Abstract
Due to its ability to track distance changes within individual molecules or molecular complexes on the nanometer scale and in real time, single-molecule fluorescence resonance energy transfer (single-molecule FRET) is a powerful tool to tackle a wide range of important biological questions. Using our recently developed single-molecule FRET assay to monitor nucleosome translocation as an illustrative example, we describe here in detail how to set up, carry out, and analyze single-molecule FRET experiments that provide time-dependent information on biomolecular processes.
1. Introduction
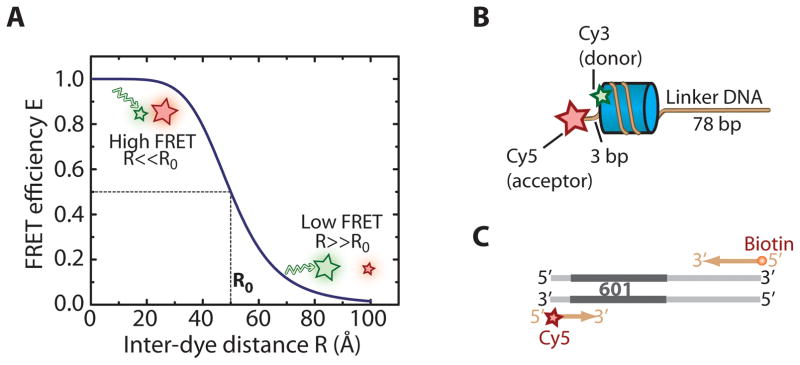
Essential cellular processes such as interactions between biomolecules or conformational changes often involve distance changes at a nanometer length scale (1–10 nm). Fluorescence resonance energy transfer (FRET) (Forster, 1949; Stryer and Haugland, 1967) is a spectroscopic technique that enables the observation of distance changes at this length scale with high sensitivity and in real time. In this technique, a donor and an acceptor fluorophore are introduced at sites whose distance is to be monitored. Upon excitation of the donor fluorophore, a fraction of its energy can be transferred to the acceptor fluorophore in a nonradiative process. The efficiency of energy transfer, E, is highly sensitive to the distance R between the two fluorescent molecules: E = 1/[1 + (R/R0)6], where R0 is the Förster radius at which E = 0.5 (Fig. 1A). FRET measurements at the single-molecule level allow the observation of dynamics on a molecular scale that would be inaccessible in ensemble measurements due to random averaging (Ha, 2001; Ha et al., 1996; Zhuang et al., 2000). The ability to monitor conformational dynamics of individual molecules in real time makes single-molecule techniques highly effective in the study of a wide range of mechanistic questions.
Figure 1.
Distance dependence of FRET efficiency and example construct design for single-molecule FRET imaging of nucleosome remodeling. (A) The FRET efficiency as a function of the distance between donor and acceptor dye molecules. The donor and acceptor fluorophores are represented as green and red stars, respectively. At the Förster distance, R0, the donor fluorophore transfers 50% of its energy to the acceptor fluorophore. (B) Example nucleosome with a single donor dye on the proximal H2A subunit. The histone octamer and the nucleosomal DNA are shown in blue and brown, respectively. The Cy5 (acceptor) and Cy3 (donor) fluorophores are depicted as red and green stars, respectively. (C) The Cy5 dye (red star) and the biotin moiety (orange dot) are included in the PCR primers used to generate the nucleosomal DNA.
Recently, several groups have reported the application of single-molecule techniques to directly observe DNA or nucleosome translocation by ATP-dependent chromatin remodeling enzymes (Amitani, Baskin, & Kowalczykowski, 2006; Blosser et al., 2009; Lia et al., 2006; Nimonkar et al., 2007; Prasad et al., 2007; Shundrovsky et al., 2006; Sirinakis et al., 2011; Zhang et al., 2006). These enzymes help regulate DNA accessibility by altering the structure of chromatin, which is based on the nucleosome as a fundamental repeating unit (Kornberg, 1974; Kornberg and Lorch, 1999). There are several subfamilies of chromatin remodeling enzymes, SWI/SNF, ISWI, CHD/Mi2, and INO80, that differ in composition and function (Becker and Horz, 2002; Clapier and Cairns, 2009; Flaus and Owen-Hughes, 2004; Narlikar et al., 2002; Smith and Peterson, 2005; Tsukiyama and Wu, 1997). Members of the ISWI subfamily reposition nucleosomes along the DNA while preserving the canonical nucleosome structure (Hamiche et al., 1999; Kassabov et al., 2002; Langst et al., 1999; Tsukiyama et al., 1999). Recently, we have used single-molecule FRET measurements to monitor in real time the translocation of individual nucleosomes by an ISWI family enzyme (Blosser et al., 2009), the ATP-dependent chromatin assembly and remodeling factor (ACF) (Ito, Bulger, Pazin, Kobayashi, & Kadonaga, 1997). These experiments allowed us to reveal previously unknown kinetic intermediates and mechanistic aspects of this remodeling enzyme (Blosser et al., 2009). The dynamic nature of chromatin structure and its regulation makes single-molecule FRET ideally suited for this type of mechanistic study. Here, we provide a detailed step-by-step guide for how to set up and carry out single-molecule FRET measurements, including experiments that require buffer exchange during data acquisition. As an illustrative example, we describe single-molecule FRET measurements on fluorescently labeled mononucleosomes during ACF-induced remodeling. With the exception of sections regarding the preparation of mononucleosomes, all protocols described here are generally applicable, with minimal adaptations, to the single-molecule FRET study of other biological systems.
2. Preparation of Fluorescently Labeled Sample for Single-Molecule FRET Imaging
The attachment sites for donor and acceptor dyes are chosen such that their distance changes are relatively large during the process under investigation. For processes that are associated with relatively small distance changes, the FRET dyes are ideally positioned at a distance close to the R0 value of the FRET pair, because at this inter-dye distance, the FRET efficiency is most sensitive to distance changes. Common labeling approaches include NHS ester or maleimide chemistry, and several different FRET dye pairs can be used (Roy, Hohng, & Ha, 2008). The labeling of nucleic acids with fluorescent dyes is generally straightforward, which greatly facilitates the study of nucleic acid–protein interactions. Because single-molecule FRET measurements of a single donor–acceptor pair yield a one-dimensional signal, finding the best labeling choice in terms of labeling sites and fluorophores is crucial in understanding conformational dynamics that typically occur in three dimensions. As an example, we describe here how to generate nucleosomes that are labeled with a Cy3–Cy5 FRET pair, but the procedures can readily be adapted to other nucleic acid and protein substrates under study.
For single-molecule measurements of chromatin remodeling, it is typically desired to carry out experiments with a homogenous population of nucleosomes where the histone octamer is initially placed at a specific location on the DNA substrate. This can be achieved by incorporating a strong nucleosome positioning sequence, such as the 601 positioning sequence (Li, Levitus, Bustamante, & Widom, 2005), into the DNA substrate used for nucleosome assembly. One should, however, be cautious about the possibility of a sequence-specific bias, and control experiments with distinct and nonrelated positioning sequences should be performed to test whether the observed phenomenon is sequence-specific. Depending on the exact mechanistic question under study, a variety of nucleosomal substrates can be used that differ in the flanking DNA length at entry and exit sides of the nucleosome, in the location of the FRET dyes on the DNA and/or the histone octamer, and in the specific composition of nucleosomal histone proteins. In our example, initially end-positioned mononucleosomes such as shown in Fig. 1B are prepared to observe the real-time dynamics of nucleosome remodeling. The double-stranded DNA molecule used to reconstitute these nucleosomes contains the FRET acceptor dye (Cy5) and a biotin moiety for immobilization at opposing ends. Histone octamers are labeled with the FRET donor dye (Cy3) on histone H2A and positioned at a defined number of base pairs (bp) away from the acceptor dye (Cy5)-labeled exit end of the DNA. As an illustrative example, we describe a construct with DNA linkers of 3 bp and 78 bp on the exit and entry sides of the nucleosome, respectively.
2.1. Preparation and purification of fluorescently labeled DNA
Nucleic acid molecules used for single-molecule FRET imaging are typically assembled by PCR or annealing reactions. This approach has the advantage that molecules such as fluorophores can be conveniently introduced by using commercially available custom-synthesized oligonucleotides that contain the desired moieties. In the example described here, the nucleosomal DNA is prepared by PCR, with the Cy5 dye and the biotin label included in custom-synthesized PCR primers (Fig. 1C).
2.1.1. Preparation of fluorophore-labeled DNA by PCR
At 4 °C, prepare an amplification mix with a total volume of 10 ml and final concentrations of 0.02 U/μl Phusion Hot Start High-Fidelity Polymerase (New England Biolabs), 200 μM of each deoxynucleotide triphosphate (dNTP), 1 × Phusion HF Buffer, 0.5 μM forward and reverse primers, 0.2 ng/μl template plasmid (containing positioning and desired flanking DNA sequences). Using a multichannel pipette, transfer 100 μl of the amplification mix into each well of a 96-well standard thin-walled PCR plate, seal the plate, and amplify the DNA in a total of 35 cycles with a final extension of 10 min.
With a multichannel pipette, combine all reactions, add 1/10 volume of 3 M sodium acetate pH 5.2 and 2.5 volumes (calculated after addition of sodium acetate) of > 95% (v/v) ethanol and precipitate the DNA overnight at − 20 °C. Pellet the DNA by centrifugation at > 14,000 × g and 4 °C for 15 min and discard the supernatant. Rinse the DNA pellet with 70% (v/v) ethanol, repeat the centrifugation, discard the supernatant, and air-dry the pellet.
Dissolve the DNA in 60 μl T50 buffer (10 mM Tris pH 7.5 and 50 mM NaCl). Incubation at 37 °C for 10 min facilitates this process. Pellet any denatured, insoluble protein by centrifugation at > 14,000 × g and room temperature for 15 min and keep the supernatant that contains the DNA. Add glycerol to a final concentration of 5% (v/v) for subsequent preparative polyacrylamide gel electrophoresis (PAGE).
2.1.2. Native PAGE purification of dye-labeled DNA
For the purification of dye-labeled DNA, pour a nondenaturing polyacrylamide, 0.5 × TBE (final concentrations 45 mM Tris–borate and 1 mM EDTA pH 8.0) gel (dimensions 20 cm × 16 cm × 3 mm). Choose the polyacrylamide percentage according to the size of the DNA sample. For example, a 5% polyacrylamide gel works well for the purification of a ~ 150–250 bp sample.
Electrophorese the gel in 0.5 × TBE at 10 V/cm and 4 °C for 1 h, rinse wells, and load sample. Resume electrophoresis for ~ 1.5 h at 4 °C. The dye label allows direct observation of DNA migration through the gel. Excise the region of the gel containing the desired DNA with a razor blade.
Transfer into 5 ml of 10 mM Tris pH 7.5 and shake at 37 °C for at least 12 h to allow the DNA to diffuse out of the gel slice and into the buffer. Take off the DNA-containing supernatant, replace with another 5 ml of 10 mM Tris pH 7.5, and repeat the incubation at 37 °C for another 12 h. Remove the supernatant and combine with the supernatant from the first incubation step.
Concentrate the DNA by adding an equal volume of 2-butanol to the combined supernatant. Mix, then separate organic and aqueous phases by brief centrifugation. Aspirate off and discard the upper organic layer. Repeat the extraction with 2-butanol until the volume of the aqueous phase is less than ~ 500 μl. Add 1/10 volume of 3 M sodium acetate pH 5.2 and 2.5 volumes (calculated after addition of sodium acetate) of > 95% (v/v) ethanol. After overnight precipitation at −20 °C, pellet the DNA by centrifugation at > 14,000 × g and 4 °C for 15 min and discard the supernatant. Rinse the DNA pellet with 70% (v/v) ethanol, repeat the centrifugation, discard the supernatant, and air-dry the pellet. Dissolve the DNA in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0).
2.2. Site-specific labeling of proteins with fluorescent dyes
A variety of different approaches for site-specific labeling of proteins have been reported (Fernandez-Suarez and Ting, 2008; Heyduk, 2002). For example, in an in vitro transcription/translation reaction, a tRNA loaded with a fluorophore-derivatized amino acid can be used to incorporate it at a specific site defined by a nonsense codon (Mendel, Cornish, & Schultz, 1995). Alternatively, peptide sequences that are specifically recognized by a fluorophore-labeled ligand can be introduced into the protein (Griffin et al., 1998, Kapanidis et al., 2001; Miller et al., 2005). Recently, various enzymes have been used to catalyze the ligation of the fluorophore to its target site (Baruah et al., 2008; Fernandez-Suarez et al., 2007; Popp et al., 2007; Tanaka et al., 2008). Further, chemoselective ligation strategies can be applied (Dawson et al., 1994; Mekler et al., 2002; Muir et al., 1998).
One of the most commonly used approaches for site-specific labeling of proteins involves using a protein without any reactive cysteine side chains other than at the site of interest, referred to as a “Cys-lite” (Rice et al., 1999) protein. A Cys-lite protein can be obtained by site-directed mutagenesis: Endogenous cysteine residues other than the target residue are mutated to nonreactive amino acid residues, and the amino acid residue at the target site is mutated to a cysteine residue. Once established that the Cys-lite mutant retains full physiological function, its cysteine thiol moiety can be derivatized by nucleophilic substitution, for example, with a monofunctional maleimide ester of a fluorescent dye. As an example for this approach, we provide here a protocol for the site-specific labeling of a core histone protein H2A with Cy3 at a single cysteine side chain (residue 120, H2A-120C) (Yang, Madrid, Sevastopoulos, & Narlikar, 2006).
At room temperature, dissolve 1 mg of lyophilized H2A-120C in 800 μl of labeling buffer (20 mM Tris pH 7.0, 7 M guanidinium HCl, 5 mM EDTA) for a protein concentration of ~ 100 μM.
In order to stabilize the free sulfhydryls, add 2 μl of 0.5 M TCEP (final concentration: 1.25 mM) and incubate for 2 h at room temperature in the dark.
Dissolve dried Cy3-maleimide ester (GE Healthcare Amersham) in anhydrous DMSO at a concentration of 100 mM, taking into account the fraction of reactive dye indicated by the manufacturer.
Add 25 μl of 100 mM Cy3-maleimide ester (~3 mM final concentration) to the histone protein and incubate for 3 h in the dark at room temperature.
Quench the labeling reaction by adding 4.6 μl of β-mercaptoethanol (final concentration: 80 mM). Set a small aliquot (~2 μl) aside for analysis via diagnostic gel.
Excess dye can be removed by dialysis, gel filtration, or the use of dye removal resin. For example, to remove excess Cy3 dye by dialysis, transfer the quenched labeling reaction into a 3-ml dialysis cassette (Slide-A-Lyzer, 7000 MWCO; Thermo Scientific) and dialyze at room temperature in the dark three times against 1000 ml of fresh dialysis buffer (20 mM Tris pH 7.0, 7 M guanidinium HCl, 1 mM DTT) for at least 3 h per iteration.
Use 1 μl of the undialyzed labeled histone sample (set aside in step 5) and dilute by a factor of 25 with water. Dilute 1 μl of the dialyzed labeled histone sample analogously. Add SDS loading dye to both dilutions and incubate at 95 °C for 2 min. In separate lanes, analyze 2–10 μl of each dilution on a 15% SDS analytical polyacrylamide gel. Quantify Cy3 intensities from the histone band in both lanes using an imaging system such as a Molecular Dynamics Typhoon. Stain the gel with SYPRO Red (Sigma-Aldrich) and quantify the protein content of both bands based on the SYPRO stain. The comparison of the SYPRO band intensities yields the concentration of the dialyzed Cy3-labeled sample, since the concentration of the undialyzed sample is known. The labeling efficiency can be roughly estimated by comparing the concentrations obtained from the SYPRO and Cy3 fluorescence signals.
Concentrate the dialyzed Cy3-labeled H2A as needed using a centrifugal filter device and use to prepare Cy3-labeled histone octamer.
2.3. Preparation of histone octamer from purified core histone proteins
Histone octamer composed of unlabeled H3, H4, H2B, and Cy3-labeled H2A subunits can be assembled by dialysis and subsequent size exclusion purification (Dyer et al., 2004; Luger et al., 1999; Shahian and Narlikar, 2012):
From each of the three lyophilized and unlabeled core histone proteins (H3, H4, H2B), make a 2 mg/ml solution in unfolding buffer (20 mM Tris pH 7.5, 7 M guanidinium hydrochloride, 10 mM DTT). Dissolve histone proteins by gently pipetting up and down. Incubate the solutions for at least 30 min to ensure complete unfolding but do not exceed an incubation time of 3 h.
Using Cy3-labeled H2A (from Section 2.2, in dialysis buffer), and the H2B, H3, and H4 solutions from step 1, prepare a mixture of the four core histone proteins with a molar ratio of 6:6:5:5 (H2A:H2B:H3:H4). The slight molar excess of H2A and H2B is used to shift the equilibrium toward more complete octamer formation. Any excess H2A/H2B dimer can be removed by subsequent size exclusion chromatography. Adjust the total protein concentration to 1 mg/ml with unfolding buffer.
Transfer the core histone mix into a dialysis bag with a 6–8 kDa molecular weight cutoff and dialyze three times against 1 l of fresh refolding buffer (2 M NaCl, 10 mM Tris pH 7.5, 1 mM EDTA, 5 mM β-mercaptoethanol). One of the dialysis steps should be carried out overnight, while the other two steps should proceed for at least 3 h each.
Recover the dialyzed sample and pellet any precipitate by ultracentrifugation at maximum speed for 20 min. Concentrate the supernatant to a volume of ~ 200 μl using a centrifugal filter device (Millipore YM-10, 10,000 NMWL). Set the filtrate and ~ 5 μl of the concentrated sample (retentate) aside for analytical purposes.
Subject the concentrated sample to size exclusion chromatography using a Superdex 200 HR 10/30 column (Amersham Pharmacia Biotech). Wash the column (typically stored in 20% (v/v) ethanol) with two column volumes of water (~50 ml) and then equilibrate with at least two column volumes of refolding buffer. Inject the sample and collect 0.5-ml fractions at a flow rate of 0.5 ml/min. The histone octamer elutes first, with a smaller elution volume than that of any excess H2A/H2B dimer. Monitor absorption at 280 nm (protein) and 550 nm (Cy3) to identify octamer peak fractions.
Analyze ~ 10 μl of the filtrate (flow-through) and 5 μl of the concentrated sample (load) from step 4 as well as 5 μl of each octamer peak fraction on a 15% SDS polyacrylamide gel. Stain the gel with SYPRO Red gel stain and combine those octamer peak fractions that display approximately equimolar quantities of all four histone core proteins.
Concentrate the pooled fractions to a concentration of at least 1 mg/ml using a centrifugal filter device (Millipore YM-10, 10,000 NMWL). To determine the final concentration more accurately, analyze several dilutions of the concentrated sample together with a series of bovine serum albumin (BSA) standards on a 15% SDS polyacrylamide gel and quantify band intensities of standards and sample upon SYPRO staining.
2.4. Nucleosome reconstitution and purification
Nucleosomes for single-molecule FRET studies can be reconstituted by combining Cy3-labeled histone octamer and Cy5-labeled DNA at a molar ratio of 6:5 using a salt gradient procedure (Dyer et al., 2004, Lee and Narlikar, 2001 and Luger et al., 1999).
For a typical small-scale reconstitution mixture of 150 μl, combine 200 pmol Cy3-histone octamer and ~ 167 pmol dye-labeled DNA in 20 mM Tris pH 7.5, 1 mM EDTA pH 8.0, 10 mM DTT, and 2 M KCl at 4 °C. Add the histone octamer last to the reconstitution mixture in order to avoid the formation of aggregates at salt concentrations < 2 M.
Prepare 250 ml of high salt buffer (10 mM Tris pH 7.5, 2 M KCl, 1 mM EDTA pH 8.0, 1 mM DTT, 0.5 mM Benzamidine) and 1000 ml of low salt buffer (10 mM Tris pH 7.5, 250 mM KCl, 1 mM EDTA pH 8.0, 1 mM DTT, 0.5 mM Benzamidine) and chill to 4 °C. At 4 °C, transfer the reconstitution mixture into a 0.5-ml dialysis cassette (Slide-A-Lyzer, 7000 MWCO; Thermo Scientific) and place the cassette into a beaker containing the high salt buffer. Under constant stirring, use a peristaltic pump setup to continually remove buffer from the dialysis beaker and to replace it with low salt buffer. Both inlet and outlet should be calibrated and adjusted to yield closely matching flow rates of ~ 215 μl/min. Perform the salt gradient dialysis at 4 °C and in the dark (to prevent photobleaching of fluorophores) for 60 h, then transfer the dialysis cassette into a beaker containing 1 l of TCS buffer (20 mM Tris pH 7.5, 1 mM EDTA pH 8.0, 1 mM DTT), prechilled to 4 °C, and dialyze for at least 2 h.
To purify nucleosomes from free DNA, prepare a 10% glycerol buffer (20 mM Tris pH 7.5, 1 mM EDTA, 0.1% NP-40, 10% glycerol) and a 30% glycerol buffer (20 mM Tris pH 7.5, 1 mM EDTA, 0.1% NP-40, 30% glycerol). Using a gradient forming instrument such as a Gradient Master (BioComp), form a linear 10–30% glycerol gradient in a 5-ml polycarbonate tube (Beckman Polyallomer No. 326819, 0.5 × 2.5″) and layer the contents of the dialysis cassette on top of the gradient. Centrifuge at 4 °C at 35,000 rpm in a SW55Ti rotor for 16.5 h.
At 4 °C, puncture the centrifuge tube with a butterfly needle ~ 1 cm from the bottom and collect 30–40 fractions of ~ 5 drops each. Analyze fractions on a 5% polyacrylamide, 0.5 × TBE gel and combine fractions that contain correctly assembled nucleosomes but no detectable free DNA. Concentrate to a volume of ~ 20–50 μl using a centrifugal filter device (Millipore YM-100, 100,000 NMWL). Upon recovery of the retentate (concentrated nucleosome preparation), store both nucleosomes and the filtrate at 4 °C in the dark. The filtrate is useful for preparing nucleosome dilutions as needed for single-molecule FRET imaging.
3. Preparing PEG-Coated Slides with Sample Chamber
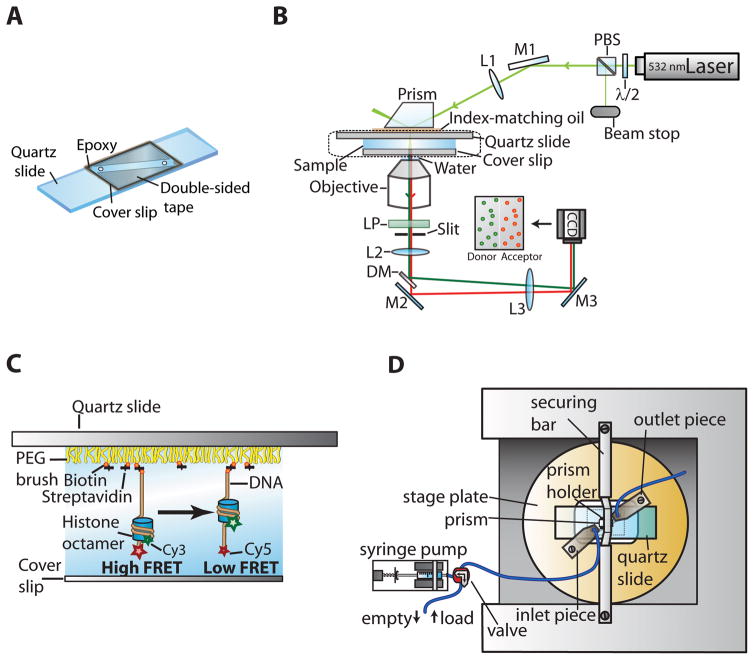
The observation of conformational dynamics at the single-molecule level often requires the ability to rapidly exchange the buffer during data acquisition. For example, an enzymatic reaction such as nucleosome remodeling can be initiated at a well-defined time point by perfusing a sample chamber (Fig. 2A) with remodeling enzyme and ATP. The quartz slide on which the sample chamber is assembled is typically passivated with a polyethylene glycol (PEG) polymer brush to minimize nonspecific protein adsorption to the quartz surface (Ha et al., 2002). A PEG mixture can be used in which a small fraction of polymer molecules is terminally coupled to a biotin moiety in order to tightly bind streptavidin, which in turn serves to immobilize the biotinylated sample.
Figure 2.
Sample chamber and optical setup for single-molecule FRET imaging with buffer exchange during data acquisition. Individual components are not drawn to scale. (A) Components used to assemble a flow chamber on a quartz slide. (B) Schematic of the optical setup for single-molecule FRET imaging using the prism-type TIR geometry. The excitation laser beam is focused into a Pellin-Broca fused silica prism with a shallow incident angle (< 23°) such that an evanescent field is created at the quartz–water interface on the quartz slide. Fluorescence emission from donor and acceptor dyes is collected by the objective, and scattered excitation laser light is removed by a long pass filter. Donor and acceptor emission is separated by a dichroic mirror. The dichroic mirror and an additional mirror are used to slightly offset the paths of donor and acceptor fluorescence such that they can be imaged onto two halves of the CCD camera. The image is reduced by a vertical slit in the imaging plane to fit onto half of the CCD chip area. λ/2, half waveplate; PBS, polarizing beam splitter; M1–M3, mirrors; L1–L3, lenses; LP, long pass filter; DM, dichroic mirror. (C) Larger view of the boxed region (dotted line) in (B). The slide is covered with a PEG brush to minimize nonspecific sticking of proteins to the quartz glass, and the sample is immobilized via a biotin–streptavidin linkage. The flow channel is formed between the quartz slide and the cover slip. (D) Top view of the microscope stage. A stage plate holds the sample cell. The prism is attached to the prism holder and placed on top of the quartz slide using a securing bar. Flow in the sample chamber is introduced with a motorized syringe pump that is connected to the inlet piece of the flow channel.
3.1. Preparing and cleaning quartz slides
For the following steps, use only MilliQ water (18.5 MΩ).
In order to assemble sample chambers, drill two holes (0.75 mm diameter) into each quartz slide as shown in Fig. 2A. The holes will serve as connection points for the inlet (connected to a syringe pump; KD Scientific) and outlet of the sample chamber.
Thoroughly scrub the slides with an aqueous slurry of Alconox detergent powder (Alconox Inc.) and rinse generously with water. Repeat the scrubbing procedure twice and then rinse with copious amounts of water. Sonicate the slides in a glass staining dish for 20 min in 1 M KOH. Next, rinse slides with water and transfer to another clean glass staining dish. Add > 95% (v/v) ethanol and sonicate for 20 min. Rinse slides with water, sonicate for 20 min in acetone, and rinse again with water.
Flame the surface to be PEG coated and imaged with a propane torch in order to minimize fluorescent background caused by residual organic molecules.
Transfer the slides into a plasma cleaner (Harrick Plasma) and treat the surface to be imaged with argon plasma.
3.2. Surface coating with PEG
Rinse a clean and dry glass staining dish with > 99.9% (w/v) acetone to remove any residual water. Transfer the slides into the glass staining dish and amino-modify them by treatment with 1% (w/v) Vectabond (Vector Laboratories) in > 99.9% (w/v) acetone for 5 min. Rinse slides by repeated dipping into a beaker with water and dry under nitrogen stream.
Apply a mixture of 20% (w/v) methoxy-PEG (Mr 5000; Nektar Therapeutics) and 0.2% biotin-PEG (Mr 5000; Nektar Therapeutics) in 0.1 M sodium bicarbonate pH 8.4 to the plasma-cleaned surface. Cover with a glass coverslip to spread PEG mixture as an even film while avoiding bubble formation. Place slides in a wet box and allow PEG coating to proceed for at least 4 h. Rinse the PEG-coated surface thoroughly with copious amounts of water and then dry the slides under nitrogen stream.
3.3. Sample chamber preparation
Attach two pieces of double-sided tape on the PEG-coated side of the quartz slide parallel to the line connecting inlet and outlet holes to form a flow channel of ~ 0.5 cm width (Fig. 2A).
Place a plasma-cleaned coverslip on top to form a sample chamber with a volume of ~ 20 μl.
Using a 5-min two-component epoxy resin, seal the boundaries between the cover slip and the quartz slide on all four sides.
PEG-coated sample chambers should be used within a few days of their assembly. Once sample has been immobilized on the slide for an experiment, PEG-coated sample chambers cannot be reused. However, the relatively expensive quartz slides can be recycled. To prepare new sample chambers, remove the coverslip, epoxy, and adhesive tape and repeat all of the above steps starting with the Alconox scrubbing. Storage of used quartz slides in water facilitates the removal of tape and epoxy residue.
4. Optical Setup and Single-Molecule FRET Data Acquisition
4.1. Optical setup
In total internal reflection (TIR) microscopy (Axelrod, 2003), an evanescent field of light is generated that decays exponentially with distance from the surface onto which the sample is immobilized. This approach greatly reduces background fluorescence, as only fluorophores within ~ 100–200 nm of the surface are excited. There are two commonly used types of TIR microscopy: objective-type TIR and prism-type TIR.
In prism-type TIR microscopy, a focused laser beam is introduced into the sample by means of a prism attached to a quartz slide surface, onto which the imaged specimen is immobilized. The fluorescence signal is collected by an objective focused on the quartz slide surface. In objective-type TIR microscopy, the excitation laser light is introduced through the microscope objective, and the specimen is immobilized on the surface of a cover glass at the focus of the objective. The fluorescence signal is collected by the same objective. Both prism-type and objective-type geometries can be used for single-molecule FRET experiments.
A prism-type TIR setup can be assembled around a commercial inverted light microscope (e.g., Olympus IX70) as shown in Fig. 2B. In this setup, the FRET donor dye molecules (Cy3) are excited by a 532-nm Nd:YAG (neodymium-doped yttrium aluminum garnet) laser (CrystaLaser). The excitation laser intensity is attenuated and polarization-cleaned by a half-wave plate and a polarizing beamsplitter. A TIR geometry is achieved by focusing the excitation beam into a Pellin-Broca fused silica prism with a shallow incident angle (< 23°). The prism holder is rigidly attached to the body of the microscope through a custom-built securing bar (Fig. 2D), which allows lateral translation of the sample on the stage while keeping the prism immobile. In order to accurately measure kinetic parameters such as transition times and rates, the temperature of the sample chamber often needs to be carefully controlled during data acquisition. This can be achieved by using a water-circulating bath that flows water at a defined temperature through tubing connected to a custom-built brass collar on the objective, a brass plate that holds the sample cell, and the metal pieces embracing the prism. The fluorescence emission from donor and acceptor molecules is collected by a water immersion objective lens (1.2 NA, 60×, Olympus) and filtered with a 550-nm long pass filter (Chroma Technology) to block scattered excitation light. Cy3 and Cy5 fluorescence signals are spectrally split by a 630-nm dichroic mirror (Chroma Technology) and imaged onto two halves of a CCD camera (Andor iXonEM+ 888). Donor and acceptor signals of the same molecule are identified by aligning these two channels using fluorescent beads whose fluorescence emission can be detected in both channels. The CCD chip is cooled to − 75 °C for acquisition of single-molecule FRET data.
In objective-type TIR microscopy, identical emission optics are used but the evanescent field is created with an oil immersion objective of high numerical aperture (for example, 1.4 NA). The excitation laser beam is focused at the rear focal plane of the objective and emerges from the objective front lens in a parallel beam. TIR at the glass–water interface on the coverslip is then achieved by steering the beam to the periphery of the objective. The fluorescence emission from sample molecules immobilized on the coverslip is collected using the same objective.
4.2. Sample immobilization and data acquisition
4.2.1. Imaging buffer and reagents
The buffer system used for single-molecule FRET imaging must not only ensure the integrity of sample and enzymes but also promote high photostability of the fluorescent dyes. Photoblinking of many fluorescent dyes is reduced in the presence of a triplet-state quenching agent such as Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid), a water-soluble derivative of vitamin E (Rasnik, McKinney, & Ha, 2006). Often, BSA is added to the imaging buffer in order to reduce nonspecific binding to the surfaces of tubes and pipette tips. In order to increase the photostability of Cy5 and Cy3 dye molecules, the sample chamber can be infused with an oxygen scavenger system to reduce photobleaching upon sample immobilization and prior to single-molecule FRET data acquisition. The oxygen scavenger system consumes molecular oxygen by the coupled enzymatic action of glucose oxidase and catalase (Benesch & Benesch, 1953). First, glucose oxidase catalyzes the reaction of molecular oxygen with β-d-glucose to produce d-glucono-1,5-lactone (which hydrolyzes to gluconic acid) and hydrogen peroxide. The second enzyme, catalase, then catalyzes the disproportionation of hydrogen peroxide into water and oxygen. The two coupled enzymatic reactions result in a net decrease of molecular oxygen concentrations. Glucose, a substrate for the glucose oxidase reaction, is provided in the imaging buffer. The enzymatic generation of gluconic acid leads to an acidification of the imaging buffer over time. As many biological samples are sensitive to changes in pH, the buffer strength of the imaging buffer must be sufficiently high to maintain appropriate pH, and the oxygen scavenger system should be added at the beginning of single-molecule FRET data acquisition. Additionally, proper sealing of the slide will minimize exposure of the sample chamber contents to air during the course of the experiment.
Prepare imaging buffer consisting of 12 mM HEPES, 40 mM Tris pH 7.5, 60 mM KCl, 0.32 mM EDTA, 3 mM MgCl2, 10% (v/v) glycerol, 10% (w/v) glucose, 0.1 mg/ml BSA (acetylated; Promega), 2 mM Trolox, and 0.02% Igepal (Sigma-Aldrich). Filter the imaging buffer with a 0.2-μm bottle filter (polyethersulfone; Nalgene) and chill to 4 °C. When stored at 4 °C, the imaging buffer can typically be used for several weeks.
To prepare a 100 × stock solution of the oxygen scavenger system (Gloxy), dissolve 10 mg of glucose oxidase in 100 μl T50 (10 mM Tris, 50 mM NaCl, pH 7.5) by gently pipetting up and down. Resuspend 20 mg/ml catalase stock (Sigma-Aldrich C100) and add 30 μl to the glucose oxidase solution. Gently mix by pipetting up and down and centrifuge at > 14,000 × g at 4 °C for 5 min. Use the supernatant as a 100 × Gloxy stock. Store the stock at 4 °C and use within a few weeks. Alternatively, flash freeze 100× Gloxy stock and store them at −80 °C.
4.2.2. Immobilization of the sample
The sample can be conveniently immobilized on the quartz slide via a biotin–streptavidin linkage (Fig. 2C). Solutions/reagents are flowed into the sample chamber (Fig. 2A) using a pipette with the pipette tip snuggly plugged into one of the two holes. If the sample chamber is already filled and its contents are to be replaced with a new solution, gently apply a Kimwipe (Kimberly Clark) to the outlet hole at the same time. This will generate capillary suction to slowly remove the sample chamber contents. Simultaneously, deliver new solution from the inlet hole with a pipette. Care must be taken to deliver new solution at the right rate so as to avoid sucking the sample chamber dry. In general, the formation of air bubbles in the sample chamber should be avoided at all times.
Apply ~ 35 μl of 0.2 mg/ml streptavidin (Molecular Probes) in T50 to the slide and incubate for ~ 2 min at room temperature to allow for quantitative binding of streptavidin to the surface-immobilized biotin moieties.
Wash out excess streptavidin by flowing ~ 100 μl of imagingbuffer through the sample cell.
To immobilize the sample, apply ~ 70 μl of an appropriate dilution of biotinylated and dye-labeled sample to the sample cell. In the case of a typical nucleosome preparation as described above, dilute by a factor of ~ 5000–10,000. Make a first 1:10 dilution with the filtrate from the concentration step and further dilutions with the imaging buffer. If the right sample dilution is unknown, start out with a dilution that likely generates too low rather than too high a sample density. Check the density on the microscope, and if necessary, increase the sample density by flowing a more concentrated dilution onto the slide. Repeat this procedure until the desired density (~1 molecule/5 μm2 imaging area) is achieved. In general, using higher sample densities is beneficial for two reasons. First, a high sample density will reduce the relative concentration of fluorescent debris molecules that may be present on the slide. Second, a higher sample density allows more single-molecule FRET traces to be analyzed per field of view so that adequate statistics can be generated more readily. Sample density should not be too high, however, since on the CCD, the images of individual fluorescent molecules need to be well separated to prevent misinterpretation of single-molecule FRET traces.
4.2.3. Collecting data for FRET histograms
Before imaging the actual sample, record images of fluorescent beads for the alignment of donor and acceptor channels as described above and in Section 5. Prepare a slide with fluorescent beads (0.1 μm TetraSpeck microspheres; Invitrogen Molecular Probes) as follows: Dilute the fluorescent beads by a factor of 100 with 4 M KCl. Assemble a flow channel on a quartz slide prepared essentially as described above, with the exception that no inlet/outlet holes need to be drilled and the channel formed by the double-sided tape and the cover slip is not sealed with epoxy glue at this point. From one edge of the channel, apply ~ 50 μl of the diluted microspheres to the sample chamber and incubate at room temperature for ~ 10 min to allow the beads to adhere to the quartz surface. Rinse channel with ~ 100 μl of 4 M KCl to remove excess beads. Should the density of adhered beads on the slide not be sufficient, repeat the initial steps with a larger concentration of microspheres. Seal all four sides of the bead slide with epoxy glue for future use.
For channel alignment, image the bead slide with 532 nm excitation in a TIR geometry as described above. Adjust the position of the lens (L1, Fig. 2B) closest to the prism to generate a beamspot that evenly illuminates the imaged area. Adjust the focal plane and record a few short movies of different imaging areas of the bead slide.
Immobilize the sample in a fresh sample chamber as described above. Prepare 100 μl of a 1:100 dilution of the 100 × Gloxy stock with imaging buffer. Flow the 1 × Gloxy dilution onto the slide and proceed immediately with the data acquisition.
Record movies from several different fields of view for construction of the FRET histogram.
4.2.4. Acquiring single-molecule FRET traces in flow experiments
In order to obtain single-molecule FRET traces where an enzymatic reaction is initiated at a well-defined starting time, flow experiments can be carried out. The buffer in the sample cell is exchanged during data acquisition using a syringe pump setup (Fig. 2D), for example, to deliver enzyme and ATP. These types of experiments are particularly well suited for the determination of kinetic properties such as transition rates and intermediate states.
Immobilize the sample on a quartz slide with sample chamber as described above. Connect the syringe to a two-way valve that will, depending on its setting, allow for flow through the inlet piece into the sample chamber or bypass the sample chamber for direct loading/emptying of the syringe (Fig. 2D). Connect one port of the valve with tubing to the inlet piece that consists of a custom-made brass plate with a rubber O-ring. Fill the syringe with ~ 300–500 μl of 1 × Gloxy dilution in imaging buffer and push a small volume through the inlet piece to remove air bubbles inside the tubing. Connect the inlet piece to the sample chamber by screwing it onto the stage such that the O-ring creates a tight connection between syringe pump tubing and the inlet of the sample chamber. Analogously, attach a custom-made brass plate with O-ring to the outlet hole of the sample cell. Direct a short piece of tubing connected to the outlet brass piece to a small disposal vessel on the microscope stage.
Using the syringe pump, infuse the sample chamber with ~ 75–100 μl of 1 × Gloxy dilution. If the seal on both inlet and outlet of the sample chamber is tight and air was properly removed from the syringe and tubing, no air bubbles should be introduced into the sample chamber during the flow.
Record a few movies from different imaging areas to construct a FRET histogram of the sample before the buffer exchange.
At 4 °C, set up the reaction mix containing all components required for enzymatic action on the sample. For example, to observe nucleosome remodeling dynamics, prepare a 400 μl remodeling mix in imaging buffer that contains 1 μM to 2 mM ATP, an equal concentration of MgCl2, 1 × Gloxy, and remodeling enzyme. Load the reaction mix into the syringe using the syringe pump motor and the valve setting that bypasses the sample chamber (Fig. 2D). Switch the syringe valve to the other setting and start the data acquisition. After 10 frames, flow the remodeling reaction mix through the sample chamber. Refocus to compensate for any slight defocusing that the flow might introduce and continue data acquisition until a large portion of the fluorescent dye molecules have bleached.
Stop data acquisition and collect movies of several different and unbleached imaging areas to construct a FRET histogram after the reaction end-point.
5. Data Analysis
We use a home-written LabView code to control the CCD camera. Fluorescence intensities are read out from the CCD chip for each pixel in each frame and stored on the hard drive in a single binary movie file.
The calculation of FRET efficiencies requires each fluorescence intensity spot in one channel to be unambiguously paired with its corresponding spot in the other channel. However, due to imperfections in the alignment of optical elements in the setup, the two halves of the CCD camera cannot be overlaid by a mere translation. Instead, an unambiguous alignment is achieved by calculating a polynomial warping map between the two channels that accounts for translocation, shear distortion, rotation, and resizing. The map coefficients are determined by analyzing a short movie (20–30 frames) of a slide with immobilized fluorescent beads that display detectable fluorescence emission in both channels. Using an IDL script, an image is calculated by averaging the first 20 frames of the movie. In this average image, the corresponding positions in donor and acceptor channel are interactively determined for three beads and used to generate an initial mapping between the two channels. The map coefficients for a more rigorous polynomial warping between the two channels are then determined by taking into account all beads on the slide.
Once a map for alignment of the two channels is determined, IDL scripts are used to analyze movie files from the single-molecule FRET experiments in a multistep procedure. First, an overlay movie is generated by mapping, frame by frame, the image of one channel onto the image of the other channel. In this overlay movie, corresponding donor and acceptor signals from the same molecule are found at the same coordinates in donor and acceptor channels. Next, to identify peaks from individual molecules, an average sum image is generated by averaging together the first 20 frames of the overlay movie. After background subtraction, the average sum image is used to identify peaks from individual molecules in either channel. Peaks are initially identified as local intensity maxima. For each peak, the average intensity of all pixels on a ring with a given radius around the maximum is calculated and compared to the maximum intensity at the peak center. Peaks are only accepted if the ratio of average ring intensity to maximum intensity is below a threshold ratio, such that accepted peaks are relatively bright inside and relatively dim outside the ring. The coordinates of all accepted peaks are stored for further analysis. The threshold ratio and ring radius for the peak finding algorithm are determined empirically. An ideal choice of parameters allows for the identification of a large number of individual molecules while overlapping peaks or dim peaks caused by fluorescent debris on the slide are excluded. Once the peak coordinates are identified based on the average sum image, the following analysis uses the overlay movie described above. In each frame of the overlay movie and at each peak position, the integrated fluorescence intensity signal of all pixels within a ring of a given, empirically determined radius is determined for each channel. This procedure yields the donor and acceptor signals for every molecule in each frame and stores them in a single traces file. The intensity information in the traces file can then be used to calculate a FRET time trace for each imaged molecule.
5.1. Calculating the FRET efficiency
The FRET efficiency is calculated from the fluorescence emission intensities measured in donor and acceptor channels. A fraction of the donor signal is expected to leak into the acceptor channel, which causes an apparent increase in the acceptor emission. Therefore, the measured raw acceptor signal IA, raw needs to be corrected by an empirically determined leakage fraction α to yield the acceptor intensity IA: IA = IA, raw − αID, where ID is the raw donor signal. The leakage fraction α is typically determined by measuring donor and acceptor signals of a molecule that is labeled with a donor dye only, but does not possess an acceptor fluorophore. The FRET efficiency can then be approximated as the ratio of acceptor intensity to the sum of acceptor and donor intensities, IA/(IA + ID). This ratio provides a good estimate of the absolute FRET value if the quantum yields and detection efficiencies of the two fluorescent dyes are very similar, which is the case for the Cy3–Cy5 FRET pair. FRET values cannot typically be used directly to extract absolute distance information, but often an experimental calibration is possible, as outlined below.
5.2. Constructing and analyzing FRET histograms
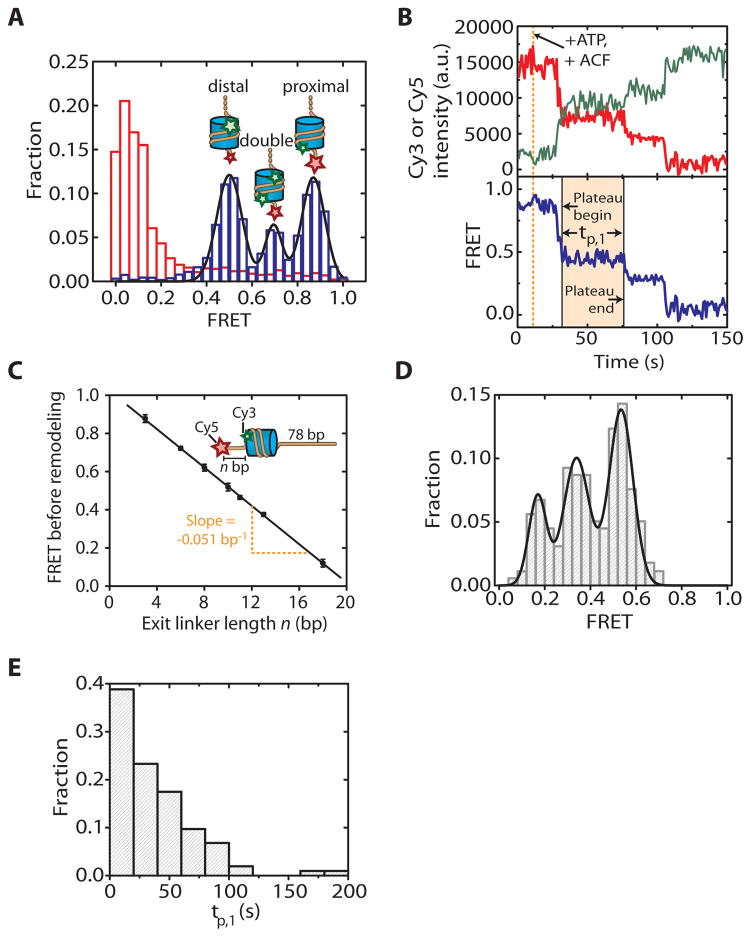
Before single-molecule FRET traces from individual molecules are analyzed, data from several imaging areas on the slide (fields of view) are combined to yield a distribution of FRET values among many molecules, usually displayed as a histogram. Using a Matlab script, the histogram is constructed by calculating the mean FRET efficiency of the first ~ 10 frames for each individual molecule in each of the imaged areas. FRET histograms are convenient for an initial analysis of a new sample because they aid in determining the different FRET populations present in the sample. In the case of only one FRET state, the FRET histogram will exhibit a single peak. The peak position is then determined in good approximation by fitting the FRET distribution to a single Gaussian. If the sample contains multiple species with different FRET values, the resulting histogram will be more complex. Such a situation could arise if multiple conformations of the same molecule or a mixture of distinct molecules with different FRET efficiencies are present in the sample. For example, the FRET histogram of nucleosomes assembled as described above and shown in Fig. 1B displays three distinct peaks (Fig. 3A). Since the FRET donor Cy3 is attached to the H2A subunit of the histone octamer, the presence of two H2A subunits in each octamer leads to three distinct populations of nucleosomes with different labeling configurations: (1) Nucleosomes with a single donor on the H2A subunit proximal to the acceptor, giving rise to a peak at high FRET, (2) nucleosomes with a single donor on the distal H2A, yielding a lower FRET level, and (3) nucleosomes with a donor dye on each of the two H2A subunits, yielding an intermediate FRET value. For this reason, the FRET histogram is best fit with three Gaussian peaks. These FRET peaks are well separated in the histogram, allowing us to clearly identify these populations and select a single population for further analysis. FRET distributions often represent a powerful tool to examine overall changes in the sample, for example, induced by enzymatic activity or by alterations in buffer composition or temperature. For example, the FRET histogram obtained from aforementioned nucleosomes displays a dramatic shift toward very low FRET values upon incubation of the slide with the chromatin remodeling enzyme ACF and ATP (Fig. 3A). Such a shift is to be expected from the remodeling activity of ACF, which centers mononucleosomes on the DNA substrate and thus reduces the FRET values by increasing the distance between donor and acceptor dyes.
Figure 3.
Example single-molecule FRET data of mononucleosomes. (A) FRET histogram constructed from many nucleosomes before (blue bars) and after (red bars) ACF-catalyzed remodeling. The three initial peaks (blue bars) arise from the three distinct donor dye labeling configurations (single proximal donor, single distal donor, proximal and distal donor present as described in the text). The black line represents a fit with three Gaussians. Upon remodeling with ACF, the FRET values are shifted toward very low values (red bars). (B) Donor fluorescence (green), acceptor fluorescence (red), and FRET (blue) traces depicting ACF-induced remodeling of an individual nucleosome with a single proximal donor fluorophore. The first intermediate FRET plateau is shaded in yellow. The duration of the first translocation pause is labeled as tp,1, and begin and end time points are indicated for the first intermediate FRET plateau. (C) Initial FRET value (before remodeling) as a function of the exit linker DNA length (n bp). The black line depicts a linear fit to the data with a slope of −0.051 ± 0.002. The deviation from the expected nonlinear dependence is caused by the flexible linkers connecting the dyes to the nucleosome or the DNA. For this ruler, data from nucleosomes with a single Cy3 dye on the proximal H2A subunit are used. (D) FRET distribution of the pauses. (E) Distribution of tp,1, the duration of the first translocation pause.
5.3. Analysis of single-molecule FRET time traces
For visual inspection of single-molecule FRET time traces, we use a Matlab script that displays the time evolution of acceptor and donor fluorescence signals as well as the FRET efficiency. In our nucleosome example, a flow experiment where the sample chamber is infused with ATP and ACF yields time traces in which the FRET efficiency decreases over time as a consequence of remodeling. In this case, the analysis can be significantly simplified by selecting time traces from nucleosomes containing a single donor on the proximal H2A (Fig. 3B). These nucleosomes display an initially high FRET value and therefore provide a large dynamic range to observe FRET decrease due to remodeling. The selection process involves the selection of traces that exhibit a mean FRET > 0.75 in the first 10 frames prior to infusion of the sample chamber with enzyme and ATP. Among these traces, those displaying a single donor bleaching step are identified. The traces selected this way comprise the entire population of nucleosomes with a single donor on the proximal H2A. Conversely, traces from nucleosomes with donor dyes on both H2A subunits, which exhibit two donor bleaching steps, are excluded.
The observed FRET efficiencies are not absolute but instead depend on the photophysical properties of the fluorophores and on the specific set of filter elements used to optically separate donor and acceptor fluorescence emission. Thus, it is typically difficult to directly convert measured FRET values into physical distances within the sample. In many cases, however, this problem can be circumvented by calibrating the experimental system with a set of samples with varying donor–acceptor distances that are known by design. In the nucleosome example, observed FRET values can be correlated to the octamer position quantitatively by calibration with a nucleosome ruler (Fig. 3C). To generate such a ruler, several ruler nucleosome constructs are prepared that differ from the construct shown in Fig. 1B only in the varying length of the exit side DNA linker that is terminally labeled with Cy5. Each ruler nucleosome therefore has a distinct but known bp distance between the Cy5 dye and the edge of the nucleosome. For each ruler nucleosome construct, a FRET histogram is obtained, and the peak position of the population with a single proximal donor dye is determined as a function of the bp distance between the Cy5 acceptor dye and the edge of the nucleosome. The calibration ruler obtained this way enables a direct correlation of measured FRET values with the bp position of the histone octamer on the DNA.
Careful analysis of single-molecule FRET time traces enables the determination of key kinetic parameters as a function of experimental conditions such as enzyme and substrate concentrations, buffer conditions, or temperature. FRET changes in single-molecule time traces sometimes proceed with pauses or plateaus at distinct FRET states. Distinct FRET states identified in time traces typically correspond to intermediate states that can provide mechanistic insight into the system under study. For example, a typical nucleosome remodeling trace (Fig. 3B) shows that translocation by ACF does not proceed at a constant rate. Instead, the FRET time trace displays periods of gradual decrease interrupted by translocation pauses at intermediate FRET values. Once characteristic intermediate FRET states have been identified at the single-trace level, they can be further analyzed by compiling information from many traces into a histogram. In the nucleosome remodeling example, such a histogram can be constructed the following way: First, a traces file is read by a Matlab script, and individual FRET time traces are displayed on screen. In traces that exhibit remodeling, the start and end time points of each intermediate FRET state are identified manually or via an automated algorithm (Fig. 3B). Next, an average plateau FRET value is calculated as the mean FRET value in the defined time interval. Average FRET plateau values from many traces and typically several remodeling experiments are then used to construct a histogram that reveals the frequency with which a given FRET plateau value occurs (Fig. 3D). In our example, the histogram displays three well-separated peaks, and their center positions are obtained by fitting the histogram with three Gaussians. By comparing with the calibration curve shown in Fig. 3C, we found that these three peaks correspond to nucleosome translocation of 7 bp (for the first peak) and 3–4 bp (for the subsequent two peaks).
The duration of characteristic phases in the time traces, such as dwell times of these FRET plateaus or the duration of the transition time between these plateaus, can be analyzed in a very similar way. In this case, start and end time points of the phases under study are determined, and a histogram is constructed from the interval durations. For example, Fig. 3E shows a dwell time histogram for the duration of the first pause (tp,1), as indicated in Fig. 3B. The shape of the dwell time distribution often can be used to infer kinetic properties of the transition. For example, if the distribution follows a single-exponential decay, the corresponding transition likely has only one rate-limiting step; conversely, a lagged exponential implies more than one rate-limiting step.
In systems with more complex dynamic transitions between multiple distinct FRET states, the identification of these states can be facilitated by applying a hidden Markov modeling routine (HaMMy) (McKinney, Joo, & Ha, 2006). In this approach, the time-binned FRET trace is modeled as a Markov process of state-to-state transitions that all display single-exponential decay kinetics.
The precise details of single-molecule FRET analysis depend on the system under study, and the analysis tools described here are intended to provide a starting point for further analysis tailored to the specific questions being addressed.
6. Summary
During the past decade, single-molecule FRET has been established as a powerful biophysical technique for the mechanistic study of a wide range of biological systems. Here we provide a detailed description of how to set up, carry out, and analyze single-molecule FRET measurements, with a particular emphasis on experiments that require a rapid buffer exchange during data acquisition. As an illustrative example, we describe the observation of nucleosome translocation by single-molecule FRET. While some of the sections regarding sample preparation are specific to this particular example, most procedures and protocols described here are readily adapted to a variety of experimental systems.
Acknowledgments
We thank William Hwang and Bryan Harada for critical reading of the manuscript. This work is in part supported by the Howard Hughes Medical Institute. X. Z. is a Howard Hughes Medical Institute Investigator. S. D. is a Merck fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
References
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Molecular Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Biophotonics, Part B. 2003;361:1–33. doi: 10.1016/s0076-6879(03)61003-7. [DOI] [PubMed] [Google Scholar]
- Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. An engineered aryl azide ligase for site-specific mapping of protein-protein interactions through photo-cross-linking. Angewandte Chemie International Edition. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annual Review of Biochemistry. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Benesch RE, Benesch R. Enzymatic removal of oxygen for polarography and related methods. Science. 1953;118:447–448. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang XW. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual Review of Biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clarklewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao YH, Chakravarthy S, Muthurajan UM, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods in Enzymology. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nature Biotechnology. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nature Reviews Molecular Cell Biology. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: Farewell to the tuna-can octamer? Current Opinion in Genetics & Development. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Forster T. Experimentelle und theoretische Untersuchung des zwischenmolekularen Ubergangs von Elektronenanregungsenergie. Zeitschrift für Naturforschung A. 1949;4:321–327. [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Heyduk T. Measuring protein conformational changes by FRET/LRET. Current Opinion in Biotechnology. 2002;13:292–296. doi: 10.1016/s0958-1669(02)00332-4. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI–containing and ATP–utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Ebright YW, Ebright RH. Site-specific incorporation of fluorescent probes into protein: Hexahistidine-tag-mediated fluorescent labeling with (Ni2+: Nitrilotriacetic acid)(n)-fluorochrome conjugates. Journal of the American Chemical Society. 2001;123:12123–12125. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Molecular and Cellular Biology. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure—Repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DFV, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Current Protocols in Molecular Biology. 2001;Chapter 21(Unit 21.6) doi: 10.1002/0471142727.mb2106s54. [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature Structural & Molecular Biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, et al. Direct observation of DNA distortion by the RSC complex. Molecular Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Chromatin. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical Journal. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- Mendel D, Cornish VW, Schultz PG. Site-directed mutagenesis with an expanded genetic-code. Annual Review of Biophysics and Biomolecular Structure. 1995;24:435–462. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- Miller LW, Cai YF, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: A flexible chemical tag. Nature Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. Journal of Biological Chemistry. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: A versatile method for protein labeling. Nature Chemical Biology. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA–translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. Journal of Molecular Biology. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and longlasting single-molecule fluorescence imaging. Nature Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahian T, Narlikar GJ. Analysis of changes in nucleosome conformation using fluorescence resonance energy transfer. Methods in Molecular Biology. 2012;833:337–349. doi: 10.1007/978-1-61779-477-3_20. [DOI] [PubMed] [Google Scholar]
- Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nature Structural & Molecular Biology. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. The EMBO Journal. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Current Topics in Developmental Biology. 2005;65(65):115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Stryer L, Haugland RP. Energy transfer—A spectroscopic ruler. Proceedings of the National Academy of Sciences of the United States of America. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. Site-specific protein modification on living cells catalyzed by Sortase. Chem Bio Chem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes & Development. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Chromatin remodeling and transcription. Current Opinion in Genetics & Development. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nature Structural & Molecular Biology. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, et al. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Molecular Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]