Abstract
Retinoblastoma (RB) is an intraocular childhood tumor which, if left untreated, leads to blindness and mortality. Nucleolin (NCL) protein which is differentially expressed on the tumor cell surface, binds ligands and regulates carcinogenesis and angiogenesis. We found that NCL is over expressed in RB tumor tissues and cell lines compared to normal retina. We studied the effect of nucleolin-aptamer (NCL-APT) to reduce proliferation in RB tumor cells. Aptamer treatment on the RB cell lines (Y79 and WERI-Rb1) led to significant inhibition of cell proliferation. Locked nucleic acid (LNA) modified NCL-APT administered subcutaneously (s.c.) near tumor or intraperitoneally (i.p.) in Y79 xenografted nude mice resulted in 26 and 65% of tumor growth inhibition, respectively. Downregulation of inhibitor of apoptosis proteins, tumor miRNA-18a, altered serum cytokines, and serum miRNA-18a levels were observed upon NCL-APT treatment. Desorption electrospray ionization mass spectrometry (DESI MS)-based imaging of cell lines and tumor tissues revealed changes in phosphatidylcholines levels upon treatment. Thus, our study provides proof of concept illustrating NCL-APT-based targeted therapeutic strategy and use of DESI MS-based lipid imaging in monitoring therapeutic responses in RB.
Keywords: cancer therapy, DESI MS imaging, nucleolin aptamer, retinoblastoma xenograft
Introduction
Retinoblastoma (RB) is a childhood eye cancer that constitutes around 3% of all childhood malignancies globally. RB leads to complete vision loss if untreated and causes mortality at advanced stages if not enucleated.1 Therefore, better treatment modalities are needed for eye salvage and good life by developing targeted therapies specific for RB cells. The retina majorly consists of glycerophospholipids which are further classified into phosphatidylethanolamines (PEs) and phosphatidylcholines (PCs).2,3 Recently, lipid imaging using desorption electrospray ionization mass spectrometry (DESI MS) has emerged as a powerful methodology to precisely measure metabolites in tissues and to study lipid changes between normal versus cancerous tissue.4 DESI MS-based lipid imaging would potentially assist to study the biology of retinal diseases.
Nucleolin (NCL) is a multifunctional protein that actively participates in the rDNA transcription, translocation of rRNA,5,6 growth, and proliferation of cells. It interacts with G-quadruplex structures in the promoter regions of c-Myc, untranslated regions (5' UTRs) of various mRNAs,7,8,9 and microprocessor machinery for the microRNA biogenesis10,11 NCL protein expression on the cell surface has been identified in many epithelial cancers.8,12,13 The presence of surface NCL in cancer cells in contrast to the non-neoplastic cells makes it a promising target for the cancer therapy.8,14,15 Aptamers are oligonucleotides developed using Systemic Evolution of Ligands by EXponential enrichment (SELEX) method. These molecules possess high affinity towards the target molecules owing to its specific three dimensional structures. Aptamers16 are utilized as diagnostic and therapeutic tools in oncology. An aptamer against NCL, NCL-APT also known as AS1411 (Antisoma, UK), is a US Food and Drug Administration (FDA)-approved NCL targeting agent. It binds to NCL on the cell surface, preferentially gets internalized, and inhibits cancer cell growth sparing normal cells17,18,19 and hence we utilized this aptamer to study its effect against RB.
DESI MS can potentially grade the stages in cancer, detect the surgical tumor margins, and study lipogenesis in tumor.4,20,21,22 DESI MS-based lipid imaging of cultured RB cells was recently optimized by spotting cells onto Whatman filter paper23 and this method was utilized for studying the aptamer siRNA chimera-mediated changes in cell lines.24 There is no information on the expression status of NCL in RB and on the use of lipid imaging to study the effect of NCL-APT in situ. In this report, we studied the role of NCL in RB and targeted it using a NCL-APT and LNA modified NCL-APT (LNA-NCL-APT). We also studied the NCL-APT influence on relevant genes, microRNAs, and lipids expressed on the RB cells using in vitro and in vivo models. We showed that DESI MS based lipid imaging can help in understanding NCL-APT treatment induced changes in RB.
Results
NCL expression in RB and uptake of NCL-APT
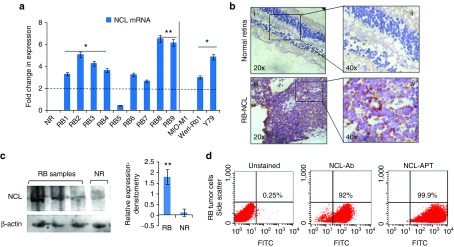
The lack of information on the status of NCL in RB intrigued us to investigate the expression of NCL on RB tumor samples and it was analyzed by qPCR, immunohistochemistry (IHC), flow cytometry, and immunoblotting. The change in NCL mRNA expression across the RB tumors normalized to normal retina varied between 0.45-and 6.56-fold with mean expression levels of 3.94 ± 1.87-fold. The samples with NCL mRNA upregulated more than twofold (marked by the threshold line) were considered as significant. The RB cell lines showed NCL mRNA upregulation of 4.8-fold in Y79 and 3.0-fold in WERI-Rb1 cells upon normalizing with MIO-M1, a nonmalignant control cell line (Figure 1a). IHC studies of NCL protein expression in 25 cases of RB primary tumor sections showed membrane/cytoplasmic positivity in the RB cells. The healthy cadaveric retinal tissues (n = 2) showed faint nuclear staining and no cytoplasmic staining (Figure 1b, i, ii). The staining pattern within the tumor was heterogeneous and on an average, 50% of cells showed positive, and in a few cases, more than 70% of cells were positive for NCL protein (Figure 1b, iii, iv).
Figure 1.
Expression of nucleolin in RB tumor samples and cell lines. (a) Fold changes in gene expression of NCL in various primary cells and cell lines. (b) Immunohistochemistry of the normal retina section (i, ii), RB tissue sections (iii, iv). (c) Expression of NCL in the cytoplasmic fraction of RB tumor tissues by immunoblotting for NCL and β-actin, graph on the right shows the densitometry analysis of tumor samples normalized to normal retina (NR). (d) Scatter plots show the expression of NCL and the NCL-APT binding to RB tumor (RB cells from the enucleated eyes). The error bar represents the SD and the * indicates significance of P < 0.05 and ** indicates significance of P < 0.001 when compared to the normal retina.
Cytoplasmic NCL protein expression levels studied by immunoblotting in both the non-neoplastic retina and RB samples showed overexpression of NCL in the RB tissues. Densitometry analysis of the blots showed significant upregulation of NCL in cytoplasm of the RB tumor cells (Figure 1c). Ten RB tumors were analyzed by flow cytometry which were found to be highly positive (>60% of cells) for NCL and the respective scatter plot is shown in Figure 1d. The overlay plot shows the expression of NCL in Y79 and WERI-Rb1 cells as analyzed by flow cytometry (Supplementary Figure S1A). AS1411 or NCL-APT has been shown to preferentially internalize into the cancer cells by utilizing the cell surface NCL. Elevated levels of NCL in the cytoplasm and cell surface of the RB tumor cells could thus aid in NCL-APT uptake and functional activity in RB tumor cells respectively. Preferential uptake of FITC labelled NCL-APT was observed in Y79 and WERI-Rb1 cells than MIO-M1 cell line (Supplementary Figure S1B). Also, the cell cycle changes analyzed post-NCL-APT treatment in cell lines showed increase in G0-G1 phase in WERI-Rb1 and arrest in S phase in Y79. MIO-M1 cells did not exert considerable changes in different cell phases (Supplementary Figure S1C). Thus, we were able to observe the differential expression of NCL in the RB cells and cell lines and NCL-APT internalization mediated cell cycle changes.
NCL-APT regulates oncogenic miRNAs in RB
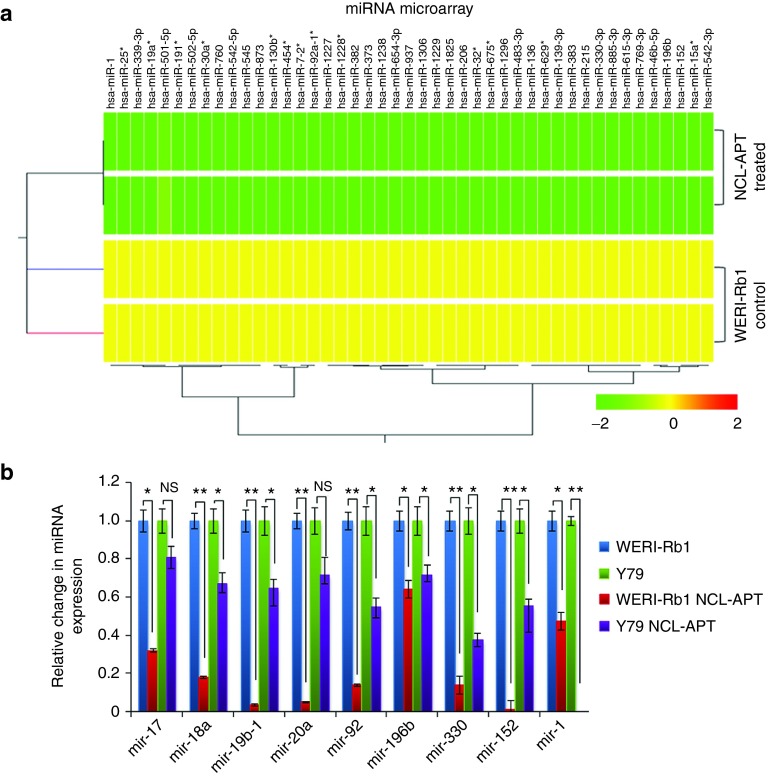
NCL is shown to involve in microprocessor machinery and regulates the DICER expression.10 As the dicer participation in microprocessor is essential for miRNA expression, we investigated the influence of the NCL-APT on miRNA expression in RB. miR-15a, miR-16, and miR-221 family were earlier reported for their altered expression upon NCL-APT treatment in breast cancer. Thus, we studied the miRNA profiles in RB cell line after 48 hours of treatment with NCL-APT.14,15 NCL-APT-treated WERI-Rb1 cells were subjected to miRNA microarray and the global miRNA expression profiling was performed. Hierarchical clustering of the miRNA showed significant (P < 0.05) downregulation of 46 miRNAs (Figure 2a).
Figure 2.
Effect of NCL-APT on miRNA expression. (a) Hierarchical clustering of the miRNAs from the WERI-Rb1 cell line and cells treated with NCL-APT. The colour range indicates the status of expression (yellow being midrange and green being downregulation). (b) Fold changes in the miRNA levels - mir-196b, mir-330, mir-152, mir-1, mir-17, mir-18a, mir-19-1, and mir-20a in Y79 and WERI-Rb1 cell lines treated with NCL-APT normalized with untreated cells.
From the differentially regulated 46 miRNAs, four miRNAs (miR-196b, miR-152, miR-1, and miR-330) and mature miRNAs of oncomir-1 cluster (miR-17–92 cluster) previously reported in RB25 were studied using qPCR for their native expression in RB cell lines by normalizing to MIO-M1 cells. The fold change in expression of the mature miRNAs of miR-17–92 cluster was similar except for miR-17 and miR-18a in both the RB cell lines, while other three miRNAs except miR-330 were highly expressed in Y79 than WERI-Rb1 (Supplementary Figure S2A). These miRNAs upon NCL-APT treatment in both the RB cell lines were found downregulated, with marked effect in WERI-Rb1 cells confirmed by qPCR (Figure 2b). Thus, the miRNA-17–92 cluster and the other miRNAs downregulated in the miRNA profiling were validated for their changes in expression in both the RB cell lines.
NCL-APT perturbs RB in vitro and in vivo: effect on miRNAs, apoptotic proteins
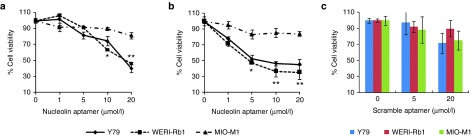
We next investigated the functional activity of the NCL-APT on the RB cell growth and proliferation. The phenotypic changes monitored post NCL-APT treatment confirmed the cell cycle blockage at S phase (Supplementary Figure S2B). The antiproliferative property of the NCL-APT was tested using dimethyl thiazolyldiphenyl tetrazolium bromide (MTT) assay in RB cell lines and MIO-M1 cell line. Various concentrations of NCL-APT (1, 5, 10, and 20 µmol/l) were studied in Y79, WERI-Rb1 and MIO-M1 cell lines and cell viabilities of 40 ± 5.0, 42 ± 1.2, and 90 ± 1.9%, respectively were obtained at 20 µmol/l by MTT assay after 48 hours of treatment. A concentration of 10 µmol/l was used to study the molecular effects of NCL-APT as the viability was 74 and 63% in Y79 and WERI-Rb1 cell lines, respectively with uncompromised viability in MIO-M1 cell line (Figure 3a). Thus the inhibition of cell division lead to cytotoxicity leading to decrease in viability of RB cell lines sparing the control retinal MIO-M1 cell line.
Figure 3.
Effect of NCL-APT on cell proliferation. NCL-APT and LNA-NCL-APT were assessed at the concentration of 1, 5, 10, and 20 μmol/l on the cell lines after 48 hours of treatment. Graph showing the percentage cell viability exhibited by the NCL-APT (a) and LNA-NCL-APT (b) on Y79, WERI-Rb1 and MIO-M1 cells at 48 hours using MTT assay. The Scrambled aptamer is treated at the concentration of 5 and 20 μmol/l. (c). Graph showing the cell viabilities upon scrambled aptamer treatment assessed by MTT assay. The mean of triplicate data ± SD are plotted and the * indicates significance of P < 0.05, ** indicates significance of P < 0.001 and NS indicates not significant.
As LNA modification of nucleic acids imparts increased serum half life, we studied the stability of LNA modified NCL-APT and found to be more stable than NCL-APT up to 120 hours under physiological pH and in serum (Supplementary Figure S3). This LNA-NCL-APT having LNA bases at three positions exhibited higher in vivo stability with unaltered cytostatic effect and was therefore included in the study.26 The antiproliferative effect of LNA-NCL-APT was assessed in Y79, WERI-Rb1, and MIO-M1 cell lines and found to have 45 ± 6.8, 35 ± 9.0, and 84 ± 5.8% of cell viability after 48 hours of treatment at the dose of 10 µmol/l. The activities were very similar not significantly different from the NCL-APT (Figure 3b). The scrambled aptamer carrying C's instead of G's in NCL-APT was tested against the RB and MIO-M1 cell lines. The scrambled aptamer did not affect significantly the cell viability of RB and MIO-M1 cell lines even at 20 µmol/l, thus confirming specificity of NCL-APT activity (Figure 3c).
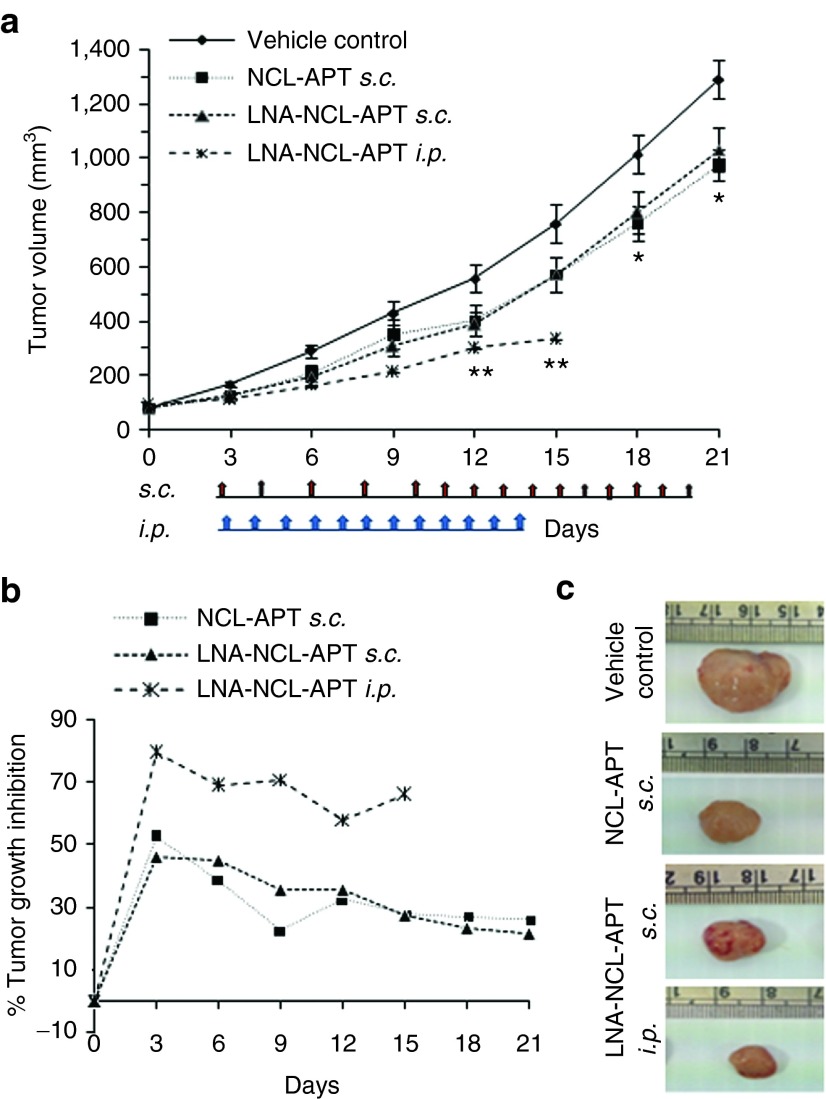
The effect of NCL-APT and LNA-NCL-APT was studied in vivo in the RB tumor using Y79 xenograft in nude mice. The Y79 xenograft model has been shown for the aggressive tumor growth characteristics and hence we used Y79 xenograft model than the WERI-Rb1-based xenograft model.27 The Y79 xenografted mice treated with NCL-APT and LNA-NCL-APT subcutaneously (s.c.) near the tumor sites showed tumor growth reduction with the difference in mean tumor volume of 380 and 440 mm3 respectively from the vehicle control group which was 1,289 mm3 (Figure 4a). The percentage tumor growth reduction/inhibition calculated were 22.2 and 26% in NCL-APT and LNA-NCL-APT treated s.c. at day 21 (Figure 4b). Notably, the effect of LNA-NCL-APT by intraperitoneal (i.p.) injection showed better tumor growth reduction of about 65% with difference in mean tumor volume of 450 mm3 at day 15. The excised tumors from respective treatments showed clear size differences between the tumors (Figure 4c).
Figure 4.
Antitumor effect of NCL-APT and LNA-NCL-APT on Y79 xenograft model. (a) Graph showing the change in percentage tumor growth inhibition of the groups injected with NCL-APT and LNA-NCL-APT subcutaneously near the tumor site and LNA-NCL-APT by intraperitoneal route. Photographs showing the representative animals (b) and excised tumors (c) showing the changes in tumor growth upon treatment by s.c. and i.p. routes. The error bar in panel A represents the SEM and the * indicates significance of P < 0.05 and ** indicates significance of P < 0.001.
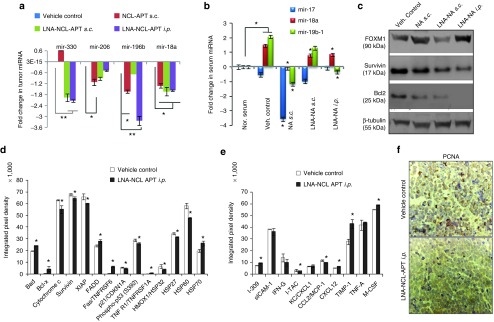
The NCL-APT and LNA-NCL-APT showed significant antitumor activity in Y79 xenograft model at day 18 and day 21. The molecular mechanism behind the tumor growth reduction was addressed by analyzing the tumor mRNA, miRNA and apoptotic protein expression in representative samples of untreated, NCL-APT and LNA-NCL-APT (s.c.) and LNA-NCL-APT (i.p.) treated xenograft tissues. Additionally, serum onco-miRNAs and cytokine levels were studied for correlating the tumor growth changes. Tumor miRNA expression was studied for the miR-17–92 cluster25,28,29 and miR-330, miR-206, miR-196b, and miR-152 that were differentially regulated by NCL-APT treatment in RB cell lines. The miR-196b was downregulated in all the treated samples followed by the miR-330, miR-206, miR-196b, and miR-18a in most of the treated samples (Figure 5a). As observed in the miRNA microarray of cells treated with NCL-APT, the above miRNAs were downregulated in the xenografted tumor tissues. This signifies the influence of NCL-APT on the miRNA changes resulting in the changes of tumor growth.
Figure 5.
Molecular changes accompanying NCL-APT and LNA-NCL-APT treatment on Y79 xenograft. (a) Graph showing the levels of Y79 xenograft tumor tissue miRNA (miR-330, miR-206, miR-196b and miR-18a) normalized to vehicle control using U6 as internal control. (b) Graph showing the levels of serum miRNA post treatment with nucleolin aptamer. Normal serum (mice without tumor) was used for normalizing the expression levels.(c) Western blotting for Bcl2, Survivin, FOXM1 proteins from xenograft tissues of the control group and treated group tissues. The levels of apoptotic markers (d) in tumor tissue and serum cytokines (e) using protein array. The integrated pixel densities of the spots were quantified using ImageJ software and plotted as graph. The error bar represents the SD and the * indicates significance of P<0.05 and ** indicates significance of P < 0.001, for panel A, treated groups were normalized to vehicle control group, for panel B - normal serum was compared to vehicle control, then the vehicle control group was used to analyze the significance in treated groups. (f) IHC for PCNA expression. The images are taken under 40X objective. The intensity of expression is indicated in the bottom left.
Serum miRNA acts as bio-marker for diagnosis and prognosis of diseases.30,31 Data from the RB patients showed that miR-17–92 cluster was overexpressed in serum, and hence changes in this serum miR-17–92 levels were of our interest.32 The relative changes in serum miRNA between normal nude mice and Y79 xenografted mice serum showed elevated levels of miR-18a and miR-19b-1 expression, followed by miR-17. The miR-17 and miR-19b-1 were downregulated in NCL-APT s.c. and LNA-NCL-APT i.p. groups, while miR-18a expression was affected at similar levels in all the treatment groups (Figure 5b). The immunoblot analysis of the apoptotic proteins, Bcl2 and survivin showed that their expressions were downregulated in all treated tissues with varying FOXM1 expression in different modes of treatments as observed by densitometry (Figure 5c). Thus the NCL-APT treatment in vitro and in vivo lead to RB growth inhibition mediated by the integrated regulation of mRNAs, oncogenic miRNAs and apoptotic markers.
LNA-NCL-APT-mediated changes in apoptotic marker and cytokine levels: an integrome analysis
The integrated changes in proteome, miRNA and gene expression upon aptamer treatment and its analysis would be able to elucidate the molecular mechanism in RB. Hence we studied the changes in the tumor-apoptotic proteins and cytokines in the xenograft treated with LNA-NCL-APT. The apoptosis markers studied by protein array from the tumor tissues treated with PBS and LNA-NCL-APT showed a significant difference in the extrinsic and intrinsic pathway mediators. There was significant decrease in XIAP, survivin, livin, cytochrome c, HSP27, clusterin and p21 with increase in the levels of Bad, Bcl-xl, Fas, TNFαR, and p53 (Figure 5d). Additionally, the cytokines expression was studied for the involvement of immune mechanisms in tumor regression. Compared to the vehicle control, the LNA-NCL-APT treated groups showed significant (*P < 0.05) increase in the expression of CXCL1, CXCL12, TIMP1, MCSF, and i-309 with decreased levels of IL2, ITAC, and CCL2. Also, there was no significant difference in expression of TNF-α and IFN-γ. Thus, there were mixed expression of the chemotactic cytokines and the mitogenic cytokines upon the LNA-NCL-APT treatment (Figure 5e). IHC studies of LNA-NCL-APT i.p. treated tissues also exhibited decreased levels of the nuclear PCNA expression (Figure 5f).
Integrated network modelling and analysis of statistically significant enriched miRNA, genes and pathways revealed key pathways were altered during NCL-APT treatment. The affected pathways were apoptosis, cancer, cell cycle, immune response, and homeostasis, harboring genes like TP53, HSPD1, Bcl2, CXCL12, AKT1, MDM2, p21, CCL2, BIRC5, and IL2. Further, the integrome network of the above affected miRNAs, mRNAs and proteins showed miR-1, miR-17, and miR-206 to be key regulators of transcriptional response and by affecting apoptotic response by NCL-APT treatment (Supplementary Figure S4).
Lipid profiling of xenograft tissues
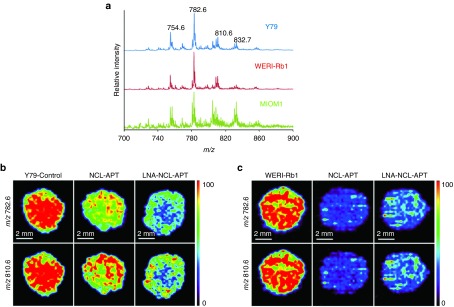
The lipid content of the cell is important in the cellular metabolism and to maintain the normal behaviour of cells. Phosphatidylcholines are the major lipid present in the cell membrane and altered PCs and choline metabolites levels leads to changes in gene expression and cell proliferation as in the case of malignancy.33 In the current work, we studied the indirect effect of NCL in PCs levels using aptamer and siRNA strategy. The mass spectra were obtained for the Y79, WERI-Rb1, and MIO-M1 cell lines (Figure 6a) and the peaks observed in DESI MS were identified as PCs by MS/MS and database search. The lipids corresponding to the different PCs are listed in Supplementary Table S1. The high intensity peaks observed in different RB cell lines were m/z 754.6 [PC(32:1)+Na]+, 782.6 [PC(34:1)+Na]+, 810.6 [PC(36:1)+Na]+, and 832.7 [PC(38:4)+Na]+ (Figure 6a). The changes in expression of lipids [PC(32:1)+Na]+ and [PC(36:1)+Na]+ differ significantly from the untreated to NCL-APT and LNA-NCL-APT treated cells of Y79 and WERI-Rb1 (m/z 782.6 and 810.6) (Figure 6b,c). Two other significant lipid peaks, m/z 754.6 [PC(34:1)+Na]+ and 832.7 [PC(38:4)+Na]+ were also studied in NCL-APT-treated RB cell lines (Supplementary Figure S5B). The effect of NCL on lipid levels was counter verified using silencing strategy. The siRNA against NCL was transfected, studied for NCL downregulation and its effect in lipid levels. Upon NCL downregulation in RB cell lines, the intensities of lipid peaks above studied were reduced (Supplementary Figure S5C). Thus, the use of NCL-APT or silencing NCL has similar effect on the predominantly identified lipids in RB cell lines studied by DESI MS.
Figure 6.
DESI MS imaging of aptamer treated RB cell lines. (a) DESI-MS spectra of Y79, WERI-Rb1 and MIO-M1 cells grown in cell. Cells were washed with 1XPBS twice and spotted on Whatmann membrane and imaged using methanol as solvent using positive mode and data acquired from 50 to 2,000 Daltons. Changes in the lipid expression accompanying the treatment with nucleolin aptamer in RB cell line, Y79 (b) WERI-Rb1 (c).
To access if there is global alteration of lipid by NCL-APT, we further evaluated the effect of both the NCL-APTs in prostate cancer cell line (PC3). The mass spectrum obtained from PC3 was presented with Y79 tumor xenograft and Y79 cells (Supplementary Figure S6A). The high intensity peaks (m/z 754.6 and 782.6) observed in PC3 cell line were affected upon APT treatment, while MIO-M1 (m/z 782.6 and 832.7) showed less intense changes by LNA-NCL-APT and not by the NCL-APT treatment (Supplementary Figure S6B, C).
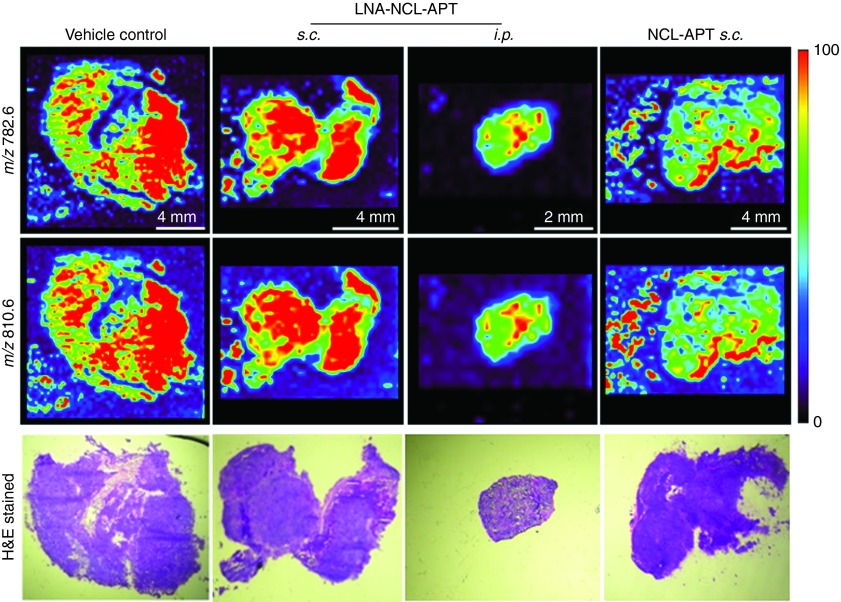
The Y79 xenografts treated with NCL-APT and LNA-NCL-APT by s.c. route and i.p. route were subjected to lipid profiling. Analysis of tumor sections showed a decrease in intensities of lipids of m/z 782.6 and 810.6 in the NCL-APT s.c. and LNA-NCL-APT i.p. treated tissues followed by LNA-NCL-APT s.c. treated tissue with insignificant changes in few areas of the tissues (Figure 7). In addition to the peaks studied above, two other lipid peaks m/z 754.6 and 832.7 studied in APT-treated xenograft tissues were found to have similar changes (Supplementary Figure S6D). Overall majority of treated tissues were identified with lesser intensities of lipids with the i.p. mode of treatment showing lowest levels of lipids. The drastic changes in lipid levels proportionately correlates with the aptamer treatment, as the inhibition of lipid synthesis or decrease in lipids levels directly affects cell proliferation. Thus, we found that blocking of NCL function using NCL-APT has affected the lipid levels in the cells, thereby regulating the cell growth.
Figure 7.
Changes in PC levels upon in vivo assessment of NCL-APT and LNA-NCL-APT. (a) DESI MS images of cryosection from the vehicle control and treatment groups. Images represent the distribution of phosphatidylcholines across the tissue sections. (b) H&E stained images of the tissue sections. Images were presented without alteration, acquired at 10X magnification.
Discussion
Our study showed the differential expression of NCL protein on the surface and cytoplasm of RB tumor cells. Targeting NCL using NCL-APT in the RB cell lines lead to downregulation of oncogenic miRNAs and lead to inhibition cell proliferation. NCL-APT and LNA modified NCL-APT-mediated inhibition of NCL, reduced phosphatidylcholine levels as confirmed by DESI MS of aptamer treated RB cells. Functional blocking of NCL from transcription and translation processes concomitantly resulted in the altered gene expression and lipid (PCs) levels. Similarly, the internalized NCL-APT leads to the cell cycle blockage due to the S phase arrest followed by increase in the G0-G1 population. These changes in cell cycle and cell proliferation inhibition are in agreement with the observations reported regarding the G-rich oligonucleotides.34,35
The maintenance of stability of oncogenic mRNAs and miRNAs is one of the essential steps in tumorigenesis. NCL plays a pivotal role in mRNA stabilization and enhanced translation in cancer.7,8 Global miRNA profiling of NCL-APT-treated RB cell line in the current study showed majorly downregulation of miRNAs with a few upregulated miRNAs confirming the importance of the NCL in the miRNA biogenesis.11 We further studied the expression of oncogenic miRNA pertaining to RB, miR-17–92 cluster and found downregulation upon treatment. The differentially expressed miRNA studied includes miR-196b, miR-373, miR-330, miR-152 and miR-1. The observation of miRNA downregulation upon NCL-APT treatment may be related to the regulation at transcription level leading to halt in the miRNA synthesis and processing. The NCL binding to the promoter G-quadruplex structure controls the transcription of c-myc gene.36 The promoter regions of these miRNA were predicted for the presence of G-quadruplex and i-tetraplex structures that could have mediated the effect.37
The tumor suppressor property of miR-152 and miR-1 in ovarian cancers38 and bladder cancer39 were reported earlier and in the current study these miRNAs were downregulated upon NCL-APT treatment, which could be attributed due to the promoter structure. NCL involvement in miRNA promoters, however, needs further investigation. The miR-330 is reported as tumor suppressor miRNA in prostate cancer, but a recent report suggested inverse role in glioblastoma.40 The oncogenic role of miR-196b has been studied in glioblastoma and its expression correlated to poor disease prognosis41 and this miRNA was upregulated in RB tumors too. Thus, the mature miRNAs of established onco-miRNA-1 cluster, miR-17, miR-18a and miR-19b and miR-196b, miR-330, miR-152, miR-206, and miR-1 were found to be downregulated in the RB cell lines upon aptamer treatment.
The in vivo studies for evaluating the anti-tumor activity of NCL-APT and LNA-NCL-APT in Y79 xenograft revealed better effect in the i.p. mode of delivery than the s.c. mode of delivery. The NCL-APT and LNA-NCL-APT both showed tumor growth inhibition with later aptamer having 4% better efficacy in tumor inhibition under s.c. mode, while the i.p. mode of LNA-NCL-APT delivery showed 65% of tumor growth inhibition. The absence of the i.p. mode of delivery for NCL-APT in the current study alleviates our ability to appreciate LNA-NCL-APT over NCL-APT. This difference in the tumor reduction suggests that the delivery mode is important to attain better therapeutic efficacy. Intraperitoneal mode has been recognized for the better absorption and availability of substance injected.42 Also NCL-APT was delivered earlier studies by both i.p. or s.c. modes in mouse and in patients by continuous infusion method.11,43 Thus, the bioavailability of the LNA-NCL-APT to the tumor could have attributed to the results observed which desires further investigation.
The mechanism behind the antitumor activity of the LNA-NCL-APT was studied by analyzing the tumor miRNAs and serum biomarkers in the i.p. mode of treatment as the efficacy is better than the s.c. mode. The miRNA expression in the treated tumor revealed that there is consistent downregulation of miR-18a and miR-196b in the treated tumors. The decreased expression of the miR-18a and miR-196b could have enhanced the antitumor activity of aptamer in RB.25 The aptamer treatment had downregulated the levels of Bcl2, survivin, and FOXM1 differentially. FOXM1 is overexpressed in cancers, controlled through miRNA expression and cell cycle changes.44 Perturbation in cell division by stathmin silencing in RB cell lines showed FOXM1 upregulation.45 FOXM1 upregulation has shown to induce genomic instability, leading to induction of p21, p53, and apoptosis of cells.46 Thus, our observation of differential regulation of FOXM1 needs further analysis. The protein array supports the apoptotic onset by the decrease of IAPs, increase in Bad and p53. The cytokine levels were also varying with some of the chemotactic attractants getting downregulated with minor or no changes in the levels of the TNF-α and IFN-γ. Thus, the multifunctional NCL protein expressed under cancerous condition modulates various activities.
DESI MS is extensively used for lipidome analysis thereby discriminating stages of cancer, surgical margins of cancerous to noncancerous regions and tumor heterogeneity.47,48 We utilized DESI MS for observing the changes in the intensity of PCs levels upon NCL-APT treatment in RB cell lines and tumor tissues. To check if NCL modulates the PCs levels in other cancers, the effect of NCL-APT was tested using prostate cancer model, PC3 cell line. We confirmed that inhibiting NCL using NCL-APT effectively brought down the PC levels in PC3 cell lines too. Thus, we intended to check this mechanism in the in vivo model and the observed results in in vivo confirm the role of the NCL in the lipid metabolism using DESI MS. This approach of using DESI MS-based lipid imaging could also monitor the therapeutic progress.
Thus, we hypothesize that perturbing NCL function using NCL-APT impaired lipid synthesis and thereby altered PC levels on the cell membrane. This could indirectly explain the mechanism behind the tumor growth inhibition in the in vivo conditions. Similar observations using conditional knock on and knock off mice for the MYCN-oncogene were obtained in the lipid profiling of hepatocellular carcinoma.49 Similarly, the importance of changes in lipids for the discrimination of the grades of astrocytoma21 evidently showed that DESI MS can be applied in cancer diagnosis. The tear fluids studied earlier had cholesterol, triglycerides, sphingomyelins, PCs, and PEs.50 Probably in future, the lipid level changes in the tear fluids could also be analyzed for studying the disease state or therapeutic progression.
To conclude, our data reports for the first time NCL overexpression in RB. The NCL-APT binds preferentially to RB cells and inhibits cell proliferation. NCL-APT treatment leads to changes in mRNA, miRNA and lipid levels resulting in effective tumor growth inhibition in vivo. The NCL-APT treatment reduced the serum onco-miRNAs. Our data also substantiates the use of intraperitoneal mode of delivery and LNA modification for the therapeutic efficacy of NCL-APT. The data confirm the effectiveness of DESI MS based lipid profiling for the monitoring of lipid level changes during treatment. Thus, further studies using orthotopic or intraocular models of RB warrant the use of aptamer in RB therapy. Future research on tear lipidome of RB using DESI MS could regard as a non-invasive method for studying the treatment mediated effects.
Materials and Methods
Cell lines and RB sample collection. Y79 and WERI-Rb1 (Riken cell bank, Japan) were cultured in RPMI 1640 medium and MIO-M1 (kind gift from Dr. G. A. Limb) in Dulbecco's modification of Eagle's medium (DMEM), respectively at 37 °C in a 5% CO2 humidified incubator. RB tumor samples were collected from the freshly enucleated eyeballs, surgically removed as a part of the treatment regimen with written consent from parents/guardian. Normal retina samples were collected from the healthy cadaveric eyes, post corneal transplantation supplied by C U Shah Eye Bank, Medical Research Foundation, Sankara Nethralaya. This study was conducted at Vision Research Foundation, Sankara Nethralaya, India, and was approved by the Vision Research Foundation Institutional Ethics Board (Ethical clearance no: 249B-2011-P). RB cells were collected in Rosewell Park Memorial Institute (RPMI) media containing 10% FBS, placed on 6-well plate, minced well, passed through 23 gauge needle and the suspended cells were washed and used for assays.
Expression of NCL in cell lines, tumors and NCL-APT uptake. The NCL expression was studied by quantitative polymerase chain reaction (qPCR), flow cytometry, IHC and western blotting on the RB tissues and normal retina. qPCR analysis was performed using SYBR green-based method. We performed flow cytometry to study the expression of surface NCL, on unfixed RB cell lines as well on RB tumor cells obtained from enucleated eyes. IHC was performed on deparaffinized tissue sections, post-antigen retrieval. Western blotting was performed on cytoplasmic lysate for NCL and β-actin. The aptamer binding and uptake was performed on live, fresh, unfixed or nonpermeabilized cells and analysed by flow cytometry.
NCL-APT treatment—miRNA microarray and miRNA regulations. The effect of NCL-APT on Y79, WERI-Rb1 and MIO-M1 cell lines were studied by treating cells with various concentrations of NCL-APT and LNA-NCL-APT (0.5 to 20 µmol/l) and scrambled aptamer (5 and 20 µmol/l) using MTT assay. The effective dose or a concentration with lesser cytotoxicity was chosen for further studies. miRNA microarrays were performed in duplicates for WERI-Rb1 untreated and NCL-APT-treated cells. Validation of the miRNAs was performed using RNU6B as normalizing control for small RNA. The aptamer-treated cells were compared to untreated cells and normalized to it. Similarly, the vehicle control samples were used for normalizing the changes observed in the aptamer treated tissue samples.
In vivo experiments: protein array and integrome analysis. Animal study was performed commercially utilizing the facility of Syngene International (Bangalore, India) and experimentations were approved by the Institutional Animal Ethics Committee (IAEC Protocol Approval No: SYNGENE/IAEC/430/10–2013). Following aseptic procedure, Y79 cells, 1 × 107 in 200 µl of serum free media containing 50% of Matrigel was injected subcutaneously in the back of 5-week-old female nude mice (Hsd: Athymic Nude-Foxn1nu/Foxn1nu). Once the tumor was palpable, the animals were randomized into four groups (n = 6) and treated with aptamers either s.c. or i.p. with 12.5 nmol/dosing for 21 days. Tumor volume and body weight were measured every 3 days. The excised treated tumor tissues were subjected for mRNA, miRNA, protein arrays. For the integrome analysis, commonly enriched biological categories were taken along with their statistical significance and differentially expressed genes and miRNA involved for downstream integrome network analysis using BridgeIsland Software (Bionivid Technology Pvt Ltd, Bangalore, India) and CytoScape V 2.8 to understand treatment specific differential regulome.
DESI MS of cell lines and xenograft tumor tissues treated with NCL-APT and LNA-NCL-APT. The effect of NCL silencing/knockdown (siNCL) and NCL-APT on the alteration in lipid profile was studied using DESI MS. A 2D DESI ion source from Prosolia (Indianapolis, IN) was coupled with Thermo Scientific LTQ XL ion trap mass spectrometer (Thermo Scientific, San Jose, CA). The treated cells (5 × 105 cells) were washed with PBS and resuspended uniformly to 15 µl 1× PBS, spotted on Whatman 42 filter paper and left for drying in laboratory condition for 5 minutes. The samples were analyzed from the filter paper by DESI MS in positive ion mode using HPLC grade methanol from Sigma-Aldrich (Bangalore, India) as solvent and conditions published earlier.23 The snap frozen tumor tissues were sectioned using a Leica cryostat and thaw mounted on glass slides and analyzed by DESI MS using the same conditions used for the cell lines. In brief, the following parameters were maintained throughout the DESI MS experiments; spray voltage: 5 kV, solvent flow rate: 5 µl/minute, nebulizer gas pressure: 150 psi, spray angle: 60° to the surface, surface to spray tip distance: 2 mm, atmospheric inlet of the mass spectrometer to spray tip distance: 3 mm. During imaging by DESI MS, a pixel size of 250 × 250 µm was chosen for both the cell lines and tissues. For MS/MS experiments, collision induced dissociation (CID) was used.
Statistical analysis. The in vitro results are represented as mean ± SD (n ≥ 3), while the in vivo results are represented as mean ± standard error of the mean (n = 6). Significance was calculated using student's t-test (paired, two-tailed) and the P values were indicated with * for P < 0.05 and ** for P < 0.001.
For detailed methods, kindly refer to Supplementary Section.
SUPPLEMENTARY MATERIAL Table S1. List of masses of the phosphatidylcholines predicted by MS/MS and database search. Figure S1. NCL expression, NCL-APT uptake and cellular effects on RB cell lines. Figure S2. miRNA expression and effect of NCL-APT on the morphology of RB cell lines. Figure S3. Stability and effect of NCL-APT and LNA-NCL-APT in cell lines. Figure S4. Schematic showing gene regulatory network upon NCL-APT treatment in RB. Figure S5. Effect of NCL-APT treatment and siRNA against NCL on lipids by DESI MS. Figure S6. Effect of NCL-APT treatment on lipids by DESI MS. Supplementary Section.
Author contributions
N.S., J.R.K., R.K.K., T.P., and S.K. designed research; P.R. and V.K. performed enucleation; N.S., B.A., and A.S. performed research; N.S., R.K.K., A.S., M.V., T.P., and S.K. analyzed data; and N.S., J.R.K., P.R., V.K., A.S., T.P., and S.K. wrote the paper.
Acknowledgments
Thanks to Sowmya Parameswaran for critical comments, proof reading of manuscript and Sukanya Mohan for technical help. Core lab facility and Department of Biochemistry of Vision Research Foundation are acknowledged for the use of flow cytometry, fluorescent microscopy and Flurochem Chemiluminescent imaging facilities. Thanks are due to the Nano Mission, Government of India for equipment support. The graduate fellowship from Deakin University (to N.S.), fellowships from CSIR and IIT Madras (to A.S.) are acknowledged. This work was supported by Department of Biotechnology, India (Grant BT/01/CE1B/11/V/16-(to S.K.)) and Australia-India Strategic Research Fund (BT/Indo-Aus/06/08/2011 (to S.K., J.R.K. and R.K.K.)). The authors declare no conflict of interest.
Supplementary Material
References
- Dimaras, H, Kimani, K, Dimba, EA, Gronsdahl, P, White, A, Chan, HS et al. (2012). Retinoblastoma. Lancet 379: 1436–1446. [DOI] [PubMed] [Google Scholar]
- Gülcan, HG, Alvarez, RA, Maude, MB and Anderson, RE (1993). Lipids of human retina, retinal pigment epithelium, and Bruch's membrane/choroid: comparison of macular and peripheral regions. Invest Ophthalmol Vis Sci 34: 3187–3193. [PubMed] [Google Scholar]
- Martinez, M, Ballabriga, A and Gil-Gibernau, JJ (1988). Lipids of the developing human retina: I. Total fatty acids, plasmalogens, and fatty acid composition of ethanolamine and choline phosphoglycerides. J Neurosci Res 20: 484–490. [DOI] [PubMed] [Google Scholar]
- Eberlin, LS (2014). DESI-MS imaging of lipids and metabolites from biological samples. Methods Mol Biol 1198: 299–311. [DOI] [PubMed] [Google Scholar]
- Cong, R, Das, S, Ugrinova, I, Kumar, S, Mongelard, F, Wong, J et al. (2012). Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res 40: 9441–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely, Y and Borowiec, JA (2000). Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol 149: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen, K, Tominaga, K, Lee, EK, Srikantan, S, Kang, MJ, Kim, MM et al. (2011). Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res 39: 8513–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen, K and Gorospe, M (2012). RNA-binding protein nucleolin in disease. RNA Biol 9: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, V and Hurley, LH (2010). The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry 49: 9706–9714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering, BF, Yu, D and Van Dyke, MW (2011). Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J Biol Chem 286: 44095–44103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri, F, Palmieri, D, De Luca, L, Consiglio, J, You, J, Rocci, A et al. (2013). In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J Exp Med 210: 951–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsioumpa, M and Papadimitriou, E (2014). Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov 9: 137–152. [DOI] [PubMed] [Google Scholar]
- Krust, B, El Khoury, D, Nondier, I, Soundaramourty, C and Hovanessian, AG (2011). Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer 11: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, H, Watanabe, T and Suganuma, M (2014). Cell-surface nucleolin acts as a central mediator for carcinogenic, anti-carcinogenic, and disease-related ligands. J Cancer Res Clin Oncol 140: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian, AG, Soundaramourty, C, El Khoury, D, Nondier, I, Svab, J and Krust, B (2010). Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS One 5: e15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megaptche, AP, Erb, U, Büchler, MW and Zöller, M (2014). CD44v10, osteopontin and lymphoma growth retardation by a CD44v10-specific antibody. Immunol Cell Biol 92: 709–720. [DOI] [PubMed] [Google Scholar]
- Bates, PJ, Laber, DA, Miller, DM, Thomas, SD and Trent, JO (2009). Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 86: 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destouches, D, El Khoury, D, Hamma-Kourbali, Y, Krust, B, Albanese, P, Katsoris, P et al. (2008). Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS One 3: e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes, EM, Teng, Y and Bates, PJ (2010). A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res 70: 8617–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calligaris, D, Caragacianu, D, Liu, X, Norton, I, Thompson, CJ, Richardson, AL et al. (2014). Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci USA 111: 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin, LS, Dill, AL, Golby, AJ, Ligon, KL, Wiseman, JM, Cooks, RG et al. (2010). Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl 49: 5953–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin, LS, Tibshirani, RJ, Zhang, J, Longacre, TA, Berry, GJ, Bingham, DB et al. (2014). Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci USA 111: 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srimany, A, Jayashree, B, Krishnakumar, S, Elchuri, S and Pradeep, T (2015). Identification of effective substrates for the direct analysis of lipids from cell lines using desorption electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 29: 349–356. [DOI] [PubMed] [Google Scholar]
- Jayashree, B, Srimany, A, Jayaraman, S, Bhutra, A, Janakiraman, N, Chitipothu, S et al. (2016). Monitoring of changes in lipid profiles during PLK1 knockdown in cancer cells using DESI MS. Anal Bioanal Chem 408: 5623–5632. [DOI] [PubMed] [Google Scholar]
- Kandalam, MM, Beta, M, Maheswari, UK, Swaminathan, S and Krishnakumar, S (2012). Oncogenic microRNA 17-92 cluster is regulated by epithelial cell adhesion molecule and could be a potential therapeutic target in retinoblastoma. Mol Vis 18: 2279–2287. [PMC free article] [PubMed] [Google Scholar]
- Roy, K, Kanwar, RK, Cheung, CHA, Fleming, CL, Veedu, RN, Krishnakumar, S, et al. (2015). Locked nucleic acid modified bi-specific aptamer-targeted nanoparticles carrying survivin antagonist towards effective colon cancer therapy. RSC Adv 5: 29008–29016. [Google Scholar]
- Chévez-Barrios, P, Hurwitz, MY, Louie, K, Marcus, KT, Holcombe, VN, Schafer, P et al. (2000). Metastatic and nonmetastatic models of retinoblastoma. Am J Pathol 157: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkrite, K, Sundby, M, Mukai, S, Thomson, JM, Mu, D, Hammond, SM et al. (2011). miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 25: 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittner, D, Lambertz, I, Clermont, F, Mestdagh, P, Köhler, C, Nielsen, SJ et al. (2012). Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat Cell Biol 14: 958–965. [DOI] [PubMed] [Google Scholar]
- Bianchi, F, Nicassio, F, Marzi, M, Belloni, E, Dall'olio, V, Bernard, L et al. (2011). A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 3: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwetha, S, Gouthamchandra, K, Chandra, M, Ravishankar, B, Khaja, MN and Das, S (2013). Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep 3: 1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beta, M, Venkatesan, N, Vasudevan, M, Vetrivel, U, Khetan, V and Krishnakumar, S (2013). Identification and insilico analysis of retinoblastoma serum microRNA profile and gene targets towards prediction of novel serum biomarkers. Bioinform Biol Insights 7: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway, ND (2013). The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit Rev Biochem Mol Biol 48: 20–38. [DOI] [PubMed] [Google Scholar]
- Edwards, SL, Poongavanam, V, Kanwar, JR, Roy, K, Hillman, KM, Prasad, N et al. (2015). Targeting VEGF with LNA-stabilized G-rich oligonucleotide for efficient breast cancer inhibition. Chem Commun (Camb) 51: 9499–9502. [DOI] [PubMed] [Google Scholar]
- Xu, X, Hamhouyia, F, Thomas, SD, Burke, TJ, Girvan, AC, McGregor, WG et al. (2001). Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J Biol Chem 276: 43221–43230. [DOI] [PubMed] [Google Scholar]
- González, V, Guo, K, Hurley, L and Sun, D (2009). Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem 284: 23622–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R, Lin, Y and Zhang, CT (2008). Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res 36(Database issue): D372–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y, Ma, N, Wang, D, Zhang, Y, Zhou, J, Wu, G et al. (2014). MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene 33: 378–386. [DOI] [PubMed] [Google Scholar]
- Yoshino, H, Chiyomaru, T, Enokida, H, Kawakami, K, Tatarano, S, Nishiyama, K et al. (2011). The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer 104: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y, Xue, Y, Ma, J, Shang, C, Wang, P, Liu, L et al. (2014). MiR-330-mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS One 9: e95060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, R, Yan, W, Zhang, G, Lv, H, Liu, Z, Fang, F et al. (2012). Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One 7: e38096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, DE, Henry, RR and Edelman, SV (1996). Acute effects of intraperitoneal versus subcutaneous insulin delivery on glucose homeostasis in patients with NIDDM. Veterans Affairs Implantable Insulin Pump Study Group. Diabetes Care 19: 1237–1242. [DOI] [PubMed] [Google Scholar]
- Rosenberg, JE, Bambury, RM, Van Allen, EM, Drabkin, HA, Lara, PN Jr, Harzstark, AL et al. (2014). A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest New Drugs 32: 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra, I (2013). FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res 119: 191–419. [DOI] [PubMed] [Google Scholar]
- Mitra, M, Kandalam, M, Sundaram, CS, Verma, RS, Maheswari, UK, Swaminathan, S et al. (2011). Reversal of stathmin-mediated microtubule destabilization sensitizes retinoblastoma cells to a low dose of antimicrotubule agents: a novel synergistic therapeutic intervention. Invest Ophthalmol Vis Sci 52: 5441–5448. [DOI] [PubMed] [Google Scholar]
- Teh, MT, Gemenetzidis, E, Chaplin, T, Young, BD and Philpott, MP (2010). Upregulation of FOXM1 induces genomic instability in human epidermal keratinocytes. Mol Cancer 9: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifa, DR and Eberlin, LS (2016). Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem 62: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata, A, Zheng, J, Ginsberg, HJ, Jaffray, DA, Ifa, DR and Zarrine-Afsar, A (2015). Contrast agent mass spectrometry imaging reveals tumor heterogeneity. Anal Chem 87: 7683–7689. [DOI] [PubMed] [Google Scholar]
- Perry, RH, Bellovin, DI, Shroff, EH, Ismail, AI, Zabuawala, T, Felsher, DW et al. (2013). Characterization of MYC-induced tumorigenesis by in situ lipid profiling. Anal Chem 85: 4259–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantamäki, AH, Seppänen-Laakso, T, Oresic, M, Jauhiainen, M and Holopainen, JM (2011). Human tear fluid lipidome: from composition to function. PLoS One 6: e19553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.