Abstract
Bioactive ergot alkaloids produced by several species of fungi are important molecules in agriculture and medicine. Much of the ergot alkaloid pathway has been elucidated, but a few steps, including the gene controlling hydroxylation of festuclavine to fumigaclavine B, remain unsolved. Festuclavine is a key intermediate in the fumigaclavine branch of the ergot alkaloid pathway of the opportunistic pathogen Neosartorya fumigata and also in the dihydrolysergic acid-based ergot alkaloid pathway of certain Claviceps species. Based on several lines of evidence, the N. fumigata gene easM is a logical candidate to encode the festuclavine-hydroxylating enzyme. To test this hypothesis we disrupted easM function by replacing part of its coding sequences with a hygromycin resistance gene and transforming N. fumigata with this construct. High pressure liquid chromatography analysis demonstrated that easM deletion mutants were blocked in the ergot alkaloid pathway at festuclavine, and downstream products were eliminated. An additional alkaloid, proposed to be a prenylated form of festuclavine on the basis of mass spectral data, also accumulated to higher concentrations in the easM knockout. Complementation with the wild-type allele of easM gene restored the ability of the fungus to produce downstream compounds. These results indicate that easM encodes an enzyme required for fumigaclavine B synthesis likely by hydroxylating festuclavine. The festuclavine-accumulating strain of N. fumigata may facilitate future investigations of the biosynthesis of dihydrolysergic acid derivatives, which are derived from festuclavine and are the basis for several important drugs.
Keywords: mycotoxin, clavines, P450 monooxygenase, gene cluster, Aspergillus fumigatus
Introduction
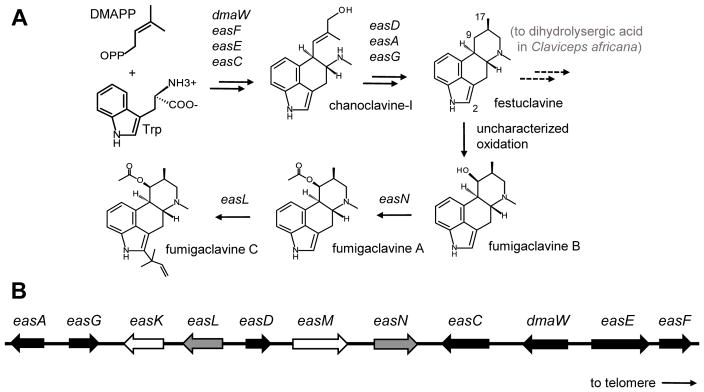
Ergot alkaloids are a large family of fungal specialized metabolites derived from prenylated tryptophan. Different branches of the ergot alkaloid pathway are found in diverse fungi including several plant-associated fungi in the Clavicipitaceae and some saprotrophs and opportunistic pathogens in the Trichocomaceae (reviewed in Gerhards et al. 2014; Robinson and Panaccione 2015; Schardl et al., 2006; Young et al 2015). Ergot alkaloids have adversely affected humans and animals when produced in association with agricultural crops (Haarmann et al. 2009; Panaccione et al. 2014) but also have been adapted and modified for clinical purposes including the treatment of dementia, Parkinson’s disease, migraines, hyperprolactinemia, and type 2 diabetes (Baskys and Hau, 2007; Perez-Lloret and Rascol, 2010; Reddy 2013; Via et al. 2010; Winblad et al. 2008). Most ergot alkaloid producers in the Clavicipitaceae produce derivatives of lysergic acid, though two species of Claviceps produce dihydroergot alkaloids (Agurell and Ramstad 1965; Barrow et al. 1974). Dihydroergot alkaloids differ from lysergic acid-derived ergot alkaloid primarily in the reduction of a particular double bond, providing them with different and valuable pharmacological properties. The ergot alkaloid producers in the Trichocomaceae, including the intensively studied Neosartorya fumigata (synonym Aspergillus fumigatus), produce the fumigaclavine class of ergot alkaloids (Fig. 1A). The fumigaclavine and dihydroergot alkaloid pathways share the simple tetracyclic ergot alkaloid festuclavine as their last common intermediate (Fig. 1A). Fungi in the Trichocomaceae modify festuclavine into fumigaclavines by hydroxylation and acetylation at carbon 2 and/or prenylation at carbon 9 (Fig. 1A), whereas the dihydroergot alkaloid-producing fungus Claviceps africana oxidizes carbon 17 to produce dihydrolysergic acid (Robinson and Panaccione 2015; Wallwey and Li 2011). Thus, festuclavine occupies an important position at the divergence point of these two branches of the ergot alkaloid pathway.
Fig 1.
Ergot alkaloid pathway and gene cluster in N. fumigata. (A) Key intermediates and products in the ergot alkaloid pathway of N. fumigata. Genes controlling steps are indicated between alkaloids. The positions of carbon molecules mentioned by number in the text are labeled in festuclavine. (B) Ergot alkaloid gene cluster of N. fumigata [redrawn from Unsöld and Li (2005); Coyle and Panaccione (2005)]. Black arrows represent genes conserved among ergot alkaloid producers in the Trichocomaceae and Clavicipitaceae and required to assemble the first tetracyclic intermediate. Arrows shaded gray represent genes with established roles in the fumigaclavine branch of the pathway and found only in ergot alkaloid clusters of certain Trichocomaceae. White arrows represent P450 monooxygenase genes that have not been functionally analyzed. The relative position of the telomere on the long arm of chromosome 2 is indicated
Many genes involved in the biosynthesis of lysergic acid-derived ergot alkaloids and the fumigaclavine class of ergot alkaloids have been characterized in recent years (reviewed in Gerhards et al. 2014; Robinson and Panaccione 2015), whereas those genes specifically involved in synthesis of dihydroergot alkaloids have not been investigated. In all cases, these ergot alkaloid synthesis (eas) genes have been found in a contiguous cluster in the genome of the producing organism (Coyle and Panaccione 2005; Lorenz et al. 2009; Schardl et al. 2013a b; Unsöld and Li 2005). In fact, no genes contributing to the production of ergot alkaloids have been found outside of an eas gene cluster. The eas cluster of the common saprotroph and opportunistic human pathogen N. fumigata has been studied extensively (Fig. 1B). Many genes in this cluster have been functionally analyzed and demonstrated to have specific roles in the synthesis of the fumigaclavines produced by N. fumigata (Fig. 1). Genes responsible for each step in the biosynthesis of the most frequently encountered N. fumigata ergot alkaloids have been identified except for one: The gene encoding the enzyme that hydroxylates festuclavine to fumigaclavine B has not been definitely identified or functionally analyzed. Two genes remain in the N. fumigata eas cluster that have not been functionally tested, and both have the capacity to encode for P450 monooxygenases—enzymes that clearly could be involved in the hydroxylation of festuclavine (Coyle and Panaccione 2005; Unsöld and Li 2005). One of these genes, easM (also called fgaP450-2), has a homolog in the eas cluster of Penicillium commune, a related fungus that produces an isomer of fumigaclavine A, whereas the other gene (easK or fgaP450-1) lacks a homolog in the P. commune eas cluster (Wallwey and Li 2011). The possibility that the products of easM and easK are functionally redundant also must be considered.
In this study, we functionally analyzed easM in N. fumigata by gene knockout and complementation followed by qualitative and quantitative analyses of ergot alkaloids. The results provide clear information on the role of easM in the ergot alkaloid pathway and provide a strain of N. fumigata that should be useful for future studies on genes involved in the synthesis of pharmaceutically important dihydroergot alkaloids.
Materials and methods
Knockout and complementation of easM
The coding sequences (including introns) of easM along with 499 bp of 5′-flanking sequences and 676 bp of 3′-flanking sequences were PCR amplified from N. fumigata Af293 genomic DNA in a reaction primed with easMF and easMR (Table 1). The reaction consisted of a 25 μL mixture containing: 1 X GoTaq Flexi buffer (Promega, Madison, WI); 1.5 mM MgCl2; 200 μM each deoxynucleotide triphosphate; 1 μM of each designated primer; and, 2.5 units GoTaq Flexi DNA polymerase (Promega, Madison, WI). The thermocycler program began with a denaturation step at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 3 min, and concluded with a final extension of 72°C for 5 min. The 3067-bp easM amplicon was then ligated into plasmid pCR 2.1 (Thermo-Fisher Scientific, Waltham, MA) and transformed into Escherichia coli. The resulting plasmid was digested with SmaI and ClaI which removed 1091 bp of sequences located near the middle of the 3,067-bp fragment of easM. The 1441-bp hygromycin resistance gene hph1 was amplified from template pCB1004 (Fungal Genetics Stock Center, Manhattan, KS) in a PCR reaction primed with SmaHygF and ClaHygR (Table 1) and utilizing the same thermocycler program described above; recognition sites for SmaI and ClaI were included in the 5′ ends of these primers to facilitate subcloning of the hph1 fragment into the gap of the easM plasmid. The insert of the resulting plasmid was amplified in a PCR primed with easMF and easMR catalyzed by Phire Hot Start II polymerase (Thermo-Fisher Scientific, Waltham, MA). The thermocycler program started with a 98 °C denaturation for 30 s, followed by 35 cycles of 98 °C for 15 s, 65 °C for 15 s, and 72 °C for 1 min. The product was then cleaned with a QIAquick column (Qiagen, Gaithersburg, MD) prior to fungal transformation.
Table 1.
PCR primers and products
| Primer combination | Primer sequence (5′ to 3′) | Product | Product length in wild type | Product length in knockout |
|---|---|---|---|---|
| easMF + easMR | GCCATTCCTCCACTCTTCAG + CCAAAGAACATTCCCAGCTC | easM | 3067 bp | 3416 bp |
| M5F + HY | ACTGCGCTACAGTCGATGTAAC + GGATGCCTCCGCTCGAAGTA | 5′ flank, ko locus | No product | 2916 bp |
| M3F + YG | GACTGCCAGTATCATCTACC + CGTTGCAAGACCTGCCTGAA | 3′ flank, ko locus | No product | 2057 bp |
| SmaHygF + ClaHygR | CATGCCCGGGGGCTTGGCTGGAGCTAGTGG + CTGTATCGATCCGTGGAGGTAATAATTGACG | hph1 | No product | 1441 bp |
Spheroplasts of N. fumigata Af293 were prepared by collecting mycelia from an overnight culture grown in malt extract broth and incubating them in 15 mL of 0.7 M NaCl containing 40 mg lysing enzyme (Sigma-Aldrich, St. Louis, MO) and 1 g VinoTaste Pro (Gusmer Enterprises Inc., Mountainside, NJ) at 37 °C for two hr. Spheroplasts were then purified and transformed with the easM-hph1 construct according to established methods (Coyle and Panaccione 2005; Coyle et al. 2010). Hygromycin-resistant transformants were transferred to malt extract agar containing 300 μg/mL hygromycin (InvivoGen, San Diego, CA) and purified to nuclear homogeneity by culturing from single conidia.
For complementation of the easM knockout, the wild-type allele of easM was amplified from N. fumigata template DNA in a reaction primed with easMF and easMR, as described above. The purified PCR product was introduced into spheroplasts of N. fumigata easM knockout by cotransformation with pBCphleo (Fungal Genetics Stock Center, Manhattan, KS) and selected on phleomycin (InvivoGen, San Diego, CA) at 400 μg/mL.
Transformants were screened for integration of the knockout construct at the native easM locus by three types of PCR assays. In the first assay, the entire easM locus was amplified from primers easMF and easMR (with conditions described above); amplification of the wild-type easM locus produced a fragment of 3067 bp, whereas integration of the knockout construct increased the length of the template and resulting product (Table 1). The other two assays were designed to amplify across the 5′-flanking junction or 3′-flanking junction of an allele resulting from homologous recombination of the knockout construct at the native copy of easM. In these assays, one primer was derived from N. fumigata DNA flanking the site of recombination at the easM locus, and the other primer was derived from the introduced hph1 gene (Table 1); thus, an amplicon can only be obtained from the knockout allele of the easM locus. PCR conditions and program were as described above for the reaction primed from easMF and easMR.
Analyses of ergot alkaloids
Fungal cultures grown on malt extract agar were extracted and analyzed by high performance liquid chromatography (HPLC) as described previously (Panaccione et al. 2012). Briefly, extracts were separated on a C18 column (Phenomenex Prodigy ODS3, 5-μm particle size, Torrence, CA) with a multilinear, binary gradient of aqueous ammonium acetate and acetonitrile and detected by fluorescence (excitation and emission at 272 nm and 372 nm, respectively). For quantitative analyses, six replicate cultures of N. fumigata Af293 (wild type), an easM knockout strain, and an easM complemented strain were grown on malt extract agar for two weeks. Cultures were extracted by removing a 50-mm2 surface area sample with the wider end of 1000-μL pipette tip and suspending it in HPLC-grade methanol for 1 hr, rotating end-over-end at 13 rpm. After centrifugation, 20 μL of the supernatant was analyzed via reverse phase HPLC as described above. Ergot alkaloids were quantified by comparing peak areas to a standard curve prepared from dihydroergotamine (Sigma, St. Louis, MO). The number of conidia in each extract was counted with the aid of a hemocytometer to provide an estimate of fungal biomass extracted, and alkaloids concentrations were normalized to conidial concentrations. Standard for chanoclavine-I was obtained from Alfarma (Prague, Czech Republic), and fumigaclavine A was purchased from Axxora (San Diego, CA). Peaks corresponding to festuclavine and fumigaclavines B and C were established by mass spectral analyses of native and de-esterified preparations (Panaccione and Coyle 2005) and were confirmed by their elimination in dmaW-knockout strains of N. fumigata (Coyle and Panaccione 2005). An uncharacterized chemical detected via HPLC and eluting from the column at 55.9 min was explored with the aid of a Thermo Fisher LCQ DecaXP Liquid Chromatography/Mass Spectrometer (LC/MS) as described by Ryan et al. (2013).
Alkaloid concentrations were analyzed statistically with JMP software (SAS, Cary, NC). We used a Brown-Forsythe test to assess inequality of group variances (with P > 0.05) then ANOVA to compare whether alkaloid quantities varied by strain (with P < 0.05). For alkaloids demonstrating a significant difference among strains in ANOVA, means were separated with a Turkey-Kramer Honestly Significant Difference test with α at 0.05.
Results
Functional analysis of easM by gene knockout
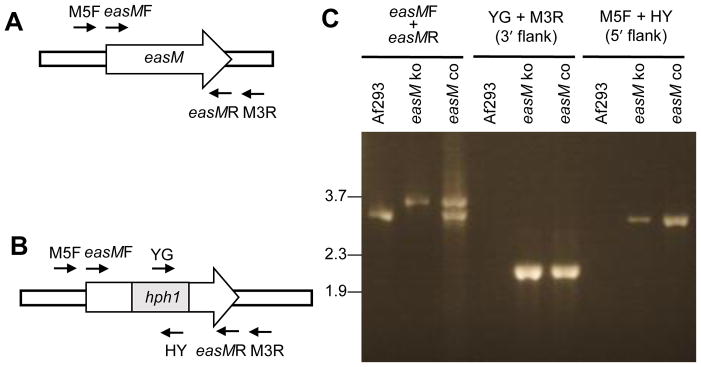
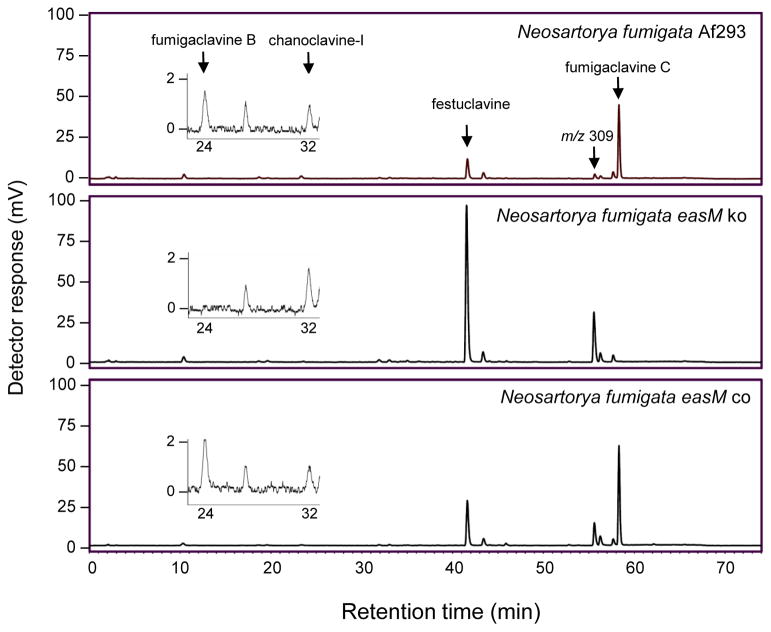
To test the hypothesis that easM is required for hydroxylation of festuclavine, easM in N. fumigata Af293 was knocked out by homologous recombination with a construct containing a mutated copy of easM. In this construct, a fragment containing 1092 bp of easM coding sequences was replaced with a 1441-bp hygromycin resistance gene cassette (hph1) (Fig. 2). Fifty-four hygromycin-resistant transformants were obtained and screened for qualitative changes in ergot alkaloid profiles by HPLC. Twelve showed an accumulation of festuclavine (the intermediate preceding the hypothesized EasM step) and loss of fumigaclavine C, the pathway end product (Fig. 3), indicative of a knockout of the gene controlling oxidation of festuclavine.
Fig 2.
Knockout of easM in N. fumigata. Schematic representation of easM locus in N. fumigata Af293 before (A) and after (B) gene knock out. Relative positions of PCR primers are indicated. Refer to Table 1 for information on primers and PCR products. hph1 represent the hygromycin phosphotransferase gene replacing part of easM. Primers M5F and M3R anneal to regions of the N. fumigata chromosome that flank the recombination site, thus when paired with primers that anneal to hph1 prime amplification only from strains in which the knockout construct has integrated at easM. (C) PCR products of N. fumigatus Af293 wild type, easM knockout (easM ko), and easM complemented (easM co) strains primed with the indicated oligonucleotides. Relative mobility of relevant fragments of BstEII-digested bacteriophage λ DNA is indicated to the left of photograph
Fig 3.
Qualitative HPLC analyses of ergot alkaloids from wild type, easM knockout (easM ko), and easM complemented (easM co) strains of N. fumigata. Data were collected with a fluorescence detector with excitation and emission settings of 272 nm and 372 nm, respectively. Peaks corresponding to ergot alkaloids are labeled only in the wild-type chromatogram. The peak with a m/z value 309.3 in LC/MS analyses, consistent with the [M+H]+ of a prenylated form of festuclavine, is labeled. The insert in each chromatogram shows a close up of time range in which fumigaclavine B and chanoclavine-I eluted
The easM locus in a putative knockout mutant was analyzed for the presence of an integrated knockout construct in three types of PCR assays. The first assay was primed with oligonucleotides flanking the intended site of integration (Table 1; Fig. 2). The fragment obtained from the easM knockout was larger than that obtained from wild-type N. fumigata due to the replacement of a portion of the easM coding sequences with a larger fragment containing the hygromycin phosphotransferase gene. The second and third assays were each conducted with one primer that flanked the 5′ or 3′ junction of the easM-hph1 construct integration site, respectively, and a second primer that annealed to hph1 (Table 1; Fig. 2). The knockout strain had bands of the predicted sizes for each screen, whereas the wild-type strain lacked a product. These data indicated that the knockout strain contained the easM-hph1 construct integrated at the native easM locus.
Complementation of easM
To demonstrate that the phenotype of the easM knockout was due to the integration at easM and not any ectopic events, the easM knockout strain was complemented in trans with the wild-type allele of easM. Complementation resulted in restoration of the wild-type chemotype (Fig. 3), indicating the change in alkaloid profile in the knockout strain resulted from inactivation of easM. Integration of the complementing easM allele (which was introduced as a separate fragment, along with co-transformed pBCphleo) was analyzed by the same set of PCR assays described above (Fig. 2). The data indicated that the complementation had not occurred by homologous recombination at the easM knockout locus..
Preliminary characterization of an additional compound affected by the easM knockout
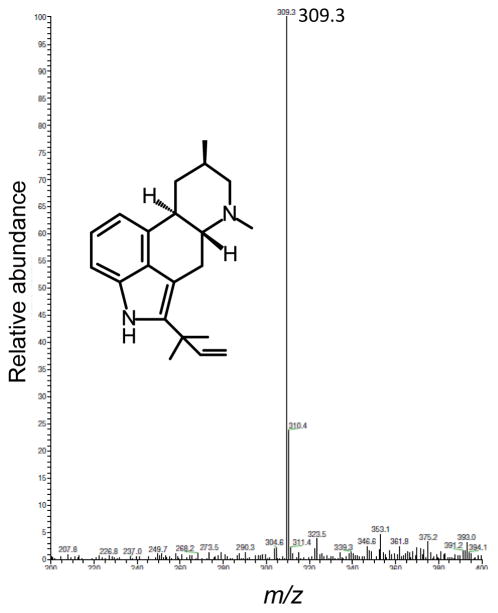
An uncharacterized chemical eluting from the column at 55.9 min also appeared to be affected by easM in that its concentration increased in the easM knockout and was reduced by complementation with the wild-type easM locus (Fig. 3). LC/MS analysis of the compound yielded a molecular ion of m/z 309.3 (Fig. 4). This molecular ion is consistent with the [M+H]+ of a prenylated form of festuclavine. Ge et al. (2009) previously reported a 2-prenylated version of festuclavine in N. fumigata that they named 9-deacetoxyfumigaclavine C. Our analyses did not allow us to determine the site of prenylation.
Fig 4.
Mass spectrum of compound accumulating to higher concentrations in the N. fumigata easM knockout strain. Mass of molecular ion [M+H]+ is indicated. Structure represents 9-deacetoxyfumigaclavine C (Ge et al. 2009), a festuclavine derivative with a molecular ion of 309.23 produced by N. fumigata
Quantitative analysis of ergot alkaloids
Whereas the data in Fig. 3 are qualitative in nature, we conducted additional alkaloid analyses on six samples each of N. fumigata Af293, an easM knockout strain, and an easM-complemented strain to compare quantities of ergot alkaloids (Table 2). Festuclavine accumulated to significantly higher concentrations in the N. fumigata easM knockout than in the wild-type or easM-complemented strains (P < 0.05). The m/z 309 compound (hypothetical prenylated festuclavine) was the second most abundant ergot alkaloid measured in the easM knockout, and its concentrations were significantly greater in the knockout compared to wild-type or easM-complemented strains (P < 0.05). The concentration of the m/z 309 compound in all three strains correlated extremely well with the concentration of its hypothetical precursor festuclavine (R2 = 0.99; P < 0.0001); in contrast, the concentration of the m/z 309 compound negatively correlated with the sum of the concentrations of all other ergot alkaloids (R2 = 0.47; P = 0.002). These data are consistent with the m/z 309 compound being a derivative of festuclavine. The early pathway intermediate chanoclavine-I also was detected in the easM knockout though in very low concentrations (Fig. 3; Table 2). The knockout accumulated more chanoclavine-I than did either the wild-type or easM-complemented strain (P < 0.05), which is reasonable considering that chanoclavine-I is found upstream of the pathway block in the easM knockout mutant (Fig. 1). Alkaloids downstream of the easM step in the pathway were not detected in the easM knockout but were measured in the wild-type and easM-complemented strains (Table 2).
Table 2.
Ergot alkaloids in wild-type and modified Neosartorya fumigata (mean amol/conidium ± standard error; n = 6)a
| Strainb | Chanoclavine I | Festuclavine | m/z 309 compoundc | Fumigaclavine B | Fumigaclavine C | total |
|---|---|---|---|---|---|---|
| Af 293 | 0.2 ± 0.03 A | 9.3 ± 2.7 A | 1.8 ± 0.5 A | 0.8 ± 0.11 A | 13.6 ± 2.2 A | 25.6 ± 2.0 A |
| easM ko | 0.5 ± 0.05 B | 30.9 ± 2.7 B | 7.7 ± 1.0 B | ndd | nd | 39.0 ± 3.7 B |
| easM co | 0.2 ± 0.03 A | 10.6 ± 1.0 A | 3.4 ± 0.7 A | 0.4 ± 0.02 B | 17.7 ± 1.4 A | 32.3 ± 2.9 AB |
values within a column that are labeled with a different letter differ significantly (α=0.05) in a Tukey-Kramer honestly significant difference test (when alkaloids were detected in all three strains) or one-way ANOVA (when alkaloids were detected in only two strains)
abbreviations: ko, knockout; co, complemented
putative prenylated festuclavine
not detected; limit of detection = 0.1 amol/spore
Since previous studies demonstrated that a small proportion of festuclavine is secreted into the medium as opposed to being retained in the colony of the fungus (Mulinti et al. 2014), we tested whether an increased proportion of the large quantity of festuclavine observed in the easM knockout was secreted. Our data indicated that the proportion of festuclavine secreted by the easM knockout (0.11) did not differ significantly (P = 0.49) from the proportion of festuclavine secreted by the wild type (0.09).
Discussion
Our data demonstrate that the putative P450 monooxygenase gene designated easM in the ergot alkaloid gene cluster encodes the enzyme that hydroxylates festuclavine to fumigaclavine B in N. fumigata. Knockout of easM eliminated the accumulation of all downstream alkaloids derived from oxidized festuclavine, including fumigaclavine B (the next intermediate in the pathway) and fumigaclavine C (the pathway end product). The intermediate fumigaclavine A did not accumulate to detectable levels in the wild-type recipient strain N. fumigata Af293 or its derivatives in this study. The lack of detectable fumigaclavine A was not surprising since N. fumigata Af293 accumulated the lowest concentration of fumigaclavine A among 12 fumigaclavine-producing isolates of N. fumigata previously surveyed (Robinson and Panaccione 2012). This particular N. fumigata isolate presumably efficiently prenylates fumigaclavine A to fumigaclavine C.
Interestingly, a compound with molecular mass and retention time predicted for a prenylated version of festuclavine (m/z 309) accumulated to significantly higher concentrations in the easM knockout as compared to wild type. Previous research indicated that the N. fumigata prenyl transferase EasL (also called FgaPT1; Unsöld and Li 2006) has a relaxed substrate specificity for certain ergot alkaloids. The primary substrate for EasL (FgaPT1) is fumigaclavine A (Unsöld and Li 2006), but Robinson and Panaccione (2014) observed accumulation of prenylated versions of the ergot alkaloids agroclavine and setoclavine in engineered strains of N. fumigata lacking fumigaclavine A. Moreover, isolates of N. fumigata that lacked a functional copy of easL failed to produce the presumed prenylated derivatives. Ge et al. (2009) characterized prenylated versions of festuclavine (9-deacetoxyfumigaclavine C) and fumigaclavine B (9-deacetylfumigaclavine C) that were prenylated at precisely the same position at which fumigaclavine A is prenylated to yield fumigaclavine C. The biosynthetic origin of these compounds was not investigated in that study, but we hypothesize that they result from prenyl transferase EasL accepting alternate ergot alkaloids as substrates. Similarly, we hypothesize that some of the large pool of festuclavine that accumulated in the easM knockout was prenylated to 9-deacetoxyfumigaclavine C by the native N. fumigata prenyl transferase EasL, which was left without its typical substrate fumigaclavine A as a result of the easM knockout. The quantitative analysis of ergot alkaloids which yielded a strong correlation of quantities of the putative prenylated form of festuclavine with festuclavine support this hypothesis.
Prior to this current study, two genes in the ergot alkaloid gene cluster of N. fumigata remained without functional analyses: easM and easK. Both of these genes are predicted to encode P450 monooxygenases (Coyle and Panaccione 2005; Unsöld and Li 2005). The possibility that easK encodes an enzyme that is functionally redundant with that encoded by easM can be excluded by our results. Whether the product of easK has a role in modifying ergot alkaloids of N. fumigata remains to be determined, but its location in the ergot alkaloid gene cluster would be unusual otherwise. One possible role for the product of easK would be hydroxylation of the N-methyl group of fumigaclavine A to fumigaclavine D, a metabolite of N. fumigata characterized recently from extracts of the fungus grown on wheat grains (Cano et al. 2013). We have not identified this metabolite in isolates of N. fumigata grown on other media in our investigations.
The genetics of the branch of the ergot alkaloid pathway leading to the biosynthesis of lysergic acid-derived ergot alkaloids is well established (Gerhards et al. 2014; Robinson and Panaccione 2015; Young et al. 2015), but the genetic differences resulting in the production of dihydrolysergic acid-derived ergot alkaloids have not been elucidated. Festuclavine is the first intermediate after the branch point in the pathway that leads to dihydrolysergic acid as opposed to lysergic acid (Barrow et al. 1974). Although many isolates of N. fumigata produce festuclavine as an intermediate in their pathway branch to fumigaclavines and variation has been documented in ergot alkaloid pathway end points among N. fumigata isolates (Robinson and Panaccione 2012), no natural isolate has been found that accumulates festuclavine as its pathway end product. A significant outcome of the present study is the engineering of a festuclavine-accumulating strain of N. fumigata which may serve as a transformation recipient in future experimental approaches to investigate the oxidation of festuclavine to dihydrolysergic acid. The determination of the biosynthetic route to dihydrolysergic acid would be significant because several important pharmaceuticals used in the treatment of dementia, migraines, hyperprolactinemia, and other ailments are semisynthetic derivatives of dihydrolysergic acid (Baskys and Hau 2007; Perez-Lloret and Rascol 2010; Reddy 2013; Via et al. 2010; Winblad et al. 2008).
Acknowledgments
We gratefully acknowledge the support of grants R15GM114774 from the National Institutes of Health, National Institute of General Medical Sciences and 2012-67013-19384 from the United States Department of Agriculture, National Institute of Food and Agriculture. This work is published the approval of the West Virginia Agriculture and Forestry Experiment Station as article number (to be supplied upon acceptance). Technology described in this article is embodied in U.S. Provisional Patent Application No. PCT/US2015/035784. We thank Paige Bragg for technical assistance.
References
- Agurell S, Ramstad E. A new ergot alkaloid from Mexican maize ergot. Acta Pharm Suecica. 1965;2:231–238. [PubMed] [Google Scholar]
- Barrow KD, Mantle PG, Quigley FR. Biosynthesis of dihydroergot alkaloids. Tet Lett. 1974;16:1557–1560. [Google Scholar]
- Baskys A, Hou AC. Vascular dementia: pharmacological treatment approaches and perspectives. Clin Interv Aging. 2007;2:327–335. [PMC free article] [PubMed] [Google Scholar]
- Cano PM, Jamin EL, Tadrist S, Bourdaud’hui P, Péan M, Debrauwer L, Oswald IP, Delaforge M, Puel O. New untargeted metabolic profiling combining mass spectrometry and isotopic labeling: Application on Aspergillus fumigatus grown on wheat. Anal Chem. 2013;85:8412–8420. doi: 10.1021/ac401872f. [DOI] [PubMed] [Google Scholar]
- Coyle CM, Panaccione DG. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl Environ Microbiol. 2005;71:3112–3118. doi: 10.1128/AEM.71.6.3112-3118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle CM, Cheng JZ, O’Connor SE, Panaccione DG. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol. 2010;76:3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge HM, Yu ZG, Zhang J, Wu JH, Tan RX. Bioactive alkaloids from endophytic Aspergillus fumigatus. J Nat Prod. 2009;72:753–755. doi: 10.1021/np800700e. [DOI] [PubMed] [Google Scholar]
- Gerhards N, Neubauer L, Tudzynski P, Li SM. Biosynthetic pathways of ergot alkaloids. Toxins. 2014;6:3281–3295. doi: 10.3390/toxins6123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarmann T, Rolke Y, Giesbert S, Tudzynski P. Ergot: from witchcraft to biotechnology. Mol Plant Pathol. 2009;10:563–577. doi: 10.1111/j.1364-3703.2009.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz N, Haarmann T, Pažoutová S, Jung M, Tudzynski P. The ergot alkaloid gene cluster: functional analyses and evolutionary aspects. Phytochemistry. 2009;70:1822–1832. doi: 10.1016/j.phytochem.2009.05.023. [DOI] [PubMed] [Google Scholar]
- Mulinti P, Allen NA, Coyle CM, Gravelat FN, Sheppard DC, Panaccione DG. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr Microbiol. 2014;68:1–5. doi: 10.1007/s00284-013-0434-2. [DOI] [PubMed] [Google Scholar]
- Panaccione DG, Coyle CM. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol. 2005;71:3106–3111. doi: 10.1128/AEM.71.6.3106-3111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaccione DG, Ryan KL, Schardl CL, Florea S. Analysis and modification of ergot alkaloid profiles in fungi. Method Enzymol. 2012;515:267–290. doi: 10.1016/B978-0-12-394290-6.00012-4. [DOI] [PubMed] [Google Scholar]
- Panaccione DG, Beaulieu WT, Cook D. Bioactive alkaloids in vertically transmitted fungal endophytes. Funct Ecol. 2014;28:299–314. [Google Scholar]
- Perez-Lloret S, Rascol O. Dopamine receptor agonists for the treatment of early or advanced Parkinson’s disease. CNS Drugs. 2010;24:941–968. doi: 10.2165/11537810-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Reddy DS. The pathophysiological and pharmacological basis of current drug treatment of migraine headache. Expert Rev Clin Pharmacol. 2013;6:271–288. doi: 10.1586/ecp.13.14. [DOI] [PubMed] [Google Scholar]
- Robinson SL, Panaccione DG. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia. 2012;104:11–310. doi: 10.3852/11-310. [DOI] [PubMed] [Google Scholar]
- Robinson SL, Panaccione DG. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Appl Environ Microbiol. 2014;80:6465–6472. doi: 10.1128/AEM.02137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SL, Panaccione DG. Diversification of ergot alkaloids in natural and modified fungi. Toxins. 2015;7:201–218. doi: 10.3390/toxins7010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KL, Moore CT, Panaccione DG. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins. 2013;5:445–455. doi: 10.3390/toxins5020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Panaccione DG, Tudzynski P. Ergot alkaloids-biology and molecular biology. Alkaloids. 2006;63:45–86. doi: 10.1016/s1099-4831(06)63002-2. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B, Panaccione DG, Schweri KK, Voisey CR, Farman ML, Jaromczyk JW, Roe BA, O’Sullivan DM, Scott B, Tudzynski P, An Z, Arnaoudova EG, Bullock CT, Charlton ND, Chen L, Cox M, Dinkins RD, Florea S, Glenn AE, Gordon A, Güldener U, Harris DR, Hollin W, Jaromczyk J, Johnson RD, Khan AK, Leistner E, Leuchtmann A, Li C, Liu JG, Liu J, Liu M, Mace W, Machado C, Nagabhyru P, Pan J, Schmid J, Sugawara K, Steiner U, Takach JE, Tanaka E, Webb JS, Wilson EV, Wiseman JL, Yoshida R, Zeng Z. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013;9:e1003323. doi: 10.1371/journal.pgen.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Young CA, Pan J, Florea S, Takach JE, Panaccione DG, Leuchtmann A. Currencies of mutualisms: sources of alkaloid genes in vertically transmitted Epichloë. Toxins. 2013;5:1064–1088. doi: 10.3390/toxins5061064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsöld IA, Li SM. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology. 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- Unsöld IA, Li SM. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem. 2006;7:158–164. doi: 10.1002/cbic.200500318. [DOI] [PubMed] [Google Scholar]
- Via MA, Chandra H, Araki T, Potenza MV, Skamagas M. Bromocriptine approved as the first medication to target dopamine activity to improve glycemic control in patients with type 2 diabetes. Diabetes Metab Syndr Obes. 2010;3:43–48. doi: 10.2147/dmsott.s9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallwey C, Li SM. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep. 2011;28:496–510. doi: 10.1039/c0np00060d. [DOI] [PubMed] [Google Scholar]
- Winblad B, Fioravanti M, Dolezal T, Logina I, Milanov IG, Popescu DC, Solomon A. Therapeutic use of nicergoline. Clin Drug Invest. 2008;28:533–552. doi: 10.2165/00044011-200828090-00001. [DOI] [PubMed] [Google Scholar]
- Young CA, Schardl CL, Panaccione DG, Florea S, Takach JE, Charlton ND, Moore N, Webb JS, Jaromczyk J. Genetics, genomics and evolution of ergot alkaloid diversity. Toxins. 2015;7:1273–1302. doi: 10.3390/toxins7041273. [DOI] [PMC free article] [PubMed] [Google Scholar]