Abstract
Mucin1 (MUC1) is an epithelial glycoprotein overexpressed in ovarian cancer and actively involved in tumor cell migration and metastasis. Using novel in vitro and in vivo MUC1-expressing conditional (Cre-loxP) ovarian tumor models, we focus here on MUC1 biology and the roles of Kras activation and Pten deletion during cell transformation and epithelial-to-mesenchymal transition (EMT). We generated several novel murine ovarian cancer cell lines derived from the ovarian surface epithelia (OSE) of mice with conditional mutations in Kras, Pten or both. In addition, we also generated several tumor-derived new cell lines that reproduce the original tumor phenotype in vivo and mirror late stage metastatic disease.
Our results demonstrate that de novo activation of oncogenic Kras does not trigger increased proliferation, cellular transformation or EMT and prevents MUC1 upregulation. In contrast, Pten deletion accelerates cell proliferation, triggers cellular transformation in vitro and in vivo and stimulates MUC1 expression. Ovarian tumor-derived cell lines MKP-Liver and MKP-Lung cells reproduce in vivo EMT and represent the first immune competent mouse model for distant hematogenous spread. Whole genome microarray expression analysis using tumor and OSE-derived cell lines reveals a 121 gene signature associated with EMT and metastasis. When applied to n=542 cases from the ovarian cancer TCGA dataset, the gene signature identifies a patient subset with decreased survival (p=0.04). Using an extensive collection of novel murine cell lines we have identified distinct roles for Kras and Pten on MUC1 and EMT in vivo and in vitro. The data has implications for future design of combination therapies targeting Kras mutations, Pten deletions and MUC1 vaccines.
Keywords: MUC1, Kras, Pten, EMT, ovarian cancer
Introduction
Ovarian cancer is often diagnosed at late stages, when the tumor has spread outside the ovaries. Loco-regional dissemination involves detachment of cancer cells from the primary tumor and attachment on the surface of surrounding anatomical sites, frequently on the omentum and peritoneum, but also on the diaphragm and small bowel mesentery1, 2. Lung metastases have been reported in up to 12% of patients3–5. As expected, disseminated stage IV ovarian cancer has poor prognosis with ten year survival rate of only 6%, in contrast to 73–92% in stage I cancer, confined to the ovaries2. It is believed that cancer dissemination may be linked, at least in part, to a process called epithelial-to-mesenchymal (EMT) transition, during which epithelial tumor cells acquire a fibroblast-like morphology, and display an invasive phenotype and increased proliferation6. Although EMT is multifactorial and has been described for several solid tumors, the process has not been captured thus far in ovarian cancer in vivo in immune competent animal models.
We focus here on the pathogenic roles of oncogenic Kras, Pten tumor suppressor and mucin 1 (MUC1) oncoprotein, a cell surface protein with roles in cell adhesion and migration7. MUC1 is a transmembrane molecule overexpressed by virtually all ovarian adenocarcinomas, regardless of histology8, 9. Although the molecule can present in several isoforms, the major full length MUC1 protein comprises an elongated extracellular region, a transmembrane domain and a cytosolic tail with signaling properties10, 11. Given the high density of negatively charged sialic acid residues that cap the branched O-linked glycosylation sites, MUC1 may aid in the initial cell detachment from the primary tumor12. In addition, due to its large extracellular domain, MUC1 may also intermediate subsequent steps, by facilitating adhesion at the metastatic site13, 14. The oncogenic function of MUC1 is attributed to its intracellular domain that contains several phosphorylation domains15. The links between MUC1 biology and the typical oncogenic or tumor suppressor pathways altered in ovarian cancer have not as yet been addressed.
High grade serous ovarian tumors are considered to arise mostly from precursor lesions in the fallopian tubes16 and are overwhelmingly characterized by TP53 mutations, present in 93% of cases17. In addition to TP53, ovarian tumors may also carry oncogenic KRAS mutations and PTEN deletion mutations18 or altered expression19, although these type of mutations are more frequent in non-serous tumors, especially endometrioid and clear cell histotypes. PTEN phosphatase acts as a repressor of the oncogenic PI3K pathway, a complex signaling network associated with membrane tyrosine kinase receptors. PTEN deletion occurs in 5% of high grade serous20, 21, 20% of clear cell and 20% of endometrioid ovarian cancer patients22. Overall, the PI3K/AKT pathway is one of the most significantly deregulated cancer associated pathways in ovarian cancer23, 24. Mutations of KRAS and PTEN have been used to model endometrioid ovarian cancer25 and have been also reported in 24.6% and 77% of endometrioid endometrial tumors, respectively emphasizing the influence of these mutations in gynecologic cancer pathogenesis17.
Here we generated several new murine ovarian cancer cell lines which express human MUC1gene as self. Using these cell lines, we elucidate the possible roles of Ras/Mek and Pten/Akt pathways in regulation of MUC1 expression during transformation and EMT in ovarian cancer cells.
Results
Kras activation and Pten loss act synergistically to increase mitosis, transformation and EMT in ovarian surface epithelial cells
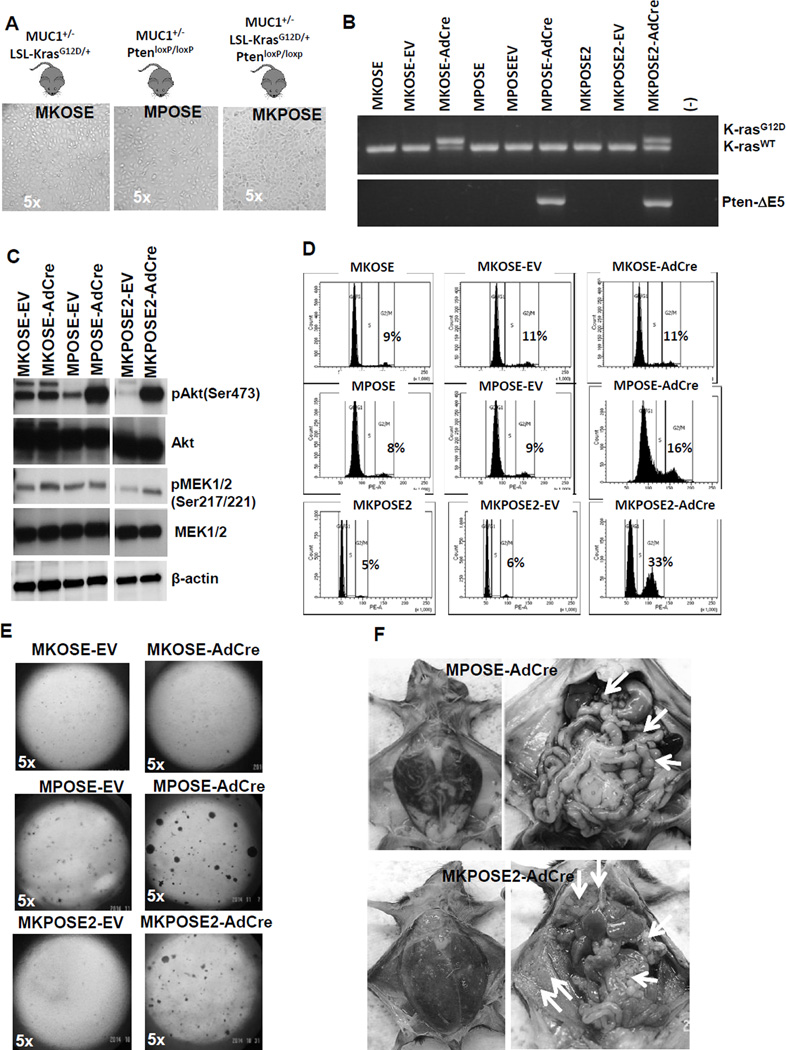
In order to test the roles of oncogenic Kras and Pi3k tumor suppressor pathways, alone or in combination, on the rate of transformation and EMT induction in ovarian epithelium, we generated a series of new ovarian cell lines, using primary ovarian surface epithelial (OSE) cells from healthy mice with conditional (Cre-loxP) genetic alterations in either oncogenic Kras, Pten tumor suppressor or both. Following OSE isolation, we established the following cell lines with silent mutations: MKOSE cells (derived from OSE of MUC1KrasG12D/+ female mice with a heterozygous, conditional KrasG12D oncogenic mutation), MPOSE cells (derived from OSE of MUC1PtenloxP/loxP female mice with homozygous, conditional Pten deletion) and MKPOSE cells (derived from OSE of MUC1KrasG12D/+PtenloxP/loxP female mice with conditional oncogenic Kras and conditional Pten deletion) (Table 1). Regardless of the originating genetic background, all primary OSE cells were immortalized at similar rates and largely maintained the cobblestone-like, epithelial morphology (Fig. 1A). To induce the mutations, we exposed the cells to AdCre, which floxes out the loxP sites from either the Kras locus (in MKOSE-AdCre cells), Pten locus (in MPOSE-AdCre) or both (in MKPOSE-AdCre) (Fig. 1B). Activation of oncogenic Kras leads to increased pMek (which acts downstream of Kras) in MKOSE-AdCre cells, while deletion of Pten (which acts as Pi3k inhibitor) increases pAkt expression in MPOSE-AdCre cells (Fig. 1C). MKPOSE-AdCre cells with simultaneous Kras activation and Pten deletion have increased levels of both pMek and pAkt (Fig. 1C). Cells exposed to no virus or to empty vector (EV) served as controls.
Table 1.
Genotype and phenotype of murine ovarian cancer cell lines
| OSE-derived cell lines |
Cell line nomenclature |
Originating mouse genotype |
Cell line genotype |
Cell line phenotype |
Soft agar colony formation |
Tumor growth in vivo (n*) |
| MKOSE | MUC1+/−LSL-KrasG12D/+ | Silenced oncogenic KrasG12D | Epithelial | No | No (0/5) | |
| MKOSE-AdCre | MUC1+/−LSL-KrasG12D/+ | Active oncogenic KrasG12D | Epithelial | No | No (0/5) | |
| MPOSE | MUC1+/−PtenloxP/loxP | Silenced Pten deletion | Epithelial | No | No (0/5) | |
| MPOSE-AdCre | MUC1+/−PtenloxP/loxP | Pten deletion | Partial EMT | Yes | Yes (6/6) | |
| MKPOSE2 | MUC1+/−LSL-KrasG12D/+PtenloxP/loxP | Silenced oncogenic KrasG12D and Silenced Pten deletion | Epithelial | No | No (0/7) | |
| MKPOSE2-AdCre | MUC1+/−LSL-KrasG12D/+PtenloxP/loxP | Active oncogenic KrasG12D and Pten deletion | Partial EMT | Yes | Yes (7/7) | |
| Tumor-derived cell lines | MKP-T | MUC1+/−LSL-KrasG12D/+PtenloxP/loxP | Kras LOH and Pten deletion | Mesenchymal | N/A | Single tumor (3/3) |
| MKP-Liver | MUC1+/−LSL-KrasG12D/+PtenloxP/loxP | Active oncogenic KrasG12D and Pten deletion | Partial EMT | N/A | Numerous tumor implants (10/10) | |
| MKP-Lung | MUC1+/−LSL-KrasG12D/+PtenloxP/loxP | Active oncogenic KrasG12D and Pten deletion | Mesenchymal | N/A | Numerous IP tumor implants and lung mets (12/12) |
Denotes number of mice with tumors of all total number of mice injected IP with the respective cell line. LOH, loss of heterozygosity; N/A, not applicable; EMT, epithelial to mesenchymal transition.
Figure 1.
Deletion of Pten tumor suppressor increases cell proliferation and induces transformation, effects that are further increased by oncogenic Kras activation. A. Ovarian surface epithelial (OSE) cells were isolated from healthy ovaries of mice with conditional mutations in Kras, Pten or both. Cells were immortalized following in vitro propagation as described in Materials and Methods. Stable lines with epithelial morphology were obtained. Images of in vitro cultured cells are shown (5× magnification).Mouse genotypes are shown above each image. B. PCR results showing floxed Kras (top) and floxed Pten (bottom). Top panel: upper band represents Cre-loxP recombination at KrasG12D locus. Lower band shows wild-type Kras. Lower panel: presence of band denotes homozygous recombination at the Pten locus (leading to Ptendel/del). C. Detection of pAkt, Akt, pMek1/2, Mek1/2 and β-actin by Western blot. D. Cell cycle analysis of PI-stained cells. Parental cells (left column) and EV-exposed cells (middle column) were used as controls. Cells exposed to AdCre (right column) show the effect on cycle due to Kras activation (upper row), Pten deletion (middle row) and combined Kras activation and Pten deletion (bottom row). Percentages of cells in G2/M phase are shown. E. Cells were plated at same density in soft agar and plates were monitored up to 20 days for colony formation. F. MPOSE-AdCre cells (upper images) and MKPOSE-AdCre cells (lower images) were injected in syngeneic mice. Mice with advanced disease were sacrificed and images of tumor burden were collected at necropsy. Left column shows abdomen distension due to ascites accumulation. Right column shows numerous tumor implants (arrows).
Loss of Pten increases mitosis while oncogenic Kras only minimally influences it (Fig. 1D and Supplementary Fig. 1). When combined, oncogenic Kras activation and Pten loss exert a synergistic effect on cell proliferation through accelerated mitosis (Fig. 1D and Supplementary Fig. 1). Growth in soft agar, a measurement of anchorage independent cell growth and cellular transformation, and ultimately a hallmark of carcinogenesis26, shows increased colony formation in cells with Pten deletion alone or in combination with Kras activation. In contrast, cells with Kras activation alone were unable to form any colonies (Fig. 1E). In line with findings from growth in soft agar, all the mice injected with MPOSE-AdCre (n= 6) and MKPOSE-AdCre (n=7) developed tumors (Fig. 1F). However none of the mice injected with MKOSE-AdCre cells (n=5), showed any signs of disease and had no macroscopic tumors at necropsy, performed 8 weeks post-tumor challenge.
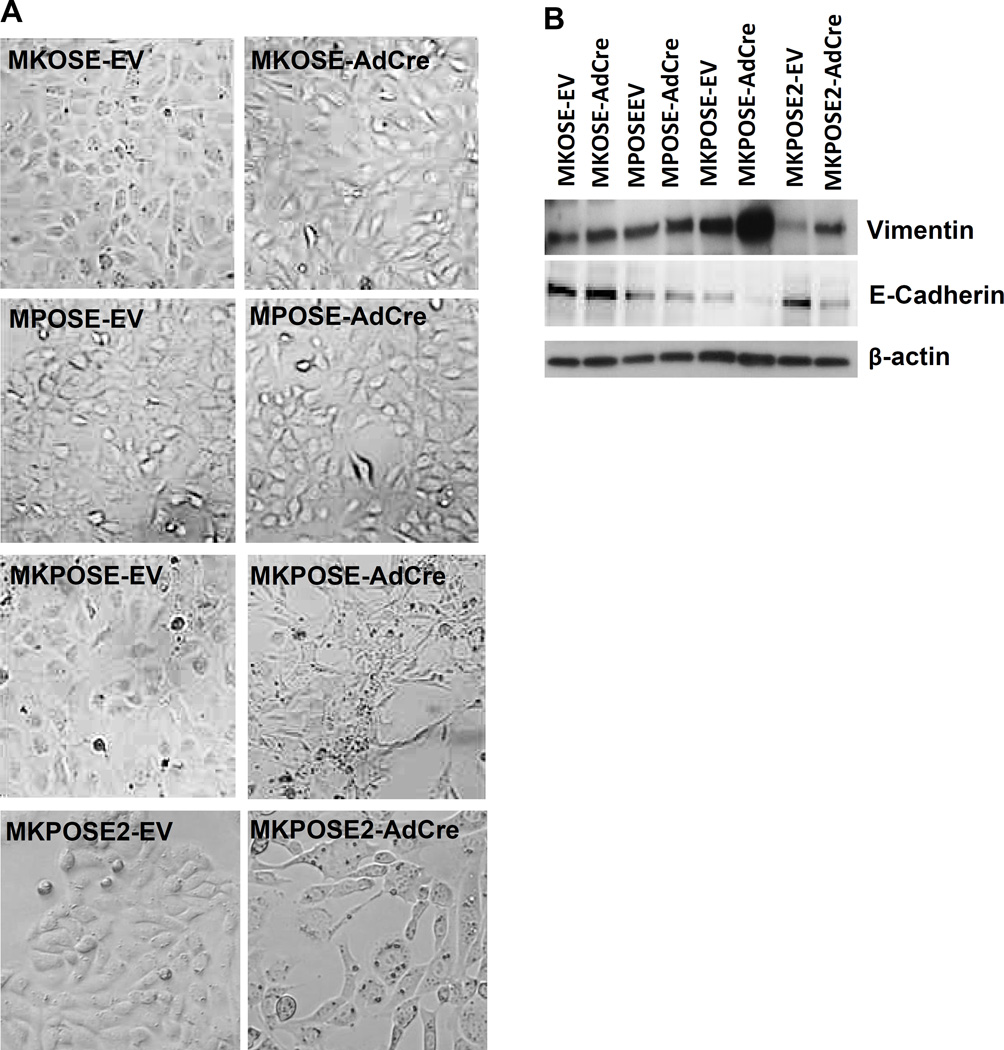
In addition to proliferation and transformation, we also assessed EMT induction, in cells with active oncogenic Kras, Pten deletion or both. MKOSE-AdCre and MPOSE-AdCre cells, cultured in vitro for the same number of passages, maintain the epithelial morphology (Fig. 2A). However, the combination of Kras activation and Pten loss in two distinct cell lines (MKPOSE-AdCre and MKPOSE2-AdCre) effectively induces EMT as demonstrated morphologically by the fibroblast-like appearance (Fig. 2A) and molecularly, by downregulation of E-cadherin and upregulation of vimentin, two molecular markers typically associated with EMT6 (Fig. 2B)Changes in vimentin and E-cadherin were minimal in MKOSE-AdCre and MPOSE-AdCre cells, reinforcing that simultaneous activation of the two pathways may synergistically induce EMT. Together, these findings demonstrate that OSE transformation can be induced in vitro by Pten loss but not oncogenic Kras and that the combination of these two pathways is synergistic for cell proliferation, transformation and EMT.
Figure 2.
Kras activation and Pten deletion triggers EMT. A. Images of OSE-derived cell lines exposed to either control adenovirus (EV, left column) or to AdCre (right column). B. Western blot identification of Vimentin (top) or E-Cadherin (middle). Beta-actin (bottom) was used as loading control.
Tumor-derived cell lines maintain the phenotype of the originating tumor
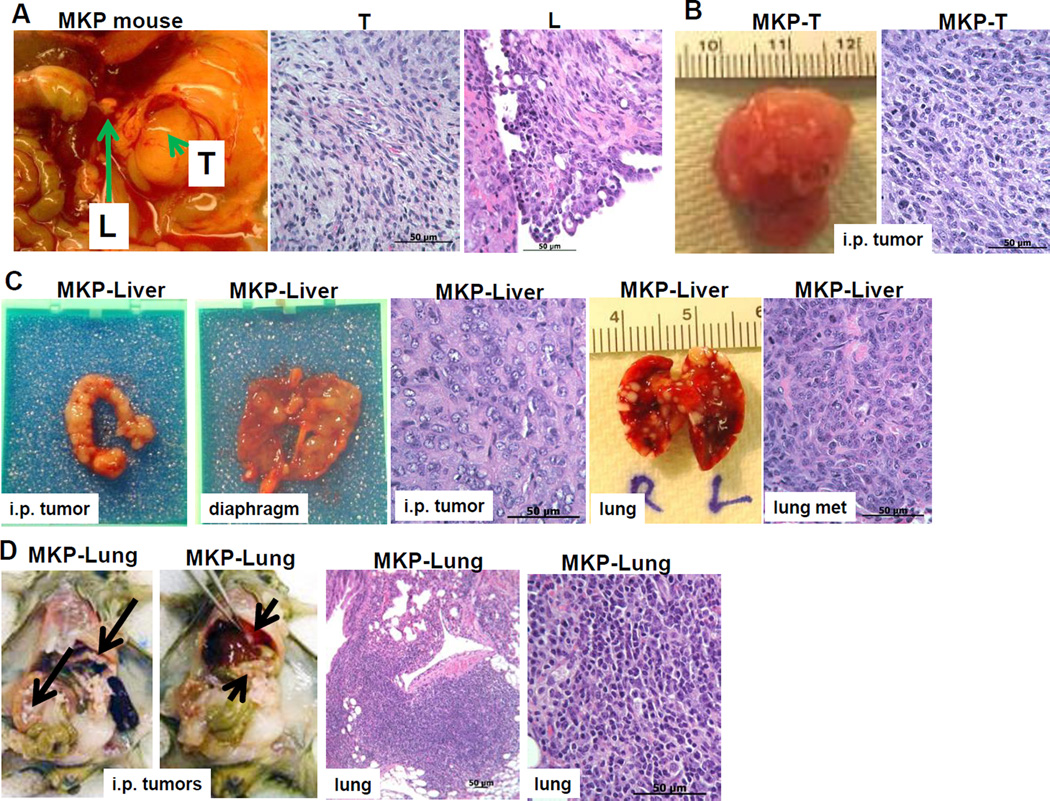
To further explore the effect of oncogenic Kras activation in conjunction with Pten deletion on EMT in vivo, we used our previously described MUC1KrasPten (Cre-loxP) conditional triple transgenic mice27. The MUC1KrasPten mice progress to human MUC1-expressing orthotopic ovarian tumors, following intrabursal injection of AdCre. At late stages the tumors spread outside of the ovaries and into the peritoneal cavity, mirroring the human disease27. Upon isolating tumors tissue from one large primary ovarian tumor and a smaller tumor implant from one of the hepatic lobes of the same tumor-bearing mouse, we generated stable cancer cell lines MKP-T and MKP-Liver, respectively (Fig. 3A). When reintroduced in the peritoneal cavity of healthy syngeneic mice the MKP-T cells grow as a single, large, localized tumor (similarly to the originating tumor), with no ascites (Fig. 3B). In contrast, the MKP-Liver cells (derived from a loco-regional implant) form tumors throughout the peritoneal cavity and on the upper abdomen (Fig. 3C). Furthermore, IP injection of MKP-Liver cells also triggers lung metastases, suggesting that in addition to loco-regional spread, these cells also have the capacity for hematogenous metastasis (Fig. 3C).
Figure 3.
De novo cell lines from primary site versus loco-regional and distant metastatic sites reflect in vivo ovarian tumor heterogeneity. A. Tumor tissue was isolated from a triple transgenic (Tg) MUC1KrasPten (MKP) mouse with late stage orthotopic ovarian tumors. The primary tumor (T, arrowhead) and a liver implant (L, arrow) were processed in vitro and two new tumor-derived cell lines were generated (MKP-T and MKP-Liver, respectively). Images of the originating T and L ovarian tumors are shown (HE staining). B. MKP-T cell line, derived from the T tumor shown in A (arrowhead) gives rise to single large tumors when injected IP in vivo in syngeneic mice (left). HE image of MKP-T tumor section following injection of 1×106 cells (right). C. MKP-Liver cells derived from the L tumor shown in A (liver implant, arrow) have epithelial morphology (left). IP injection of 4×106 MKP-Liver cells in syngeneic mice triggers multiple loco-regional tumor implants outside of the genital tract, on the diaphragm. MKP-Liver tumors also metastasize to the lungs. D. Tumor tissue from one of the lung metastases triggered by MKP-Liver cells was used to generate the MKP-Lung cell line. IP injection of 4×105 MKP-Lung cells triggers multiple tumor implants in the peritoneal cavity and on the diaphragm (arrows, left two panels). HE images of a lung metastasis are shown at two magnifications (right two panels).
Tumor tissue from one of the lung metastases was used to generate the MKP-Lung cell line (Fig. 3D). When injected IP in syngeneic mice, the MKP-Lung cells develop as IP tumors with lung metastases, in each of the recipient mice (Fig. 3D), suggesting that their molecular programming consistently leads to loco-regional plus hematogenous spread, mirroring stage IV human disease.
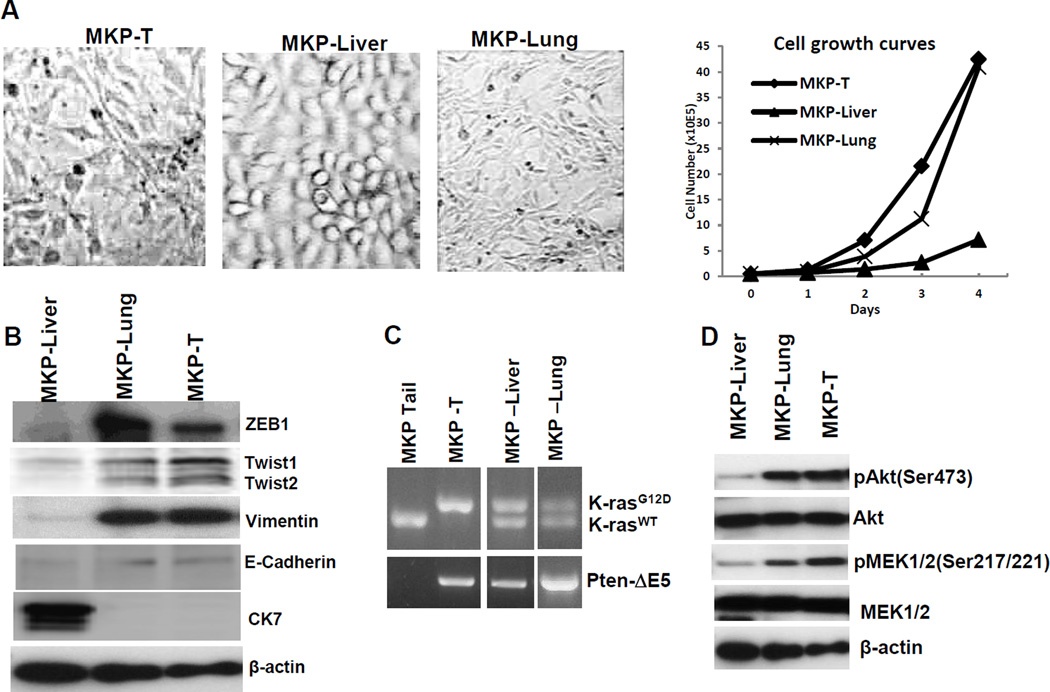
In line with the aggressive behavior in vivo, the most rapidly dividing MKP-T and MKP-Lung cells also show morphological (Fig. 4A) and molecular signs of EMT with high vimentin and low E-cadherin (Fig. 4B). Furthermore, MKP-T cells show loss of heterozygosity at the Kras locus (Kras LOH) (Fig. 4C) and have highest pMEK levels (Fig. 4D).
Figure 4.
Kras and Pten effects on EMT. A. Morphology and growth curves of MKP-T, MKP-Liver and MKP-Lung cells. B. Western blot for EMT associated markers. From top to bottom: ZEB1, Twist1, Twist 2, Vimentin, E-cadherin, cytokeratin 7 (CK7). Beta actin was used as loading control. C. PCR to identify floxed KrasG12D (top lane, top panel) and floxed Pten which results in exon 5 deletion (bottom panel). D. Western blot to identify phosphorylation downstream of PI3K (pAkt) and downstream of Kras (pMek1/2). Beta-actin serves as loading control.
In MKP-Liver cells derived from a loco-regional (liver) implant, E-cadherin is expressed at low levels while cytokeratin 7 remains high and vimentin upregulation has not occurred yet, suggesting a partial EMT, and explaining their potential for loco-regional dissemination. In line with their ability for systemic metastasis and aggressive growth, the MKP-Lung cells are clearly positive for EMT (with increased vimentin, loss of E-cadherin and cytokeratin) and have an accelerated doubling time in vitro and in vivo.
Overall, these in vivo experiments with new ovarian cancer cell lines derived from tumor implants with different anatomical locations demonstrate that similarly to patients, mice with orthotopic tumors show high degree of intra-tumor heterogeneity and that cells from various locations reproduce the tumor growth characteristics of the originating tumor and their loco-regional or distant mode of spread.
Kras activation and Pten deletion inversely modulate MUC1 protein expression
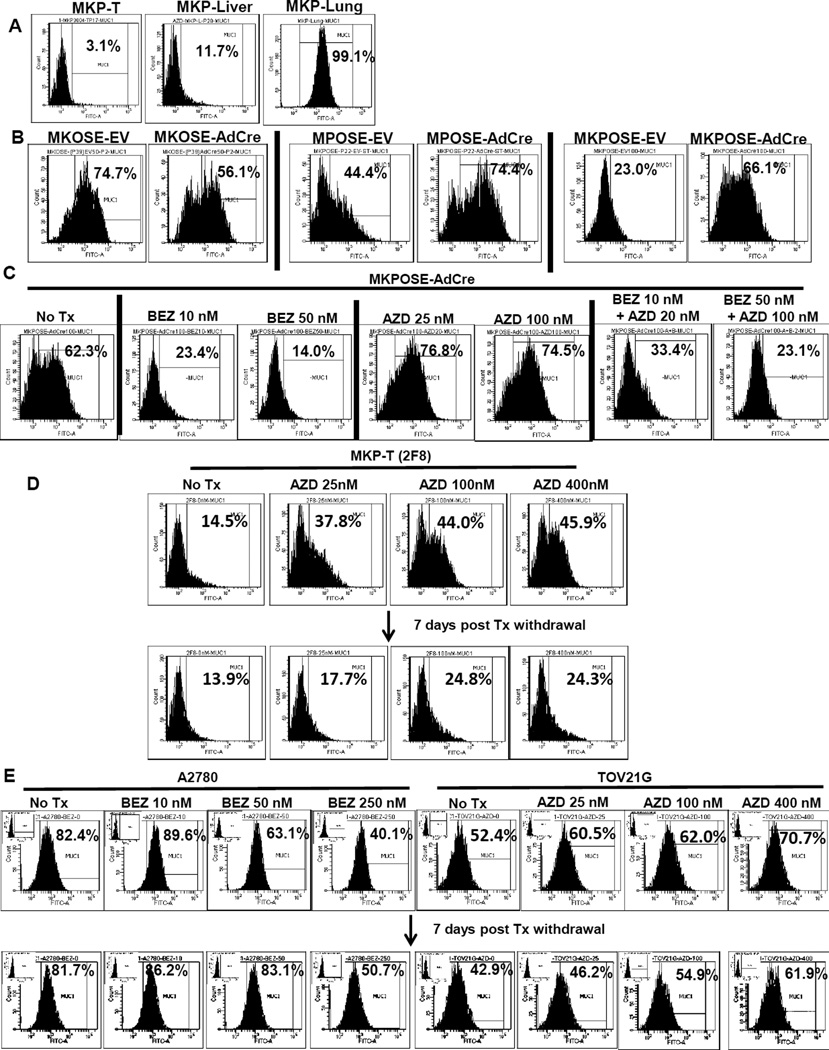
Having established the EMT profile in vitro and metastatic properties of MKP-T, MKP-Liver and MKP-Lung cells in vivo we asked next if and how MUC1 protein expression changes during EMT and metastasis. Interestingly, MUC1 expression was lowest in MKP-T cells, intermediate in MKP-Liver and highest in MKP-Lung, suggesting a direct correlation with the ability to disperse loco-regionally or to distant sites (Fig. 5A). Notably however, MKP-T cells with EMT have lowest MUC1 levels and grow as single tumors (Fig. 3A and 4C), in contrast to the expectation for local and/or distant dissemination and high MUC1 expression. Nevertheless, MKP-T cells show Kras LOH, raising the possibility that oncogenic Kras inhibits MUC1 expression. In line with this hypothesis, oncogenic Kras activation in MKOSE cells causes a significant drop in percentages of MUC1 positive cells (Fig. 5B). In contrast, Pten deletion triggers increased MUC1 production in MPOSE-AdCre and MKPOSE-AdCre cells, suggesting that Pten deletion alone or in conjunction with oncogenic Kras leads to MUC1 upregulation (Fig. 5B). These results are further supported by in vitro treatment with pathway-specific chemical inhibitors, currently used in clinical trials28, 29. Treatment of MKPOSE cells with PI3K inhibitor BEZ235 results in decreased MUC1 expression (Fig. 5C) while exposure to the pMEK inhibitor AZD6244, triggers the opposite effect, leading to increased percentage of MUC1 positive cells (Fig. 5C). When BEZ235 and AZD6244 were used in combination, the overall inhibitory effect on MUC1 expression was preserved, although the drop in MUC1 was less pronounced than for BEZ235 alone, likely due to the counteracting effect from AZD6244 (Fig. 5C).
Figure 5.
Activated Kras and Pi3k pathways have opposing effects on MUC1 protein levels in vitro. A. Flow cytometry measurement of MUC1 on tumor –derived cell lines MKP-T, MKP-Liver and MKP-Lung. PercentageMUC1 positive cells are shown. B. Cell surface MUC1 levels OSE-derived cell lines after Kras activation (left two panels), Pten deletion (middle two panels) and Kras activation plus Pten deletion (right two panels). Cells exposed to AdCre were compared to control empty vector (EV) treated cells. Percentage MUC1 positive cells are shown. C. Cell surface MUC1 in MKPOSE-AdCre after exposure to either BEZ235 (BEZ), AZD6244 (AZD) or both (BEZ + AZD) at two different drug concentrations. DMSO only treated cells (No Tx) served as controls. Percentages of MUC1 positive cells determined six days post treatment are shown. D. Upper row: cell surface MUC1 expression following exposure of MKP-T-2F8 cells to increasing concentration of AZD6244, after six days in culture. Lower row: MUC1 expression seven days after drug withdrawal. E. MUC1 expression at baseline and after four days exposure to BEZ235 (of A2780 cells, left 4 panels) or AZD6244 (of TOV21G cells, right 4 panels). MUC1 levels at day 7 following drug removal are shown in the corresponding lower histograms. In all assays, gates to determine MUC1 + cells were set according to isotype control (IC) antibody. Examples for IC histograms are shown in insets..
Furthermore, we tested the effect of AZD6244 on MUC1 expression in MKP-T cells (clone 2F8) with Kras LOH and high pMek levels. MUC1 levels significantly increase upon treatment with AZD6244 (Fig. 5D). The effect is largely reversed following drug withdrawal although the frequency of MUC1 positive cells remained higher among treated cells compared to cells that were never exposed to the inhibitor, seven day post-treatment.
The pathway-specific effects on MUC1 were mirrored in two human ovarian cancer cell lines (A2780 and TOV21G), with defined PTEN and KRAS mutations30. The A2780 carry PTEN deletion and have high baseline MUC1. Exposure to BEZ235 triggers dose-dependent inhibition of MUC1 expression. In contrast, exposure of Kras mutant TOV21G ovarian cancer cells led to MUC1 upregulation. One week post-drug withdrawal there is a trend to reverse MUC1 expression although pre-treatment levels are not fully achieved, especially at higher doses (Fig. 5E).
Collectively, these results from mouse and human ovarian cancer cell lines demonstrate that oncogenic Kras activation and Pten deletion inversely modulate MUC1 expression in ovarian cancer.
Gene signature of EMT associates with decreased overall survival in ovarian cancer patients
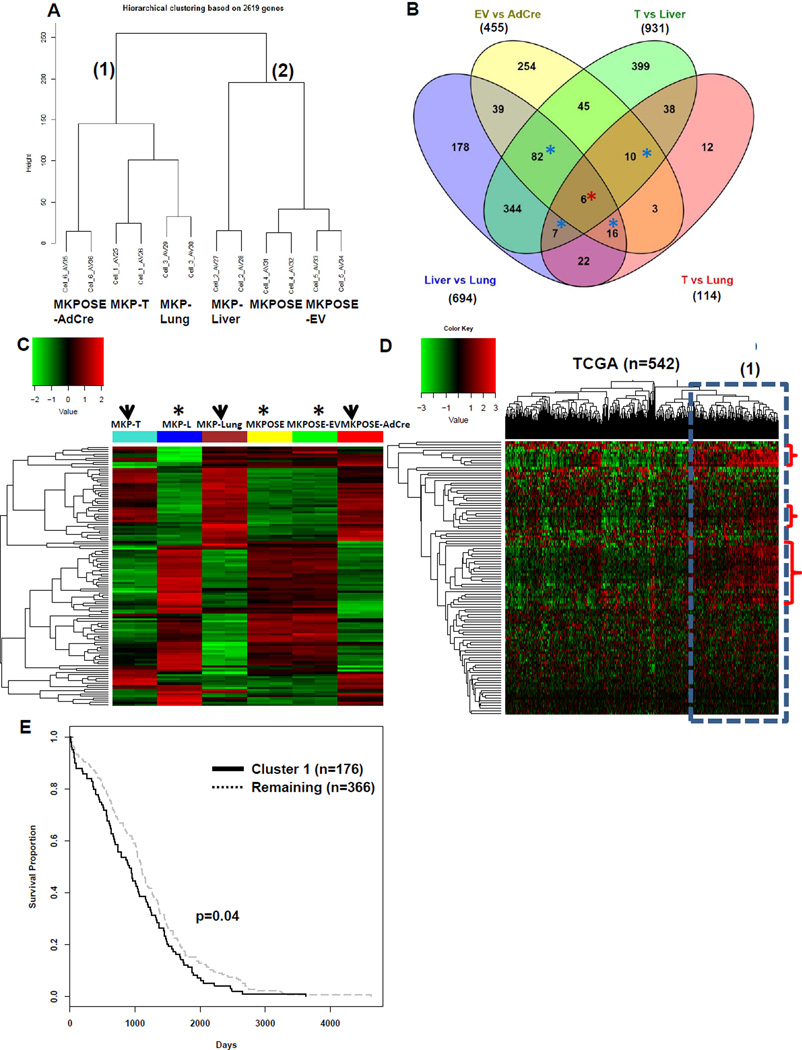
The ovarian cancer models described here recapitulate EMT in vitro and in vivo. To explore gene expression changes associated with this phenotype we performed whole genome microarray expression analysis using MouseWG-6 v2.0 Expression BeadChip. Unsupervised cluster analysis shows that MKPOSE-AdCre, MKP-T and MKPOSE-Lung (all with mesenchymal appearance, aggressive cell proliferation and positive for EMT molecular markers) cluster together, separate from epithelial MKPOSE and MKPOSE-EV cells and MKP-Liver cells with partial EMT, suggesting a genotypic and phenotypic equivalence (Fig. 6A).
Figure 6.
EMT-associated gene signature identifies patients with reduced survival. A. Unsupervised cluster analysis of microarray results from 6 murine cancer cell lines. Each cell line was run in duplicate., Three of the cell lines were derived from ovarian tumors (MKP-T, MKP-Liver and MKP-Lung) and three were derived from primary OSEs (MKPOSE, MKPOSE-EV and MKPOSE-AdCre). The six lines fall into two main clusters: MKPOSE-AdCre, MKP-T and MKP-Lung are in cluster 1 while MKP-Liver, MKPOSE and MKPOSE-EV are in cluster 2. B. Venn diagram showing the number of differentially expressed (DE) genes (q<0.001) that resulted from comparisons of MKPOSE-EV vs MKPOS-AdCre; MKP-T vs MKP-Liver; MKP-T vs MKP-Lung and MKP-Liver vs MKP-Lung. A total of n=6 DE genes are common for the four comparison (red asterisk) A total of n=115 genes were common to three group comparisons (blue asterisks). Collectively, the common DE genes (n=121) are listed in Supplemental Table 2. C. Heat map using the n=121 common murine genes in epithelial cell lines (asterisks) and in cell lines positive for EMT (arrowheads).. D Heat map of n=542 patients with high grade serous tumors using microarray gene expression data. Patient clusters are shown on top. Gene clusters are shown along the vertical axis. The highlighted patient cluster (dotted line) shows gene expression changes similar to those seen in mouse cell lines with EMT (upregulated genes, accolade). E Overall survival analysis and Kaplan Meier curve of ovarian cancer patients in cluster 1 shown in panel D (bold line, 176 patients) versus the remaining of the TCGA patient cohort (dotted line, 366 patients) p =0.04 (log-rank test).
We identified differentially expressed (DE) genes (q<0.001), using the following four comparisons: MKPOSE-EV versus (vs) MKPOSE-AdCre (n=455 DE genes), MKP-T vs MKP-Liver (n=931 DE genes), MKP-T vs MKP-Lung (n=114 DE genes) and MKP-Liver vs MKP-Lung (n=694 DE genes). All genes are listed Supplementary Table 1. Total DE gene numbers and overlapping gene numbers are shown in Fig 6B. The lowest number of DE genes (n=114) were between T and Lung cells, further supporting their similar phenotype, positive for EMT. Of all DE genes obtained from the above analyses, six genes were common to all four group comparisons (Fig. 6B, red asterisk) while additional n=115 genes were DE in three group comparisons (Fig 6B, blue asterisks). Heatmap of the n=121 common = genes (DE in three or four group comparisons, red and blue asterisks, Fig 6B and Supplementary Table 2), reveals that most genes follow a similar pattern of expression in the three cell lines positive for EMT (MKP-T, MKP-Lung and MKPOSE-AdCre, Fig 6C, arrowheads). Cells with no or partial EMT (MKPOSE, MKPOSE-EV and MKP-Liver) also show gene profiles highly similar to each other (Fig. 6C, asterisks). Ingenuity Pathway Analysis (IPA) reveals cellular movement as the top molecular and cellular dysregulated function (Supplementary Table 3).
Given the aggressive in vivo behavior in preclinical models of cells positive for EMT, we postulated next that the common genes identified above (n=121) may also identify tumors with increased aggressive behavior in ovarian cancer patients. To test for this, we applied the murine gene signature to the ovarian cancer TCGA microarray dataset of 542 patients. Of the 121 genes generated from our microarray approach, only 108 human genes could be retrieved from the TCGA dataset, due to gene filtering. The resulting heatmap using n=108 human genes revealed one patient cluster (Fig. 6D) with gene upregulations (cluster 1, red accolades) similarly found in mouse cells with EMT (MKP-T, MKP-Lung and MKPOSE-AdCre). Genes upregulated in patients (Fig. 6D, cluster 1) and EMT+ murine cell lines (Fig. 6C, arrowheads) are shown in Supplementary Table 4. In line with our hypothesis, patients in cluster 1 show a modest but significant (P=0.04) decrease in survival compared to the remaining patients. Overall, these results demonstrate that the EMT-related gene signature obtained from our in vivo and in vitro preclinical studies reveals similar gene expression changes in patients with more aggressive ovarian tumors and decreased survival.
DISCUSSION
Epithelial to mesenchymal transition in ovarian cancer has been difficult to reproduce in vivo in models that mimic the human disease. This is partly due to limited availability of ovarian cancer cell lines from human or mouse primary tumors. Until recently, only 26 human ovarian cancer cell lines from repositories like ATCC and ECACC have been publically available31. Many of these lines have been derived from ascites and appear to lose the characteristics of the original tumors. Using a novel in vitro culture system Ince et al recently generated 25 new human ovarian cancer cell lines that preserve the phenotype of original tumors31. The collection of murine ovarian cancer cell lines that can be used in immune competent mice to mirror primary or recurrent ovarian cancer is even more limited, with OSE-derived ID8 and IG10 cells with no well-defined mutations being predominantly used32–34. We report here a highly versatile collection of novel mouse OSE-derived cell lines featuring a unique combination of conditional or constitutive mutations in Kras, Pten or both. Although ovarian tumors may originate, at least in part in tubal epithelia, our novel cell lines could further support the emerging view that tumors sharing specific genomic traits could be studied together, regardless of the cell/organ of origin35, 36. Using cells from genetically engineered mice we demonstrate contrasting roles for Kras and Pten on cell proliferation, cellular transformation and MUC1 expression. Most importantly, using in vivo orthotopic tumor models we generated several additional tumor-derived cell lines that mirror the phenotype of the originating tumor. Moreover, the MKP-Lung cells that consistently metastasize to the lungs upon IP injection establish the first immune competent in vivo model for distant hematogenous spread in ovarian cancer.
Our recently reported triple transgenic orthotopic mouse model demonstrates that when conditional oncogenic Kras (LSL-KrasG12D/+) and Pten deletion (PtenloxP/loxP) are simultaneously triggered in OSE cells, the mice progress to human MUC1 overexpressing ovarian tumors27. However, these studies do not address the individual contributions of Kras and Pten to MUC1 expression. Here, we report that conditional activation of oncogenic Kras leads to diminished cell surface MUC1 expression and pathway inhibition with AZD6244 leads to dose dependent increase in MUC1 expression. Potential consequences of these findings are twofold: first, MUC1 acts as an oncogene (reviewed in37) and any molecular events leading to its upregulation may provide cells during treatment with AZD6244 additional MUC1-induced survival benefit. Although MUC1 protein levels return upon drug withdrawal, reversal is not complete and selection of MUC1-high clones with survival advantage may be possible. Second, understanding AZD6244-induced effect on MUC1 upregulation can impact the design of treatments in combination with MUC1 immune therapies. MUC1 is a tumor associated antigen expressed by virtually all adenocarcinomas. MUC1 overexpression initiates EMT in pancreatic cancer via increased interactions with b-catenin and can modulate transcription of genes downstream of Erk in breast cancer38, 39. Different types of adenocarcinomas are currently tested for response to AZD6244 (Selumetinib) in clinical trials40–42. In light of our results, it would be important to determine if and how soluble, tumor-derived MUC1 levels change (as determined in the clinic by the CA15-3 test, largely expected to regress during tumor involution) in patients treated with Mek/Erk inhibitors and to determine influences of these changes on de novo MUC1-specific antibody and/or cellular responses. Most importantly, we postulate that MUC1 vaccines given soon after AZD6244 administration may show synergistic anti-tumor effects, due to higher MUC1-expressing targets. In contrast to Kras activation, deletion of Pten (triggering PI3K activation and increased pAkt levels) leads to MUC1 upregulation while pathway inhibition with BEZ235 reverses the effect. Following a similar syllogism, we argue that combination therapies that would include MUC1 vaccines need to explore the correct timing in order to avoid low protein levels in target tumor cells.
Based on its pro-migration and pro-adhesion properties, we expected MUC1 to be overexpressed in cells with confirmed morphological and molecular EMT characteristics. In line with this hypothesis, epithelial-looking cells (like MKOSE, MPOSE or MKPOSE) express intermediate MUC1 levels and MUC1 was upregulated during EMT in vitro (MKPOSE-AdCre) and in vivo (MKP-Lung). Nevertheless, the mesenchymal-looking MKP-T cells have low MUC1, although this may be due to Kras LOH which likely hindered MUC1 increase.
In vivo selection of MKP-Lung tumor cells represents the first capture of distant metastasis and EMT in vivo in an ovarian cancer model. Remarkably, the MKP-Lung cells consistently follow the same pattern of migration to the lung post-IP injection, suggesting an imprinted ability for hematogenous spread, a measurement of aggressive in vivo biology, and offering an unprecedented opportunity to profile genome-wide gene expression changes associated with EMT and metastasis in ovarian cancer. To test translatability of our findings to the human disease we queried the TCGA and postulated that some of genes associated with EMT could be used as negative prognostic indicators in patients with ovarian cancer. Because the TCGA dataset contains almost exclusively patients with late stage tumors (and thus impeding comparisons according to disease stage) we focused on correlations of gene expression and overall survival. Patients with gene expression changes similar to those found in the EMT positive murine cell lines survived significantly less than the remaining patients. Further refinement of this gene list to identify tumors with aggressive biology is currently ongoing.
In summary, we have generated new ovarian cancer preclinical models using a series of cell lines with versatile mutations and different in vitro and in vivo tumor phenotypes. These models demonstrate distinct roles of Kras and Pten in ovarian cancer biology and can serve a wide range of applications, from screening new Kras and/ or Pten inhibitors, to exploring EMT induction in vivo and in vitro. In addition, our findings open unparalleled opportunities for further studies on the crosstalk between oncogenic MUC1, Kras and Pten tumor suppressor. As MUC1 is overexpressed in several other solid tumor types driven by Kras and/or Pten (like pancreas, lung, colon etc.)43, these results impact future research in ovarian as well as non-ovarian adenocarcinomas and can guide future combination therapies with small drug inhibitors and MUC1 vaccines.
Materials and Methods
Generation of ovarian cell lines from tumor tissues and ovarian surface epithelium (OSE)
MUC1KrasPten triple transgenic mice were injected with AdCre under the bursa of the left ovary and tumors were recovered after 8–10 weeks, as previously described27. Fresh tumor tissue was collected at necropsy, cut into 3–5 mm pieces, then digested with 0.1% Trypsin, 0.02% EDTA (Gibco) at 37 °C for 2 hours. Recovered cells were cultured in DMEM (Corning Cellgro), supplemented with 10% FBS (Cellgro), 10,000 U/L penicillin, 10,000 g/L streptomycin, 2 mM L-glutamine, 1% non-essential amino-acids, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol (all from Sigma). OSE cell lines were generated in vitro according to Roby et al32 until stable cell lines were obtained. Genomic DNA from the established cell lines was extracted with AllPrep DNA/RNA/Protein Mini Kit (QIAGEN) according to manufacturer’s instructions. PCR analysis with specific primers (Integrated DNA Technologies), as previously described27 were used to confirm the genotypes of the cell lines containing specific genetic traits (LSL-KrasG12D/+, PtenloxP/loxP, MUC1) and Cre-loxP recombination post-exposure to AdCre in vitro or in vivo.
For preparing growth curves of cell lines, cells were seeded in 24-well plates in triplicates and cell numbers were counted at five different time points.
Authentication of all OSE-derived cell lines described here was performed through genomic PCR for the presence of MUC1 and loxP cassettes at the (murine-specific) Kras and/or Pten loci before and after AdCre infection. Verification was performed for each experiment described herein. Surveillance for mycoplasma was performed periodically using mycoplasma PCR detection kit (Sigma).
In vitro cell transformation assay and in vivo tumor growth
ECM570 Cell Transformation Detection Assay kit (Millipore) was used for in vitro colony formation analysis according to the protocol provided. Tumors cells (5×105 to 2×106) were suspended in PBS and injected intraperitoneally (ip) or subcutaneously (sc) into syngeneic (129 S4/SvJae) MUC1.Tg mice. Mice were sacrificed according to IACUC protocol for tumor burden.
Flow cytometry
Cells were stained with FITC-CD227 (BD, 559774, 1:10) or isotype control (BD, 555748) in 1% BSA/PBS on ice for 40min, and the data were acquired with LSR II (BD) and analyzed with FACSDiva (BD).
Propidium iodide (BD) was used for cell cycle analysis.
Western-blot
Cell pellets were lysed with RIPA (Pierce) plus Halt™ protease inhibitor cocktail, Halt™ Phosphatase inhibitor cocktail and 0.05M EDTA (Thermo Scientific); 10–20 µg of protein lysates were loaded to 4–20% Mini-Protean precast gel (BIO-RAD) and transferred to nitro-cellulose membranes (BIO-RAD). Membranes were incubated in blocking buffer (1% BSA, Fisher Scientific), 0.05% Tween-20) at room temperature for 1 hour and then blotted with primary antibodies at 4°C, overnight. Primary antibodies to the following antigens were used: MUC1 (BD, 550488), cytokeratin 7 (ab181598), E-cadherin (ab133597), vimentin (ab8978) (all from Abcam), pan Akt (4691S), pAkt (4060S), Mek1/2 (8727S), pMek1/2(9154S) (all from Cell Signaling), Twist (Santa Cruz,sc-15393), ZEB1 (Novus Biologicals, NBP1-05987), and β-actin (Sigma, A1978). Goat-anti-mouse (170–6516) or goat-anti-rabbit (170–6515) IgG-HRP (BIO-RAD) were 1:3000 to 1:30000 diluted in blocking buffer. Immun-Star WesternC kit (BIO-RAD) or SuperSignal West Dura kit (Thermo Scientific) were used to develop the membranes and images were taken by Chemidoc XRS darkroom system (BIO-RAD).
Inhibition of Kras and/or Pi3k pathways
Mek1/2 inhibitor AZD6244 and PI3K/mTOR dual-inhibitor BEZ235 (Selleck Chemicals LLC) were dissolved in DMSO (Sigma) and used at various dilution, as indicated. DMSO-only treated cells were used as controls.
Histopathology
Tumor tissues were harvested and fixed in 10% buffered formalin (Fisher Scientific) for 24 hours, stored in 70% ethanol for 2 to 3 days and subsequently embedded in paraffin. Four micron sections were cut and the gross histopathology was assessed by hematoxylin/eosin (HE) staining.
Microarray analysis
Total RNAs were isolated from the cell lines using AllPrep DNA/RNA/Protein Mini Kit (Qiagen). Each cell line had RNAs isolated from two different times (passages) in culture. RNA samples were sent to Genomics and Proteomics Core Laboratories, University of Pittsburgh for MouseWG-6 v2.0 Expression BeadChip (Illumina) analysis. Raw data from scanned microarray chips were retrieved with background corrected using Illumina BeadStudio (as tab-delimited files) software. The lumi package in Bioconductor44 was used to preprocess the raw data. For quality control, we kept probes with detection p-values less than 0.05 in at least one third of all samples to ensure the remaining probes were truly detected. To minimize the effect of unwanted technical variation between RNA samples, the raw data was quantile normalized. In addition, the data was also log2 transformed to stabilize the variance. Heatmap of sample correlation was plotted to identify problematic samples and batch effects.
After gene annotation, the Linear Model for Microarray Data (LIMMA) in Bioconductor45 was used to identify differentially expressed genes between two specified cell lines. We used Benjamini–Hochberg procedure to control for false discovery rate. Differentially expressed genes were loaded to Ingenuity Pathway Analyses (IPA) for functional analysis. We also plotted the expression heatmap for a pre-specified list of genes that are associated with epithelial to mesenchymal transition (EMT).
To analytically characterize samples across the cell lines, we carried out unsupervised clustering. We performed feature filtering by removing genes with means or standard deviations under the median of all the genes. Based on the remaining 2619 expressed (with high mean values) and informative (with large standard deviations) genes, hierarchical clustering with Ward linkage was applied. All statistical programming was implemented in R (code available upon request). The complete microarray data sets are available in the Gene Expression Omnibus database under accession number GSE69544.
The TCGA Ovarian Cancer mRNA expression data was retrieved from the Pittsburgh Genome Resource Repository (PGRR: http://www.pgrr.pitt.edu/pgrr); 542 samples with available survival information were used. We plotted the heatmap for the selected 121 genes and a hierarchical clustering with Ward linkage was applied. A cluster of patients was identified from the heatmap and survival analysis was performed. Kaplan-Meier curve and log-rank test was used to compare survival distributions.
Supplementary Material
Acknowledgments
This study was partly supported by the Department of Defense (DOD) Ovarian Cancer Academy Award W81XWH-10-1-0525 and NIH/NCI R01 CA163462 (to AMV).
Footnotes
Conflicts of interest.
Dr Vlad’s work has been funded by the NIH. All other authors declare no conflict of interest.
This manuscript presents original research, has not been previously published, and is not being considered for publication elsewhere.
Ethical standards
All animal studies have been approved by University of Pittsburgh Institutional Animal Care and Use Committee (IACUC), according to the Guide for the Care and Use of Laboratory Animals from the National Research Center of the National Academies.
References
- 1.Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nature reviews Clinical oncology. 2015;12:239–245. doi: 10.1038/nrclinonc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 3.Cormio G, Rossi C, Cazzolla A, Resta L, Loverro G, Greco P, et al. Distant metastases in ovarian carcinoma. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2003;13:125–129. doi: 10.1046/j.1525-1438.2003.13054.x. [DOI] [PubMed] [Google Scholar]
- 4.Dauplat J, Hacker NF, Nieberg RK, Berek JS, Rose TP, Sagae S. Distant metastases in epithelial ovarian carcinoma. Cancer. 1987;60:1561–1566. doi: 10.1002/1097-0142(19871001)60:7<1561::aid-cncr2820600725>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 5.Kerr VE, Cadman E. Pulmonary metastases in ovarian cancer. Analysis of 357 patients. Cancer. 1985;56:1209–1213. doi: 10.1002/1097-0142(19850901)56:5<1209::aid-cncr2820560542>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Davidson B, Trope CG, Reich R. Epithelial-mesenchymal transition in ovarian carcinoma. Frontiers in oncology. 2012;2:33. doi: 10.3389/fonc.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng J, Wang L, Chen H, Li L, Ma Y, Ni J, et al. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer metastasis reviews. 2013;32:535–551. doi: 10.1007/s10555-013-9423-y. [DOI] [PubMed] [Google Scholar]
- 8.Dong Y, Walsh MD, Cummings MC, Wright RG, Khoo SK, Parsons PG, et al. Expression of MUC1 and MUC2 mucins in epithelial ovarian tumours. The Journal of pathology. 1997;183:311–317. doi: 10.1002/(SICI)1096-9896(199711)183:3<311::AID-PATH917>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Ma J, Liu F, Yu Q, Chu G, Perkins AC, et al. Expression of MUC1 in primary and metastatic human epithelial ovarian cancer and its therapeutic significance. Gynecologic oncology. 2007;105:695–702. doi: 10.1016/j.ygyno.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Vlad AM, Kettel JC, Alajez NM, Carlos CA, Finn OJ. MUC1 immunobiology: from discovery to clinical applications. Advances in immunology. 2004;82:249–293. doi: 10.1016/S0065-2776(04)82006-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Vlad A, Milcarek C, Finn OJ. Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms. Cancer immunology, immunotherapy : CII. 2013;62:423–435. doi: 10.1007/s00262-012-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, Hayashi T, et al. Overexpression of MUC1 reconfigures the binding properties of tumor cells. International journal of cancer Journal international du cancer. 2001;94:783–791. doi: 10.1002/ijc.1554. [DOI] [PubMed] [Google Scholar]
- 13.Besmer DM, Curry JM, Roy LD, Tinder TL, Sahraei M, Schettini J, et al. Pancreatic ductal adenocarcinoma mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer research. 2011;71:4432–4442. doi: 10.1158/0008-5472.CAN-10-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. The Journal of biological chemistry. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 15.Carson DD. The cytoplasmic tail of MUC1: a very busy place. Science signaling. 2008;1:35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- 16.Kindelberger DW, Lee Y, Miron A, Hirsch MS, Feltmate C, Medeiros F, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: Evidence for a causal relationship. The American journal of surgical pathology. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuuchi H, Mori Y, Sato K, Kamiya H, Okamura N, Nasim S, et al. High incidence of point mutation in K-ras codon 12 in carcinoma of the fallopian tube. Cancer. 1995;76:86–90. doi: 10.1002/1097-0142(19950701)76:1<86::aid-cncr2820760112>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.Roh MH, Yassin Y, Miron A, Mehra KK, Mehrad M, Monte NM, et al. High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23:1316–1324. doi: 10.1038/modpathol.2010.119. [DOI] [PubMed] [Google Scholar]
- 20.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. International journal of cancer Journal international du cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 21.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nature genetics. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 22.Obata K, Morland SJ, Watson RH, Hitchcock A, Chenevix-Trench G, Thomas EJ, et al. Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors. Cancer research. 1998;58:2095–2097. [PubMed] [Google Scholar]
- 23.Huang J, Zhang L, Greshock J, Colligon TA, Wang Y, Ward R, et al. Frequent genetic abnormalities of the PI3K/AKT pathway in primary ovarian cancer predict patient outcome. Genes, chromosomes & cancer. 2011;50:606–618. doi: 10.1002/gcc.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nature medicine. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- 26.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. Journal of visualized experiments : JoVE. 2014:e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budiu RA, Elishaev E, Brozick J, Lee M, Edwards RP, Kalinski P, et al. Immunobiology of human mucin 1 in a preclinical ovarian tumor model. Oncogene. 2013;32:3664–3675. doi: 10.1038/onc.2012.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future medicinal chemistry. 2011;3:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 30.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature communications. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ince TA, Sousa AD, Jones MA, Harrell JC, Agoston ES, Krohn M, et al. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nature communications. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roby KF, Taylor CC, Sweetwood JP, Cheng Y, Pace JL, Tawfik O, et al. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 33.Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, et al. Epithelial ovarian cancer experimental models. Oncogene. 2014;33:3619–3633. doi: 10.1038/onc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasan N, Ohman AW, Dinulescu DM. The promise and challenge of ovarian cancer models. Translational cancer research. 2015;4:14–28. doi: 10.3978/j.issn.2218-676X.2015.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horlings HM, Flanagan AM, Huntsman DG. Categorization of cancer through genomic complexity could guide research and management strategies. J Pathol. 2015;236:397–402. doi: 10.1002/path.4542. [DOI] [PubMed] [Google Scholar]
- 36.Horlings HM, Shah SP, Huntsman DG. Using Somatic Mutations to Guide Treatment Decisions: Context Matters. JAMA Oncol. 2015;1:275–276. doi: 10.1001/jamaoncol.2015.35. [DOI] [PubMed] [Google Scholar]
- 37.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nature reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. The Journal of biological chemistry. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 40.Jain N, Curran E, Iyengar NM, Diaz-Flores E, Kunnavakkam R, Popplewell L, et al. Phase II study of the oral MEK inhibitor selumetinib in advanced acute myelogenous leukemia: a University of Chicago phase II consortium trial. Clin Cancer Res. 2014;20:490–498. doi: 10.1158/1078-0432.CCR-13-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kandil E, Tsumagari K, Ma J, Abd Elmageed ZY, Li X, Slakey D, et al. Synergistic inhibition of thyroid cancer by suppressing MAPK/PI3K/AKT pathways. J Surg Res. 2013;184:898–906. doi: 10.1016/j.jss.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 42.Paolo M, Assunta S, Antonio R, Claudia SP, Anna BM, Clorinda S, et al. Selumetinib in advanced non small cell lung cancer (NSCLC) harbouring KRAS mutation: endless clinical challenge to KRAS-mutant NSCLC. Rev Recent Clin Trials. 2013;8:93–100. doi: 10.2174/15748871113089990047. [DOI] [PubMed] [Google Scholar]
- 43.Iwanaga K, Yang Y, Raso MG, Ma L, Hanna AE, Thilaganathan N, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer research. 2008;68:1119–1127. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 45.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.