Abstract
In the recent years, the possibility of utilizing extracellular vesicles for drug delivery purposes has been investigated in various models, suggesting that these vesicles may have such potential. In addition to the choice of donor cell type for vesicle production, a major obstacle still exists with respect of loading the extracellular vesicles efficiently with the drug of choice. One of the proposed solutions to this problem has been drug loading by electroporation, where small pores are created in the membrane of the extracellular vesicles, hereby allowing for free diffusion of the drug compound into the interior of the vesicle. We investigated the utility of adipose-derived stem cells (ASCs) as an efficient exosome donor cell type with a particular focus on the treatment of glioblastoma multiforme (GBM). In addition, we evaluated electroporation-induced effects on the ASC exosomes with respect to their endogenous potential of stimulating GBM proliferation, and morphological changes to single and multiple ASC exosomes. We found that electroporation does not change the endogenous stimulatory capacity of ASC exosomes on GBM cell proliferation, but mediates adverse morphological changes including aggregation of the exosomes. In order to address this issue, we have successfully optimized the use of a trehalose-containing buffer system as a way of maintaining the structural integrity of the exosomes.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-016-9952-7) contains supplementary material, which is available to authorized users.
Keywords: Extracellular vesicles, Exosomes, Drug delivery, Adipose-derived stem cells, Electroporation, Trehalose, Glioblastoma multiforme
Introduction
Exosomes are a class of extracellular vesicles (EVs) that are secreted by most cell types of the body able to transport biologically active cargo between cells, including protein, lipids, mRNA and miRNA (Andaloussi et al. 2013). They are 30–120 nm in diameter and have a complex surface structure composed of special lipids and transmembranous proteins (especially the tetraspanins, CD9, CD63, and CD81), which facilitate specific interaction and fusion with the recipient cells (Vlassov et al. 2012).
Exosomes have received a lot of attention with respect to drug delivery since Alvarez-Erviti et al. (2011) showed that these nanoparticles could deliver small interfering RNA (siRNA) across the blood–brain barrier (BBB) to the brain parenchyma (Alvarez-Erviti et al. 2011). Exosomes derived from immature dendritic cells were engineered to express a targeting ligand on the extracellular domain of the exosome-associated protein, Lamp2b. This was followed by electroporation-induced loading with siRNA against the housekeeping gene, GAPDH, and the Alzheimer’s disease-associated gene, BACE1. Injection of these exosomes into mice resulted in a brain-specific knockdown of gene expression, illustrating the potential of modulating the endogenous exosomes to function as drug delivery vehicles (Alvarez-Erviti et al. 2011).
The potential of using exosomes to deliver therapeutics was also shown in GBM, where anti-miRs against the oncogenic miRNA, miR-9, could be loaded into exosomes derived from mesenchymal stem cells (MSC), and mediate miRNA knockdown in glioblastoma multiforme (GBM) cells in vitro. The knockdown of miR-9 was followed by an increase in sensitivity to temozolomide, the most widely used chemotherapeutic drug in the treatment of GBM (Munoz et al. 2013). MSC exosomes could also transfer miR-143 to GBM cells in vitro, which resulted in decreased cell viability (Lee et al. 2013). In addition, MSC exosomes enriched in miR-146b could reduce GBM tumor size after intratumoral injection, hereby showing that exosomes may have a great potential as drug delivery vehicles in GBM, and that mesenchymal stem cells may be used as an efficient producer of these particles (Katakowski et al. 2013).
We recently scrutinized all available literature pertaining to the field of exosome-based drug delivery (Johnsen et al. 2014). Through this work, several factors were identified as being important to address when designing exosomes for drug delivery purposes. These factors included, for example, the correct choice of donor cell, because the exosomes derived for drug delivery should be devoid of any stimulating properties on the immunological system as well as on the disease, in which they are used (Tian et al. 2013). The donor cell should also be an efficient producer of exosomes to obtain clinical scale quantities of exosomes for human use (Chen et al. 2011; Yeo et al. 2013). To secure specific action of the delivered drug, exosomes should be endowed with a targeting ligand to facilitate transport into a particular tissue or area of disease (Alvarez-Erviti et al. 2011; Ohno et al. 2012; Tian et al. 2013). This can be achieved by bioengineering exosome surface proteins to express a targeting peptide, but also by adsorbing proteins, such as whole antibodies and single-chain variable fragments, onto the exosome surface (Bryniarski et al. 2013). Efficient loading of therapeutically active cargo into the exosomes is a crucial requirement for successful development of an exosome-based drug delivery system. Several methods have been proposed, including electroporation of the exosomes and transfection of the donor cell with the cargo of choice. Although new methods are being described, most of the currently used loading methods still have problems that need to be solved (Kooijmans et al. 2013; Fuhrmann et al. 2015). For example, electroporation is known to adversely affect the structural integrity of the exosomes, whereas transfection of the donor cell is a time-consuming process with numerous steps that need quality assurance (Ohno et al. 2012; Hood et al. 2014). In addition, the choice of cargo and administration routes is also important to consider in order to optimize the efficiency of the resulting treatment (Johnsen et al. 2014).
In the present study, we investigated whether electroporation of adipose-derived stem cell (ASC) exosomes induced any aberrant effects on the normal interaction observed between ASC exosomes and GBM cancer cells. In addition, we tested whether an electroporation buffer based on the disaccharide trehalose could keep the structural integrity of exosomes released from ASCs and subjected to electroporation, hereby maintaining a favourable size distribution of the exosomes for drug delivery purposes.
Materials and methods
Cell culture
Human adipose-derived stem cells (ASCs; passages 5–8; Rasmussen et al. 2011; Yang et al. 2012) were thawed by standard procedures, seeded and cultured in T175 culture flasks in Dulbecco’s Modified Eagle’s Medium (DMEM) F12 (Lonza, Vallenbaek, Denmark) containing 10 % fetal calf serum (FCS; Gibco, Life Technologies, Carlsbad, CA, USA) and 1 % penicillin/streptomycin (pen/strep; Gibco, Life Technologies). The ASCs were incubated in a 37 °C incubator with 5 % CO2 and grown to 70–80 % confluence before being used for exosome production and isolation.
Isolation of exosomes
Production of conditioned medium (CM) was initiated by administrating DMEM F12 (Lonza) containing 10 % FCS (Gibco, Life Technologies) and 1 % pen/strep (Gibco, Life Technologies) to a 70–80 % confluent ASC culture. Prior to the production of CM, the FCS was depleted of exosomes by ultracentrifugation at 100,000g for 18 h followed by filtration through a 0.22 μm filter. The CM was harvested after 24 h of incubation and exosomes were isolated by two different procedures. Implications of the choice of isolation protocol for obtaining exosomes and other extracellular vesicles (with respect to yield and purity) have recently been reviewed (Gudbergsson et al. 2015).
The first isolation method utilized the Total Exosome Isolation Kit (Life Technologies), which contained a polyethylene glycol (PEG) precipitation reagent that enabled pelleting of nanoparticles with low-speed centrifugation. The harvested CM was spun at 2000 g for 30 min and filtered through a 0.22 μm filter to clear the medium from any debris, apoptotic bodies and larger nanoparticles. The filtered supernatant was mixed with the Total Exosome Isolation Reagent (2:1) and incubated under agitation at 4 °C overnight. After incubation, exosomes were pelleted by centrifugation at 10,000g for 65 min and resuspended in either phosphate-buffered saline (PBS)(Life Technologies) or trehalose pulse medium (TPM). Trehalose pulse medium has recently been shown to maintain exosomes structural integrity of melanoma exosomes (Hood et al. 2014). The yield of exosomes was determined by nanoparticle tracking analysis (NTA)(see below) and the Pierce BCA Protein Assay Kit (Life Technologies).
In the second isolation method, exosomes were isolated by sequential ultracentrifugation. Briefly, CM was cleared from live and dead cells by centrifugation at 300 and 2000g for 10 min. An additional centrifugation step at 10,000g for 30 min was performed to clear the CM from any cell debris. The supernatant was filtered through 0.22 µm filters, and then subsequently exposed to two steps of ultracentrifugation for 90 min at 100,000g with an intermediate washing step. The resulting pellets were resuspended in either PBS (Life Technologies) or TPM and analysed by NTA.
Nanoparticle tracking analysis
Size determination and concentration measurements of the ASC exosomes were performed on a NanoSight LM10-HS (NanoSight Ltd, Amesbury, United Kingdom) equipped with a 638 nm laser. The samples were diluted to a volume of 1 mL in clean PBS (Life Technologies) or TPM prior to analysis. The diluted sample was loaded into the sample chamber using a sterile syringe until the liquid reached the chamber outlet. To avoid particle drift during analysis, the outlet was sealed with Parafilm. The loaded sample chamber was mounted on the microscope, and the location for analysis was determined by identifying the region of intense diffraction of the laser beam, called “thumbprint”. The relevant point of analysis was found by moving the visual field to the right from the “thumbprint”, hereby avoiding disturbance of the data by interference from the “thumbprint” light scatter.
The obtained images were visually evaluated, and the gain and shutter time were adjusted to allow for smaller particles to be detected. Afterwards, the NTA 2.2 software was used to track all particles in the visual field throughout the captured video. All measurements were performed at a temperature in a range of 24–26 °C, determined with a thermometer attached to the sample chamber. The background of all video data was automatically extracted prior to the particle tracking. Size distribution diagrams, mean/mode size values and standard deviations were calculated within the NTA 2.2 software and used for downstream analysis.
Atomic force microscopy
Atomic force microscopy (AFM) was performed to characterize the shape and size of the exosomes. Purified exosomes were diluted 1:50 in sterile PBS (Life Technologies) and adsorbed onto freshly cleaved mica sheets. The adsorbed samples were rinsed thoroughly in deionized water and dried at ambient temperature for 30 min before analysis. AFM was carried out on a NTEGRA Aura (NT MDT, Moscow, Russia) operating in tapping mode using Olympus OMSCL240TS cantilevers (nominal spring constant 1.8 N/m, nominal tip radius <10 nm). Images were acquired in areas of 2, 4 and 10 µm2, typically at a resolution of 1024 × 1024 pixels. All data were processed using the AFM software, Gwyddion (Czech Metrology Institute, Prague, Czech Republic).
Transmission electron microscopy and immunogold labelling
Exosomes isolated from ASCs were placed as a 10 μL drop of exosome suspension on a freshly glow-discharged 400-mesh copper grid, negative stained by adding three drops of 1 % phosphotungstic acid (PTA)(pH 7.0) and incubated 15 s per drop. The exosome preparations were analysed by transmission electron microscopy (TEM) using a JEM 1010 (JEOL Ltd., Tokyo, Japan) and images were obtained using an electron sensitive Olympus KeenView CCD camera (Kamiina-gun, Nagano, Japan). For immunogold labelling, a 10 μL drop of exosome suspension was placed on a freshly glow-discharged 400-mesh nickel grid and washed 3 x 5 min in two drops of PBS. All samples were blocked with one drop 0.5 % ovalbumin in PBS. The grids were transferred to one drop of a primary antibody cocktail against known exosome-associated surface markers (CD9, CD63 or CD81; 1:250; LifeSpan BioSciences, Inc., Seattle, WA, USA) and incubated for 45 min at 37 °C. After incubation, the grids were washed in three drops of PBS and incubated with a goat-anti-mouse secondary antibody conjugated to 15 nm colloidal gold particles in 1 % cold-fish gelatine for 45 min at 37°C. Washing with three drops of PBS followed the incubation with secondary antibody. To remove any unbound colloidal gold, the grids were washed in two drops of 1 % cold-fish gelatine (5 min per wash) followed by two drops of PBS and one drop of ultrapure water. Lastly, the grids were negative stained with one drop of 1 % PTA and analysed by TEM using a JEM 1010 (JEOL Ltd., Japan).
CFSE proliferation assay
The GBM cell line, U87 (ATCC (Manassas, VA, USA); Cat. No. HTB-14), and the in-house generated GBM stem cell line, C16 (Pilgaard et al. (submitted)), were seeded into T25 culture flasks at a density of 2000 cells/cm2 in DMEM F12 (Lonza, Vallenbaek, Denmark) containing 10 % FCS and 1 % pen/strep, and allowed to set for 24 h. After this initial incubation, the growth medium was discarded, the cells washed twice in PBS, and stained with 10 µM carboxyfluorescein succinimidyl ester (CFSE; Cell Trace CFSE Cell Proliferation Kit, Life Technologies) in PBS for 15 min. The CFSE solution was discarded and pre-heated DMEM F12 (Lonza) containing 10 % exosome-free FCS and 1 % penicillin/streptomycin (pen/strep) was added to each T25 culture flask. This growth medium also included 15 µg/mL ASC exosomes, 15 µg/mL electroporated ASC exosomes (400 V, 125 µF) or no ASC exosomes to enable analysis of the proliferation-stimulating capacity of ASC exosomes. After an additional 30 min of incubation, which allowed for acetate hydrolysis of the CFSE, “Day 0” samples were detached with trypsin, washed twice in PBS, fixed in 3.7 % paraformaldehyde for 15 min, and stored at 4 °C until flow cytometric analysis. Samples were also prepared after 2 and 4 days of growth. The fixed cells were pelleted and stained in a 1:5000 dilution of Hoeschst 33342 (Thermo Scientific, Waltham, MA, USA) for 15 min. A maximum of 25,000 events were acquired on a MoFlo Astrios Cell Sorter (Beckman Coulter Inc., Fullerton, CA, USA) depending on the sample cell density, and the resulting data was analysed using the FlowJo 10 software (Tree Star Inc., Ashland, OR, USA).
Electroporation of ASC exosomes
To evaluate electroporation as a method of cargo loading into exosomes, ASC exosomes isolated by Total Exosome Isolation reagent or ultracentrifugation were resuspended in the electroporation buffers, cytomix electroporation buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM KPO4, 25 mM HEPES, 2 mM EGTA and 5 mM MgCl2, adjusted to pH 7.6 with KOH) or trehalose pulse medium (TPM; 50 mM trehalose (Sigma-Aldrich, Cat. No. T0167) in PBS). Electroporation was performed on a GenePulser II Electroporation System with capacitance extender (Bio-Rad, Hercules, CA, USA) using 0.4 cm cuvettes with aluminium electrodes. The volume of electroporated samples had a volume between 200 and 450 µL (containing a maximum of 100 µg total exosome protein) with settings of 400 V and 125 µF. The electroporated samples were subsequently analysed by NTA, AFM and TEM as described earlier.
RNA electroextraction assay
RNA release was measured to investigate whether electroporation induces pore formation in ASC exosomes. The maximal absorbance of RNA is at a wavelength of 260 nm, which differs from proteins having maximal absorbance at 280 nm. However, since proteins also absorb light at lower levels at 260 nm, and therefore able to shield light from the intravesicular RNA, any released RNA would be detected as an increase in absorbance at 260 nm (Hood et al. 2014). It follows that such an increase in RNA absorbance could only (in theory) be due to electroextraction (pore formation) of the ASC exosomes. Exosomes isolated by ultracentrifugation (100 µL) was diluted in TPM to a final volume of 450 µL. The absorbance of each sample was measured on a GENESYS 10 Spectrophotometer (Thermo Scientific) in quartz cuvettes (enabling absorbance measurements at 260 nm). Afterwards, each sample was transferred into a 0.4 cm electroporation cuvette with aluminium electrodes and electroporated at 400 V and 125 µF for 3 ms. The electroporated samples were then transferred back to the quartz cuvette for re-measurement of absorbance at 260 nm. The measured absorbance for each sample was noted, and the RNA release compared to an RNA standard curve. The data were analysed using a paired T test in Prism 5.
Results
Adipose-derived stem cells are efficient producers of extracellular vesicles (EV)
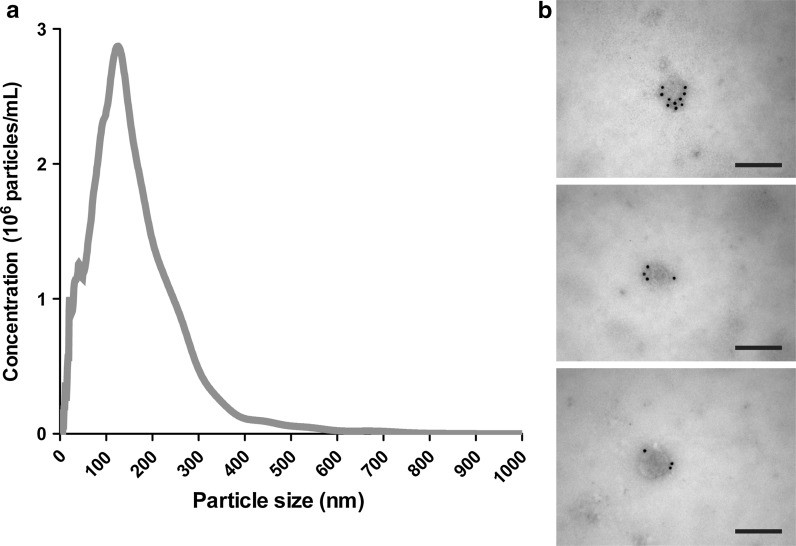
EVs (term used until identification of exosome characteristics) were isolated from ASCs either with the Total Exosome Isolation Kit or sequential ultracentrifugation. With NTA, the EVs secreted from these cells were examined with regards to the size distribution and particle yield. ASCs produced EVs within a size range normally defined as exosomes (30–120 nm), although EVs within a size range regarded as shedding microvesicles (approx. > 150 nm) could also be detected. It should be noted that the distinction between different types of EVs cannot solely be based on their size distribution, as some level of diameter overlap will be expected (Gudbergsson et al. 2015). The total yield of EVs from the CM was found to be high, averaging around 6 × 1010 particles/T175 culture flask (Fig. 1a). Transmission electron microscopy showed the presence of particles with a spherical shape with diameters ranging between 40 and 200 nm. Examination with immunogold labelling showed that the particles stained positive for a cocktail of antibodies against the exosome surface markers, CD9, CD63 or CD81 (Fig. 1b).
Fig. 1.
Characterization of ASC extracellular vesicles. a Nanoparticle tracking analysis of ASC extracellular vesicles isolated with Total Exosome Isolation Kit revealed a size distribution of the particles encompassing diameters of both exosomes and other extracellular vesicles. b Immunogold labelling using a cocktail of gold-conjugated antibodies targeting the exosome surface markers CD9, CD63 or CD81 (all panels) showed that the analysed extracellular vesicles were exosomes. Scale bars depict 200 nm
Electroporation of adipose-derived stem cell exosomes induces aggregation
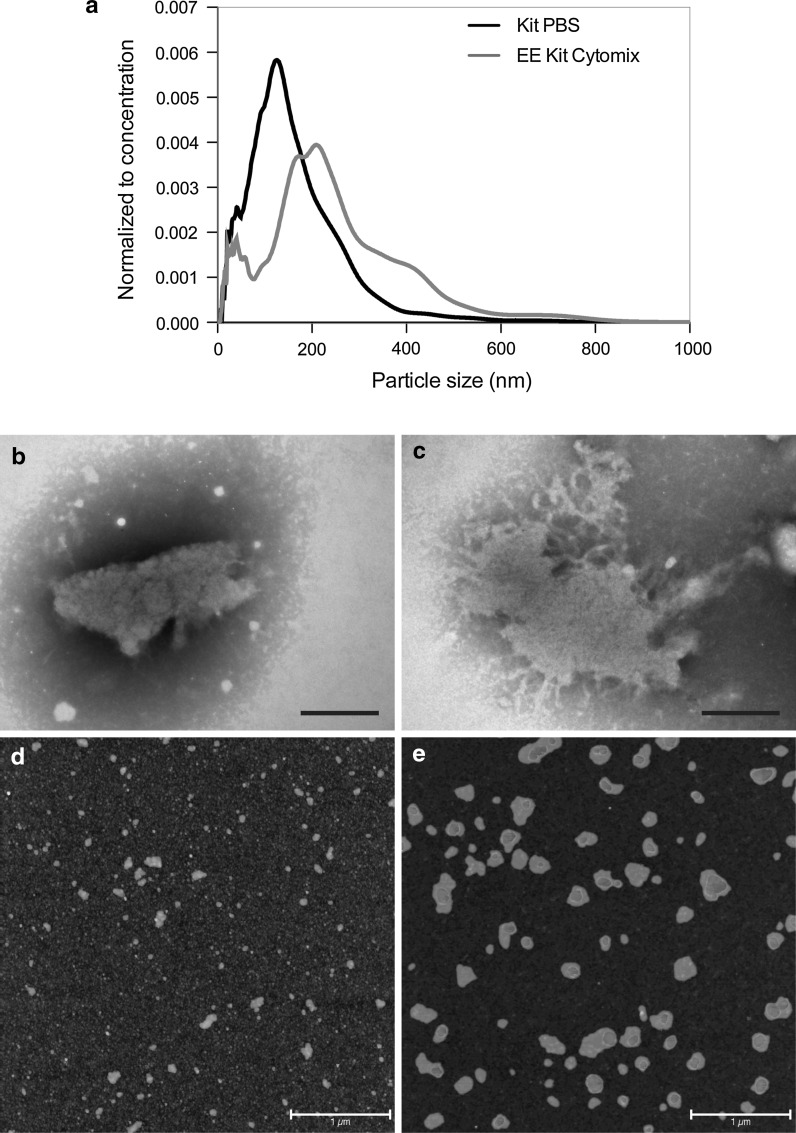
To investigate whether electroporation induced any aberrant effects, ASC exosomes were resuspended in cytomix buffer (an optimized cell transfection buffer), because it contains several constituents that should aid in avoiding changes to the exosomes or therapeutic cargo. The ASC exosomes were isolated with Total Exosome Isolation reagent followed by electroporation in the cytomix buffer. The electroporation mediated a shift in the size distribution of the exosomes, suggestive of a significant formation of aggregates following electroporation compared to non-electroporated exosomes (Kolmogorov–Smirnov test, p < 0.0001; Fig. 2a). The results from NTA were underscored by TEM imaging, which confirmed the presence of large aggregates, but also showed that the aggregates appeared to be membranous structures, as would be expected for exosome aggregates (Fig. 2b, c). AFM imaging also showed the presence of large aggregates in the electroporated samples (Fig. 2d, e). While the non-electroporated exosomes presented with diameters between 75 and 100 nm (Fig. 2d), the diameters of the electroporated exosomes were in a range of 100–500 nm (Fig. 2e). In addition, the high level of background observed in the non-electroporated exosome samples (possibly due to co-isolated protein aggregates) had disappeared after electroporation (Fig. 2d, e).
Fig. 2.
Electroporation induces aggregation of ASC exosomes. a In cytomix buffer, electroporation induced a right shift in the size distribution of the ASC exosomes. b, c Visualization of the electroporated samples with TEM showed the presence of large aggregates of membranous material. Scale bars depict 200 nm. d, e Atomic force microscopy analysis was performed on normal and electroporated ASC exosomes. Small particles of 75–100 nm could be observed in the normal exosome samples (d), whereas larger aggregate-like structures of 100–500 nm could be observed in the electroporated exosomes samples (e). All exosomes were isolated with the Total Exosome Isolation Kit. Scale bars depict 1 µm
The proliferation-stimulating properties of adipose-derived stem cell exosomes remain unaltered after electroporation
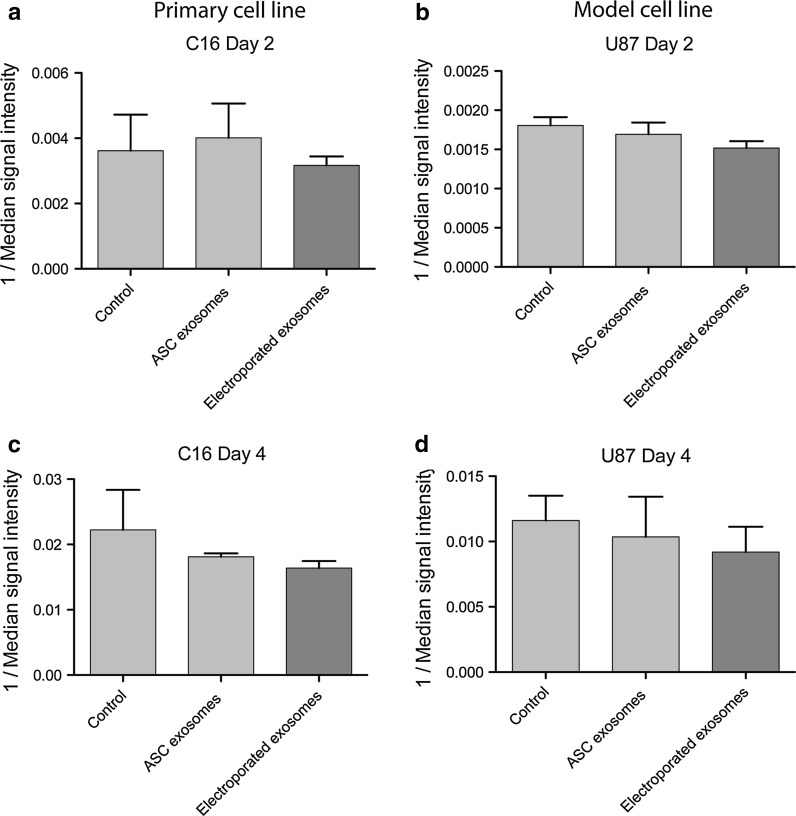
ASC exosomes (with and without electroporation) were applied to two GBM cell lines, U87 and C16. Both cell lines showed a reduction of median signal intensity over time, consistent with the known division of total CFSE signal during proliferation (Fig. 3). After 2 days of culture, no proliferation-inducing effects could be detected after administration of any of the ASC exosomes to any of the cell lines (Fig. 3a, b). The same pattern could be observed after 4 days of culture (Fig. 3c, d). In fact, a tendency towards ASC exosomes having a small inhibitory effect on the cancer cell proliferation after 4 days was seen, but this was not statistically significant. Furthermore, no significant difference in the effect on GBM cell proliferation could be detected between ASC exosomes and electroporated ASC exosomes.
Fig. 3.
Flow cytometry analysis of GBM cell proliferation stimulated by electroporated and non-electroporated ASC exosomes. Cells were stained with CFSE and incubated for 2 or 4 days. Only singlets were gated and analysed in the C16 and U87 samples. a, c Quantification of the median fluorescent signal intensity over time showed that neither ASC exosomes nor electroporated ASC exosomes stimulated proliferation in C16 cells on day 2 or 4. b, d ASC exosomes or electroporated ASC exosomes did not have any proliferation-inducing effect on U87 cell growth. No statistically significant difference could be observed between the effects of normal and electroporated ASC exosomes
Addition of trehalose pulse medium does not change the size of adipose-derived stem cell exosomes
To reduce the formation of aggregates during the electroporation process, we tested whether a trehalose-based electroporation buffer could keep the structural integrity of ASC exosomes after electroporation. Before electroporating ASC exosomes in the trehalose-based buffer, NTA was performed to observe whether the addition of trehalose to the samples induced any size-modifying effect in itself. This analysis did not detect any significant differences in the mean size of the particles or in the fraction of particles within the size range of exosomes isolated either by use of Total Exosome Isolation reagent or by sequential ultracentrifugation (Supplementary Figure 1).
Trehalose pulse medium decreases the formation of exosome aggregates after electroporation
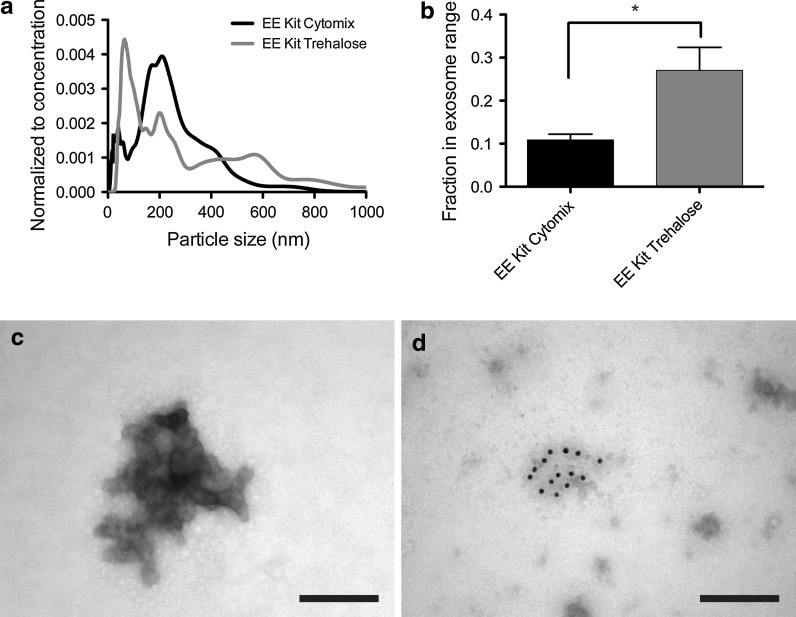
ASC exosomes (isolated with Total Exosome Isolation reagent) were resuspended in the TPM and electroporated. The resulting samples were analysed by NTA, which showed that electroporation in this medium gave rise to a large population of particles within the size range of exosomes (30–120 nm)—a population that had been lost with electroporation in cytomix buffer (Fig. 4a). Quantification of the fraction of particles within the exosome size range showed a statistically significant increase from 10 to 30 % with the change in electroporation buffer (Mann–Whitney test, p = 0.012; Fig. 4b). However, even though a large fraction of smaller particles had been retained, the samples still presented small peaks at higher diameters, implying a continued presence of exosome aggregates in the samples. This observation was confirmed with TEM, which could identify these larger aggregated structures together with some smaller particles, thus reflecting the NTA results (Fig. 4c). Immunogold labelling showed that some of these aggregates were positive for the exosome surface markers CD9, CD63 or CD81, suggesting that these aggregates were originally ASC exosomes (Fig. 4d).
Fig. 4.
Electroporation of ASC exosomes in trehalose pulse medium (TPM) reduces the level of aggregation. a Addition of trehalose to the electroporation buffer shifted the size distribution back to encompass a large population of particles within the exosome size range. b The fraction of particles within the exosome size range increased significantly after electroporation in TPM compared to cytomix buffer (Mann–Whitney test, p = 0.012). c TEM analysis of the ASC exosomes electroporated in TPM confirmed the sustained presence of larger aggregates in the resulting samples, although to a smaller extent compared to electroporation in cytomix buffer. d The aggregates stained positive with an antibody cocktail for the exosome surface markers, CD9, CD63 or CD81, suggesting them to be of exosomal origin. EE: Electroporated exosomes. Scale bars depict 200 nm
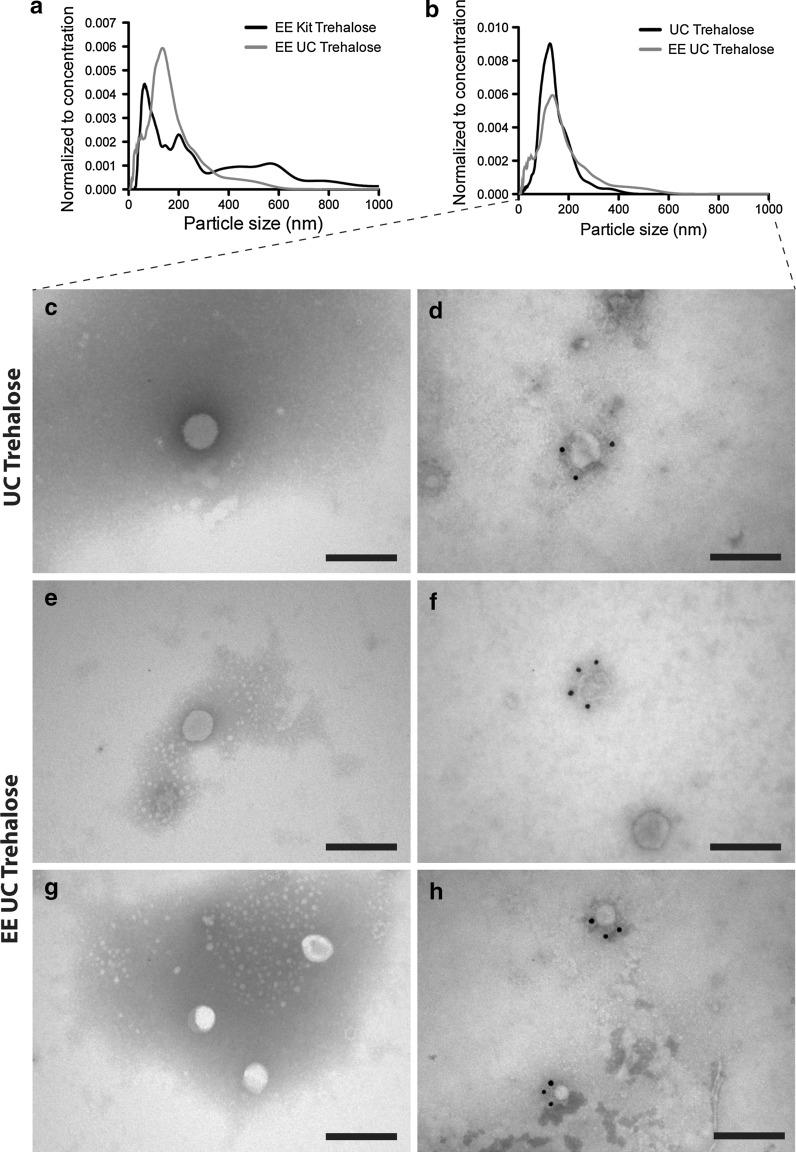
Electroporation of ultracentrifuged exosomes in trehalose pulse medium does not modulate the size distribution
Since sequential ultracentrifugation is known to yield exosome samples with higher purity than commercial isolation kits (Witwer et al. 2013; Gudbergsson et al. 2015), we wanted to assess whether such high purity could avoid the continued aggregation after electroporation. The ultracentrifuged ASC exosomes were resuspended in TPM, electroporated, and analysed by NTA. The results showed that when comparing these samples to the ones isolated with Total Exosome Isolation reagent, the ultracentrifuged and electroporated exosomes presented with a size distribution that did not include any larger aggregates (Fig. 5a). The fraction of particles within the exosome size range did not change significantly. TEM analysis of the samples showed that exosome aggregates could not be detected to the same extent, whereas smaller particles (exosomes) were abundant (Fig. 5c, d). These smaller particles were also positive for the exosome surface markers CD9, CD63 or CD81 (Fig. 5e, f). In addition, a comparison between the ultracentrifuged ASC exosomes electroporated in TPM and non-electroporated ultracentrifuged ASC exosomes was made. This comparison showed that electroporation did not induce any major changes to the size distribution of the exosomes isolated with ultracentrifugation (Fig. 5b).
Fig. 5.
Electroporation in trehalose pulse medium (TPM) retains the size distribution of ASC exosomes isolated by sequential ultracentrifugation. a The electroporated ASC exosomes isolated by ultracentrifugation (blue line) presented with a size distribution devoid of large aggregates compared to those isolated with Total Exosome Isolation Kit (black line). b Comparing the electroporated ASC exosomes isolated with sequential ultracentrifugation (blue line) and non-electroporated control exosomes (black line) revealed similar size distribution in TPM. c, d TEM showed that non-electroporated ASC exosomes with a round morphology and reactivity against CD9, CD63 or CD81 (antibody cocktail). e, g Electroporated ASC exosomes also presented with a round morphology and diameters around 100 nm. f, h The electroporated ASC exosomes were also positive with an antibody cocktail for the exosome surface markers, CD9, CD63, and CD81 (both panels). All ASC exosomes were isolated by sequential ultracentrifugation. Scale bars depict 200 nm. EE: Electroporated exosomes. UC: Exosomes isolated by sequential ultracentrifugation
RNA electroextraction as a measure of pore formation in the exosome membrane during electroporation
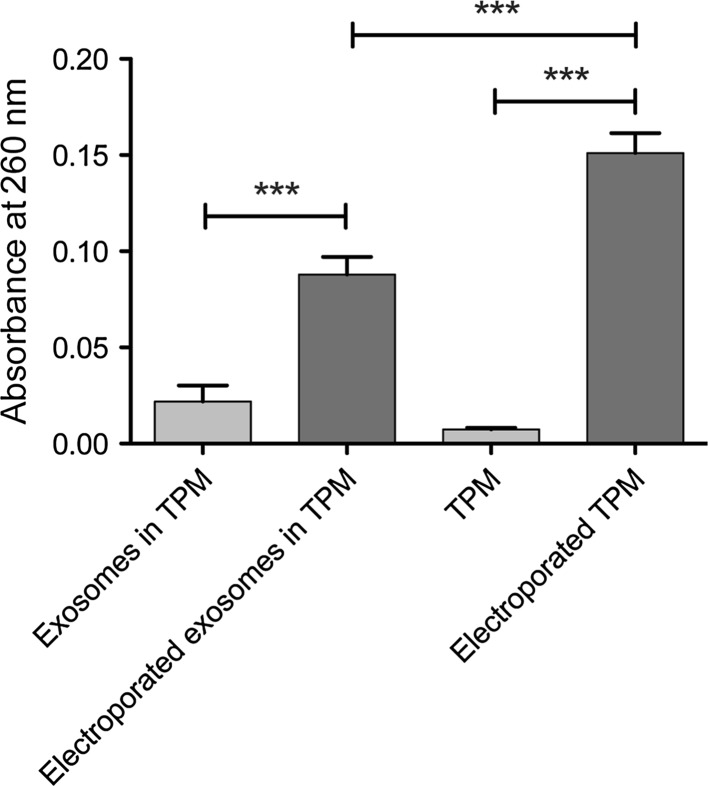
With optimized parameters for the electroporation to keep the structural integrity of the ASC exosomes, we wanted to assess whether these parameters also theoretically allowed for loading of therapeutic cargo into the exosome lumen. This was achieved by measuring the release of RNA (assessed by absorbance at 260 nm) after electroporation, hereby illustrating the formation of pores in the exosome membrane as recently suggested (Hood et al. 2014).
ASC exosomes were resuspended in TPM and the absorbance at 260 nm was measured before electroporation to allow for pairwise comparison. The samples were electroporated, and the absorbance of the samples was re-measured. The results from this experiment showed RNA release from the ASC exosomes with an almost absolute increase in absorbance observed for each sample (Fig. 6). The difference in absorbance before and after electroporation was found to be statistically significant (paired T test, p < 0.0001), suggesting that electroporation induced pore formation in the ASC exosomes. However, in subsequent control experiments, a much larger increase in absorbance at 260 nm could be observed when pure TPM was electroporated without the presence of exosomes, suggesting spectral interference of trehalose that overshadows the signal of RNA released from the exosome interior (Fig. 6). Electroporation of cytomix buffer did not produce the same increase in absorbance (data not shown). It therefore seems that when analysing electroporation in trehalose-containing buffer conditions, this method is inadequate.
Fig. 6.
RNA electroextraction on ASC exosomes as a measure of pore formation. RNA was measured as the absorbance at 260 nm. After electroporation, an increase in absorbance could be detected, suggesting the release of a stable amount of RNA from the ASC exosomes. Trehalose pulse medium without exosomes was also electroporated, yielding an even higher increase in absorbance at 260 nm. Quantification of the mean absorbance before and after electroporation showed a statistically significant increase in absorbance after electroporation (paired T test, ***p < 0.0001)
Discussion
Exosomes from ASCs are poorly characterized compared to exosomes from other types MSCs. ASCs are known to produce exosomes with a size distribution comparable to the one presented in the present study, and the exosomes are able to exhibit beneficial effects in Alzheimer’s disease and angiogenesis (Katsuda et al. 2013; Lopatina et al. 2014). Furthermore, they promote migration of breast cancer cells, and are known to communicate with GBM cells (Lee et al. 2013; Lin et al. 2013). Even though the combined effect of the results presented above may suggest a pro-tumorigenic profile, ASC exosomes were recently shown to mediate successful delivery of therapeutic miRNA to glioma cells (Lee et al. 2013). In the present study, ASC exosomes were shown to be devoid of any proliferation-stimulating properties after 4 days on two different GBM cell lines, including a cancer stem cell line, underscoring their relevance with respect to safety in delivering therapeutics to GBM tumors. However, for ASC (or any other cell type’s) exosomes to be used in any sort of therapy, one would also need to include a thorough assessment of how the cell culture conditions affect the production and characteristics of the secreted exosomes (Gudbergsson et al. 2015). For example, several types of cell stress, such as hypoxia, cell activation and high glucose, are known to interfere with the released quantity of exosomes, but also with their composition of proteins and RNA, whereas the effects of other components of cell culture, like the surface structure of the culture dish, remain completely unknown (de Jong et al. 2012; King et al. 2012). Electroporation of the ASC exosomes did not mediate any adverse effects with respect to modulation of the proliferative capacity of GBM cells, but based on our data it cannot be ruled out that the electroporation could modulate other parameters related to the ASC exosome interplay with GBM cells.
Exosome-based drug delivery is a rapidly expanding field of research, which is reflected in the frequent release of new publications regarding this subject (Johnsen et al. 2014). One of the main issues being addressed in these papers is the way of loading the exosomes with therapeutic cargo and reaching superior therapeutic effects compared to synthetic liposomes (Smyth et al. 2015; Fuhrmann et al. 2015). As described in Johnsen et al. (2014), the method of electroporation has been questioned due to the fact that exosomes as well as siRNA cargo tend to aggregate in the process (Kooijmans et al. 2013; Johnsen et al. 2014). In the study by Kooijmans et al. (2013), siRNA was found to accumulate in the exosomes with an efficiency similar to that previously reported (Alvarez-Erviti et al. 2011; Kooijmans et al. 2013). However, by thorough analysis of the loaded siRNA, large aggregates were observed, suggesting that the loading efficiency quantified only by total siRNA fluorescence was erroneous, because the loaded siRNA would be devoid of any functionality (Kooijmans et al. 2013). One could therefore speculate that electroporation is inapplicable for the loading of therapeutic cargo, and that a method like transfection of the exosome donor cell would be more appropriate. However, for exosomes to be clinically relevant as drug delivery vehicles, and become a part of the concept of personalized medicine, there must be a high throughput element to allow for proper loading efficiency within a relevant time frame.
Electroporation has been used to load various types of exogenous cargo into exosomes. Even though one study suggested that electroporation is an inappropriate method for loading of interfering RNA molecules (as described above), such loading has been achieved in several studies, showing efficient knockdown of GAPDH, BACE1, MAPK, RAD51, and RAD52 (Alvarez-Erviti et al. 2011; Wahlgren et al. 2012; Shtam et al. 2013; Kooijmans et al. 2013). Mimics of miR-155 was also loaded into exosomes by electroporation (Momen-Heravi et al. 2014). Moreover, small molecule drugs like the chemotherapeutic compound, doxorubicin, were loaded into exosomes by electroporation, resulting in a decrease of doxorubicin-mediated side effects while still providing therapeutic efficacy in a breast cancer model (Tian et al. 2013). In the present study, optimization of the electroporation process was performed to ensure the structural integrity of the exosomes. Based on initial data showing that exosomes aggregated in the cytomix electroporation buffer, a new buffer system based on the disaccharide trehalose was introduced. The addition of trehalose to the electroporation buffer allowed the exosomes to keep their pre-electroporation diameter, which was reflected in the size distribution of the exosomes being unchanged after electroporation.
Trehalose is a disaccharide consisting of two glucopyranose subunits adjoined by an α-(1 → 1) linkage. It is found in organisms like insects, yeast and fungi; organisms that tend to accumulate trehalose under stressful conditions like anhydrobiosis, heat shock or osmotic stress to a greater degree than any other known sugars. Several studies have shown that trehalose could stabilize membranes and proteins under unfavourable conditions (Pereira et al. 2004). Based on these characteristics, trehalose has been examined for its favourable effects on human cells and biostructures undergoing stress, e.g. during freezing, lyophilisation and electroporation (Mussauer et al. 2001; Crowe 2007). For example, trehalose was found to increase the recovery rate of lyophilized liposomes in the size range of 50–100 nm, whereas smaller (25 nm) or larger (200–400 nm) liposomes could not be recovered to the same extent. Interestingly, the best recovered size range of liposomes reflects the size range of exosomes, and therefore, trehalose may be an appropriate additive for the storage buffer of exosomes, since the results of the present study showed that the exosome size range was unaffected by this additive (Crowe and Crowe 1988; Chen et al. 2010). Moreover, the recovery and viability of cells subjected to electroporation for transfection was significantly higher when performed in a trehalose-containing buffer (Mussauer et al. 2001).
The mechanism by which trehalose interacts with a phospholipid bilayer membrane is still debated. Evidence shows that trehalose is able to replace water at the membrane to some extent and associate with the head groups of the phospholipids. This is proposed to keep the spacing between the individual lipids during dehydration, possibly explaining the favourable effects of trehalose in lyophilisation. Several studies have shown that trehalose is able to form hydrogen bonds with the phosphate groups of membranes, hereby mediating the replacement of water molecules—a process that is distinct from monosaccharides in stressful conditions (Pereira et al. 2004; Villarreal et al. 2004; Pereira and Hünenberger 2006; Kapla et al. 2015). Other studies support another hypothesis, where trehalose acts in a vitrification process resulting in mechanical protection of the membrane (Koster et al. 1994; Kapla et al. 2013, 2015). Either way, trehalose is able to stabilize lipid membranes under stress, e.g. during electroporation, and to prevent fusion of lipid bilayers, which are favourable characteristics for cargo loading process as shown by this study and others (Pereira et al. 2004; Hood et al. 2014). Whether the results presented in this study and by others can be extrapolated onto other disaccharides needs further investigations (Hood et al. 2014).
Electroporation is most often used for transfecting cells with DNA vectors. For these vectors to be introduced into the cell cytoplasm, the membrane must allow for the creation of small openings. These small openings (pores) are around 1 nm in diameter, and are only present within the short time frame (millisecond range) of electroporation. The formation of pores is a result of the current in the solution, and of secondary effects such as heating, affecting the membrane integrity (Weaver 1993). Transport of genetic material in the opposite direction (out of cells) was shown as a way of extracting plasmids from cyanobacteria (Moser et al. 1995). In the present study, this approach was used to determine whether genetic material could be released from the exosomes during electroporation as a measure of pore formation. Intriguingly, the increase in absorbance measured after electroporation was almost absolute, suggesting that a specifically defined amount of RNA can be present inside an exosome. However, since our control experiments showed that a larger increase in absorbance could be observed by electroporating pure TPM without the presence of exosomes, it is up for debate whether this assay is useful for determining RNA release from exosomes in trehalose-containing buffers. We hypothesize that it may be necessary to include an additional isolation step after the electroporation, from which RNA quantification can be performed.
Conclusion
The field of exosome-based drug delivery is still rapidly expanding, and the increasing amounts of evidence suggest that the future will see a clinical perspective of this concept. In the current state, however, the available knowledge and the associated methodology are insufficient to reach maximum potential in a clinical setting. In this study, some of the problems in the methodology associated with exosome-based drug delivery were addressed with a particular focus on an application in GBM. Electroporation was found to induce aggregation of ASC exosomes, although this did not have any effects on the stimulating properties of ASC exosomes on GBM cell proliferation. The structural integrity of ASC exosomes could be maintained after electroporation in an optimized buffer system containing the disaccharide, trehalose. In addition, without electroporation, trehalose did not interfere with the particle size, suggesting that this sugar may be a relevant additive for the storage of exosomes. Lastly, the presence of trehalose did not seem to hinder the formation of pores in the membrane during electroporation when measuring the absorbance at 260 nm. However, whether the changes in absorbance stem solely from RNA release from the exosomes could not be unambiguously proven in our experiments, since we observed spectral interference from the TPM. Further studies are thus still needed to validate the dynamics of nucleic acid movement over the exosome membrane during electroporation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to acknowledge laboratory technician Rikke Sophie Holm Kristensen, Aalborg University for her excellent technical assistance. Furthermore, Andreas Rasmussen, Laboratory of Stem Cell Research, Aalborg University is acknowledged for his kind help and facilitation of electroporation. This work was supported by Spar Nord Fonden. Kasper Bendix Johnsen is supported by the Novo Scholarship Programme (Novo Nordisk, Denmark).
Abbreviations
- AFM
Atomic force microscopy
- ASC
Adipose-derived stem cells
- BBB
Blood–brain barrier
- CD
Cluster of differentiation
- CFSE
Carboxyfluorescein succinimidyl ester
- CM
Conditioned medium
- EE
Electroporated exosomes
- ESCRT
Endosomal sorting complex required for transport
- FCS
Fetal calf serum
- GBM
Glioblastoma multiforme
- miRNA
MicroRNA
- mRNA
Messenger RNA
- MSC
Mesenchymal stem cell
- MVB
Multivesicular bodies
- NTA
Nanoparticle tracking analysis
- PEG
Polyethylene glycol
- Pen/strep
Penicillin/streptomycin
- PTA
Phosphotungstic acid
- RNA
Ribonucleic acid
- siRNA
Small interfering RNA
- TEM
Transmission electron microscopy
- TPM
Trehalose pulse medium
- UC
Ultracentrifugation
References
- Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Bryniarski K, Ptak W, Jayakumar A, et al. Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J Allergy Clin Immunol. 2013 doi: 10.1016/j.jaci.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Han D, Cai C, Tang X. An overview of liposome lyophilization and its future potential. J Control Release. 2010;142:299–311. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Chen TS, Arslan F, Yin Y, et al. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. J Transl Med. 2011;9:47. doi: 10.1186/1479-5876-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM. Factors affecting the stability of dry liposomes. Biochim Biophys Acta. 1988;939:327–334. doi: 10.1016/0005-2736(88)90077-6. [DOI] [PubMed] [Google Scholar]
- Crowe JH. Trehalose as a “chemical chaperone”: fact and fantasy. Adv Exp Med Biol. 2007;594:143–158. doi: 10.1007/978-0-387-39975-1_13. [DOI] [PubMed] [Google Scholar]
- de Jong OG, Verhaar MC, Chen Y, et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:569. doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi SE, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013 doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Fuhrmann G, Serio A, Mazo M, et al. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J Control Release. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- Gudbergsson JM, Johnsen KB, Skov MN, Duroux M. Systematic review of factors influencing extracellular vesicle yield from cell cultures. Cytotechnology. 2015 doi: 10.1007/s10616-015-9913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Anal Biochem. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen KB, Gudbergsson JM, Skov MN, et al. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Kapla J, Wohlert J, Stevensson B, et al. Molecular dynamics simulations of membrane–sugar interactions. J Phys Chem B. 2013;117:6667–6673. doi: 10.1021/jp402385d. [DOI] [PubMed] [Google Scholar]
- Kapla J, Engström O, Stevensson B, et al. Molecular dynamics simulations and NMR spectroscopy studies of trehalose–lipid bilayer systems. Phys Chem Chem Phys. 2015;17:22438–22447. doi: 10.1039/C5CP02472B. [DOI] [PubMed] [Google Scholar]
- Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuda T, Tsuchiya R, Kosaka N, et al. Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes. Sci Rep. 2013;3:1197. doi: 10.1038/srep01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijmans SAA, Stremersch S, Braeckmans K, et al. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release. 2013 doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Koster KL, Webb MS, Bryant G, Lynch DV. Interactions between soluble sugars and POPC (1-palmitoyl-2-oleoylphosphatidylcholine) during dehydration: vitrification of sugars alters the phase behavior of the phospholipid. Biochim Biophys Acta. 1994;1193:143–150. doi: 10.1016/0005-2736(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Lee HK, Finniss S, Cazacu S, et al. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget. 2013;4:346–361. doi: 10.18632/oncotarget.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang S, Zhao RC. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol Cell Biochem. 2013 doi: 10.1007/s11010-013-1746-z. [DOI] [PubMed] [Google Scholar]
- Lopatina T, Bruno S, Tetta C, et al. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhance their angiogenic potential. Cell Commun Signal. 2014;12:26. doi: 10.1186/1478-811X-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F, Bala S, Bukong T, Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine. 2014 doi: 10.1016/j.nano.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D, Zarka D, Hedman C, Kallas T. Plasmid and chromosomal DNA recovery by electroextraction of cyanobacteria. FEMS Microbiol Lett. 1995;128:307–313. doi: 10.1111/j.1574-6968.1995.tb07541.x. [DOI] [PubMed] [Google Scholar]
- Munoz JL, Bliss SA, Greco SJ, et al. Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussauer H, Sukhorukov VL, Zimmermann U. Trehalose improves survival of electrotransfected mammalian cells. Cytometry. 2001;45:161–169. doi: 10.1002/1097-0320(20011101)45:3<161::AID-CYTO1159>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Ohno S-I, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microrna to breast cancer cells. Mol Ther. 2012 doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Lins RD, Chandrasekhar I, et al. Interaction of the disaccharide trehalose with a phospholipid bilayer: a molecular dynamics study. Biophys J. 2004;86:2273–2285. doi: 10.1016/S0006-3495(04)74285-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Hünenberger PH. Interaction of the sugars trehalose, maltose and glucose with a phospholipid bilayer: a comparative molecular dynamics study. J Phys Chem B. 2006;110:15572–15581. doi: 10.1021/jp060789l. [DOI] [PubMed] [Google Scholar]
- Rasmussen JG, Frøbert O, Pilgaard L, et al. Prolonged hypoxic culture and trypsinization increase the pro-angiogenic potential of human adipose tissue-derived stem cells. Cytotherapy. 2011;13:318–328. doi: 10.3109/14653249.2010.506505. [DOI] [PubMed] [Google Scholar]
- Shtam TA, Kovalev RA, Varfolomeeva EY, et al. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth T, Kullberg M, Malik N, et al. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–155. doi: 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2013 doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- Villarreal MA, Díaz SB, Disalvo EA, Montich GG. Molecular dynamics simulation study of the interaction of trehalose with lipid membranes. Langmuir. 2004;20:7844–7851. doi: 10.1021/la049485l. [DOI] [PubMed] [Google Scholar]
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wahlgren J, Karlson TDL, Brisslert M, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JC. Electroporation: a general phenomenon for manipulating cells and tissues. J Cell Biochem. 1993;51:426–435. doi: 10.1002/jcb.2400510407. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:18389. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Pilgaard L, Chase LG, et al. Defined xenogeneic-free and hypoxic environment provides superior conditions for long-term expansion of human adipose-derived stem cells. Tissue Eng Part C Methods. 2012;18:593–602. doi: 10.1089/ten.tec.2011.0592. [DOI] [PubMed] [Google Scholar]
- Yeo RWY, Lai RC, Zhang B, et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.