Abstract
Adeno-associated viral (AAV) vectors are considered as one of the most promising delivery systems in human gene therapy. In addition, AAV vectors are frequently applied tools in preclinical and basic research. Despite this success, manufacturing pure AAV vector preparations remains a difficult task. While empty capsids can be removed from vector preparations owing to their lower density, state-of-the-art purification strategies as of yet failed to remove antibiotic resistance genes or other plasmid backbone sequences. Here, we report the development of minicircle (MC) constructs to replace AAV vector and helper plasmids for production of both, single-stranded (ss) and self-complementary (sc) AAV vectors. As bacterial backbone sequences are removed during MC production, encapsidation of prokaryotic plasmid backbone sequences is avoided. This is of particular importance for scAAV vector preparations, which contained an unproportionally high amount of plasmid backbone sequences (up to 26.1% versus up to 2.9% (ssAAV)). Replacing standard packaging plasmids by MC constructs not only allowed to reduce these contaminations below quantification limit, but in addition improved transduction efficiencies of scAAV preparations up to 30-fold. Thus, MC technology offers an easy to implement modification of standard AAV packaging protocols that significantly improves the quality of AAV vector preparations.
Keywords: AAV vectors, DNA impurities, Minicircle, Vector production, Vector quality
Introduction
Initiated more than two decades ago, human gene therapy has entered clinical reality with the first advanced therapeutic medicinal product on the European market and a number of clinical trials achieving therapeutic efficacies comparable or even better than standard treatment.1 A vector system based on the nonpathogenic, replication-deficient adeno-associated virus (AAV) contributed to this success in particular by mediating long-term transgene expression in postmitotic tissues such as liver, muscle, eye, and brain.2,3,4,5
AAV vectors are composed of a nonenveloped protein capsid defining tissue preference and antigenic reactivity and a DNA vector genome delivering the transgene expression cassette (TEC), i.e., the gene/s of interest including control elements. The vector genome contains at either end inverted terminal repeats (ITRs) representing the viral origin of replication and the packaging signals. It can be designed either as a single-stranded DNA providing sense and antisense versions of the TEC on separate molecules (single-stranded AAV vectors (ssAAV)) or on a single molecule, separated by an additional ITR (self-complementary AAV vectors (scAAV)).6
AAV vectors transduce both dividing and terminally differentiated cells, and lack an intrinsic integrase activity, thereby lowering the risk of insertional mutagenesis. A remarkable feature of AAV is the impressive particle stability against temperature and shifts in pH,7,8 which allows for sophisticated purification protocols. However, despite extensive efforts to improve the AAV vector manufacturing process, production of vector preparations that reach the level of purity requested by regulatory authorities remains a challenge.9,10
For vector production, transient plasmid transfection protocols in mammalian cell lines are commonly employed. While so-called AAV vector plasmids provide the TEC flanked by the ITRs, thus serving as template for vector genome replication, AAV helper plasmids introduce viral replication and packaging proteins (Rep proteins), viral capsid proteins (VP1, VP2, and VP3) as well as the assembly activating protein (AAP).11 The third “component” essential for AAV vector production are helper virus functions, as AAV relies on the assistance of unrelated viruses, such as members of the herpes virus or adenovirus (Ad) family, for replication and particle production. These functions are either provided by helper virus infection,12 by addition of a further helper plasmid encoding for helper virus function13 or by combination of the latter with the AAV open reading frames on a single AAV/Ad helper plasmid.14 Vector particle production takes place in the cell nucleus where vector genomes are transferred in a Rep-dependent process into preassembled capsids.15 Vector-producing cells are lysed 48–72 hours post-transfection, followed by nuclease treatment and density gradient and/or chromatography-based purification of the vector containing cell lysate.16 If required, centrifugal or gel filtration is included as final polishing step.17 Apart from AAV vector particles carrying the desired TEC, however, empty particles as well as DNA impurities containing AAV-, genomic-, or antibiotic resistance-specific information are present in vector preparations.9,10,18,19,20,21 In the interest of vector safety, transfer of such impurities should be avoided, as empty particles may induce anti-AAV immune responses,22 while unintended transfer of viral genetic information may result in production of AAV or helper virus proteins. In contrast to viral promoters, prokaryotic promoter sequences are nonfunctional in mammals. However, transfer of plasmid backbone sequences should also be avoided, since they contain motifs that are recognized by the cell-autonomous immune system and are therefore prone to induce inflammatory responses or gene silencing.23,24 In addition, antibiotic resistance genes may get integrated into the host genome,19 thus bearing the risk to come under the control of eukaryotic promoters.
While antibiotic resistance genes constitute a minor population in viral vector preparations, nonviral vectors traditionally consist of plasmids and therefore always transfer antibiotic resistance genes and further backbone sequences upon administration.25 In order to avoid the above mentioned challenges and to thus improve safety of nonviral vectors as well as efficiency and duration of cell modification, the minicircle (MC) technology was developed.26,27,28,29 MCs are circular DNA expression cassettes which do not contain functional or coding prokaryotic sequences. They originate from parental plasmids harboring at least the selection marker, an origin of replication and two recombination sites flanking the TEC. Following amplification in Escherichia coli, this parental plasmid is split enzymatically by a cis-recombination reaction resulting in two circular supercoiled and monomeric molecules: a miniplasmid with all the unwanted bacterial sequences and the MC containing the sequences of interest and a small additional sequence (here 213 bp), termed sequence for chromatography, affinity and recombination (SCAR), which represents one recombination sequence and a tag for affinity purification.25,30,31
Given the lack of functional prokaryotic sequences in MC, we reasoned that the MC technology might be an easy and straightforward strategy for the production of AAV vector preparations devoid of antibiotic resistance genes. We therefore developed AAV vector plasmids and a combined AAV/Ad helper plasmid as MC constructs and compared them to standard packaging plasmids regarding efficiency of ssAAV and scAAV vector production and vector purity. This side-by-side comparison revealed that both strategies did not differ in total particle yield, but in quality. Specifically, MC-based vector preparations consisted of a higher number of TEC-containing particles (packaging efficiency). In addition, exchanging plasmids for MCs resulted in vector preparations with superior transduction efficiency, in particular in case of scAAV vector preparations. In line with earlier reports9,19,21 on vector plasmids being the main source for DNA impurities, replacing just the AAV vector plasmid by its MC equivalent was sufficient to reduce the frequency of particles containing packaged plasmid backbone sequences up to two orders of magnitude. However, aiming to also avoid the minor contribution of helper plasmids, both AAV vector and helper plasmids have to be replaced by MCs.
Results
Prokaryotic sequences in AAV vector preparations cannot be removed by standard purification methods
We examined ssAAV vector preparations purified by standard protocols for the presence of plasmid backbone sequences using the ampicillin resistance gene (ampR) bla encoding for TEM-1 β-lactamase as marker, as it is contained in our vector and helper plasmids. Briefly, AAV serotype 2 (AAV2) vector preparations were produced by plasmid transfection of HEK293 using a standard plasmid packaging system.13 Following discontinuous iodixanol density gradient centrifugation (IDGC) of Benzonase-treated cell lysates, vector preparations #1 and #2 were purified by affinity chromatography utilizing an anti-AAV single-domain antibody (AVB column) followed by centrifugal filtration (Amicon tubes). Vector preparation #3 was instead further purified by affinity chromatography harnessing the heparin binding ability of AAV2, followed by gel filtration. Aliquots of the vector preparations after density gradient centrifugation, chromatography and filtration were analyzed by quantitative polymerase chain reaction (qPCR). The quantity of vector genomes was determined by primers specific for the enhanced green fluorescent protein (eGFP) gene, the TEC intended to be packaged, while primers specific for ampR were employed to indicate the presence of plasmid backbone sequences. As expected, the majority of sequences (> 98% for all preparations) in our purified preparation matched the intended transgene (Figure 1a). However, in line with previous studies,9,19,20,21 we also detected a substantial number of ampR sequences. The analysis of density-gradient purified vectors revealed ampR-specific DNA sequences in all preparations, ranging from 0.5% (vector preparation #3) to 1.8% (vector preparation #2) relative to TEC (eGFP) sequences (Figure 1a). None of the employed subsequent purification steps succeeded in removing these undesired prokaryotic sequences from the vector preparations (ampR concentrations relative to TEC: vector preparation #1: IDGC 1.3% versus AC-AVB 1.1%; vector preparation #2: IDGC 1.8% versus AC-AVB 1.7%, vector preparation #3: IDGC 0.5% versus AC-Heparin 0.6%).
Figure 1.
Antibiotic resistance gene (ampR) is tightly associated with adeno-associated viral (AAV) vector particles. (a) Quantification of DNA sequences contained in AAV vector preparations. Lysates of vector-producing cells were purified by discontinuous iodixanol density gradient centrifugation (IDGC). The 40%-phases of three different vector preparations—containing AAV vector particles16—were further purified by either affinity chromatography using AVB column (AC-AVB) or Heparin column (AC-Heparin), followed by centrifugal filtration (CF) or gel filtration (GF). At each step of purification, total DNA from aliquots was isolated and analyzed by qPCR using indicated primers. (b) Benzonase protection assay. Two aliquots of AAV vector preparation #3 encoding for eGFP were spiked with 400 ng of a plasmid encoding for the kanamycin/neomycin resistance gene (kanR). One aliquot was treated with Benzonase as described in Materials&Methods section. Total DNA of the treated and an untreated second aliquot was isolated and analyzed by qPCR using indicated primers (Supplementary Table S1). qPCR analyses described in (a) and (b) were performed three times independently.
In order to investigate whether these DNA impurities are protected by the viral capsid and thus tightly associated or encapsidated, we performed a Benzonase protection assay. We therefore spiked aliquots of vector preparation #3 with 400 ng of a plasmid containing the kanamycin/neomycin resistance gene cassette (kanR). One aliquot was subjected to Benzonase treatment followed by isolation of total DNA from the treated and untreated aliquot, and qPCR analyses (Figure 1b). Benzonase treatment neither affected the quantity of TEC nor ampR-specific sequences. In contrast, Benzonase treatment resulted in the removal of > 99.9% of input control plasmid sequences. Accordingly, TEC and ampR sequences are not accessible to the nuclease and thus protected by the capsid from enzymatic digestion. Therefore, the prokaryotic DNA contained in AAV vector preparations is Benzonase-resistant and cannot be removed by standard purification methods.
Development of MC constructs of AAV vector and AAV/Ad helper plasmids
In order to apply the MC technology, recombination sequences flanking the TEC and thus separating backbone sequences from those that should later be contained in the MC construct had to be introduced. We therefore cloned the whole vector genome of a standard ssAAV vector plasmid, pAAV-ssGFP (pGFP in ref. 32), encoding for two TECs (Supplementary Figure S1a) flanked by ITRs of AAV2 into our parental plasmid (PP) between the two minicircle-recombination sites. Subjecting plasmid preparations to L-Arabinose induced a ParA resolvase-mediated recombination event through which two circles were produced, one of them being the MC with the ITR2-flanked vector genome and the 213 bp SCAR sequence. This MC, in the following termed MC.AAV-ssGFP (Supplementary Figure S1c), was separated from the second circle, the miniplasmid, by affinity chromatography. Final quality control confirmed that MC.AAV-ssGFP was a supercoiled and homogenous monomeric product (Supplementary Figure S2). Likewise, the combined AAV/Ad helper plasmid pDP2rs (Supplementary Figure S1b) encoding for the structural and nonstructural proteins of AAV2 and adenoviral helper functions33 was used as source to clone the PP for production of MC.DP2rs (Supplementary Figure S1d), the MC construct that ought to replace the combined AAV/AdV helper plasmid pDP2rs. MC.DP2rs was produced and characterized as described for MC.AAV-ssGFP.
MC constructs are efficient substitutes for plasmids in AAV vector production
In order to investigate whether MCs can substitute plasmids in AAV vector production and whether indeed both, vector and helper plasmids have to be replaced, we performed a side-by-side comparison of the four possible combinations of MCs and plasmids (Table 1). The respective vector preparations were produced in HEK293 cells and purified by discontinuous iodixanol density gradient centrifugation of Benzonase-treated cell lysates. The 40% phase of the gradient was isolated and then characterized by qPCR, Western Blotting and enzyme-linked immunosorbent assay (ELISA) using transgene-specific primers,22 capsid protein specific antibody B1 (ref. 34) and capsid specific antibody A20 (ref. 35), respectively. With all four combinations, AAV vector preparations of comparable yield could be obtained (Table 1). Specifically, for all preparations comparable physical particle titers (capsids per ml) (analysis of variance (ANOVA), not significant (n.s.)) and an identical capsid composition (data not shown) was determined. Quantitative PCR analyses further revealed that the capsids contained vector genomes (TEC) and that MC-based vector preparations yielded a genomic particle titer, i.e., Benzonase-resistant TEC-containing particles per ml, which did not differ negatively from those obtained by the dual plasmid packaging system (ANOVA; Tukey post hoc tests versus dual plasmid, n.s.). On the contrary, the highest genomic particle titers were measured for those preparations that were produced using MC.AAV-ssGFP revealing that MC.AAV-ssGFP served as efficient template for vector genome replication, although the ITRs are separated by only 213 bp (Supplementary Figure S1c).
Table 1. Characterization of AAV2-ssGFP.

As volume-independent measure, we then calculated the packaging efficiency. The latter is defined as ratio of physical (capsid) titer to genomic (= TEC) particle titer (vp:vg). A ratio of 50 and below is judged as a wild-type phenotype,36 while higher values indicate that either replication of vector genomes and/or the packaging process itself occurred inefficiently. In none of the cases in which plasmids were exchanged by MC, a significant negative impact on packaging efficiency was observed (ANOVA; Tukey post hoc tests versus dual plasmid, n.s.). On the contrary, the lowest value, i.e., the highest packaging efficiency, was determined for preparations in which MC.AAV-ssGFP was used (Table 1). Thus, replacing packaging plasmids by MC constructs results in AAV vector preparations that were at least comparable in titer and in packaging efficiency to preparations obtained by standard packaging plasmids.
Next, we incubated the cervix carcinoma cell line HeLa, which is highly permissive for AAV2, with a serial dilution of the ssAAV vector preparations, followed by assessment of eGFP-expressing cells by flow cytometric measurements 48 hours post-transduction (p.t.). We thereby determined the transducing titer of each preparation (Table 1). Again, we did not observe a significant negative impact on AAV vector production in cases in which either one or both packaging plasmids were exchanged by MCs (ANOVA; Tukey post hoc tests). On the contrary, MC-based preparations tended to show a higher transducing titer with preparations produced by combining pDP2rs and MC.AAV-ssGFP being superior to the dual MC system (ANOVA; Tukey post hoc tests, P < 0.001). However, in a subsequently performed repetition experiment in which we specifically compared ssAAV vector preparations produced by MC.AAV-ssGFP either in combination with pDP2rs or with MC.DP2rs, transducing titers did not differ significantly (data not shown).
As volume-independent measure, we then calculated the transduction efficiency, which indicates the number of particles per cell required to obtain an infectious unit. A ratio below 104 is defined as wild-type phenotype36 and was reached by all preparations. Thus, ssAAV vector preparations produced by transfection of MCs demonstrated an at least comparable overall yield and transduction efficiency compared to ssAAV vector preparations produced by transfection of vector and AAV/Ad helper plasmids.
Vector plasmids are the main source of prokaryotic sequences in vector preparations
In standard AAV vector preparations, ampR sequences are present which can neither be removed by nuclease treatment of purified preparations nor by applying further purification steps (Figure 1; refs. 9,20). In line, analyzing our side-by-side produced vector preparations—in which lack of free nucleic acid had been confirmed by Benzonase protection assay (Supplementary Table S2)—revealed the presence of DNA impurities in preparations produced with the dual plasmid strategy. Specifically, we determined 3.2 and 4.6 × 109 ampR-specific sequences per ml (Figure 2). This corresponds to approximately one ampR sequence per 150 particles or an approximate 1:40 ratio of ampR- to TEC-containing particles. Replacing the helper plasmid by MC.DPrs had no beneficial effect (ANOVA; Tukey post hoc test versus dual plasmid, n.s.). In contrast, replacing the vector plasmid by MC.AAV-ssGFP reduced the number of ampR-containing and thus falsely packaged particles by more than two orders of magnitude. Under these conditions, the ratio of ampR to TEC-containing particles was reduced to less than 1:6,000 (ANOVA; Tukey post hoc test versus dual plasmid, P = 0.0021). A further decrease in the frequency of ampR sequences was observed in preparations produced by the dual MC approach for which the frequency of ampR particles was lowered to 0.004% or less relative to TEC (ANOVA; Tukey post hoc test versus dual plasmid, P < 0.001). Thus, by replacing packaging plasmids for respective MC constructs, ampR DNA impurities can be reduced to background levels.
Figure 2.
Replacing vector plasmid by minicircle construct significantly decreases the amount of encapsidated ampR DNA particles. DNA isolated from vector preparations was quantified for ampR sequences by qPCR using specific primers (Supplementary Table S1). # < Limit of quantification. All analyses were performed in parallel for all vector preparations. All analyses were performed three times independently. Differences in ampR content between preparations were assessed using ANOVA (P < 0.0001) and subsequent Tukey post hoc tests.
Proximity to ITRs rather than specific sequence elements is decisive for packaging of backbone sequences
As cause for ampR DNA impurities, presence of a weak packaging signal was proposed.18 To test this hypothesis and to gain insight into the mechanism of false packaging, we analyzed our dual plasmid-based preparations for the presence of further plasmid backbone sequences. Specifically, we chose the bacterial origin of replication (ori) and the f1 origin of replication, which are neighboring the left and the right ITR, respectively (Supplementary Figure S1a). We again performed qPCR analyses using target sequence specific primers (Supplementary Table S1). Interestingly, we found that both sequences were present with a frequency that correlated with the findings for ampR: we detected 2.0 to 2.3% and 2.5 to 3.0% of ori- and f1 ori-containing relative to TEC-containing particles, respectively (Table 2). Thus, prokaryotic sequences located in cis to the ITRs are packaged independent of a specific sequence element.
Table 2. Quantification of prokaryotic DNA in the dual plasmid preparations by qPCR.

Based on this result, we wondered whether also the short prokaryotic noncoding SCAR sequence which remains present in MC constructs becomes encapsidated (Supplementary Figure S1c). qPCR quantification showed that this DNA sequence is indeed being packaged into viral capsids when MC.AAV-ssGFP, containing this sequence in cis, was used for vector production. Up to 1.3% of SCAR relative to TEC-containing particles were found, with no significant difference in content between vectors produced with either MC.DP2rs or pDP2rs (Table 3, t-test n.s.) further confirming that backbone sequences are packaged independent of a specific motif.
Table 3. Quantification of SCAR sequences in AAV vector preparations by qPCR.

Self-complementary AAV vector preparations produced by MC constructs show improved transduction efficiencies and contain no ampR DNA impurities
Given the preferred use of scAAV vectors for in vivo applications and thus the obvious need to establish also for this vector type an easy to implement strategy to avoid unintended transfer of functional prokaryotic sequences, we next developed an MC construct as substitute for a scAAV vector plasmid. As source, we decided for pAAV-scGFP (pscAAV/EGFP in ref. 38 (Supplementary Figure S1e)) that encodes for eGFP and results in production of self-complementary vector genomes due to the deletion of the terminal resolution site (trs) of the left ITR sequence.6 The respective MC construct (MC.AAVscGFP, Supplementary Figure S1f) was designed and produced as described for MC.AAVssGFP.
Again, we performed a side-by-side comparison of all four possible combinations of vector (pAAV-scGFP, MC.AAV-scGFP) and helper constructs (pDP2rs and MC.DP2rs) and analyzed the vector preparations following standard purification by ELISA and qPCR (Table 4). In line with our results for the ssAAV vector preparations (Table 1), MC constructs were as efficient as plasmid-based counterparts in particle production (vp per ml) (ANOVA n.s.). Replacing plasmids by MC constructs, however, beneficially impacted on genomic (TEC) particle titer and packaging efficiency, although statistical significance was not reached (ANOVA, Tukey post hoc tests). We observed the most remarkable differences between MC-based and plasmid-based scAAV vector preparations, in the biological activity of the preparations: we determined up to 30-fold improved transducing titers on HeLa (ANOVA, Tukey post hoc tests versus dual plasmid, P < 0.001) and as a consequence a significantly higher transduction efficiency (vp:tu) for vector preparations produced with MCscGFP.
Table 4. Characterization of self-complementary vectors.

We then determined the amount of DNA contaminations, again using the ampR sequence as marker. Surprisingly, we observed that scAAV vector preparations produced by the dual plasmid system contained a significantly higher amount of ampR sequences than ssAAV vector preparations, relative to TEC. Specifically, we measured 4.8 × 109 to 1.1 × 1010 ampR-specific sequences per ml (Figure 3). This corresponds to approximately one ampR sequence per 100 particles or an approximate 1:3 ratio of ampR to TEC-containing particles. Replacing just the helper plasmid by the respective MC construct (MC.DP2rs) again had no beneficial effect (ANOVA; Tukey post hoc test versus dual plasmid, n.s.), while, solely by replacing the vector construct, ampR-containing particles were decreased to 0.2% or less relative to TEC (ANOVA; Tukey post hoc test versus dual plasmid, P < 0.001). Replacing both vector and helper plasmid by MCs resulted in scAAV vector preparation free of ampR-containing particles (Figure 3, below limit of quantification) providing further proof that packaging of functional prokaryotic sequences can be avoided by employing the MC technology, and that the helper plasmid contributes to packaging of DNA impurities in AAV vector preparations.
Figure 3.
Replacement of both plasmids by minicircle results in ampR-free vector preparations. DNA isolated from vector preparations was quantified for ampR sequences by qPCR using specific primers (Supplementary Table S1). # < Limit of quantification. Analysis was performed in parallel for all vector preparations. Differences in ampR content between preparations were assessed using ANOVA (P < 0.0001) and subsequent Tukey post hoc tests.
Overall, these results allow us to conclude that replacing plasmids for MC constructs significantly improves the biological activity and the quality of scAAV vector preparations.
Discussion
Viral vectors including AAV are developed as tools for efficient transfer of genes of interest into cells either ex vivo or in vivo to provide a novel treatment option, or to contribute to answering basic biological questions. In line with previous reports, we observed that AAV vector preparations produced by transient transfection of HEK293 cells do contain DNA other than the intended TEC as genetic payload (Figure 1; refs. 9,10,18,19,20,21). Benzonase protection assays conducted on purified vector preparations indicated that the DNA impurities are protected from nuclease digest, pointing toward their encapsidation or at least to a close enough association with the viral capsid to protect them from degradation. In ssAAV vector preparations, the TEC represent the main population (>98%), but up to 1.8% of plasmid backbone sequences were detected in our preparations that underwent relatively elaborate purification protocols (Figure 1). These values as well as those determined for our side-by-side comparison are in the lower range of those which have been reported (1–8%) elsewhere for ssAAV.9,10,19,21 In contrast, we found that scAAV vector preparations, which are preferentially used for in vivo gene transfer and modification of primary cells,6 do contain a much higher amount of encapsidated prokaryotic sequences (up to 26.1%, Figure 3).
Such sequences are transferred in vivo upon administration of the vector preparations and are delivered into target tissue, presumably with half-lives comparable to AAV vectors delivering the intended TEC.19 Although Hauck and colleagues provided evidence that these sequences are not being transcribed,21 it cannot be excluded that they trigger immune responses. The latter have been reported to limit long-term expression of TEC delivered by nonviral vectors.39,40 Furthermore, the long-term consequences of co-delivered antibiotic resistance genes cannot be predicted. Accordingly, it is highly desirable to remove or at least extensively reduce these elements in vector preparations. As removal of DNA impurities from a vector preparation appears impossible, we here report a strategy that precludes packaging of these sequences in the first place. Precluding the packaging of plasmid backbone sequences, while following the transient transfection protocol for AAV vector production, has become possible through the invention of MC. The initial goal of the MC technology was the removal of antibiotic resistance sequences from therapeutic nonviral vectors.26,27,28,29 Due to their significantly reduced size compared to plasmids and the absence of CpG motifs, MCs achieve an increased transgene expression level.25 As they are considered as substantial improvement in nonviral vector technology they are currently employed in a number of in vivo studies.41
Here, the vector plasmid for ss- and scAAV and the combined AAV/AdV helper plasmid were successfully produced as so-called parental plasmids from which respective MC constructs (Supplementary Figure S1) could be produced by a standard purification process.31 A side-by-side comparision revealed that MC constructs are at least as potent as plasmids in AAV vector production as each component of the standard dual plasmid system could be replaced by a MC without impairing total genomic particle yield, capsid titer, and genomic titer (Tables 1 and 4). Vectors produced by transfection of MC.AAV-ssGFP or MC.AAVscGFP and either helper (MC.DP2rs or pDP2rs) yielded preparations in which packaging of functional prokaryotic sequences was significantly reduced or even avoided and thus in preparations with improved packaging efficiency compared to the dual plasmid system (Tables 1 and 4). Furthermore, regarding transducing titers and transduction efficiency, MC.AAV-ssGFP-based vector preparations did not differ for the worse (Table 1), while MC.AAV-scGFP-based vector preparations significantly outperformed vector preparations produced by the standard dual plasmid transfection system (Table 4).
Using the MC constructs as tools (Figures 2 and 3), we obtained data supporting Chadeuf and coworkers, who provided evidence for the vector plasmid being the main source of antibiotic resistance gene sequences found in AAV vector preparations, while the helper plasmid contributes, though only to a minor extent.19
To better understand what causes the packaging of plasmid backbone sequence, we quantified our ssAAV preparations not only for the presence of ampR, but also for the presence of ori and f1 ori located in the plasmid backbone neighboring either ITR (Supplementary Figure S1a). We found that f1 ori and pUC ori are present in AAV vector preparations to a similar extent as ampR (Table 2). Also in this case the vector plasmid was identified as main source, as their presence was significantly decreased when pAAV-ssGFP was replaced by MC.AAV-ssGFP (data not shown). Thus, it is unlikely that a specific sequence contained in ampR fosters its encapsidation. The more intriguing explanation would be that sequences of the plasmid backbone are being packaged by virtue of location in cis to the ITRs, as a side product during vector genome rescue/replication from a circular plasmid (see below). In line with this assumption, we observed that the SCAR also becomes encapsidated. This sequence is the sole non-vector genome element in MC.AAV-ssGFP (Supplementary Figure S1c). It does not contain a Rep-binding site or functional element, but is surrounded by the two ITR sequences.
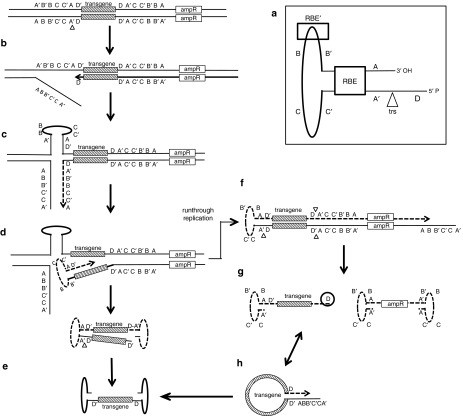
The ITRs consist of a 125-nucleotide palindrome with six segments (A-A', B-B', C-C') and a 20-nucleotide internal D sequence (Figure 4a). Between the A segment and the D sequence resides AAV's origin of replication, the terminal resolution site (trs), that is recognized by the viral Rep proteins.42 In order to replicate integrated AAV proviruses or—in case of vector production—to replicate AAV vector genomes provided on vector plasmids, viral/vector genome templates have to be rescued. The rescue process is postulated to be initiated by Rep-mediated nicking of one strand of the duplex DNA at the trs (Figure 4b).37 The partially single-stranded GC rich palindromic sequence of the ITR then forms a T-shaped hairpin (HP) (Figure 4c)—stabilized by Rep binding to a Rep-binding element (RBE') at the apex of the HP structure43—and serves as primer for genome replication. Replication initially ensues along the displaced strand. Upon reaching the 3'-end, Rep helicase activity resolves the duplex, which allows the newly synthesized ITR to fold upon itself and serve as primer for replication—this time—into the AAV vector genome (Figure 4d). At the second ITR, the same event takes place, thus generating a linear AAV genome with intact ITRs that can serve as further template for viral/vector genome replication (Figure 4e).44 If the trs of the second ITR is not nicked prior to arrival of the replication complex, replication proceeds beyond the ITR structures, producing a vector genome-plasmid backbone molecule (Figure 4f). This duplex DNA molecule can be processed—again Rep-dependent—into two single-stranded DNA molecules,44 but none with two intact ITRs. Specifically, the vector genome containing the TEC possesses one intact ITR and the D sequence of the second ITR, while the plasmid backbone sequence contains one ITR or, if replication covers the entire plasmid backbone, two ITRs, both without the D sequence (Figure 4g). The defective ITR of the vector genome can be repaired through a panhandle intermediate involving base pairing of the two D sequences flanking the vector genome by using the intact ITR as template (Figure 4h).46 While the presence of one HP structure is sufficient to rescue AAV vector genomes and to induce replication, efficiency of vector genome replication and in particular packaging of vector genomes into viral capsids depends on the presence of the D sequence in cis.47,48 Based on the latter observation, Wang and coworkers postulated the D sequence to function as actual packaging signal for AAV.48 Thus, the presence of the D sequence—independent of the pathway followed for its production (Figure 4)—is the likely reason for the so obviously preferred packaging of the TEC-containing vector genomes.47,48,49
Figure 4.
Model of proposed rescue mechanism. Circular plasmid genome is represented as linearized. (a) Schematic representation of AAV2 ITR. The triangle represents the nicking activity of Rep at the trs. (b) Rep nicks at trs and creates a single-strand break. (c) The template strand folds into a hairpin (HP) conformation thus enabling replication along the displaced strand. (d) The newly generated inverted terminal repeat (ITR) folds into the HP conformation which allows for replication into the vector genome sequence. (e) Upon the nicking event at the trs of the second ITR, the single-stranded AAV vector genome is created. (f) If the second ITR is not nicked before the arrival of the replication machinery a large replicon encompassing TEC and backbone sequences is generated. (g) The TEC genome can be rescued by Rep nickase activity which then creates the TEC with one intact ITR and an additional D sequence and the prokaryotic backbone sequences with two ITRs lacking the D sequence. (h) The defect ITR of the TEC genome can be repaired by a panhandle mechanism, thus generating an intact AAV vector genome. RBE: Rep protein binding element, trs: terminal resolution site. Figure adapted from Ward et al.44
To generate vector plasmids harboring scAAV vector genomes, the trs of the left ITR, including the D sequence, is deleted. Our data demonstrate that this modification of the ITR structure in pAAV-scGFP considerably increases the encapsidation of prokaryotic sequences (Figure 3). According to our model, this increase is the product of a run-through replication that occurs more frequently in scAAV compared to ssAAV vector production as specific nicking of Rep at the trs of the modified ITR does not occur (Figure 4f–h).
Lecomte and coworkers quantified DNA impurities in a ssAAV vector preparation with next-generation sequencing and reported a clear ranking with vector plasmid backbone being the most frequent one (0.84–5.97%), followed by the helper plasmid (0.01–0.08%) and human genome sequences (0.04–0.30%).9 In line, we (Figures 2 and 3; Tables 2 and 3) and others19 observed a striking preference for backbone sequences derived from the vector plasmid (or the corresponding MC construct). Since plasmid backbone sequences of vector plasmids become rescued and equipped with HP structures (Figure 4g), thereby gaining the ability to serve as templates for replication, it is reasonable to speculate that they become exponentially amplified during vector production. As Rep proteins bind to RBE' at the apex of the HP structure43 as well as to the pores at the fivefold symmetry axis of the capsid through which newly produced genomes are channeled,15,50 HP-flanked plasmid backbone containing sequences may become connected and eventually packaged. In support of this model Chadeuf and coworkers isolated ITR-plasmid junctions from AAV vector stocks and AAV-transduced tissue that showed a preferential retention of the A region containing the RBS in the otherwise damaged ITRs. However, they also observed partially retained D-sequences, including the trs,19 which argues for mechanisms other than those postulated in Figure 4 through which undesired DNA sequences additionally become equipped with signals that foster packaging.45,46 Such events may explain how sequences others than those contained in the vector plasmid become targets for encapsidation.9 Additionally, it is possible that the helicase activity of Rep leads to unspecific packaging events, even in absence of DNA replication.
In one of our single-stranded dual MC vector preparations, an ampR signal corresponding to a mean of 6,500 sequences per µl remained detectable (Figure 2). While we judge this signal as background as it exceeds our qPCR background level by less than 1,000 particles, it nevertheless prompted us to analyze the MC preparations which were used for packaging of ssAAV using ampR-specific primers. While MC.ssGFP contained only marginal amounts of ampR-specific sequences (<0.0005%, relative to TEC), we found ampR particles ranging from 0.03–0.14% (relative to rep) in different batches of MC.DP2rs. As MC constructs contain neither an antibiotic resistance gene sequence nor—with exception of the SCAR sequence, which contains significantly less CpG and has been shown to be safe for in vivo applications25,51—any sequence that is not intended by the researcher to be transferred, the findings highlight the requirement of sophisticated production procedures including single-use material and preferably a dedicated facility to generate MC preparations free of any kind of contamination.
The second issue concerns the elaborate production process itself. Up-scaling of the MC production and purification to produce large batches of AAV vectors may present a challenge for the combined AAV/Ad helper constructs owing to their size and the high quantities required for vector production. This limitation, however, could be overcome by shifting to the triple transfection strategy, where AAV and Ad helper functions are provided on separate and thus smaller plasmids. MCs from these plasmids are smaller in size and, at least at present, easier to produce.
In summary, we here report that MC technology offers an elegant and potent strategy to avoid unintended transfer of functional prokaryotic plasmid backbone sequences and thus to improve safety of in vivo gene transfer. MC constructs were at least equally efficient in ssAAV and scAAV production compared with the dual plasmid strategy, while the biological activity, in particular in case of scAAV vector preparations, was increased.
Materials and Methods
Cell lines and plasmids. The human embryonic kidney cell line HEK293 (ATCC CRL-1573) and the human cervix epitheloid cell line HeLa (ATCC CCL2) were maintained in Dulbecco's modified Eagle's medium with GlutaMAX-I (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (Invitrogen), 100 IU/ml of penicillin (Invitrogen) and 100 µg/ml of streptomycin (Invitrogen). HEK293 cells were used for viral vector production, while HeLa cells served as model cell line to determine transducing titers of AAV vector preparations. As plasmids, pAAV-ssGFP (pGFP in ref. 32), pAAV-scGFP (pscAAV/EGFP in ref. 38) and pDP2rs were used. Plasmid pDP2rs is a combined AAV and adenovirus helper plasmid providing AAV2 rep, cap, and AAP sequences as well as Ad E2A, E4, and VA helper virus functions plus a red fluorescent protein gene.33
Minicircle constructs. MC producer Escherichia coli K12 bacteria were cultivated in LB-medium without antibiotics at 37 °C in a preculture followed by further cultivation in a Sartorius-Stedim bioreactor Biostat C plus (Sartorius-Stedim, Guxhagen, Germany) with 10 l or 20 l working volume, for approximately 15–20 hours. MC constucts were generated in two major steps, as previously published.30,31 Specifically, ParA resolvase recombinase expression was induced at an OD600 » 4 by adding L-arabinose. After 1 hour of further growth, cells were harvested by centrifugation and freezing. The recovered “recombination product” (RP) consists of the MC and the miniplasmid (MP). The content of both DNA populations was analysed by agarose gel electrophoresis or by a dielectrophoresis based continuous-flow nanosorter as also published earlier.30,31 The In-Process-Control showed for all different preparations the presence of the two circular and supercoiled DNA molecules with a slightly increased amount of MP, which replicates further within the producer cell. The total productivity was in the range of a standard DNA production harvested at the same OD value (0.5 mg/g wet weight biomass). MC DNA was recovered from RP using affinity chromatography based on interaction of lactose operon (LacO) with repressor of lactose operon (LacI) as previously described.52 In-Process-Control showed that just the MC binds to the chromatography matrix, while the undesired MP is not binding. The resuting MC fraction was precipitated and resuspended in water for injection at 1 mg/ml and the product QC was performed (see Supplementary Figure S2).
AAV vector production. An AAV vector and a Helper construct (either as MC or plasmid) were transfected into HEK293 cells using the CaCl2 method at a molar ratio of 1:1.3 (ssAAV: total DNA 37.5µg per 15 cm petri dish: 30µg pDP2rs, 7.5µg pAAV-ssGFP; scAAV: total DNA 46.2µg per 15 cm petri dish: 38.7µg pDP2rs, 7.5µg pAAV-scGFP. To replace plasmid by equimolar amount of MC, the amount of respective MC DNA was calculated according to molecular mass). Forty-eight hours post-transfection, cells were harvested and pelleted by low-speed centrifugation. Cell pellets were subjected to three rounds of freeze-thaw for lysis. Subsequently, preparations were treated with 50 U/ml Benzonase nuclease (Merck Millipore, Darmstadt, Germany) at 37 °C for 30 minutes. Cellular debris was removed by low-speed centrifugation. The supernatant was loaded onto an iodixanol step gradient.16 The 40% phase was isolated and characterized (see below). Affinity chromatography purifications were performed using HiTrap Heparin HP or AVB Sepharose HP column (GE Healthcare, Freiburg, Germany). Ultrafiltration was performed using either Amicon Ultra-15 centrifugal filter units (Merck Millipore, Darmstadt, Germany) or Sephadex G50 packed illustra NICK column (GE Healthcare).
Characterization of vector preparations. Capsid titers of vector preparations were determined by enzyme-linked immunosorbent assay using the AAV2 capsid-specific antibody A20 (Progen, Heidelberg, Germany). Capsid composition was analyzed by western blotting using capsid protein specific antibody B1 (kindly provided by Martin Müller, DKFZ, Heidelberg, Germany) and peroxidase-conjugated donkey anti-mouse IgG. Transducing titers were determined by transduction of HeLa cells with a serial dilution of the vector preparations and determination of percentage of transgene-expressing cells by flow cytometry 48 hours p.t. (FACS Canto II, BD Biosciences, Franklin Lakes, NJ).53 Viral vector genomic particles (genomic titers) were determined by isolating DNA from vector preparations according to the DNeasy kit protocol (Qiagen, Hilden, Germany), followed by real time LightCycler (LC) PCR (Roche Diagnostics, Mannheim, Germany) using TEC-specific primers.22 DNA impurities were quantified by qPCR using primers specified in Supplementary Table S1. Specificity of amplification was confirmed by melting curve analysis. Sensitivity of qPCR measurements was determined for each primer pair (Supplementary Table S1). For each DNA sequence, the background level of the qPCR quantification was determined and subtracted from the qPCR results.
Statistical analyses. Data are presented as mean ± standard error. Quantitative data was log2-transformed and tested with t-test or ANOVA, followed by Tukey test. Calculated P values below 0.05 were considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Schematic representation of AAV vector and AAV/Ad helper plasmids and thereof derived MC constructs. Figure S2. Quality control of MC DNA. Table S1. Primers for qPCR analyses. Table S2. Benzonase protection assay of a vector preparation. Table S3. Significance values of Tukey post hoc analyses for ssAAV vectors. Table S4. Significance values of Tukey post hoc comparison of means for scAAV vectors.
Acknowledgments
This work was supported by grants from the German Federal Ministry of Education and Research (BMBF) within the program Nano4Life, grant 13149063, the program BioChancePLUS, grant 0313749 to M.S., Federal Ministry of Economics and Technology grant KF2429612AJ3 to M.S. and H.B., as well as by the Center for Molecular Medicine Cologne (CMMC) of the University of Cologne to H.B. (CMMC-C1). M.S., M.S., and C.K. are employed at PlasmidFactory (Bielefeld, Germany).
Supplementary Material
References
- Naldini, L (2015). Gene therapy returns to centre stage. Nature 526: 351–360. [DOI] [PubMed] [Google Scholar]
- Brimble, MA, Reiss, UM, Nathwani, AC and Davidoff, AM (2016). New and improved AAVenues: current status of hemophilia B gene therapy. Expert Opin Biol Ther 16: 79–92. [DOI] [PubMed] [Google Scholar]
- Wang, D, Zhong, L, Nahid, MA and Gao, G (2014). The potential of adeno-associated viral vectors for gene delivery to muscle tissue. Expert Opin Drug Deliv 11: 345–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx, M, Dutheil, N, Bezard, E and Dehay, B (2014). Systemic gene delivery to the central nervous system using Adeno-associated virus. Front Mol Neurosci 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, LS and Vandenberghe, LH (2015). Promising and delivering gene therapies for vision loss. Vision Res 111(Pt B): 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, DM (2008). Self-complementary AAV vectors; advances and applications. Mol Ther 16: 1648–1656. [DOI] [PubMed] [Google Scholar]
- Rayaprolu, V, Kruse, S, Kant, R, Venkatakrishnan, B, Movahed, N, Brooke, D et al. (2013). Comparative analysis of adeno-associated virus capsid stability and dynamics. J Virol 87: 13150–13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, M, Lins, B, Mietzsch, M, Heilbronn, R, Van Vliet, K, Chipman, P et al. (2014). A simplified purification protocol for recombinant adeno-associated virus vectors. Mol Ther Methods Clin Dev 1: 14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecomte, E, Tournaire, B, Cogné, B, Dupont, JB, Lindenbaum, P, Martin-Fontaine, M et al. (2015). Advanced Characterization of DNA Molecules in rAAV Vector Preparations by Single-stranded Virus Next-generation Sequencing. Mol Ther Nucleic Acids 4: e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, J (2014). Product-related impurities in clinical-grade recombinant AAV vectors: characterization and risk assessment. Biomedicines 2: 80–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag, F, Schmidt, K and Kleinschmidt, JA (2010). A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA 107: 10220–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolling, F and Samulski, RJ (1995). AAV as a viral vector for human gene therapy. Generation of recombinant virus. Mol Biotechnol 3: 9–15. [DOI] [PubMed] [Google Scholar]
- Xiao, X, Li, J and Samulski, RJ (1998). Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D, Kern, A, Rittner, K and Kleinschmidt, JA (1998). Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther 9: 2745–2760. [DOI] [PubMed] [Google Scholar]
- King, JA, Dubielzig, R, Grimm, D and Kleinschmidt, JA (2001). DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20: 3282–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin, S, Byrne, BJ, Mason, E, Zolotukhin, I, Potter, M, Chesnut, K et al. (1999). Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 6: 973–985. [DOI] [PubMed] [Google Scholar]
- Strobel, B, Miller, FD, Rist, W and Lamla, T (2015). Comparative analysis of cesium chloride- and iodixanol-based purification of recombinant adeno-associated viral vectors for preclinical applications. Hum Gene Ther Methods 26: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, JF (2008). Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther 15: 840–848. [DOI] [PubMed] [Google Scholar]
- Chadeuf, G, Ciron, C, Moullier, P and Salvetti, A (2005). Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol Ther 12: 744–753. [DOI] [PubMed] [Google Scholar]
- Allay, JA, Sleep, S, Long, S, Tillman, DM, Clark, R, Carney, G et al. (2011). Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum Gene Ther 22: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, B, Murphy, SL, Smith, PH, Qu, G, Liu, X, Zelenaia, O et al. (2009). Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol Ther 17: 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hösel, M, Broxtermann, M, Janicki, H, Esser, K, Arzberger, S, Hartmann, P et al. (2012). Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors. Hepatology 55: 287–297. [DOI] [PubMed] [Google Scholar]
- Bauer, S, Kirschning, CJ, Häcker, H, Redecke, V, Hausmann, S, Akira, S et al. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA 98: 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde, SC, Pringle, IA, Abdullah, S, Lawton, AE, Davies, LA, Varathalingam, A et al. (2008). CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol 26: 549–551. [DOI] [PubMed] [Google Scholar]
- Kobelt, D, Schleef, M, Schmeer, M, Aumann, J, Schlag, PM and Walther, W (2013). Performance of high quality minicircle DNA for in vitro and in vivo gene transfer. Mol Biotechnol 53: 80–89. [DOI] [PubMed] [Google Scholar]
- Darquet, AM, Cameron, B, Wils, P, Scherman, D and Crouzet, J (1997). A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther 4: 1341–1349. [DOI] [PubMed] [Google Scholar]
- Kreiss, P, Cameron, B, Darquet, AM, Scherman, D and Crouzet, J (1998). Production of a new DNA vehicle for gene transfer using site-specific recombination. Appl Microbiol Biotechnol 49: 560–567. [DOI] [PubMed] [Google Scholar]
- Bigger, BW, Tolmachov, O, Collombet, JM, Fragkos, M, Palaszewski, I and Coutelle, C (2001). An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J Biol Chem 276: 23018–23027. [DOI] [PubMed] [Google Scholar]
- Chen, ZY, He, CY, Ehrhardt, A and Kay, MA (2003). Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther 8: 495–500. [DOI] [PubMed] [Google Scholar]
- Mayrhofer, P, Blaesen, M, Schleef, M and Jechlinger, W (2008). Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. J Gene Med 10: 1253–1269. [DOI] [PubMed] [Google Scholar]
- Rischmüller A, Viefhues M, Dieding M, Blaesen M, Schmeer M, Baier R et al. (2013). In: Minicircle and Miniplasmid DNA Vectors. The Future of Nonviral and Viral Gene Transfer. Schleef M (ed.). Wiley-Blackwell, Weinheim, Germany. PP. 71–91. [Google Scholar]
- Hacker, UT, Gerner, FM, Büning, H, Hutter, M, Reichenspurner, H, Stangl, M et al. (2001). Standard heparin, low molecular weight heparin, low molecular weight heparinoid, and recombinant hirudin differ in their ability to inhibit transduction by recombinant adeno-associated virus type 2 vectors. Gene Ther 8: 966–968. [DOI] [PubMed] [Google Scholar]
- Grimm, D, Kay, MA and Kleinschmidt, JA (2003). Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther 7: 839–850. [DOI] [PubMed] [Google Scholar]
- Wobus, CE, Hügle-Dörr, B, Girod, A, Petersen, G, Hallek, M and Kleinschmidt, JA (2000). Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74: 9281–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D, Kern, A, Pawlita, M, Ferrari, F, Samulski, R and Kleinschmidt, J (1999). Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther 6: 1322–1330. [DOI] [PubMed] [Google Scholar]
- Kern, A, Schmidt, K, Leder, C, Müller, OJ, Wobus, CE, Bettinger, K et al. (2003). Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol 77: 11072–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister, JR and Muzyczka, N (1999). Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol 73: 9325–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, UT, Wingenfeld, L, Kofler, DM, Schuhmann, NK, Lutz, S, Herold, T et al. (2005). Adeno-associated virus serotypes 1 to 5 mediated tumor cell directed gene transfer and improvement of transduction efficiency. J Gene Med 7: 1429–1438. [DOI] [PubMed] [Google Scholar]
- Chen, ZY, He, CY, Meuse, L and Kay, MA (2004). Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther 11: 856–864. [DOI] [PubMed] [Google Scholar]
- Tan, Y, Li, S, Pitt, BR and Huang, L (1999). The inhibitory role of CpG immunostimulatory motifs in cationic lipid vector-mediated transgene expression in vivo. Hum Gene Ther 10: 2153–2161. [DOI] [PubMed] [Google Scholar]
- Gaspar, V, de Melo-Diogo, D, Costa, E, Moreira, A, Queiroz, J, Pichon, C et al. (2015). Minicircle DNA vectors for gene therapy: advances and applications. Expert Opin Biol Ther 15: 353–379. [DOI] [PubMed] [Google Scholar]
- Daya, S and Berns, KI (2008). Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21: 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, JH, Zolotukhin, S and Muzyczka, N (1996). Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol 70: 1542–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, P, Elias, P and Linden, RM (2003). Rescue of the adeno-associated virus genome from a plasmid vector: evidence for rescue by replication. J Virol 77: 11480–11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt, FC and Samulski, RJ (2010). Creating a novel origin of replication through modulating DNA-protein interfaces. PLoS One 5: e8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski, RJ, Srivastava, A, Berns, KI and Muzyczka, N (1983). Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33: 135–143. [DOI] [PubMed] [Google Scholar]
- Wang, XS, Ponnazhagan, S and Srivastava, A (1995). Rescue and replication signals of the adeno-associated virus 2 genome. J Mol Biol 250: 573–580. [DOI] [PubMed] [Google Scholar]
- Wang, XS, Ponnazhagan, S and Srivastava, A (1996). Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol 70: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, P and Berns, KI (1991). In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol 218: 791–804. [DOI] [PubMed] [Google Scholar]
- Bleker, S, Sonntag, F and Kleinschmidt, JA (2005). Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol 79: 2528–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrels, W, Talluri, TR, Ziegler, M, Most, I, Forcato, DO, Schmeer, M et al. (2016). Cytoplasmic injection of murine zygotes with Sleeping Beauty transposon plasmids and minicircles results in the efficient generation of germline transgenic mice. Biotechnol J 11: 178–184. [DOI] [PubMed] [Google Scholar]
- Schleef, M, Blaesen, M, Schmeer, M, Baier, R, Marie, C, Dickson, G et al. (2010). Production of non viral DNA vectors. Curr Gene Ther 10: 487–507. [DOI] [PubMed] [Google Scholar]
- Büning, H, Perabo, L, Coutelle, O, Quadt-Humme, S and Hallek, M (2008). Recent developments in adeno-associated virus vector technology. J Gene Med 10: 717–733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.