CA and Remien, RH (2014). Optimizing ART adherence: update for HIV treatment and prevention. Curr HIV/AIDS Rep 11: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detels, R, Muñoz, A, McFarlane, G, Kingsley, LA, Margolick, JB, Giorgi, J et al. (1998). Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. Multicenter AIDS Cohort Study Investigators. JAMA 280: 1497–1503. [DOI] [PubMed] [Google Scholar]
- Pérez-Hoyos, S, del Amo, J, Muga, R, del Romero, J, García de Olalla, P, Guerrero, R et al.; GEMES (Spanish Multicenter Study Group of Seroconverters) (2003). Effectiveness of highly active antiretroviral therapy in Spanish cohorts of HIV seroconverters: differences by transmission category. AIDS 17: 353–359. [DOI] [PubMed] [Google Scholar]
- Siliciano, JD and Siliciano, RF (2013). Recent trends in HIV-1 drug resistance. Curr Opin Virol 3: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, SG (2011). HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med 62: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, M, Carrington, M, Winkler, C, Huttley, GA, Smith, MW, Allikmets, R et al. (1996). Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273: 1856–1862. [DOI] [PubMed] [Google Scholar]
- Dragic, T, Litwin, V, Allaway, GP, Martin, SR, Huang, Y, Nagashima, KA et al. (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381: 667–673. [DOI] [PubMed] [Google Scholar]
- Liu, R, Paxton, WA, Choe, S, Ceradini, D, Martin, SR, Horuk, R et al. (1996). Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86: 367–377. [DOI] [PubMed] [Google Scholar]
- Glass, W.G.,McDermott, D.H., Lim, J.K., Lekhong, S., Yu, S.F., Frank, W.A., Pape,J., Cheshier, R.C., and Murphy, P.M.(2006). CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med. 203, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani, AP and Novembre, J (2005). The evolutionary history of the CCR5-Delta32 HIV-resistance mutation. Microbes Infect 7: 302–309. [DOI] [PubMed] [Google Scholar]
- Allers, K, Hütter, G, Hofmann, J, Loddenkemper, C, Rieger, K, Thiel, E et al. (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117: 2791–2799. [DOI] [PubMed] [Google Scholar]
- Hütter, G, Nowak, D, Mossner, M, Ganepola, S, Müssig, A, Allers, K et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360: 692–698. [DOI] [PubMed] [Google Scholar]
- Henrich, TJ, Hanhauser, E, Marty, FM, Sirignano, MN, Keating, S, Lee, TH et al. (2014). Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich, TJ, Hu, Z, Li, JZ, Sciaranghella, G, Busch, MP, Keating, SM et al. (2013). Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis 207: 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, P and June, C (2011). Chemokine receptor 5 knockout strategies. Curr Opin HIV AIDS 6: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiem, HP, Jerome, KR, Deeks, SG and McCune, JM (2012). Hematopoietic-stem-cell-based gene therapy for HIV disease. Cell Stem Cell 10: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L, Krymskaya, L, Wang, J, Henley, J, Rao, A, Cao, LF et al. (2013). Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther 21: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didigu, CA, Wilen, CB, Wang, J, Duong, J, Secreto, AJ, Danet-Desnoyers, GA et al. (2014). Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood 123: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, DA, Brennan, AL, Jiang, S, Binder-Scholl, GK, Lee, G, Plesa, G et al. (2013). Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas, P, Stein, D, Tang, WW, Frank, I, Wang, SQ, Lee, G et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov, FD, Miller, JC, Lee, YL, Beausejour, CM, Rock, JM, Augustus, S et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- Yi, G, Choi, JG, Bharaj, P, Abraham, S, Dang, Y, Kafri, T et al. (2014). CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids 3: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel, S, Jarjour, J, Astrakhan, A, Adey, A, Gouble, A, Duchateau, P et al. (2014). megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res 42: 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather, BD, Romano Ibarra, GS, Sommer, K, Curinga, G, Hale, M, Khan, IF et al. (2015). Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med 7: 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour, J, West-Foyle, H, Certo, MT, Hubert, CG, Doyle, L, Getz, MM et al. (2009). High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res 37: 6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y, Khan, IF, Boissel, S, Jarjour, J, Pangallo, J, Thyme, S et al. (2014). Progressive engineering of a homing endonuclease genome editing reagent for the murine X-linked immunodeficiency locus. Nucleic Acids Res 42: 6463–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörner, A, Björndal, A, Albert, J, Kewalramani, VN, Littman, DR, Inoue, R et al. (1999). Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol 73: 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo, MT, Gwiazda, KS, Kuhar, R, Sather, B, Curinga, G, Mandt, T et al. (2012). Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods 9: 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornalusse, GG, Mummidi, S, Gaitan, AA, Jimenez, F, Ramsuran, V, Picton, A et al. (2015). Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor. Proc Natl Acad Sci USA 112: E4762–E4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme, SB, Jarjour, J, Takeuchi, R, Havranek, JJ, Ashworth, J, Scharenberg, AM et al. (2009). Exploitation of binding energy for catalysis and design. Nature 461: 1300–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, EE, Wang, J, Miller, JC, Jouvenot, Y, Kim, KA, Liu, O et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, M, Dean, M, Martin, MP and O'Brien, SJ (1999). Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet 8: 1939–1945. [DOI] [PubMed] [Google Scholar]
- Osborn, MJ, Webber, BR, Knipping, F, Lonetree, CL, Tennis, N, DeFeo, AP et al. (2016). Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol Ther 24: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba, R, Bosch, A and Chillon, M (2005). Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther 12 (suppl. 1): S18–S27. [DOI] [PubMed] [Google Scholar]
- Stilwell, JL and Samulski, RJ (2004). Role of viral vectors and virion shells in cellular gene expression. Mol Ther 9: 337–346. [DOI] [PubMed] [Google Scholar]
- Seiler, MP, Cerullo, V and Lee, B (2007). Immune response to helper dependent adenoviral mediated liver gene therapy: challenges and prospects. Curr Gene Ther 7: 297–305. [DOI] [PubMed] [Google Scholar]
- Almåsbak, H, Rian, E, Hoel, HJ, Pulè, M, Wälchli, S, Kvalheim, G et al. (2011). Transiently redirected T cells for adoptive transfer. Cytotherapy 13: 629–640. [DOI] [PubMed] [Google Scholar]
- Kordelas, L, Verheyen, J, Beelen, DW, Horn, PA, Heinold, A, Kaiser, R et al.; Essen HIV AlloSCT Group (2014). Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med 371: 880–882. [DOI] [PubMed] [Google Scholar]
- Hildinger, M, Dittmar, MT, Schult-Dietrich, P, Fehse, B, Schnierle, BS, Thaler, S et al. (2001). Membrane-anchored peptide inhibits human immunodeficiency virus entry. J Virol 75: 3038–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, N, Dorgham, K, Levacher, B, Burlion, A, Gorochov, G and Marodon, G (2014). Targeting both viral and host determinants of human immunodeficiency virus entry, using a new lentiviral vector coexpressing the T20 fusion inhibitor and a selective CCL5 intrakine. Hum Gene Ther Methods 25: 232–240. [DOI] [PubMed] [Google Scholar]
- Perez, EE, Riley, JL, Carroll, RG, von Laer, D and June, CH (2005). Suppression of HIV-1 infection in primary CD4 T cells transduced with a self-inactivating lentiviral vector encoding a membrane expressed gp41-derived fusion inhibitor. Clin Immunol 115: 26–32. [DOI] [PubMed] [Google Scholar]
- Egelhofer, M, Brandenburg, G, Martinius, H, Schult-Dietrich, P, Melikyan, G, Kunert, R et al. (2004). Inhibition of human immunodeficiency virus type 1 entry in cells expressing gp41-derived peptides. J Virol 78: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstein, O, Boyd, M, Millington, M, Impey, H, Boyer, J, Howe, A et al. (2014). Preclinical safety and efficacy of an anti-HIV-1 lentiviral vector containing a short hairpin RNA to CCR5 and the C46 fusion inhibitor. Mol Ther Methods Clin Dev 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L, Patel, B, Ghanem, MH, Bundoc, V, Zheng, Z, Morgan, RA et al. (2015). Novel CD4-based bispecific chimeric antigen receptor designed for enhanced anti-HIV potency and absence of HIV entry receptor activity. J Virol 89: 6685–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu, GK, Sango, K, Selliah, N, Ma, Q, Skowron, G and Junghans, RP (2013). Anti-HIV designer T cells progressively eradicate a latently infected cell line by sequentially inducing HIV reactivation then killing the newly gp120-positive cells. Virology 446: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton, N, Verrier, F, Debré, P and Gorochov, G (1998). Characterization of T cell-expressed chimeric receptors with antibody-type specificity for the CD4 binding site of HIV-1 gp120. Eur J Immunol 28: 4177–4187. [DOI] [PubMed] [Google Scholar]
- Romeo, C and Seed, B (1991). Cellular immunity to HIV activated by CD4 fused to T cell or Fc receptor polypeptides. Cell 64: 1037–1046. [DOI] [PubMed] [Google Scholar]
- Gartner, S, Markovits, P, Markovitz, DM, Kaplan, MH, Gallo, RC and Popovic, M (1986). The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233: 215–219. [DOI] [PubMed] [Google Scholar]
- Popovic, M, Gartner, S, Read-Connole, E, Beaver, B, and Reitz, M (1988). Cell Tropism and expression of HIV-1 isolates in natural targets. In: Girard M and Valette L (eds). Retroviruses of Human AIDS and Related Animal Diseases. Fondation Marcel Merieux (Lyon); Pasteur Vaccins (Marnes-la-Coquette): Lyon; Marnes-la-Coquette. pp. 275. [Google Scholar]
- Yuan J,Wang J,Crain K, Fearns C, Kim KA, Hua KL, Gregory PD, Holmes MC,Torbett BE(2002). Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol. Ther. 20(4): 849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain, C, Lee, B, Vakili, J, Doranz, BJ, Govaerts, C, Migeotte, I et al. (1999). Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J Biol Chem 274: 18902–18908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.