Abstract
The Escherichia coli Tsr protein contains a periplasmic serine-binding domain that transmits ligand occupancy information to a cytoplasmic kinase-control domain to regulate the cell's flagellar motors. The Tsr input and output domains communicate through sequential conformational changes transmitted through a transmembrane helix (TM2), a five-residue control cable helix at the membrane-cytoplasm interface, and a four-helix HAMP bundle. Changes in serine occupancy are known to promote TM2 piston displacements in one subunit of the Tsr homodimer. We explored how such piston motions might be relayed through the control cable to reach the input AS1 helix of HAMP by constructing and characterizing mutant receptors that had one-residue insertions or deletions in the TM2-control cable segment of Tsr. TM2 deletions caused kinase-off output shifts; TM2 insertions caused kinase-on shifts. In contrast, control cable deletions caused kinase-on output, whereas insertions at the TM2-control cable junction caused kinase-off output. These findings rule out direct mechanical transmission of TM2 conformational changes to HAMP. Instead, we suggest that the Tsr control cable transmits input signals to HAMP by modulating the intensity of structural clashes between out-of-register TM2 and AS1 helices. Inward displacement of TM2 might alter the sidechain environment of control cable residues at the membrane core-headgroup interface, causing a break in the control cable helix to attenuate the register mismatch and enhance HAMP packing stability, leading to a kinase-off output response. This helix-clutch model offers a new perspective on the mechanism of transmembrane signaling in chemoreceptors.
Keywords: bacterial chemotaxis, piston model, control cable, HAMP domain, dynamic-bundle model
Graphical Abstract
Introduction
All organisms use transmembrane signal transduction to monitor and respond to environmental stimuli. In motile bacteria and archaea, chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) convert information about external attractant and repellent levels into signals that control locomotor behavior. The extensively studied MCPs of Escherichia coli serve as important models for understanding transmembrane signal transduction (see [1; 2; 3; 4] for recent reviews).
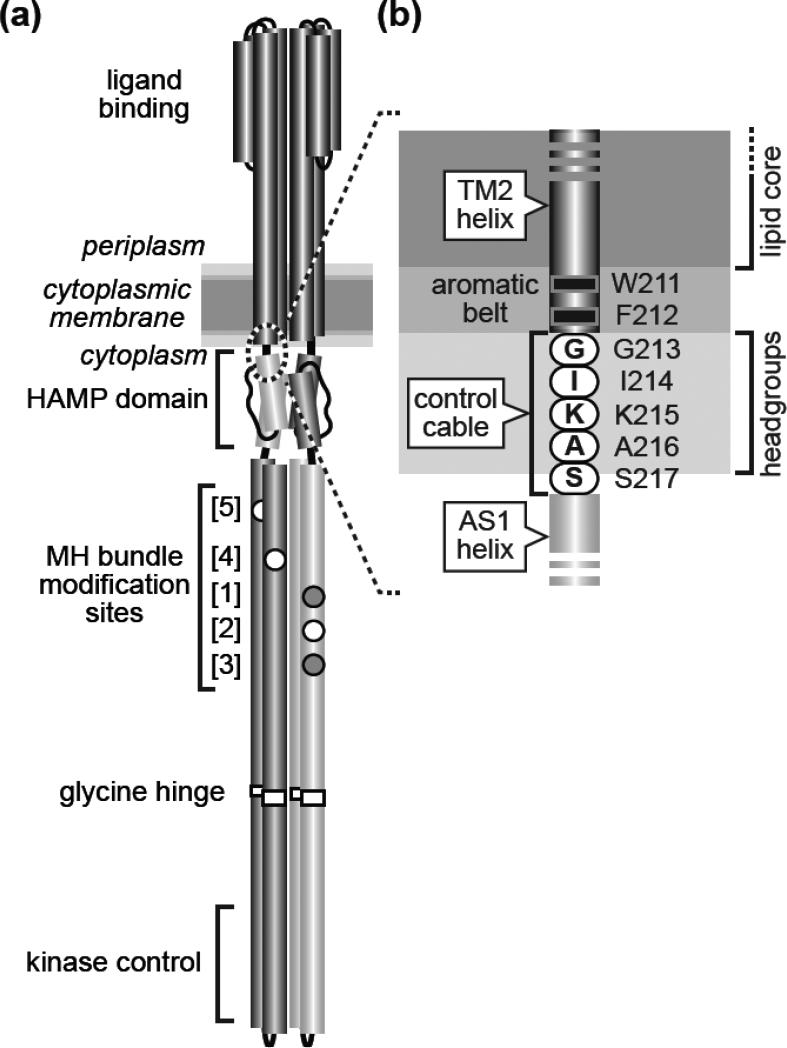
E. coli has four MCP species (Tsr, Tar, Tap, Trg) that detect various small molecule ligands. They have similar functional architectures: mainly α-helical protomers of ~550 residues organized as homodimers with a ligand-sensing periplasmic domain connected via a transmembrane helix in each subunit to a cytoplasmic signal-processing domain (Fig. 1a). The cytoplasmic portion of MCP molecules contains a membrane-proximal HAMP domain, a sensory adaptation domain containing sites for reversible covalent modifications, and a hairpin tip that regulates the activity of a histidine autokinase, CheA. CheA phosphoryl groups are, in turn, donated to the CheY response regulator, whose phosphorylated form binds to the base of flagellar motors to initiate random directional changes in cell swimming trajectory. Whenever the cell happens to head up an attractant gradient, the increasing ligand concentration causes the receptor to down-regulate CheA activity, extending up-gradient swimming runs.
Fig. 1. Tsr structural features and transmembrane signaling models.
(a) The Tsr homodimer [1]. Cylindrical segments represent α-helices, drawn approximately to scale. Each Tsr subunit contains five adaptation sites within a methylation-helix (MH) bundle. Sites 2, 4, and 5 (white circles) are translated as glutamic acid residues, the substrate for CheR methylation. Sites 1 and 3 (gray circles) are translated as glutamine residues that can be deamidated to glutamic acid residues by CheB, making them competent for subsequent methylation. This study focuses on the Tsr region at the membrane-cytoplasm interface (dashed circle), enlarged in (b).
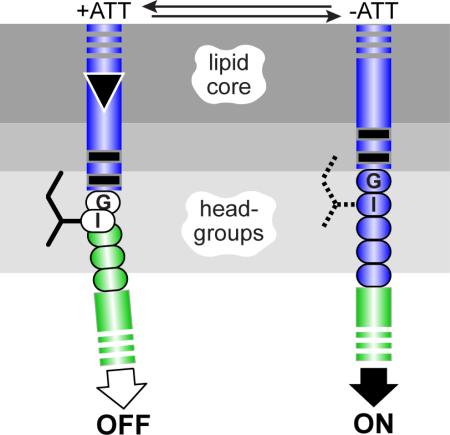
(b) The Tsr control cable. Detail of the dashed region in (a). The aromatic residues at the C-terminus of the TM2 transmembrane helix and the following control cable residues are shown at their probable positions relative to the hydrophobic (lipid core) and polar (headgroup) membrane regions [14].
Sensory adaptation plays a critical role in the MCP-mediated chemotactic behavior of E. coli. The sensory adaptation enzymes, CheR (MCP methyltransferase) and CheB (MCP methylesterase and deamidase) modify specific residues in the four-helix methylation bundle of the receptor molecule. The overall methylation state of the receptor matches the level of its chemoeffector in the environment. Thus, MCP modification state serves as a memory store for detecting temporal changes in chemoeffector levels as the cell swims through spatial gradients. Ligand occupancy changes update the modification record by adjusting the CheR/CheB substrate properties of MCP molecules. Attractant ligands shift receptor molecules to a kinaseoff state that serves as a substrate for CheR-mediated reactions; reduced ligand occupancy shifts receptors to a kinase-on state that serves as a substrate for CheB-mediated reactions.
The mechanism of transmembrane signaling by E. coli MCPs has been explored most extensively with the Tar (aspartate-sensing), Tsr (serine-sensing), and Trg (ribose/galactose-sensing) receptors and the current mechanistic picture is a montage derived from all three (see [5; 6; 7; 8] for reviews). Hereafter, residue names and coordinates for Tsr, the subject of the studies in this report, will be used to describe structure-function features important for transmembrane signaling.
In Tsr homodimers, two membrane-spanning segments flank the serine-binding portion of each subunit: an N-terminal TM1 helix and a TM2 helix that connects to the AS1 helix of the HAMP domain (Fig. 1b). TM2 comprises 19 mostly hydrophobic residues (A192-V210) that embed in the membrane lipid core [9; 10] and two aromatic residues (W211, F212) at its cytoplasmic end that partition at the core-headgroup interface [11; 12; 13]. A five-residue control cable joins the TM2 and HAMP AS1 helices and mediates their signaling transactions [14; 15; 16].
Studies of the Tsr control cable led us to a working model of transmembrane signaling in which the control cable mediates structural interactions between the mismatched registers of the TM2 and AS1 helices [14; 15]. For example, a proline replacement at any position in the Tsr control cable except its N-terminal G213 abolishes stimulus responses, suggesting that control cable helicity is important to the signal transmission mechanism [15]. Moreover, charged amino acid replacements at the I214 position interfere with signal transmission, implying that interaction of less polar sidechains at that position with the membrane interfacial environment might assist stimulus-induced shifts in receptor signal state [14]. We reasoned that a control cable with high helix potential might enable TM2 to destabilize packing of the HAMP bundle, whereas a control cable with reduced helicity or a distinct helix break might enhance HAMP packing [14]. Various proposed mechanisms of HAMP action, including the gearbox [17], scissors [18; 19], and dynamic-bundle [20] models, all predict that altered packing arrangements of the HAMP bundle produce changes in CheA output activity.
Considerable evidence supports the view that piston motions of the TM2 helix normal to the plane of the membrane initiate transmembrane signal transmission in MCPs [8; 21; 22; 23; 24; 25]. Attractant stimuli, for example, promote inward piston displacements of 1-2 Å in one subunit of the receptor dimer [21; 26; 27; 28]. The resultant structural asymmetry that impinges on the control cable might be transmitted directly to the AS1 helix of HAMP, as proposed by the gearbox and scissors models, or it could somehow modulate control cable helicity to influence HAMP packing, as specified in the dynamic-bundle model. To explore the TM2-control cable transmission mechanism in further detail, we constructed and characterized a series of mutant Tsr receptors with changes in the length and helical register of these signaling elements. Our study revealed evidence for a structural and signaling change at the junction of the TM2 and control cable helices that argues against direct mechanical transmission of TM2 motions through the control cable.
Results
Creating Tsr structural changes that mimic transmembrane signals
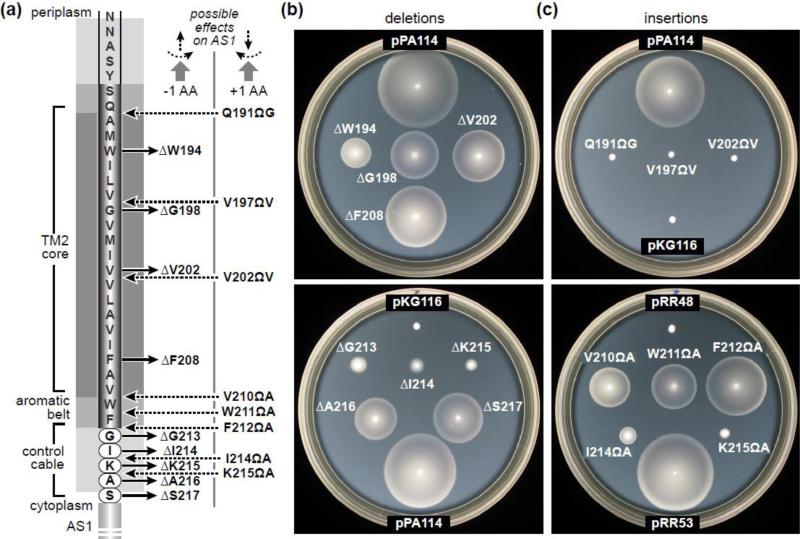
We constructed a series of Tsr mutants, each with a one-residue deletion or insertion in the TM2-control cable region (Fig. 2A). The TM2 residues embedded in the lipid core most likely have an alpha-helical secondary structure [10]. Except for an overall hydrophobic character, there appears to be little or no sidechain specificity to their signaling function [29]. Thus, removing one residue from the TM2 helix could shorten its membrane-spanning length by as much as 1.5 Å and rotate its cytoplasmic end by up to ~100° counter-clockwise, viewed in the N to C direction (Fig. 2A). Adding a residue to TM2 should cause comparable structural changes in the opposite direction.
Fig. 2. One-residue deletions and insertions in the TM2-control cable region of Tsr.
(a) Mutant derivatives characterized in this study. Shadings for various membrane regions correspond to those used in Fig. 1. “Possible effects on AS1” indicates the structural forces expected at the AS1 helix if the TM2-AS1 segment were a continuous α-helix. Viewed from the membrane toward AS1, one-residue deletions should cause outward displacement and counter-clockwise rotation; one-residue insertions should cause inward displacement and clockwise rotation.
(b) Chemotactic behaviors mediated by Tsr deletion mutants. Derivatives of plasmid pPA114 were tested for ability to support chemotaxis of receptor-less host strain UU2612 on tryptone semi-solid agar containing 25 μg/ml chloramphenicol and 0.6 μM salicylate. Plates were photographed after incubation at 30°C for 7-8 hours.
(c) Chemotactic behaviors mediated by Tsr insertion mutants. Derivatives of plasmid pRR53 were tested for ability to support chemotaxis of receptor-less host strain UU2612 on tryptone semi-solid agar containing 100 μg/ml ampicillin and 100 μM IPTG. Plates were photographed after incubation at 30°C for 7-8 hours.
Sidechain character is evidently more important in the vicinity of the aromatic belt [11; 12; 13] and the first two residues of the control cable [14; 15; 30; 31]. Moreover, although the control cable seems to have helical character, modulated changes in its helix potential may play a role in signal transmission [14; 31]. Thus, the structural changes caused by one-residue insertions or deletions of belt or control cable residues could differ from their TM2 counterparts and might be confounded by sidechain-specific interactions with the membrane core-headgroup transition zone (Fig. 2A). Accordingly, to investigate control cable length effects on signaling without sidechain-specific complications, we also constructed Tsr receptors in which the control cable comprised five consecutive serine residues (the wild-type length), or four or six serine residues, corresponding, respectively, to a one-residue deletion or insertion in the all-serine control cable.
Chemotactic behaviors supported by the mutant receptors
Mutations were constructed in tsr expression plasmids pRR53 (IPTG-inducible) and pPA114 (salicylate-inducible). We use the following notations to designate the resultant Tsr structural changes: e.g., ΔW194 = deletion of residue 194; e.g., Q191ΩG = insertion of a glycine residue immediately C-terminal to residue 191. All mutant receptors exhibited native steady-state levels (Table S1), so any functional alterations they might have cannot be due to reduced expression or elevated instability. When tested in strain UU2612, an otherwise receptor-less host that contains the CheR and CheB enzymes of the sensory adaptation system, many of the mutant Tsr plasmids mediated demonstrable serine chemotaxis in soft agar plate assays (Fig. 2B and 2C; Table S1). Deletions in TM2 and in the HAMP-proximal residues of the control cable retained Tsr function, whereas insertions in those regions did not. However, insertions in the aromatic belt retained Tsr function, whereas deletions in the belt-proximal control cable residues did not. The different functional consequences of comparable lesions in different locations suggest that the TM2-control cable segment of Tsr may have several structurally distinct components.
Assessing the signaling properties of the mutant receptors
MCP molecules approximate two-state signaling devices with kinase-off (OFF) and kinaseon (ON) output states. Chemoeffector ligands and adaptational modifications produce stimulus responses and subsequent sensory adaptation by shifting the equilibrium distribution of ON and OFF receptor complexes. Structural interactions between the HAMP domain and adjoining input (TM2-control cable) and output (MH bundle) elements are key to receptor signal-state control. Whether those control interactions obey two-state or more graded structural mechanisms remains an open question, but for simplicity we use two-state terminology to describe the mutant signaling behaviors and then revisit this mechanistic issue in the Discussion.
We determined the signal output shifts of mutant Tsr receptors with an in vivo kinase assay based on Förster resonance energy transfer (FRET) [32; 33; 34]. In brief, this assay monitors phosphorylation-dependent interactions between CheY tagged with YFP (the FRET acceptor) and its phosphatase, CheZ, tagged with CFP (the FRET donor). The FRET signal provides a measure of receptor-controlled CheA autokinase activity, the source of CheY phosphoryl groups. The kinase inhibition responses to serine stimuli provide three important signaling parameters: K1/2, the serine concentration that inhibits 50% of the kinase activity; the Hill coefficient, reflecting response cooperativity; and the maximal kinase activity of the receptor signaling complexes.
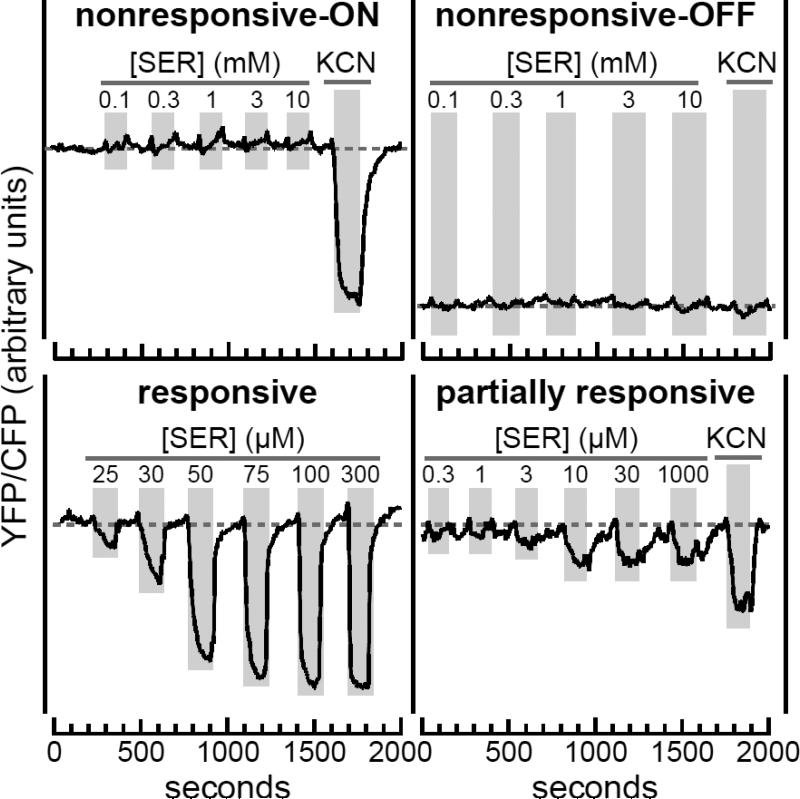
We first expressed mutant Tsr plasmids in FRET reporter strain UU2567, which lacks the CheR and CheB adaptation proteins. In this host all Tsr molecules retain their initial QEQEE residue pattern at the five modification sites in each protomer (see Fig. 1A). Homogeneous populations of wild-type Tsr receptors in the QEQEE state exhibit moderate serine sensitivity (K1/2 ~15 μM) and high response cooperativity (Hill ~15) [14; 34; 35]. Mutant Tsr receptors exhibited one of four response patterns (Fig. 3; Table S2). (i) Some receptors were nonresponsive to even very high levels of serine, but had wild-type kinase activity, revealed by KCN treatment, which depletes cellular ATP, the phosphodonor for CheA autophosphorylation [34]. (ii) Other nonresponsive receptors evinced no kinase activity. These two classes of nonresponsive receptors respectively represent strongly ON-shifted and strongly OFF-shifted signal outputs. (iii) Some mutant receptors responded to serine stimuli in similar fashion to the wild type. (iv) Other receptors were only partially responsive, showing slower, less complete control of kinase activity (Fig. 3; Table S2).
Fig. 3. Stimulus response patterns of mutant receptors in an adaptation-deficient host.
Mutant derivatives of plasmid pPA114 and pRR53 in strain UU2567 (CheR− CheB−) were tested for serine responses in FRET kinase assays. The YFP/CFP ratios reflect the level of receptor-controlled CheA kinase activity. Four different mutant receptor response patterns were seen in the UU2567 host:
upper left: Nonresponsive-ON receptors (ΔK215 shown) exhibited no activity changes in response to addition and removal of serine (SER; vertical gray bars), but activated CheA kinase, as evidenced by the response to KCN, which depletes cellular ATP, the phosphodonor for the CheA autophosphorylation reaction [34].
upper right: Nonresponsive-OFF receptors (ΔF208 shown) exhibited no serine or KCN responses and had low baseline values, indicating no receptor-stimulated kinase activity.
lower left: Responsive receptors (ΔS217 shown) exhibited kinase inhibition responses that saturated at high serine concentrations.
lower right: Partially responsive receptors (cc-6S shown; see Fig. 5) exhibited slow changes in kinase activity in response to both serine addition and removal (compare to the responsive example). Receptors of this type produced some kinase activity, but only inhibited a fraction of that activity at saturating stimulus levels.
We next examined the signaling properties of the mutant Tsr plasmids in FRET reporter strain UU2700, which contains the CheR and CheB adaptation enzymes. CheR converts E residues at receptor modification sites to glutamyl methyl esters (Em), shifting output toward the ON state. CheB deamidates sites with a Q residue and demethylates Em sites, creating E residues that shift output toward the OFF state. In cells with both adaptation enzymes, the receptor population is heterogeneously modified, with an average modification state that offsets ambient chemoeffector levels. In strain UU2700 wild-type Tsr produces sensitive serine responses (K1/2 ~0.5 μM) with modest cooperativity (Hill ~2.5) [14; 34; 35]. As expected, mutant receptors that mediated serine responses in the adaptation-incompetent host became more sensitive to serine in the adaptation-competent host (Table S2).
We found that most mutant receptors that were nonresponsive in UU2567 became responsive in UU2700, consistent with the fact that many of those receptors produced chemotactic behaviors on soft agar in an adaptation-competent host (Fig. 2B & 2C). Two general response patterns emerged under action of the sensory adaptation system (Fig. 4). Some previously nonresponding, kinase-ON receptors remained less sensitive than wild type, but had wild-type cooperativities; others had wild-type sensitivities, but significantly reduced response cooperativities. Similarly, some previously nonresponding, kinase-OFF receptors regained wild-type response sensitivity and cooperativity, whereas others had wild-type sensitivities, but significantly reduced response cooperativities.
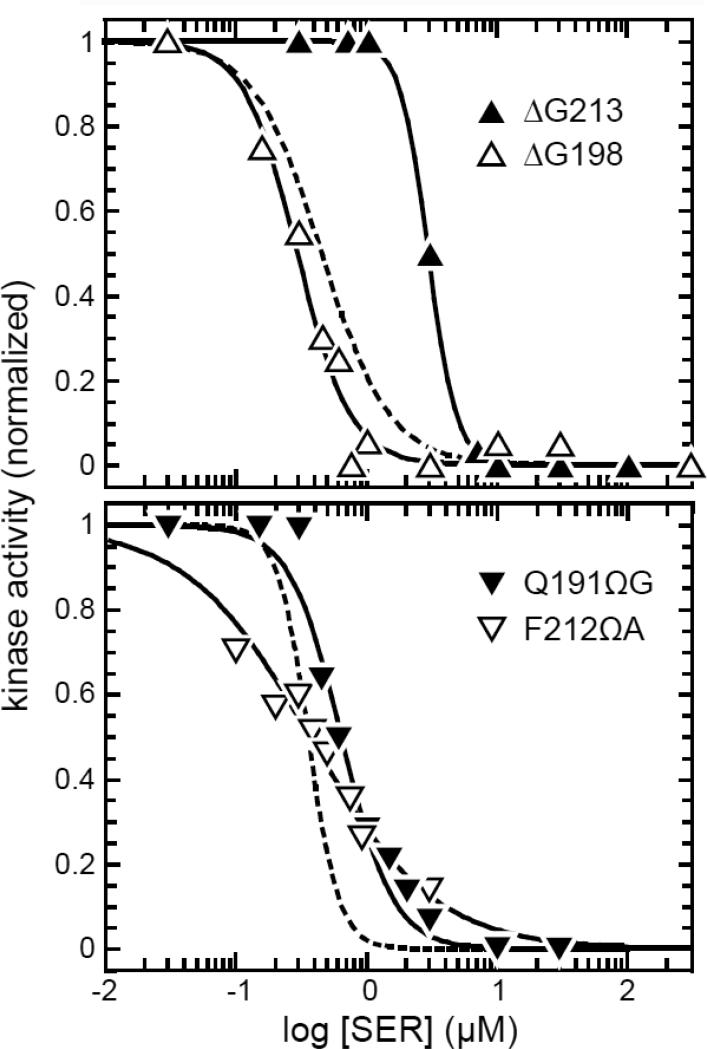
Fig. 4. Signaling by nonresponsive mutant receptors in an adaptation-proficient host.
Mutant derivatives of plasmids pPA114 (upper panel) and pRR53 (lower panel) were examined in strain UU2700 (CheR+ CheB+) by FRET kinase assay. The dashed lines show the Hill fits (with data points omitted) for the dose-response of the corresponding wild-type Tsr plasmid in strain UU2700. Solid lines (with data points) are Hill fits for the indicated Tsr mutants. Black symbols denote receptors with nonresponsive-ON behaviors in an adaptation-deficient host; white symbols denote receptors with nonresponsive-OFF behaviors in an adaptation-deficient host (see Fig. 3). K1/2 and Hill coefficient values were [upper panel: 0.5 μM, 1.7 (pPA114 Tsr-wt); 3.0 μM, 4.0 (ΔG213); 0.3 μM, 2.1 (ΔG198)]; [lower panel: 0.4 μM, 3.7 (pRR53 Tsr-wt); 0.8 μM, 0.9 (G191ΩG); 0.4, 0.8 (F212ΩA)].
All FRET response parameters of the mutant receptors are listed in Table S2.
Adaptational modification of the mutant receptors
CheR acts on kinase-OFF receptor conformation(s); CheB acts on kinase-ON conformation(s) [14]. Thus, the CheR/CheB substrate properties of a mutant receptor molecule reflect the conformational properties of its methylation helix bundle. We expressed mutant Tsr proteins in strain UU2632 (CheR+ CheB−) and in strain UU2611 (CheR− CheB+) to determine if they were subject to CheR or CheB modification, respectively. Adaptational modifications of Tsr subunits were detected as small electrophoretic mobility shifts in denaturing polyacrylamide gels [36]. The mutant receptors fell into three classes with respect to these modification tests (Table S1): (i) Most mutant receptors were substrates for both CheR and CheB modification(s); (ii) a few were modified by CheB but not by CheR; (iii) others were substrates for neither enzyme.
Signaling shifts of the mutant receptors
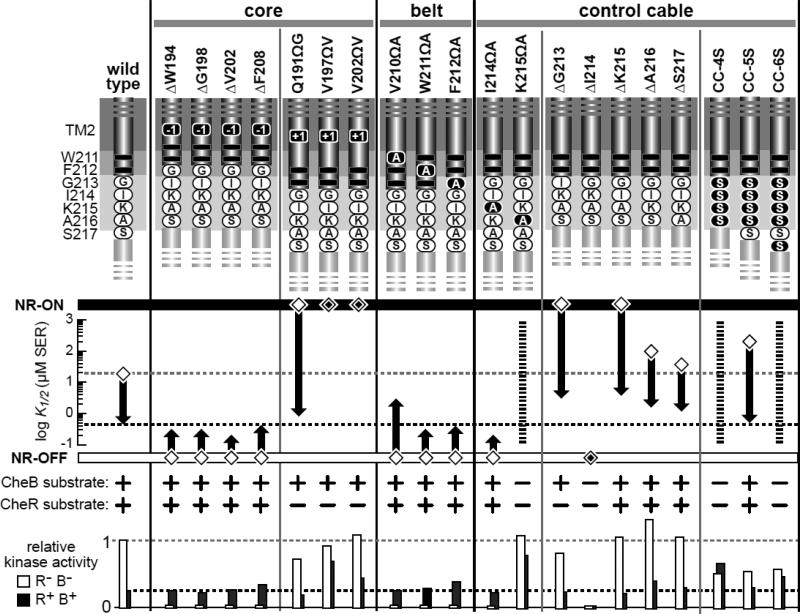
The FRET response parameters and modification properties of the mutant receptors are summarized in Fig. 5. Four different one-residue deletions in TM2 caused OFF-shifted outputs that responded to adaptational control. Those mutant receptors were substrates for both CheR and CheB modifications and regained a wild-type level of kinase activity in the adaptation-competent host. Three different one-residue insertions in TM2 caused ON-shifted outputs, two of which remained locked in the ON state in the adaptation-competent host. All three mutant receptors were modified by CheB but not by CheR. In contrast, three different one-residue insertions in the adjacent aromatic belt region produced OFF-shifted signaling properties like those of TM2 deletions.
Fig. 5. Signaling properties of Tsr mutants with TM2 or control cable alterations.
The K1/2 values for FRET kinase responses in UU2567, the adaptation-deficient host, are indicated with diamond symbols; K1/2 values for responses in UU2700, the adaptation-proficient host, are indicated with thick vertical arrows that begin at the UU2567 response and end at the serine sensitivity in UU2700. White diamonds enclosing a smaller black diamond denote locked-ON (NR-ON) or locked-OFF (NR-OFF) behaviors that are refractory to sensory adaptation control. Abilities of the mutant receptors to undergo adaptational modification in strain UU2611 (CheB+) or strain UU2632 (CheR+) are shown below the K1/2 scale. Kinase activities produced by the mutant receptors in host strains UU2567 (CheR− CheB−) and UU2700 (CheR+ CheB+) are indicated relative to that of wild-type Tsr in UU2567. Dashed horizontal lines show the wild-type kinase values in each host for comparison purposes. Broken vertical lines for the K215ΩA, CC-4S, and CC-6S receptors denote partially responsive behaviors of indeterminate serine sensitivity in both adaptation-deficient (UU2567) and adaptation-proficient (UU2700) host strains.
The signaling properties of control cable mutants were more variable (Fig. 5). The I214ΩA receptor had OFF-shifted output subject to adaptational control, whereas the K215ΩA receptor exhibited partial responses to serine in both FRET hosts and was not modified by either adaptation enzyme. Four of five one-residue deletions in the control cable produced ON-shifted output subject to partial (ΔG213) or full (ΔK215, ΔA216, ΔS217) adaptational control. The ΔI214 receptor, by contrast, was locked in the OFF state and was not a substrate for either CheR or CheB modification. The disparate signaling properties of the control cable mutants may reflect structural influences of particular sidechains when shifted to an adjacent residue position or when a neighboring residue is removed (Fig. 5).
The three Tsr mutants with all-serine control cables confirm that sidechain character influences the signaling consequences of control cable length changes (Fig. 5). The receptor with a five-serine control cable had near-normal signaling properties, indicating that control cable function tolerates a serine sidechain at all residue positions. However, the four-serine and sixserine control cables caused similar aberrant behaviors: partial responsiveness, no adaptational control, and no modification by CheB or CheR (Fig. 5). These findings demonstrate that a one-residue length change in the control cable severely impairs signal transmission, but that native control cable sidechains can moderate those defects.
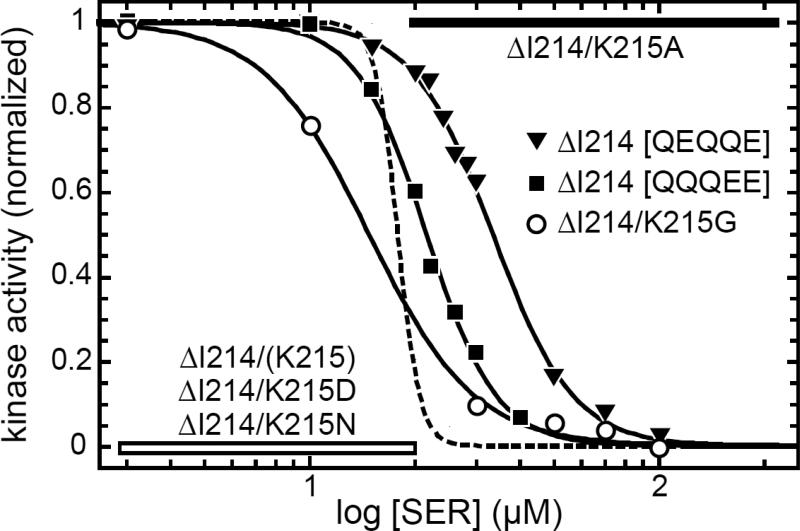
To explore the ability of different sidechains to enhance or suppress the signaling consequences of control cable length changes, we focused on the ΔI214 receptor, whose properties were very different from the other four control cable deletion mutants (Fig. 5). This control cable residue is known to play a critical role in transmembrane signaling in Tsr [14]. We suspected that the locked-OFF behavior of the ΔI214 receptor might be due to relocation of K215 to the 214 residue position. Perhaps in the context of the neighboring G213 residue that shift allows the positively-charged lysine sidechain to influence control cable structure through an interaction with the anionic membrane headgroups. This scenario predicts that replacing the K residue in the control cable of the ΔI214 receptor should alter its signaling properties. We made several such replacements and tested their signaling effects with FRET kinase assays in strain UU2567 (CheR− CheB−) to avoid any adaptational modification effects (Fig. 6). An A residue at the 214 position shifted ΔI214 output from locked-OFF to locked-ON, whereas a G residue restored responsiveness (Fig. 6). However, a D or N replacement failed to change the locked-OFF behavior of the ΔI214 receptor, suggesting that there might be several structural ways in which the particular sidechain at control cable residue 214 causes OFF output.
Fig. 6. Signaling properties of TsrΔI214 derivatives in an adaptation-deficient host.
Mutant derivatives of plasmid pPA114 were examined by FRET kinase assay in strain UU2567 (CheR− CheB−). The dashed line shows the Hill fit (with data points omitted) for the dose-response of wild-type Tsr. Solid lines (with data points) are Hill fits for the indicated Tsr-ΔI214 derivatives. K1/2 and Hill coefficient values for these serine-responsive receptors were: 18 μM, 14 (Tsr-wt); 15 μM, 2.9 (ΔI214/K215G); 22 μM, 4.3 (ΔI214 [QQQEE]); 34 μM, 3.8 (ΔI214 [QEQQE]). The parental mutant receptor [ΔI214/(K215)] and two of its derivatives (K215D and K215N) were locked-OFF (horizontal white bar); the K215A derivative was locked-ON (horizontal black bar).
The OFF signaling conformation of the ΔI214 receptor is not a substrate for CheR modification; the accessibility or conformation of its adaptation sites must differ in some way from those of the native OFF state. To ask whether higher methylation states of the ΔI214 receptor could shift it toward kinase-ON output, we created ΔI214 derivatives with E to Q replacements at various adaptation sites. In the wild-type Tsr receptor, a Q residue at an adaptation site mimics methylation effects, shifting output toward the kinase-ON state [34; 35]. We found that derivatives of the ΔI214 receptor with one additional Q site (QQQEE or QEQQE) shifted output toward the ON state and became serine-responsive in the UU2567 host (Fig. 6). These results indicate that modification-dependent output controls still operate normally in the ΔI214 mutant receptor, despite its CheR-refractory character.
Discussion
Binding of a single ligand molecule by the Tsr and Tar receptors induces relative rotation of the two receptor subunits around the occupied binding site and piston motions of one TM2 transmembrane helix [23; 25]. Because ligand binding is negatively cooperative, chemoeffector stimuli cause asymmetric signal inputs to the cytoplasmic HAMP domain. However, symmetric structural changes, generated by insertion or deletion of a residue in TM2 or the adjoining control cable, also shift receptor output, indicating that induced HAMP asymmetry is not critical to the transmembrane signaling mechanism. Rather, structural inputs to one or both of the AS1 helices probably modulate output kinase activity in the same manner by influencing the overall conformation [17; 18] or packing stability [20] of the HAMP bundle.
Signaling consequences of one-residue additions or deletions in the TM2 helix
One-residue additions or deletions should cause both length and register changes in the TM2 helix. The maximum magnitudes of these structural changes should be ~1.5 Å in length, comparable to stimulus-induced piston displacements, and ~100° rotations in helical register. The spring-like properties of alpha helices and the aromatic residues that position the cytoplasmic end of TM2 at the core-headgroup interface [11; 12; 13], could conceivably dampen both sorts of TM2 structural changes. Although we do not know the resultant magnitude of these structural changes, a priori, one-residue insertions might mimic the piston motion that accompanies an attractant stimulus (kinase-off), whereas one-residue deletions might mimic a repellent stimulus (kinase-on). This was clearly not the case: For the portion of TM2 embedded in the lipid core, one-residue deletions shifted Tsr output toward a kinase-off state, whereas one-residue insertions shifted output strongly toward a kinase-on state (Fig. 7). The [−1] receptors remained substrates for both CheR and CheB modifications, whereas the [+1] receptors served as substrates for CheB, but not for CheR (Fig. 5; Fig. 7). Thus, the TM2 [±1] receptors evidently underwent structural changes sufficient to alter signal transmission, but the sign of their mutant output was not consistent with piston displacement effects.
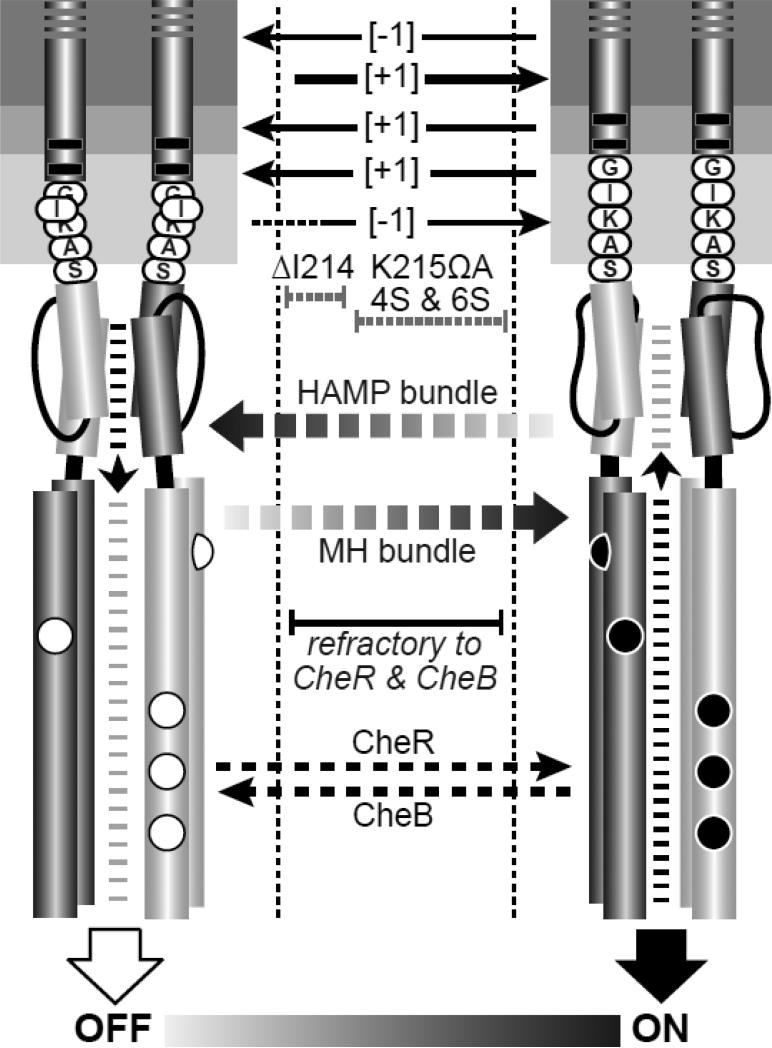
Fig. 7. Mechanistic summary of mutant receptor behaviors.
The dynamic-bundle model of HAMP input-output signaling proposes a series of meta-stable conformational states, produced through opposing structural interactions of the HAMP and MH bundles, that range between kinase-off and kinase-on states. In the full-off state (OFF), HAMP is stably packed and the MH bundle is loosely packed. During sensory adaptation, the CheR methyltransferase acts on loosely packed modification sites of OFF-state receptors to shift their output toward the ON state. In the full-on state (ON), HAMP is loosely packed and the MH bundle is stably packed. The CheB enzyme acts on stably packed modification sites of ON-state receptors to shift their output toward the OFF state. MH bundles with intermediate stabilities along the OFF-ON conformational landscape are not substrates for either adaptation enzyme. Structural alterations in the TM2-control cable that destabilize the HAMP OFF and/or ON state can trap the receptor within this intermediate regime, preventing modification by one or both sensory adaptation enzymes.
We suggest that the overall signal shifts of TM2 [±1] mutant receptors are due mainly to changes in TM2 helix register rather than to any piston movements they might have. Viewed in the N to C direction along the TM2 helix, counter-clockwise rotation [−1] evidently shifts output toward a kinase-off state and clockwise rotation [+1] shifts output toward a kinase-on state (Fig. 7). If these changes in TM2 register propagate directly to the AS1 helix of HAMP, they could favor alternative packing arrangements of the HAMP bundle. The gearbox model, for example, proposes that the HAMP bundle has two stable packing arrangements, designated x-da and a-d, that differ by 26° counter-rotations of its four helices [17]. However, in the context of the gearbox model, clockwise rotation of the HAMP AS1 helix favors a-d packing, which has been assigned to the OFF state [34; 37]. Similarly, counter-clockwise rotation favors the x-da arrangement, which has been assigned to the ON state [34; 37]. We conclude that the helix rotations in TM2 [±1] mutants do not elicit the signaling shifts predicted by the HAMP gearbox model.
A structural-signaling shift at the aromatic belt - control cable junction
The signaling shifts caused by TM2 deletions and insertions changed dramatically at the core-headgroup interfacial region. One-residue insertions adjacent to or between the aromatic belt residues (V210ΩA, W211ΩA, F212ΩA) or in the adjoining control cable (I214ΩA) shifted output toward the OFF state and preserved substrate properties for both CheR and CheB (Fig. 7). Conversely, four of five one-residue deletions in the control cable shifted output toward the ON state. The ΔK215, ΔA216, and ΔS217 receptors were good substrates for both adaptation enzymes, but the signaling properties of the ΔS217 receptor were most similar to the wild type (Fig. 5). In contrast, deletion of G213 at the N-terminus of the control cable produced the most strongly ON-shifted behavior, evidenced by inability to serve as a CheR substrate (Fig. 5). The decline in severity of the control cable [−1] signal shifts with distance of the deleted residue from the aromatic belt implicates the junction of the control cable and aromatic belt as the likely site of the structural features responsible for signal reversal. These mutant behaviors imply that piston displacements and changed registers in the TM2 helix are not transmitted as such to AS1, but rather converted to another signaling conformation through interaction of Tsr structural elements with the membrane-cytoplasm interface. This important insight argues against transmembrane signaling mechanisms, for example those proposed by the crankshaft-gearbox [17] and pushrod-scissors [18] models, that invoke direct transmission of TM2 structural changes to AS1. A continuous, structurally rigid TM2-AS1 connection, as required by those models, would not produce the signal reversal observed at the cytoplasmic end of TM2.
Signaling consequences of control cable length changes
Tsr with a five-residue all-serine control cable had ON-shifted output in the QEQEE modification state, but exhibited wild-type signaling behavior and supported robust chemotaxis in an adaptation-competent background (Fig. 5; Table S1). However, unlike [±1] changes in the native control cable, Tsr molecules with a four- or six-residue all-serine control cable failed to support chemotaxis and were not subject to modification by either the CheB or CheR sensory adaptation enzymes (Fig. 5). Those mutant receptors produced some kinase activity, but down-regulated only part of that activity in response to serine stimuli (Fig. 5). We attribute the 4S and 6S signaling defects to structural changes that destabilize the native OFF and ON output conformations that serve as substrates for CheR and CheB (Fig. 7). Such defects might confine the interacting HAMP and methylation helix bundles to a subset of conformations between the OFF and ON signaling states (Fig. 7). The existence of additional conformational states along an OFF-ON structural continuum is central to the dynamic-bundle model of HAMP signaling [16], but not predicted by discrete two-state models, such as the gearbox [17] or scissors [18] proposals for HAMP operation.
Like the 6-residue all-serine control cable, the K215ΩA control cable caused partial serine responses (Fig. 7). However, unlike the 4-residue all-serine control cable, one-residue deletions in the native control cable produced ON-shifted, serine responsive, behavior (Fig. 5; Fig. 7). We ascribe these differences to the native residues still present in the [−1] control cables, particularly G213 and I214, which adjoin the aromatic belt and are known to play important signaling roles [14; 15]. Deletion of G213 shifts I214 to the 213 position and prevents the receptor from accessing the CheR substrate state (Fig. 5). Deletion of I214 shifts K215 to the 214 position and locks output in a kinase-off state. The ΔI214 receptor appears to be trapped in signaling conformations intermediate to the native ON and OFF states (Fig. 7) because amino acid replacements at its lysine residue, or an E to Q change at an adaptation site, can restore its kinase activity and serine responsiveness (Fig. 6).
A helix-clutch model of transmembrane signaling
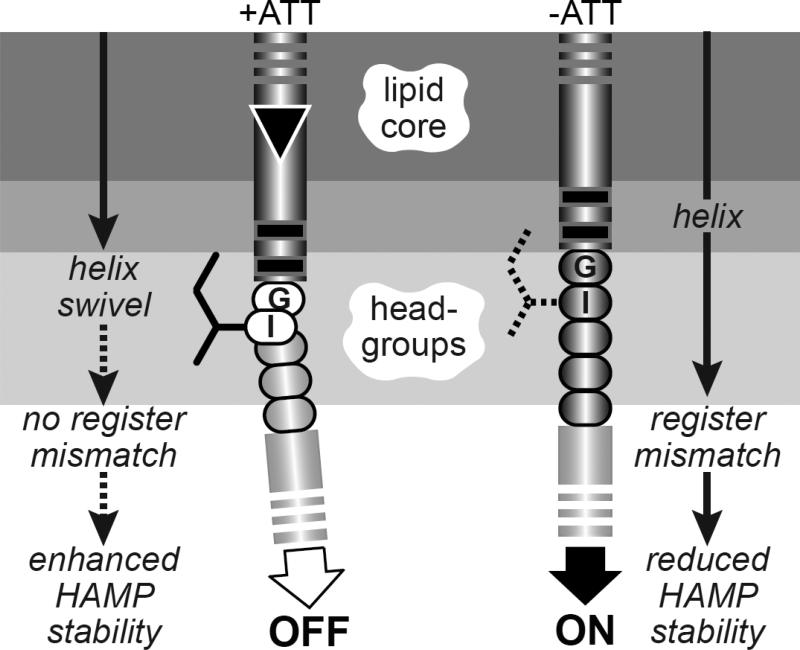
The different signaling behaviors produced by control cables of the same length, either one residue shorter or longer than the wild-type, indicate that the control cable most likely has a malleable structure subject to influence by the flanking TM2 and AS1 helices and by its sidechain interactions with the membrane interfacial environment. Previous studies of both Tar and Tsr suggested that the control cable has helical character that changes in response to stimulus inputs [14; 15; 31]. The structural factors that most likely modulate signal transmission through the control cable helix are piston displacements of the TM2 helix, the packing stability of the HAMP bundle, and the mismatched registers of the TM2 and AS1 helices resulting from the 5-residue control cable helix that joins them. In the context of the dynamic-bundle model of HAMP signaling, we suggest that inward piston displacements disengage a structural clutch at the TM2 aromatic belt to promote a bend or helix break in the first few control cable residues (Fig. 8). The resultant helix swivel in turn reduces the intensity of the register mismatch between TM2 and AS1, enhances HAMP packing, and shifts output to the OFF state (Fig. 8). Engaging the structural clutch that joins the TM2 and control cable helices increases the register mismatch, destabilizes HAMP packing, and shifts output to the ON state.
Fig. 8. Helix-clutch model for transmembrane signaling by Tsr.
A portion of the TM2-control cable-AS1 segment in one Tsr subunit is pictured. Dark- or light-gray shaded segments are proposed to have α-helical secondary structure. Sidechains of control cable residues (only the I214 sidechain is shown) are proposed to interact with the membrane environment, but not with other residues in Tsr or in other proteins. A stable helical connection between TM2 and the AS1 helix of HAMP destabilizes packing of the HAMP bundle, favoring kinase-on output. The inward TM2 piston displacement (black triangle) promoted by binding of an attractant ligand to the periplasmic sensing domain perturbs the membrane environment of the I214 sidechain, creating a kink or break in helicity that alleviates the register mismatch between TM2 and AS1, thereby enhancing HAMP bundle packing to promote kinase-off output.
This helix-clutch model best explains the disparate signaling consequences of one-residue insertions or deletions at different points in the TM2-AS1 segment of the Tsr molecule. Helix length changes on the input side of the clutch probably fail to mimic piston displacements because of their attendant rotational component. Indeed, the signal shifts caused by TM2 [±1] lesions are consistent with their expected effects on the TM2-AS1 register mismatch. One-residue insertions would exacerbate the mismatch, producing the observed kinase-on shifts; one-residue deletions would reduce the mismatch, producing the observed kinase-off shifts. In contrast, shortened control cables generally shifted output toward the kinase-on state. Although a 4-residue control cable should reduce the TM2-AS1 register mismatch, the overall ON-shifted behavior of [−1] control cable mutants might arise from increased structural tension on the AS1/AS1' helices of HAMP caused by a shortened connection to the TM2/TM2' membrane helices. With no native residue sidechains in the vicinity of the clutch, the 4-residue all-serine control cable might destabilize all HAMP conformations, producing partially responsive behavior. Six-residue control cables should exacerbate the TM2-AS1 register mismatch, but on the output side of the clutch their HAMP-destabilizing effects could be sufficiently severe to cause partially responsive behavior in some [+1] mutants (K215ΩA, 6S; Fig. 5; Fig. 7).
The structural changes produced by TM2 piston displacements at the proposed helix clutch and their trigger mechanism remain to be determined. Neither the aromatic belt residues (W211, F212 in Tsr; W209, Y210 in Tar) nor the key control cable residues (G213, I214 in Tsr; G211, I212 in Tar) are essential for transmembrane signaling, provided that the receptor's overall signaling poise remains in a responsive structural range, for example, through appropriate adaptational modifications [11; 12; 13; 14; 15; 30; 31]. Further structure-function studies of the helix-clutch residues should provide important insights into the mechanism of transmembrane signaling in chemoreceptors.
Materials and Methods
Bacterial strains
Strains were derivatives of E. coli K12 strain RP437 [38]; their relevant genotypes are: UU1250 [Δaer-1 ygjG::Gm Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] [39]; UU2610 [Δaer-1 ygjG::Gm Δ(tar-cheB)4346 Δtsr-5547 Δtrg-4543] [40]; UU2611 [Δaer-1 Δ(tar-cheR)4283 Δtsr-5547 Δtrg-4543] [40]; UU2612 [Δaer-1 Δ(tar-tap)4530 Δtsr-5547 Δtrg-4543] [40]; UU2632 [Δaer-1 Δ(tar-tap)4530 ΔcheB4345 Δtsr-5547 Δtrg-4543] [40]; UU2567 [Δ(tar-cheZ)4211 Δtsr-5547 Δaer-1 Δtrg-4543] [34]; UU2700 [Δ(cheY-cheZ)1215 Δ(tar-tap)4530 Δtsr-5547 Δaer-1 Δtrg-4543] [34].
Plasmids
Plasmids used in the study were: pKG116, a derivative of pACYC184 [41] that confers chloramphenicol resistance and has a sodium salicylate-inducible expression/cloning site [42]; pPA114, a relative of pKG116 that carries wild-type tsr under salicylate control [39]; pRZ30, a derivative of pKG116 that expresses CheY-YFP and CheZ-CFP fusion proteins under salicylate control [34]; pRR48, a derivative of pBR322 [43] that confers ampicillin resistance and has an expression/cloning site with a tac promoter and an ideal (perfectly palindromic) lac operator under the control of a plasmid-encoded lacI repressor, inducible by IPTG [44]; pRR53, a derivative of pRR48 that carries wild-type tsr under IPTG control [44]; and pVS88, a plasmid that expresses CheY-YFP and CheZ-CFP fusion proteins under IPTG control [33].
Construction of TM2 and control cable mutants
Mutations in plasmids pPA114 and pRR53 were generated by QuikChange™ PCR mutagenesis, using site-specific primers, and verified by sequencing the entire tsr coding region, as previously described [39; 45].
Chemotaxis assays
Mutant tsr plasmids carried in strain UU2612 were assessed for ability to support chemotaxis on tryptone soft agar plates [46] containing appropriate antibiotics (ampicillin [50 Δg/ml] or chloramphenicol [12.5 Δg/ml]) and inducers (100 ΔM IPTG or 0.6 ΔM sodium salicylate). Plates were incubated at 30°C and 32.5°C for 7 to 10 h and at 24°C for 15-20 hours.
Expression levels and modification patterns of mutant Tsr proteins
Cells harboring pRR53 derivatives were grown in tryptone broth containing 50 Δg/ml ampicillin and 100 ΔM IPTG; cells harboring pPA114 derivatives were grown in tryptone broth containing 12.5 Δg/ml chloramphenicol and 0.6 ΔM sodium salicylate. Expression levels of mutant proteins were determined in strain UU2610 (R−B−) in which receptor molecules have a uniform modification state. Strains UU2611 (R−B+) and UU2632 (R+B−) were used to assess the CheR and CheB substrate properties of mutant Tsr proteins. Cells were grown at 30°C to mid-exponential phase, and 1-ml samples were pelleted by centrifugation, washed twice with KEP (10 mM K-PO4, 0.1 mM K-EDTA, pH 7.0), and lysed by boiling in sample buffer [36]. Tsr bands were resolved by electrophoresis in 11% polyacrylamide gels containing sodium dodecyl sulfate and visualized by immunoblotting with a polyclonal rabbit antiserum raised against Tsr residues 290-470 [47].
In vivo FRET CheA kinase assay
The experimental system, cell sample chamber, stimulus protocol, and data analysis followed the hardware, software, and methods described by Sourjik et al. [33] with minor modifications [34]. Cells containing a FRET reporter plasmid (pRZ30 or pVS88) and a compatible tsr expression plasmid (pRR53 or pPA114 derivative) were grown at 30°C to mid-exponential phase in tryptone broth, washed, attached to a round coverslip with polylysine, and mounted in a flow cell [48]. The flow cell and all motility buffer test solutions [KEP containing 10 mM Na lactate, 100 μM methionine, and various concentrations of serine] were maintained at 30°C throughout each experiment. Cells were illuminated at the CFP excitation wavelength and light emission detected at the CFP (FRET donor) and YFP (FRET acceptor) wavelengths with photomultipliers. The ratio of YFP to CFP photon counts reflects CheA kinase activity and changes in response to serine stimuli [32; 33]. In some extended experiments differential rates of YFP and CFP bleaching caused a slow decline in YFP/CFP values. In such cases, a linear fit of YFP/CFP versus time was used to correct for baseline drift, similar to the approach used by Meir et al. [49]. Fractional changes in kinase activity versus applied serine concentrations were fitted to a multi-site Hill equation, yielding two parameter values: K1/2, the attractant concentration that inhibits 50% of the kinase activity; and the Hill coefficient, reflecting the extent of cooperativity of the response [33; 50].
Protein modeling and structural display
Atomic coordinates for the Tsr HAMP domain were generated from the Af1503 HAMP coordinates (PDB accession number 2ASW) [45]. Coordinates for the TM bundle of S. typhimurium Tar were based on the modeled TM structure of Trg [10] and provided by Dr. Gerald Hazelbauer (U. Missouri). Structure images were prepared with MacPyMOL software (http://www.pymol.org).
Supplementary Material
Highlights.
How do chemoreceptors transmit stimulus information across the cytoplasmic membrane?
One-residue length changes in a transmembrane helix cause receptor output shifts.
Similar lesions cause reversed signal outputs in the cytoplasmic control cable helix.
Input signals modulate control cable helicity to elicit receptor output responses.
Acknowledgements
This work was supported by research grant GM19559 from the National Institute of General Medical Sciences. DNA sequencing and primer synthesis were carried out by the Protein-DNA Core Facility at the University of Utah, which receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute. We thank Dr. Smiljka Kitanovic for sharing several of the mutant plasmids characterized in this study.
Abbreviations
- MCP
methyl-accepting chemotaxis protein
- FRET
Förster resonance energy transfer
- IPTG
isopropyl-Δ-D-thiogalactopyranoside
- AS1
AS1', N-terminal HAMP helices
- MH1
MH1', MH2, MH2', methylation helices
- TM1
TM1', TM2, TM2', transmembrane helices
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falke JJ, Piasta KN. Architecture and signal transduction mechanism of the bacterial chemosensory array: Progress, controversies, and challenges. Curr. Opin. Struc. Biol. 2014;29:85–94. doi: 10.1016/j.sbi.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 2012;24:262–8. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–51. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falke JJ. Piston versus scissors: chemotaxis receptors versus sensor His-kinase receptors in two-component signaling pathways. Structure. 2014;22:1219–20. doi: 10.1016/j.str.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high- performance signaling in networked arrays. Trends. Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends. Biochem. Sci. 2001;26:257–265. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldog T, Hazelbauer GL. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 2004;13:1466–1475. doi: 10.1110/ps.04648604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peach ML, Hazelbauer GL, Lybrand TP. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 2002;11:912–923. doi: 10.1110/ps.4640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry. 2005;44:1268–77. doi: 10.1021/bi048969d. [DOI] [PubMed] [Google Scholar]
- 12.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 13.Adase CA, Draheim RR, Manson MD. The residue composition of the aromatic anchor of the second transmembrane helix determines the signaling properties of the aspartate/maltose chemoreceptor Tar of Escherichia coli. Biochemistry. 2012;51:1925–32. doi: 10.1021/bi201555x. [DOI] [PubMed] [Google Scholar]
- 14.Kitanovic S, Ames P, Parkinson JS. A Trigger Residue for Transmembrane Signaling in the Escherichia coli Serine Chemoreceptor. J. Bacteriol. 2015;197:2568–79. doi: 10.1128/JB.00274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitanovic S, Ames P, Parkinson JS. Mutational analysis of the control cable that mediates transmembrane signaling in the E. coli serine chemoreceptor. J. Bacteriol. 2011;193:5062–5072. doi: 10.1128/JB.05683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 17.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 18.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 22.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: A model. Proc. Natl. Acad. Sci. USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D, Ma X, Tu Y, Lai L. Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Scientific reports. 2015;5:8640. doi: 10.1038/srep08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park H, Im W, Seok C. Transmembrane signaling of chemotaxis receptor Tar: insights from molecular dynamics simulation studies. Biophys. J. 2011;100:2955–2963. doi: 10.1016/j.bpj.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall BA, Armitage JP, Sansom MS. Transmembrane helix dynamics of bacterial chemoreceptors supports a piston model of signalling. PLoS Comput Biol. 2011;7:e1002204. doi: 10.1371/journal.pcbi.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughson AG, Lee GF, Hazelbauer GL. Analysis of protein structure in intact cells: crosslinking in vivo between introduced cysteines in the transmembrane domain of a bacterial chemoreceptor. Protein Sci. 1997;6:315–22. doi: 10.1002/pro.5560060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chervitz SA, Lin CM, Falke JJ. Transmembrane Signaling by the Aspartate Receptor: Engineered Disulfides Reveal Static Regions of the Subunit Interface. Biochemistry. 1995;34:9722–33. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 1995;270:24043–53. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama IN, Mikawa YG, Maruyama HI. A model for transmembrane signalling by the aspartate receptor based on random-cassette mutagenesis and site-directed disulfide cross-linking. J. Mol. Biol. 1995;253:530–546. doi: 10.1006/jmbi.1995.0571. [DOI] [PubMed] [Google Scholar]
- 30.Adase CA, Draheim RR, Rueda G, Desai R, Manson MD. Residues at the cytoplasmic end of transmembrane helix 2 determine the signal output of the TarEc chemoreceptor. Biochemistry. 2013;52:2729–38. doi: 10.1021/bi4002002. [DOI] [PubMed] [Google Scholar]
- 31.Wright GA, Crowder RL, Draheim RR, Manson MD. Mutational analysis of the transmembrane helix 2-HAMP domain connection in the Escherichia coli aspartate chemoreceptor Tar. J. Bacteriol. 2011;193:82–90. doi: 10.1128/JB.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:363–91. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 34.Lai RZ, Parkinson JS. Functional suppression of HAMP domain signaling defects in the E. coli serine chemoreceptor. J. Mol. Biol. 2014;426:3642–55. doi: 10.1016/j.jmb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han XS, Parkinson JS. An unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J. Bacteriol. 2014;196:641–9. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Ferris HU, Dunin-Horkawicz S, Mondejar LG, Hulko M, Hantke K, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19:378–85. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol. Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gosink KK, Buron-Barral M, Parkinson JS. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 2006;188:3487–3493. doi: 10.1128/JB.188.10.3487-3493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolivar F, Rodriguez R, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization of new cloning vehicles. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 44.Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkinson JS. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ames P, Parkinson JS. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg HC, Block SM. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 1984;130:2915–20. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 49.Meir Y, Jakovljevic V, Oleksiuk O, Sourjik V, Wingreen NS. Precision and kinetics of adaptation in bacterial chemotaxis. Biophys. J. 2010;99:2766–74. doi: 10.1016/j.bpj.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.