Abstract
The regulation of function of endothelial cell–cell junctions is fundamental in sustaining vascular integrity. The polycistronic microRNA (miR) complexes containing miR-23a-27a-24-2, and 23b-27b-24-1 are predicted to target the majority of major endothelial junctional proteins. We focus on miR-23a and miR-23b, and investigate the functional effects of these miRs on junctions. While miR-23a and 23b only differ by 1 nucleotide (g19) outside the seed region and thus are predicted to have the same targets, they function differently with miR-23a inhibiting permeability and miR-23b inhibiting angiogenesis. Both miRs target the junctional attractive molecule (tight junction protein 2) ZO-2 and the repulsive molecule junctional adhesion molecule C (JAM-C), although the inhibition of JAM-C by miR-23a is more profound than by miR-23b. The difference in potency is attributable to differences at g19 since a mutation of the t17, the g19 binding site of miR-23b in the 3′UTR of JAM-C restores identity. We also show that the pattern of expression of miR-23a and miR-23b and their targets are different. Thus, the paralogues miR-23a and miR-23b can have profoundly different effects on endothelial cell function due at least partially to selective effects on target proteins and differences in expression patterns of the miRs. This work exposes a hitherto unappreciated complexity in therapeutically targeting miRs.
Keywords: miR-23-27-24, endothelial cells, cell-cell junctions, miR-23
Introduction
The sustenance of normal vascular permeability is one of the chief functions of endothelial cells (ECs) and is achieved mainly by the regulation of tightness of endothelial cell–cell junctions. Junctional integrity is maintained by a balance of adhesive junctional proteins, such as VE-Cadherin, ZO-1, ZO-2, and repulsive proteins such as JAM-C, SEMA-6A, and semaphorin.1,2,3,4,5 The coordination of regulation of expression and signaling through these proteins is very important to allow transient changes as seen in acute inflammation and prevent chronic changes evident in almost all inflammatory and degenerative diseases. Targeting such junctional proteins is a possible approach for development of therapeutics particularly for diseases where vascular leak is central to the underlying pathology.
MiRNAs (miRs) are a highly conserved class of endogenous small noncoding RNAs (20–25 nucleotides), which regulate genes at the post-transcriptional stage of expression.6 MiRs have been shown to be dysregulated and contribute to the initiation and development of many diseases.7,8 MiRs are able to inhibit the post-transcriptional expression of multiple genes or gene families involved in development or complex cellular functions,9 and are thus well suited to coordinate the function of cell junctions.
The complexity and importance of these interactions was highlighted by our recent description of miR-27a as a potent regulator of the proadhesive protein VE-Cadherin,10 while others have demonstrated miR-27 regulation of the antiadhesive proteins SEMA-6A and semaphorin.5,11,12 This contrasting function led to the development of an RNA-derived drug that specifically targets the miR-27a-VE-Cadherin interaction and has potent antipermeability properties and has suitable drug characteristics.10
MiR-27 is part of a poly-cistronic group of cotranscribed miRs, including 23 and 24. There are two copies of this miR cistron in the genome, miR-23a-27a-24 being intergenic with its own promoter on chromosome 19 and miR-23b-27b-24, being intronic on chromosome 9. This organization has been highly preserved in vertebrates and also in fish, albeit less closely clustered.13,14 Members of the cluster have been described to have functions in several organ/developmental systems, including in cancer,15 the central nervous system 16 and vascular organs.17,18 Most recently the cluster has been shown to regulate T cell differentiation.19 In the vascular system there has been a linkage primarily to angiogenesis, cardiac development and apoptosis.5,11,20,21,22 However bioinformatic analysis of the potential targets of these miRs finds a concentration of molecules expressed in adherens and tight junctions,13 and in particular, we note representation of almost all known members of endothelial cell–cell junctions, giving rise to the possibility that endothelial junctional regulation is a chief function of these coevolved set of miRs.
Mature sequences of miR-23a and miR-23b have identical seed sequences and only differ by one nucleotide in the nonseed region. As a result, in silico target analysis (Targetscan, DIANA lab, and www.microRNA.org) has suggested they share the same putative target genes and thus are likely to have similar biological functions. Despite reports showing distinct function between miRs with the same seed sequence, few studies have focused on the possible difference in the biological effects or the targets of these miRs.23,24,25 By using both gain- and loss-of-function approaches, show that in ECs, miR-23a and miR-23b do not behave in the expected identical manner. Overexpression of miR-23a inhibits EC permeability whereas miR-23b increases permeability. Moreover, miR-23b but not miR-23a overexpression regulates angiogenesis. Importantly, such differences are also seen in vivo. Further, we demonstrate that these biological differences likely are attributed to the differential regulation of miR-23a and miR-23b on EC junctional molecules in either a direct or indirect manner. Together our findings provide substantial evidence for the evolution of a vascular permeability miR operon, and demonstrate the differing potent effects of miR-23s on their target genes and diverse role on EC function. The findings have both physiological and clinical relevance at a time where miRNAs and their inhibitors are entering the clinic.
Results
The miR-23-27-24 cistron targets endothelial junctional proteins
We examined the predicted targets of the individual members of the miR-23-27-24 cistron. 2,517 genes are predicted (by TargetScan 6.2) to be targets of at least one member of miR-23-27-24 cluster. A gene set enrichment analysis (GSEA) revealed that many of the predicted targets (see Supplementary Table S1) are involved in the adherens junctions pathway (P = 9.67 e−18) with a highly significant False Discovery Rate q-values of 8.01 e−16.
All of the 17 major junctional and 18 downstream signaling or junctional regulating proteins (Figure 1a) known to play important roles in regulating EC junctions,3,5,26,27,28,29,30,31,32,33 are potential targets of at least one member of miR-23-27-24 clusters. Thus, it is highly likely that the whole cistron has a major role in governing endothelial function.
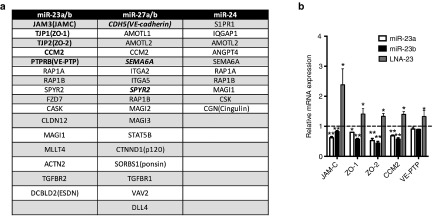
Figure 1.
Regulation of predicted targets by miR-23-27-24 cluster. (a) Predicated and validated targets of miR-23~miR-27~miR-24 clusters known to be present in tight and adherens junctions in ECs. miR-23 includes miR-23a and miR-23b. miR-27 includes miR-27a and miR-27b. Validated targets are in bold. (b) Relative mRNA expression of the predicted miR-23 targets JAM-C, VE-PTP, CCM2, ZO-1 and ZO-2 in HUVEC transfected with miR-23a or miR-23b mimic, LNA to miR-23 or controls. Expression levels were measured by qRT-PCR with results of mRNA normalized to β-actin. mean ± SEM; n = 3. *P < 0.05; **P < 0.005. LNA, locked nucleic acid; HUVEC, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Of the predicted EC junctional targets of miR-23-27-24, three (VE-cadherin, SEMA6A, Sprouty2) have been confirmed by our group10 and by others.24 Here we focus on targets of miR-23 namely the expression of five of the most conserved (ZO-1, ZO-2, JAM-C, VE-PTP, and CCM2). Human umbilical vein endothelial cells (HUVEC) (Figure 1b) or human cerebral microvascular endothelial cell line (hCMEC/D3) (see Supplementary Figure S1a, b) transfected with either locked nucleic acid (LNA) to miR-23 or mimics of miR-23a or miR-23b. Mimics to miR-23a and miR-23b were able to specifically increase miR-23a and miR-23b expression respectively (see Supplementary Figure S2a), whereas with the LNA, both miR-23a and miR-23b was reduced to a similar level (see Supplementary Figure S2b), The manipulation of endogeneous miR-23a/b led to appropriate up or down regulation of all five genes, as measured by polymerase chain reaction (PCR). Therefore there are at least eight major EC junctional molecule targeted, either directly or indirectly, by the miR-23-27-24 cistron5,10,11,22,29,34(bold in Figure 1a).
Taken together, these data indicate that miR-23-27-24 targets are specifically highly enriched in EC tight and adherens junctions. We thus propose that the miR-23-27-24 cluster serves as a vascular cell–cell junctional operon. It should be noted that the miRs target both attractive and repulsive proteins, thus there is likely to be a fine tuning in the release of these miRs or access to their respective targets to coordinate the opening and closing of cell–cell junctions.
Expression patterns of miR-23a and miR-23b in angiogenesis
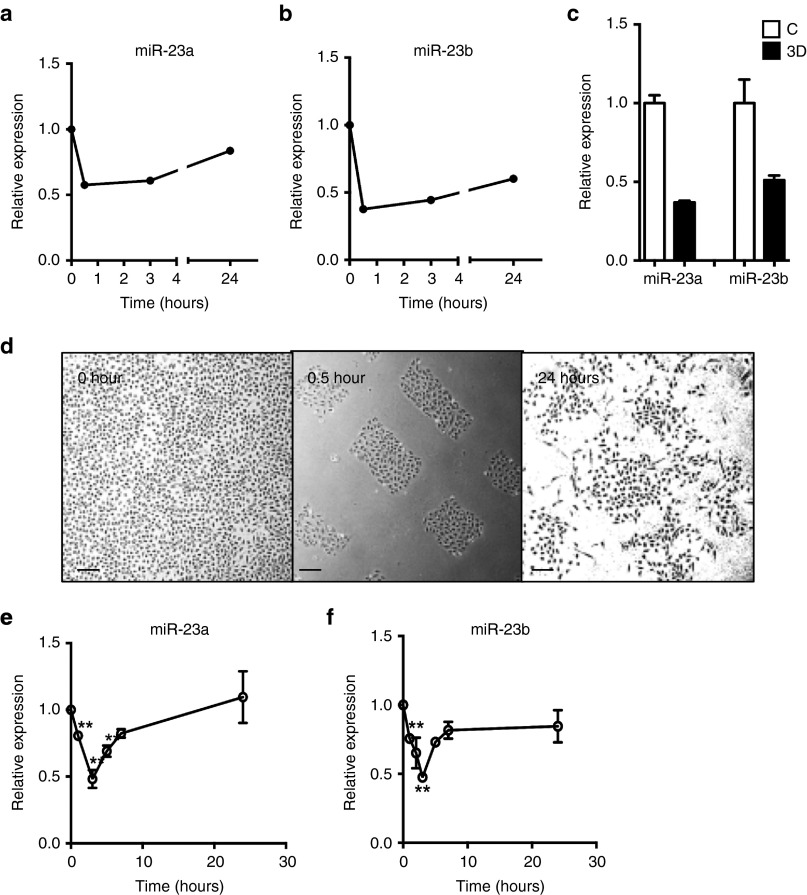
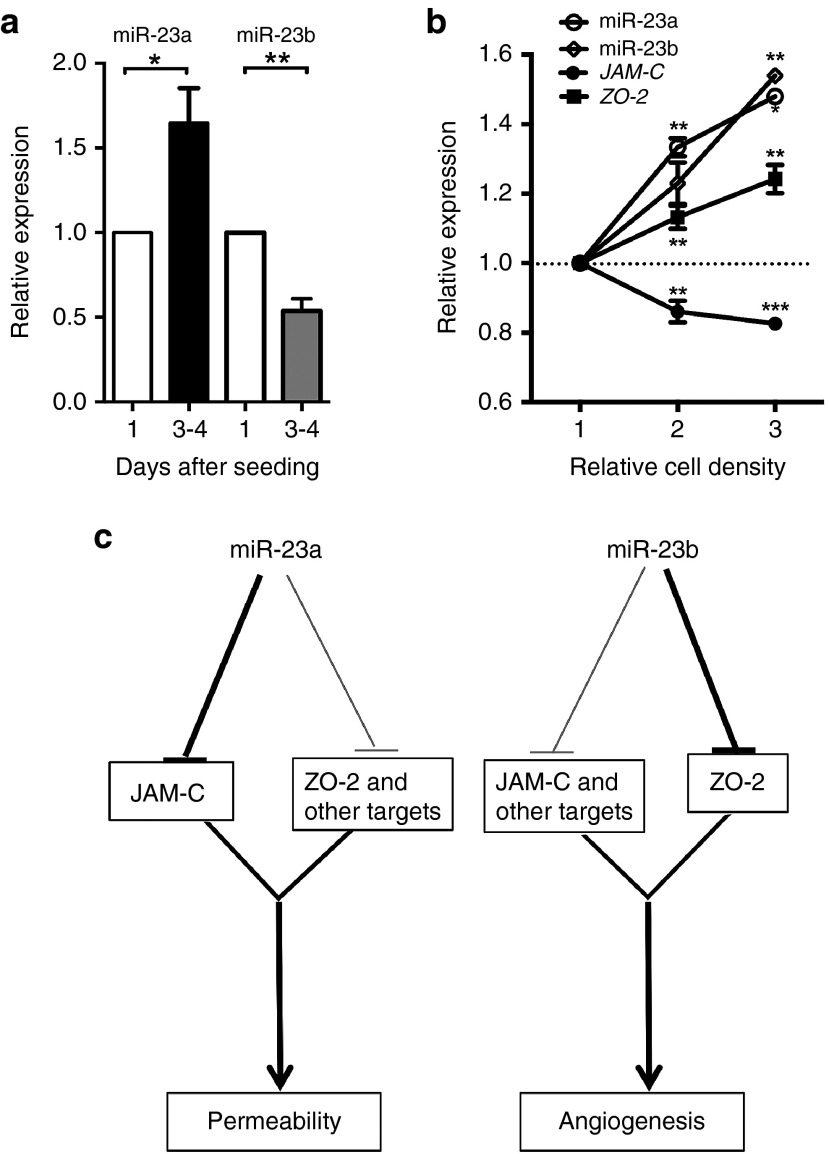
The miR-23-27-24 cluster was originally identified using microarray analysis (GSE50437) and found to be regulated in ECs during in vitro 3-dimensional (3D) collagen angiogenesis assay. Both miR-23a and miR-23b showed a rapid and sustained downregulation in their expression (Figure 2a,b). This microarray data for miR23 was validated in the 3D collagen angiogenesis assay by quantitative PCR (Figure 2c). The pattern is similar to that we have previously observed with miR-27a.10 Since the earliest events in in vitro angiogenesis are cell movement and alignment, the rapid decrease in miR maybe associated with loss of cell–cell interactions or cell migration. To test this we measured miR expression in migrating ECs, in a modified “scratch assay”, from a wounded edge of a confluent monolayer (Figure 2d). Both miR-23a and miR-23b were rapidly down-regulated within 30 minutes of wounding and also at times that reflect the migration stage. Normal levels of expression of both miRs are achieved by 6 hours and times thereafter (Figures 2e,f). These data indicate that miR-23a and miR-23b were both down-regulated in the early stage of angiogenesis, times associated with loss of cell–cell interactions and the induction of migration.
Figure 2.
Expression pattern of miR-23a and miR-23b during angiogenesis. (a) Expression of miR-23a and (b) miR-23b in EC during in vitro 3D collagen angiogenesis assay measured by microarray. (c) Confirmation of microarray by real-time PCR. C = nonangiogenic, 3D = in vitro 3D collagen angiogenesis assay. mean ± SEM; n = 2. (d) Morphology of EC either before wounding (0 hour) and 0.5 and 24 hours after wounding. Representative of three experiments. Scale bar = 100 μm. Quantification of miR-23a (e) and miR-23b(f) after scratch (mean ± SEM; n = 3). Data is compared with the levels in the confluent cells. Expression levels were measured by qRT-PCR with results of microRNA normalized to U48. *P < 0.05; ***P < 0.001; ****P < 0.0001 by unpaired two-tailed t-test. qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
MiR-23a and miR-23b differentially regulate angiogenesis
To test whether the down-regulation of miR-23 family is essential to the angiogenic process, we manipulated the levels of the miRs and investigated changes in the early phases of an angiogenesis assay.
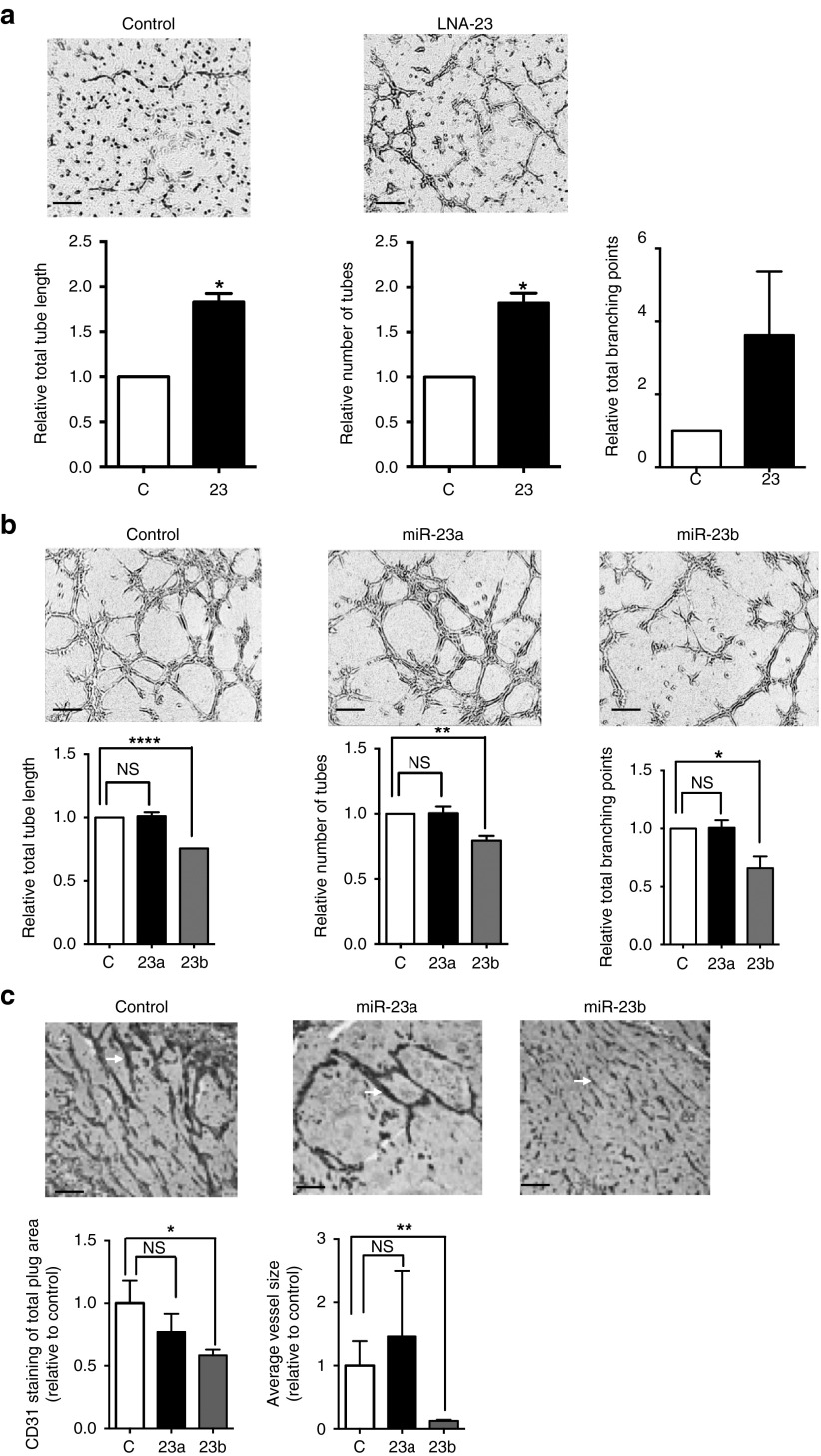
The knockdown of miR-23a and miR-23b by LNA-23 significantly enhanced tube formation on Matrigel, as quantified by tube length, tube number and number of branching points (Figure 3a). These results indicate that down-regulation of miR-23 promotes angiogenesis in vitro. Next, we investigated the role of each of the individual miRs in angiogenesis. Since LNAs against the individual members could not be designed to achieve specificity, we used mimics of each of these transfected into ECs. Cells were seeded onto Matrigel 48 hours after transfection. Different cell concentrations were used in order to see enhancement or inhibition of tube formation (See “Materials and Methods”). Overexpression of miR-23b, but not miR-23a significantly impaired the formation of capillary-like structures in ECs causing a decrease of total tube length, total tube numbers, and total branching points (Figure 3b). This was interesting as miR-23a and miR-23b are predicted to have the same targets due to their identical seed sequence and were similarly downregulated in angiogenesis (Figure 2a,b). The difference in the regulation of angiogenesis between miR-23a and miR-23b also occurs in vivo. Matrigel plugs containing either miR23a, miR-23b or control miR mimic were injected into mice and the neovascularization quantified by total vessel area and average vessel size, as determined by immunohistochemical analyses of CD31 expression (Figure 3c). MiR-23b containing plugs significantly inhibited the angiogenic response whereas the miR-23a mimic treated mice did not show any significant change compared with control. Taken together, these data indicate that miR-23b but not miR-23a regulates angiogenesis.
Figure 3.
Mir-23b but not and miR-23a regulates EC tube formation. (a) LNA-23 increased HUVEC tubule formation on Matrigel. (b) Over-expression of miR-23b but not miR-23a inhibits tubule formation on Matrigel. Cells were monitored and photos taken every hour, for 7 hours. Representative images all taken at the same time point for each experiment are shown with a scale bar = 100 μm. Images analysis and quantification were performed by Wimasis software (Wimasis GmbH). Graphs of tube length, number and branching points show the mean ± SEM. *P < 0.05; **P < 0.005; ****P < 0.0001 by unpaired two-tailed t-test. n = 3 independent EC lines. (c) Angiogenic effects of miR-23a and miR-23b in mice. Mice received subcutaneous injection of 0.5 ml of a Matrigel mixture containing FGF-2, Heparin, and mimics. The animals were killed and gels dissected 2 weeks later. 5 μm sections were stained for CD31 (n = 3–4 for each plug) and a representative image is shown. Pixel histogram quantitation of percentages of vessel area (CD31 positive staining) and average vessel size with miR-23a, miR-23b or control mimic is shown. Each group contained 3–4 animals. Scale bar = 200 μm, *P < 0.05, **P < 0.01. NS = not significant. LNA, locked nucleic acid; HUVEC, human umbilical vein endothelial cells.
MiR-23a and miR-23b regulate vascular permeability differently
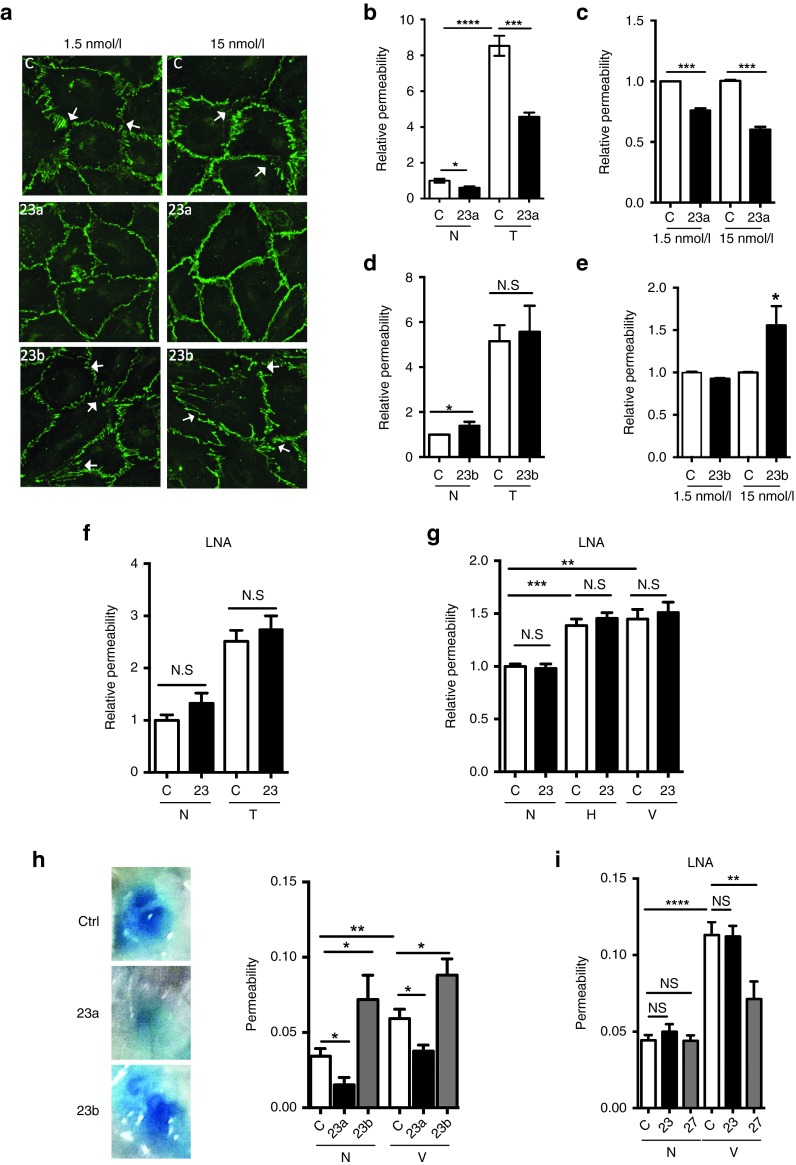
Changes in vascular permeability are associated with pathological angiogenesis.35,36 We investigated whether miR-23a and miR-23b impacted on permeability. Since EC junctions are directly involved in regulating vascular permeability, we first examined the EC junction pattern, as shown by VE-cadherin staining, after miR-23 mimics treatment. In a loosely packed HUVEC monolayer, the control cells showed open zipper–like staining for VE-cadherin, whereas miR-23a mimic treated cells showed a smoother and continuous VE-cadherin staining at both 1.5 nmol/l and 15 nmol/l, indicating tightly apposed junctions (Figure 4a). In contrast, more gaps and wider zipper-like staining for VE-cadherin between cells were observed in miR-23b mimic treated cells particularly in the cells that received 15 nmol/l of 23b mimic (Figure 4a, white arrows). These data indicate miR-23a and miR-23b may have opposite roles in the regulation of cellular junctions and hence permeability, where miR-23a may inhibit permeability while miR-23b may promote permeability.
Figure 4.
MiR-23a inhibits and miR-23b enhances EC permeability. Cells were stimulated with 0.1–0.2 U/ml of thrombin, 10 µmol/l of histamine or 50 ng/ml of human VEGF165 for 30 minutes as indicated for in vitro experiments. (a) 48 hours after 1.5 or 15 nmol/l of miR-23a or miR-23b mimic treatment, HUVEC were stained for VE-cadherin (green). White arrows = examples of changed adherens junctions (VE-cadherin-staining). (b,d) Permeability measured in control or miR-23a (b) or miR-23b (d)-mimic–transfected cells without (N) or with thrombin stimulation (T). n = 4 experiments. (c,e) Permeability measured at basal level in control or cells transfected with 1.5 or 15 nmol/l miR-23a-mimic (c) or miR-23b (e). n = 3 experiments, where values are normalized to control (C). (f,g) Permeability was measured without (N) or with thrombin (T, n = 6 experiments), histamine (H, n = 4 experiments) or VEGF (V, n = 5 experiments) stimulation in control or miR-23 LNA transfected EC. (h) The Miles assay was performed with 4 µg of control mimic or miR-23a or miR-23b mimic injected intradermally into the back of the mice. 24 hours later, 200 μl 0.5% Evan's Blue was injected intravenously. After 30 minutes, 10 ng VEGF (V) or phosphate-buffered saline (N) was given into the same site as the mimic. Mice were sacrificed 30 minutes later and the dye was extracted from skin samples and quantified; # mice are: N+C, n = 10; N+23a, n = 6; N+23b, n = 5; V+C, n = 12, V+23a, n = 11; V+23b, n = 6. Representative photos of lesions are given. (i) The Miles assay was performed with 4 µg of control LNA, LNA-23 or LNA-27 without (N) or with VEGF stimulation (V) with set up given as in f above. # mice are: N+C, n = 16; N+23, n = 16; N+27, n = 8; V+C, n = 16, V+23, n = 16; V+27, n = 8. All graph represents mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001 by unpaired two-tailed t-test. LNA, locked nucleic acid.
Later, we investigated the effects of miR-23a and miR-23b using in vitro permeability assays. HUVEC were transfected with miR mimics or LNA-23 and 24 hours later, plated on permeable membranes for another 24 hours to allow the development of cell junctions. Thrombin was used to induce permeability and extent of permeability measured by the passage of fluorescein isothiocyanate (FITC)-dextran through the monolayer. Basal and thrombin stimulated permeability was inhibited by mimic 23a overexpression (Figure 4b). The inhibition by miR-23a was independent of the amount of mimic transfected since inhibition was seen at both 1.5 nmol/l and 15 nmol/l (Figure 4c). In contrast miR-23b overexpression did not inhibit permeability rather showing a significant increase in basal permeability with the higher dose of 15 nmol/l of mimic 23b (Figure 4d,e). A slight trend to increase thrombin-induced permeability was seen in some experiments but was not consistent using mimic 23b transfected cells (Figure 4d). These data are consistent with the effect of miR-23a and miR-23b on the pattern of EC junctions (Figure 4a). In comparison, LNA to miR-23 (inhibiting both 23a and b) did not significantly change either basal nor thrombin (Figure 4f), histamine or vascular endothelial growth factor (VEGF) (Figure 4g) stimulated permeability. Since we showed that LNA was able to down-regulate the endogenous expression of miR23a/b and also to enhance EC tube formation, the LNA was indeed functional. Therefore, the lack of effectiveness in changing permeability is likely due to opposing roles of miR-23a and b in regulating EC permeability.
The effect of the mimics on permeability in vivo was assessed by the Miles assay.10 Mice were injected intradermally with 4 µg of the miR-23 mimics, LNA-23 or controls and 24 hours later assessed for response to VEGF. Mir-23a mimic significantly inhibited both the basal permeability and the VEGF-induced increase in vascular permeability (Figure 4h). MiR-23b mimic actually increased permeability consistent with that seen in vitro (Figure 4d,e). LNA to miR-23 did not alter vascular permeability either basal or VEGF-stimulated permeability (Figure 4i). However, LNA to miR-27 inhibited VEGF-induced permeability as we have shown previously (Figure 4i).10 Thus, miR-23a and miR-23b inhibited permeability in opposite manner as shown both in vitro and in vivo.
JAM-C and ZO-2 are direct targets of miR-23a and miR-23b
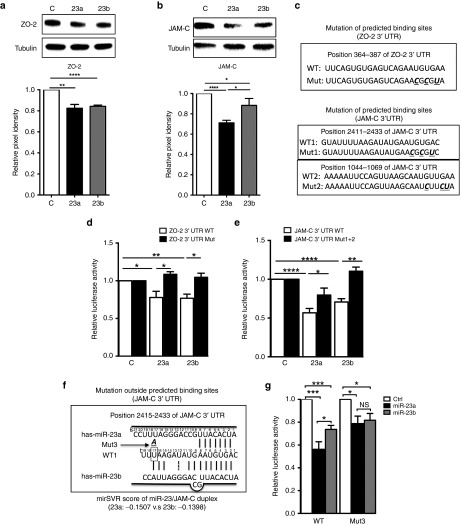
Our functional data shows that miR-23a and miR-23b regulate permeability and angiogenesis differently. In order to elucidate the molecular basis of miR-23a/b-mediated regulation of EC function we chose to analyze the responses of endogenous ZO-2 and JAM-C to miR-23. These were chosen as the miR binding sites in their 3′UTR are of high affinity and conserved and these targets have been shown to influence angiogenesis and permeability.2,3
MiR-23 binding sites are the only miR binding sites in the 3′UTR of JAM-C and one of the only two in ZO-2 3′UTR as predicted by Targetscan (see Supplementary Figure S3a,b). HUVECs were transfected with miR-23a, miR-23b, or control mimics and harvested for protein analysis 48 hours after. Overexpression of either miR-23a or miR-23b inhibited ZO-2 to a similar extent (Figure 5a). Overexpression of miR-23a and miR-23b inhibited JAM-C expression although the extent of inhibition of JAM-C by miR-23a mimic was consistently and significantly greater than that seen by miR-23b (Figure 5b). The difference became greater when the transfection dose decreased from 15 nmol/l to 1.5 nmol/l (see Supplementary Figure S4a). The miR-23a/b complementary target site in JAM-C and ZO-2 3'UTR are different between human and mouse (see Supplementary Figure S3a,b). However, mouse and human EC had a similar pattern of response to miR23a and miR-23b in target preference. Overexpression of either miR-23a or miR-23b in mouse brain microvascular ECs inhibited zo-2 mRNA to a similar extent, but miR-23a inhibited jam-c expression to a greater extent especially at lower doses (see Supplementary Figure S4b) similar to that in human ECs (Figure 1b). Thus, miR-23a/b regulate in a similar fashion the expression of the proadhesive protein ZO-2, but miR-23a inhibits the propermeability protein JAM-C consistently and more significantly than miR-23b. Interestingly miR-23a overexpression inhibits permeability (Figure 4h) consistent with its inhibition of JAM-C. Although the degree of inhibition of target proteins was not high, the results are consistent with what we have previously observed for miR-27a targeting VE-cadherin10 and also consistent with the biology of EC junctions, where small changes in the junctional proteins can have profound functional effects.
Figure 5.
MiR-23 directly target ZO-2 and JAM-C. (a) overexpression of miR-23a and miR-23b inhibited ZO-2 protein expression to a similar extent, n = 4 independent experiments.(b) overexpression of miR-23a inhibited JAM-C to a greater extent than miR-23b overexpression, n = 7 independent experiments. Representative Western blot is shown in upper panel and quantified by ImageJ as Pixel density. α-Tubulin was used as a loading control. (c) Putative miR-23 binding site and mutation within the ZO-2 and JAM-C 3′UTR. Nucleotides that were mutated are given in bold, underlined and italic ones in Mut. (d,e) Luciferase reporter assays, in which HEK293T cells were cotransfected with reporter constructs containing full length 3′UTR of the wild-type ZO-2, JAM-C mRNA (WT) or mutated miR-23 binding sites (Mut), together with miR-23a, miR-23b or control mimics. n = 3 independent experiments. (f) Schematic alignment between mature miR-23a/b and JAM-C 3′UTR shown at www.microrna.org. g, guide miRNA; t, target mRNA. Site t17 (in the rectangle) within WT1 was mutated to A (bold, italic and underlined) to become Mut3. (g) Luciferase reporter assays, in which HEK293T cells were cotransfected with reporter constructs containing full length 3′UTR of the wild-type JAM-C (WT), or single nucleotide mutation outside binding sites (Mut3), together with miR-23a, miR-23b or control mimics. n = 7 independent experiments. Data in all graph shown as mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001.
To confirm that ZO-2 and JAM-C were direct targets we constructed luciferase reporter plasmids containing the complete wild-type 3′UTR of ZO-2 and JAM-C mRNA as well as mutants containing mutation sites in the predicted miR-23a/b binding sites (Figure 5c). These vectors were cotransfected into HEK293T cells with either miR-23a/b mimic or negative control. Firefly vector was also cotransfected as an internal control. Both miR-23a and miR-23b mimic significantly inhibited ZO-2 (Figure 5d) and JAM-C (Figure 5e) compared with the negative control and mutation of the predicted binding sites reversed the loss of luciferase activity (Figure 5d,e). Collectively, our data suggested that miR-23a and miR-23b directly targeted JAM-C and ZO-2.
A single nucleotide difference outside the seed sequence of miR-23s is responsible for differential regulation of JAM-C
The inhibition of luciferase activity of ZO-2 is similar between miR-23a and miR-23b (Figure 5d). However, the inhibition of luciferase activity of JAM-C by miR-23a was significantly stronger than by miR-23b (Figure 5e). This is consistent with the greater inhibition of protein expression of JAM-C by miR-23a compared with miR-23b (Figure 5b). Furthermore, this difference was maintained even after the mutation of the two most conserved binding sites (1,044 and 2,415), suggesting the difference did not arise from miR-23 seed sequence binding (Figure 5e). An increase in the dose of mimic (miR-23a or miR-23b) still showed miR-23a has a more pronounced inhibitory effect than miR-23b on JAM-C (not shown).
Since nucleotide g19 (guide strand19) is the only difference between miR-23a and miR-23b, we examined the effect this difference had on binding characteristics. The miR-23a/ZO-2 alignment is the same as miR-23b/ZO-2 alignment on the conserved 366 site, with an identical mirSVR score of −0.9603 (www.microrna.org). However, the miR-23a/JAM-C alignment is different to miR-23b/JAM-C alignment on the well-conserved 2415 site (WT1) with mirSVR scores of −0.1507 versus −0.1398 respectively. This suggests that miR-23a has a better base-pair binding than miR-23b to JAM-C 3′UTR. Indeed, as shown in Figure 5f, g19 “A” in miR-23b pairs with t17 “U”, with an accompanying loop-out of residues g9 and g10 to allow an additional pairing of t9 and t10 with g11 and g12. This altered configuration of miR-23b/JAM-C spanning nucleotides 9–12 is likely to cause topological constraints within its nucleic acid-binding binding channel of Ago2. The extension of complete pairing beyond the seed region has been shown, in structural studies, to cause major changes in Kd.37 To examine whether these differences contributed to stronger effects of miR-23a on JAM-C, we mutated t17“U” in JAM-C 3′UTR site to “A” (Figure 5f). The mutation (Mut3) abolished the difference in binding to JAM-C 3′UTR between miR-23a and miR-23b, although both miR-23a and miR-23b still significantly inhibited the luciferase activity (Figure 5g). Thus together, these experiments show that seed sites are not the only sequences contributing to target repression, with nucleotides outside the seed sequence also contributing additional affinity.
Physiological regulation of miR-23 paralogues
Since miR-23a and miR-23b potentially can have different targets, it may suggest that they will show different patterns of regulation during the different stages of angiogenesis. Therefore, we measured endogenous expression of miR-23a/b, JAM-C, and ZO-2 in HUVECs under different conditions. When HUVECs were grown to confluence, miR-23a was significantly and consistently up-regulated on day 3 and 4 whereas miR-23b was down-regulated (Figure 6a). In contrast both showed a parallel up-regulation when cells were plated at different densities (Figure 6b). The increase in miR-23a and miR-23b upon plating at different densities was accompanied by a decrease of JAM-C but not of ZO-2 (Figure 6b). Thus, both miR-23a and miR-23b and their targets ZO-2 and JAM-C are differentially regulated in expression depending on context.
Figure 6.
Physiological regulation of miR-23 paralogues. (a) Expression of miRNA-23a and miR-23b in 3–4 days postseeding. Data are normalized to levels on Day 1 after seeding, which did not differ from Day 0. (b) Expression of miR-23a, miR-23b, and mRNA for JAM-C and ZO-2 24 hours after seeding at different density. Density 1, 2, and 3 corresponds to 1 × 105, 2 × 105, and 3 × 105 HUVECs per well of six-well plate. Expression levels were measured by qRT-PCR, with the results of microRNA normalized to U48 and mRNA normalized to β-actin. Graph represents mean ± SEM; n = 5 independent experiments. *P < 0.05, **P < 0.01. (c) Schematic of proposed function of miR23a and miR-23b in the regulation of EC function. LNA, locked nucleic acid; HUVEC, human umbilical vein endothelial cells; PCR, polymerase chain reaction.
Discussion
The tight control of vascular permeability is one of the chief functions of the endothelial lining of blood vessels. Abnormal vascular leakage contributes to many pathophysiological events including all forms of ischemia, trauma, inflammatory and degenerative diseases.10,38,39,40 EC–EC junctions are formed by at least 17 proteins, uniting in either adherens or tight junctions. These in turn are controlled by intracellular signals generated by agonists such as histamine, VEGF and thrombin, by the balance of expression of proadhesive and antiadhesive proteins and by the macromolecular assembly of the components. It has been anticipated that such a complex cellular system is subject to fine tuning and overall orchestration by microRNAs, since they are known to have special roles in complex cellular and developmental systems.5,11,20,21,22
The miR-23-27-24 cluster has been noted to have a diverse set of effects on the cardiovascular system and been linked to individual targets of single miRs in this cluster.18,41 While this has been a significant advance, an additional challenge is to understand how this highly conserved cistron orchestrates these cardiovascular effects. We and others have noted that bioinformatics analysis has revealed a highly significant concentration of targets related to adherens junctions, tight junctions and transendothelial migration, of members of the cistron.13 We have previously validated VE-cadherin being a target of miR-27a10 and now we show that an additional five junctional proteins are regulated by miR-23. Two of these five proteins are confirmed to be direct targets of miR-23. In total, 3 of 17 major junctional molecular (18%) and 8 of 35 junctional regulating proteins (23%) are validated. Statistical analysis demonstrated that this enrichment in EC junctional related genes was extremely unlikely to have accumulated by chance (P = 2.87 e−6, q = 5.31 e−3), thus suggesting very strongly that this conserved group of miRs has evolved to orchestrate (among other things) the function of EC junctions. The paradoxical situation however arises, as the proteins regulated often have opposing effects. For example, VE-cadherin and ZO-2 keep junctions intact,1,2 while JAM-C and SEMA-6A have repulsive properties.3,12 Thus, questions arise as to how the balance between these influences is regulated, and indeed whether individual miRs can be therapeutic targets having opposing functional targets. We have previously addressed the latter issue where we used the novel Blockmir technology to specifically and successfully target miR-27a-VE-cadherin interaction.10
With an identical seed sequence, it was somewhat surprising that the two miRs demonstrated robust differences on two important EC functions, angiogenesis and vascular leak. A similar difference between these isoforms was also noted in their effect on cytotoxic T cells where only miR-23a suppressed function although miR-23a and miR-23b had similar expression pattern.25 We have observed a difference in the effect of miR-27a/b on VE-cadherin (data not shown), and others have of the miR-126a/b-Pak1 interaction.24 The complexity of miR effects arises not only because the same miR can target genes with opposing functional effects, but also because some targets (or long noncoding RNAs) can also act as decoys altering the availability of miRs42,43,44,45 and in some instances the star strand (with a complementary sequence to the miR) can become functional.46,47,48,49 We have here investigated the molecular basis of these differences in miR-23 on the vascular system by choosing two of its predicted targets, the attractive protein ZO-2 and repulsive protein JAM-C. The choices were based on the tightness of binding of miR-23 to the 3′UTR of these genes (both bound at least 7/8 of the seed region in both human and mouse Supplementary Figure S3), the uniqueness of binding (being either the only (JAM-C) or one of two miRs (ZO-2) and the fact that these targets have been linked to regulation of angiogenesis and permeability.2,3 We confirmed that JAM-C and ZO-2 are direct targets of miR-23a/b, but miR-23a had more significant effects on JAM-C expression than did miR-23b (Figure 5). The single difference predicted a differential pairing (Figure 5f) with a loop out in the original residue 9 and 10, and thus extending complementarity to t9 and 10 (with now g11 and 12). A point mutation in JAM-C changing g19 from A to U abolished the difference in function between the two miRs, whereas mutating the seed region reversed the effect of miR-23b, while maintaining the differential effect of miR-23a and miR-23b. These results are consistent with recent structural and functional studies37 showing contributions from nonseed sequences in miR function and affinity for Ago2. The complementary target site in mouse and human differs by several nucleotides outside the seed region. However, g19 is the critical nucleotide in this region to confer target selection as demonstrated by mutations at this site. We also show that the two paralogues can be expressed differentially. As EC approach confluence there is an increase in expression of miR-23a but a decrease of miR-23b, and there is also a divergence in the expression of the two targets JAM-C and ZO-2. In other cells and stimulation the paralogues are differentially expressed49 and in a number of normal tissue (miRGator v3.0) and tumors.13 Thus, these experiments demonstrate that these two miRs can preferentially target different genes, that they can have different functional consequences depending on their target genes and that this differential target specificity, together with a differential expression of the miRs may have a major role in their functional effects (schematic of proposed functions is given in Figure 6c). Such differences are likely to have general applicability across many miR paralogues. Thus, therapeutic manipulation may need to address these issues.
Materials and Methods
Cell culture. HUVECs were isolated and cultured as previously described10 and used between passage two and four. The umbilical cords were anonymous (donors nonidentifiable) and informed consent was given for their use. HEK293T were maintained in Dulbecco's modified essential medium (Life technologies. Carlsbad, CA) supplemented with 10 % fetal calf serum. The mouse brain EC line MVEC/B3 cell line was cultured in Roswell Park Memorial Institute 1640 (RPMI) 1640 (Life technologies) supplemented with 10% fetal calf serum.50 HCMEC/D3 cell line was kept in Endothelial Basal Medium supplemented by bullet kit (Lonza, Basel, Switzerland).
Transfection with miRNA mimics and LNA. HUVECs were seeded at 3 × 105 cells per 25 cm2 flask and grown overnight. Prior to performing all in vitro experiments, we have confirmed the high level of transfection efficiency (>90%) by using microRNA mimic transfection control- Dy547 labeled (Thermo Scientific Dharmacon) and fluorescence labeled LNA inhibitor (Exiqon, Vedbaek, Denmark). Mimics or control (miRIDIAN microRNA mimics, Dharmacon, Lafayette, CO) of miR23a or miR23b were transfected at final concentration of 15 or 1.5 nmol/l for in vitro functional assays. LNA-modified antisense oligonucleotide miRNA inhibitor or scramble control (Exiqon) were used for inhibition of miR23a/b at final concentration of 15 nmol/l. All transfections were performed with HiPerFect transfection reagent (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Scratch assay. HUVEC cells were grown to 100% confluency on gelatin-coated six-well plates. Scratches across the HUVEC monolayer were then made using a 200 µl pipet tip. Cells were harvested for RNA extraction during an indicated time course.
In vitro permeability assay. In vitro permeability assay was performed in a 24-well tissue culture plate with 12 cell culture inserts as previously described. In brief, 24 hours after transfection, transfected HUVECs were plated onto fibronectin-coated 0.3-µm pores inserts (Transwell; Corning Costar, Acton, MA) at 1 × 105 cells per insert in HUVEC medium. The cell monolayer formed in 24 hours. Cells were stimulated with 0.1–0.2 U/ml thrombin (Sigma, St. Louis, MO), 10 µmol/l histamine (Sigma), or 50 ng/ml human VEGF165 (R&D systems, Minneapolis, MN) for 30 minutes if required. FITC-conjugated dextran was added to the upper chamber, and medium was taken from the lower chamber (20 µl) over the following 60 minutes. The amount of fluorescence was measured using a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany) at an excitation wavelength of 485-12 and emission wavelength of 520P. The FITC-dextran fluorescence intensity over that of controls was used as a measure of the permeability of EC monolayers.
Miles assay for in vivo permeability measurement. Miles assay was employed for measuring in vivo permeability.10 In brief, 4 µg of the miR-23a mimic or LNA-23 or corresponding control was injected intradermally into the back of 8 week-old C57BL/6J-Tyrc-2J/J mice. During intradermal injection, mice were anaesthetized using isoflurane/oxygen (induced at 4% and maintained at 1–2% isoflurane vaporizer during treatment). After 24 hours, 200 µl of 0.5% Evans blue dye was injected intravenously, followed by injection of phosphate-buffered saline or 10 ug of VEGF (R&D systems) intradermally into the same site as the mimic 30 minutes later. After another 30 minutes, mice were sacrificed by cervical dislocation. For measurement, a biopsy of the injection site was taken, the dye eluted in formamide overnight at 56°C with shaking. After centrifugation, the amount of dye was measured at 620 nm using a POLARstar Omega microplate reader (BMG Labtech).
In vitro 3D collagen angiogenesis assay. This was similar to that described elsewhere.51 HUVEC were isolated from umbilical cords and cells were cultured under normal conditions. Cells were then detached and stimulated with an anti-α2β1 antibody (AC11) for 7 minutes and then with phorbol-12-myristate-13-acetate (PMA) for 1 minute. Cells were plated onto 3D collagen for the appropriate time. Capillary tube formation occurred over the following 24 hours.
In vitro Matrigel capillary tube formation assay. The tube formation capability of ECs was evaluated by the Matrigel angiogenesis assay.10 Briefly, HUVECs (3.5 × 104 cells per well for mimic treated cells; 2.5 × 104 cells for LNA experiments in order to see inhibition and enhancement respectively) were plated in a 96-well plate precoated with 100 µl Matrigel (BD Biosciences, Bedford, MA) per well 48 hours post-transfection. Cells were monitored and photos were taken every hour, for 7 hours and then repeated after 18 hours. Image analysis and quantification was performed using the WimTube software (Wimasis GmbH, Munich, Germany) to evaluate the generation of new vessels base on parameters including total tubes, tube length and total branching points. The result was normalized to control.
In vivo Matrigel plug angiogenesis assay. The Matrigel plug assay was performed as we have previously described.10 Six to eight week-old female C57BL/6 mice were injected subcutaneously with 500 µl of Matrigel (BD) premixed with 60 U/ml heparin and 400 ng/ml FGF-2 (Sigma), 15 µg control or miR-23a or miR-23b mimic. After 14 days, the mice were killed, the Matrigel plugs were isolated, resected and fixed in 10% paraformaldehyde. 4 µm cross-sections were stained with CD31 monoclonal Ab (mAb, Caltag Medsystems, Buckingham, UK) to detect ECs of blood vessels in the Matrigel by using immunohistochemistry. The extent of angiogenesis was quantified as the blood vessel density by visually counting the total number of CD31-positive vessels per microscopic field.
Immunofluorescence and confocal microscopy. Localization studies were performed as we have previously described.10 In brief, 1.5 × 104 HUVECs were plated onto fibronectin coated labtek slides, LabTek slides (Thermo Fisher Scientific, Rochester, NY) for 24 hours, followed by treated with miR23a/b mimics or control as described in the method of transfection. 48 hours later, cells were fixed with a 50–50 mixture (v/v) of −20°C acetone – methanol for 20 minutes. Cells were then permeabilized by application of 0.1% Triton X-100 in 2% bovine serum albumin (Sigma). Incubation with the primary antibody (VE-cadherin, goat anti rabbit, Santa Cruz Biotechnology, Santa Cruz, CA) was followed by a secondary antibody Alexa 594 donkey anti goat, Life technologies). Localization of VE-cadherin was viewed under a Leica SP5 confocal microscopy (Leica Microsystems, Wetzlar, Germany). Images were analyzed by using Fiji software (Version 2.2.2-rc-34/1.50a).
Luciferase reporter assay. HEK293T cells were transfected with Renilla luciferase reporter constructs containing miR-23 binding sites (JAM-C 3′UTR WT or mutated sites, Switchgear genomics, Carlsbad, CA), along with pGL3 control vector, which contained the firefly luciferase reporter gene (Promega, Madison, WI) and 30 nmol/l of miR-23a/b mimics or control. Luciferase activity was quantitated 24 hours later using Dual-Luciferase Reporter Assay System (Promega) on a POLARstar Omega microplate reader (BMG Labtech) according to the manufacturer's recommendations. Three independent experiments were performed and assayed in triplicate per group. Data represent the S.E for three experiments. Relative luciferase activity (Renilla/firefly) was expressed in relative luminescence units and plotted.
RNA preparation and quantification of gene expression using quantitative Reverse Transcription PCR. Total RNA, including small RNA fraction was extracted from ECs using Trizol10 For validation of miR-23 family expression from microarray, HUVEC were harvested, stimulated with PMA and AC11 and lysed (as control) or replated after stimulation onto a 3D collagen gel (3D) for 30 minutes. RNA was isolated for each condition and two steps real-time PCR was used to measure miRs. First, 1 μg of RNA was reverse transcribed using microRNA-specific primers from Applied Biosystems. Second, real-time PCR was performed in combination with predesigned primers for miR-23a/b and RNU48 or RNU6 small nuclear RNA (snRU48 and snRNU6B reference gene). For mRNA quantification, cDNA were made by using reverse transcription kit followed by TaqMan probe based real-time PCR (Life technologies). The relative RNA amount was calculated with the 2-ΔΔCT method. All reactions were performed in quadruplicates and repeated in three biological replicates using Applied Biosystems 7900HT machine (Life technologies).
Preparation of cell extracts and Western blotting. Western blot analysis was performed as previously described.10 Whole-cell lysates were prepared using lysis buffer (1 mol/l TrisHCl, pH 7.5, with 1% NP-40, 5 mol/l NaCl, 200 mmol/l EGTA, 500 mmol/l NaF,100 mmol/l Na4P2O7 containing 1× protease inhibitor cocktail (Sigma)). We used NP-40, a milder, nonionic detergents in the lysis buffer for isolating cytoplasmic proteins although it does not lyse nuclear membranes. Proteins concentrations were quantified using Bradford reagent (BioRad, Hercules, CA) and bovine serum albumin as standards. Equal amounts of protein were loaded onto a 4–15% precast polyacrylamide gel (BioRad) and separated by electrophoresis in the running buffer, and then transferred to PVDF (polyvinyl difluoride) membranes in transferring buffer, blocked with 5% skim milk powder in PBS-Tween 20 (0.05%). The membranes were probed with a JAM-C antibody (mouse monoclonal antihuman, Santa Cruz), ZO-2 (Cell Signaling, Danvers, MA), β-actin, α-tubulin (Santa Cruz) overnight at 4 °C or for 1 hour at room temperature, followed by corresponding HRP-conjugated antibody (1:5,000; Amersham, UK) for 1 hour at room temperature. Proteins were visualized using ECL or ECL Plus Western blotting Detection Reagents (GE Healthcare, Little Chalfont, UK) on ChemiDoc MP system (Bio-rad). Densitometric analysis was performed using ImageJ version 1.48, NIH, USA software.
Target prediction. The predicted targets of individual members of the miR-23-27-24 cistron were analyzed by TargetScan (Release 6.2). Modified gene set enrichment analysis (GSEA) was used to assess functional significance at the level of sets of genes as previous described.52 All of these potential targets were compared with the “c2_all” collection of curated gene sets from the Molecular Signatures Database (GSEA Version 2.0.14 Broad Institute, Cambridge, MA), consisting of 1,077 genesets corresponding to BIOCARTA, KEGG, and REACTOME biological pathways. The False Discovery Rate q-values is deemed significant value at < 0.05.
Statistical analysis. Data were presented as mean ± SEM and plotted using GraphPad Prism 5 software. For statistical comparison of two groups, unpaired, two-tailed Student's t-test was used. A value of P < 0.05 was considered statistically significant.
Ethics statement
Animal and Human Ethics approval were obtained from the NSW Local Sydney Health District Ethics Committees. All animal procedures confirm the guideline from National Health and Medical Research Council of Australia. This study was conducted in accordance with the Declaration of Helsinki.
SUPPLEMENTARY MATERIAL Table S1. Targetscan 6.2 or Diana predicated targets of miR-23~miR-27~miR-24 clusters known to be present in KEGG adherens junctions pathway. Figure S1. Quantification of miR-23a and its predicated targets in human cerebral microvascular endothelial cell line treated with miR-23a mimic and LNA to miR-23. Figure S2. Quantification of miR-23a/b in ECs treated with microRNA mimics and LNAs. Figure S3. TargetScan 6.2 predictions of interactions between microRNA and 3′-untranslated region of the JAM-C (JAM3) or ZO-2 (TJP2) mRNA. Only conserved sites for miRNA families broadly conserved among vertebrates were considered for prediction. Figure S4. Differential regulation of JAM-C and ZO-2 by miR-23a and miR-23b.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia, Program Grant #349332 and the National Heart Foundation (G 11S 5855). J.L. is a NHMRC/Gustav Nossal Postgraduate Scholarship holder. J.R.G. holds the Wenkart Chair of the Endothelium at the Centenary Institute and the Sydney Medical School, University of Sydney. The authors declare no conflicts of interest.
Supplementary Material
References
- Vestweber, D (2008). VE-cadherin: the major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler Thromb Vasc Biol 28: 223–232. [DOI] [PubMed] [Google Scholar]
- Tsukita, S, Katsuno, T, Yamazaki, Y, Umeda, K, Tamura, A and Tsukita, S (2009). Roles of ZO-1 and ZO-2 in establishment of the belt-like adherens and tight junctions with paracellular permselective barrier function. Ann NY Acad Sci 1165: 44–52. [DOI] [PubMed] [Google Scholar]
- Li, X, Stankovic, M, Lee, BP, Aurrand-Lions, M, Hahn, CN, Lu, Y et al. (2009). JAM-C induces endothelial cell permeability through its association and regulation of β3 integrins. Arterioscler Thromb Vasc Biol 29: 1200–1206. [DOI] [PubMed] [Google Scholar]
- Serini, G, Maione, F, Giraudo, E and Bussolino, F (2009). Semaphorins and tumor angiogenesis. Angiogenesis 12: 187–193. [DOI] [PubMed] [Google Scholar]
- Zhou, Q, Gallagher, R, Ufret-Vincenty, R, Li, X, Olson, EN and Wang, S (2011). Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA 108: 8287–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Negrini, M, Ferracin, M, Sabbioni, S and Croce, CM (2007). MicroRNAs in human cancer: from research to therapy. J Cell Sci 120(Pt 11): 1833–1840. [DOI] [PubMed] [Google Scholar]
- Ventura, A and Jacks, T (2009). MicroRNAs and cancer: short RNAs go a long way. Cell 136: 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia, I and Miska, EA (2005). MicroRNA functions in animal development and human disease. Development 132: 4653–4662. [DOI] [PubMed] [Google Scholar]
- Young, JA, Ting, KK, Li, J, Moller, T, Dunn, L, Lu, Y et al. (2013). Regulation of vascular leak and recovery from ischemic injury by general and VE-cadherin-restricted miRNA antagonists of miR-27. Blood 122: 2911–2919. [DOI] [PubMed] [Google Scholar]
- Urbich, C, Kaluza, D, Frömel, T, Knau, A, Bennewitz, K, Boon, RA et al. (2012). MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 119: 1607–1616. [DOI] [PubMed] [Google Scholar]
- Dhanabal, M, Wu, F, Alvarez, E, McQueeney, KD, Jeffers, M, MacDougall, J et al. (2005). Recombinant semaphorin 6A-1 ectodomain inhibits in vivo growth factor and tumor cell line-induced angiogenesis. Cancer Biol Ther 4: 659–668. [DOI] [PubMed] [Google Scholar]
- Liang, T, Yu, J, Liu, C and Guo, L (2014). An exploration of evolution, maturation, expression and function relationships in mir-23 ∼ 27 ∼ 24 cluster. PLoS One 9: e106223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds, E, Kloosterman, WP, Miska, E, Alvarez-Saavedra, E, Berezikov, E, de Bruijn, E et al. (2005). MicroRNA expression in zebrafish embryonic development. Science 309: 310–311. [DOI] [PubMed] [Google Scholar]
- Acunzo, M, Romano, G, Palmieri, D, Laganá, A, Garofalo, M, Balatti, V et al. (2013). Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci USA 110: 8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, ST, Huang, Y, Zhang, L, Heng, MY, Ptácek, LJ and Fu, YH (2013). MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci USA 110: 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra, R, Adlakha, YK, Hariharan, M, Scaria, V and Saini, N (2009). Upregulation of miR-23a-27a-24-2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One 4: e5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra, R, Dubey, R and Saini, N (2010). Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer 9: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S, Wu, CJ, Yasuda, T, Cruz, LO, Khan, AA, Lin, LL et al. (2016). miR-23∼27∼24 clusters control effector T cell differentiation and function. J Exp Med 213: 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, A, Lozano, E, Daimi, H, Esteban, FJ, Crist, C, Aranega, AE et al. (2011). MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc Res 89: 98–108. [DOI] [PubMed] [Google Scholar]
- Ruan, W, Xu, JM, Li, SB, Yuan, LQ and Dai, RP (2012). Effects of down-regulation of microRNA-23a on TNF-α-induced endothelial cell apoptosis through caspase-dependent pathways. Cardiovasc Res 93: 623–632. [DOI] [PubMed] [Google Scholar]
- Biyashev, D, Veliceasa, D, Topczewski, J, Topczewska, JM, Mizgirev, I, Vinokour, E et al. (2012). miR-27b controls venous specification and tip cell fate. Blood 119: 2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovat, F, Fassan, M, Gasparini, P, Rizzotto, L, Cascione, L, Pizzi, M et al. (2015). miR-15b/16-2 deletion promotes B-cell malignancies. Proc Natl Acad Sci USA 112: 11636–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J, Li, WQ, Li, Q, Li, XQ, Zhang, JT, Liu, GQ et al. (2011). Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res 108: 201–209. [DOI] [PubMed] [Google Scholar]
- Lin, R, Chen, L, Chen, G, Hu, C, Jiang, S, Sevilla, J et al. (2014). Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest 124: 5352–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana, E, Tournier-Lasserve, E and Weinstein, BM (2009). The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev Cell 16: 209–221. [DOI] [PubMed] [Google Scholar]
- Dejana, E (2004). Endothelial cell-cell junctions: happy together. Nat Rev Mol Cell Biol 5: 261–270. [DOI] [PubMed] [Google Scholar]
- Nagashima, Y, Oosako, T, Takase, Y, Ejiri, A, Watanabe, O, Kobayashi, H et al. (2010). Observation of beat oscillation generation by coupled waves associated with parametric decay during radio frequency wave heating of a spherical tokamak plasma. Phys Rev Lett 104: 245002. [DOI] [PubMed] [Google Scholar]
- Nie, L, Guo, X, Esmailzadeh, L, Zhang, J, Asadi, A, Collinge, M et al. (2013). Transmembrane protein ESDN promotes endothelial VEGF signaling and regulates angiogenesis. J Clin Invest 123: 5082–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, PA, Dudek, SM, Chiang, ET and Garcia, JG (2005). Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P1 receptor, PI3 kinase, Tiam1/Rac1, and alpha-actinin. FASEB J 19: 1646–1656. [DOI] [PubMed] [Google Scholar]
- Bratt, A, Birot, O, Sinha, I, Veitonmäki, N, Aase, K, Ernkvist, M et al. (2005). Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem 280: 34859–34869. [DOI] [PubMed] [Google Scholar]
- Lee, HJ, Cho, CH, Hwang, SJ, Choi, HH, Kim, KT, Ahn, SY et al. (2004). Biological characterization of angiopoietin-3 and angiopoietin-4. FASEB J 18: 1200–1208. [DOI] [PubMed] [Google Scholar]
- Broermann, A, Winderlich, M, Block, H, Frye, M, Rossaint, J, Zarbock, A et al. (2011). Dissociation of VE-PTP from VE-cadherin is required for leukocyte extravasation and for VEGF-induced vascular permeability in vivo. J Exp Med 208: 2393–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira Tojais, N, Peghaire, C, Franzl, N, Larrieu-Lahargue, F, Jaspard, B, Reynaud, A et al. (2014). Frizzled7 controls vascular permeability through the Wnt-canonical pathway and cross-talk with endothelial cell junction complexes. Cardiovasc Res 103: 291–303. [DOI] [PubMed] [Google Scholar]
- Dvorak, HF, Brown, LF, Detmar, M and Dvorak, AM (1995). Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am J Pathol 146: 1029–1039. [PMC free article] [PubMed] [Google Scholar]
- Dvorak, HF, Detmar, M, Claffey, KP, Nagy, JA, van de Water, L and Senger, DR (1995). Vascular permeability factor/vascular endothelial growth factor: an important mediator of angiogenesis in malignancy and inflammation. Int Arch Allergy Immunol 107: 233–235. [DOI] [PubMed] [Google Scholar]
- Schirle, NT, Sheu-Gruttadauria, J and MacRae, IJ (2014). Structural basis for microRNA targeting. Science 346: 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, Y, Sharma, HS, Pettersson, A and Cervos-Navarro, J (1992). Release of endogenous neurochemicals may increase vascular permeability, induce edema and influence cell changes in trauma to the spinal cord. Prog Brain Res 91: 197–203. [DOI] [PubMed] [Google Scholar]
- Cotran, RS and Majno, G (1964). The delayed and prolonged vascular leakage in inflammation. I. Topography of the leaking vessels after thermal injury. Am J Pathol 45: 261–281. [PMC free article] [PubMed] [Google Scholar]
- Ohya, Y and Guth, PH (1988). Ethanol-induced gastric mucosal blood flow and vascular permeability changes in the rat. Dig Dis Sci 33: 883–888. [DOI] [PubMed] [Google Scholar]
- Lin, Z, Murtaza, I, Wang, K, Jiao, J, Gao, J and Li, PF (2009). miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA 106: 12103–12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, J, Zhu, J and van Oudenaarden, A (2007). MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y, Wang, B, Jiang, F, Wang, D, Liu, H, Yan, Y et al. (2013). A feedback loop consisting of microRNA 23a/27a and the β-like globin suppressors KLF3 and SP1 regulates globin gene expression. Mol Cell Biol 33: 3994–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, CM, Wang, RY, Saijilafu, Jiao, ZX, Zhang, BY and Zhou, FQ (2013). MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev 27: 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev, B, Stanley, P and Papalopulu, N (2012). MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep 2: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal, M, Xu, J, Seitz, H, Weng, Z and Zamore, PD (2010). Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, JS, Phillips, MD, Betel, D, Mu, P, Ventura, A, Siepel, AC et al. (2011). Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, K, Phillips, MD, Tyler, DM, Duan, H, Chou, YT and Lai, EC (2008). The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363.18376413 [Google Scholar]
- Wu, X, Bhayani, MK, Dodge, CT, Nicoloso, MS, Chen, Y, Yan, X et al. (2013). Coordinated targeting of the EGFR signaling axis by microRNA-27a*. Oncotarget 4: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assaad, F, Wheway, J, Mitchell, AJ, Lou, J, Hunt, NH, Combes, V et al. (2013). Cytoadherence of Plasmodium berghei-infected red blood cells to murine brain and lung microvascular endothelial cells in vitro. Infect Immun 81: 3984–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, JR, Matthias, LJ, Meyer, G, Kaur, P, Russ, G, Faull, R et al. (1993). Regulation of in vitro capillary tube formation by anti-integrin antibodies. J Cell Biol 121: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J, Sze, DM, Brown, RD, Cowley, MJ, Kaplan, W, Mo, SL et al. (2010). Clonal expansions of cytotoxic T cells exist in the blood of patients with Waldenstrom macroglobulinemia but exhibit anergic properties and are eliminated by nucleoside analogue therapy. Blood 115: 3580–3588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.