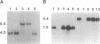Abstract
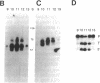
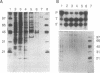
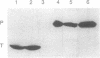
RNase P from the mitochondria of Saccharomyces cerevisiae was purified to near homogeneity > 1800-fold with a yield of 1.6% from mitochondrial extracts. The most abundant protein in the purified fractions is, at 105 kDa, considerably larger than the 14-kDa bacterial RNase P protein subunits. Oligonucleotides designed from the amino-terminal sequence of the 105-kDa protein were used to identify and isolate the 105-kDa protein-encoding gene. Strains carrying a disruption of the gene for the 105-kDa protein are viable but respiratory deficient and accumulate mitochondrial tRNA precursors with 5' extensions. As this is the second gene known to be necessary for yeast mitochondrial RNase P activity, we have named it RPM2 (for RNase P mitochondrial).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akaboshi E., Guerrier-Takada C., Altman S. Veal heart ribonuclease P has an essential RNA component. Biochem Biophys Res Commun. 1980 Sep 30;96(2):831–837. doi: 10.1016/0006-291x(80)91430-8. [DOI] [PubMed] [Google Scholar]
- Baer M., Altman S. A catalytic RNA and its gene from Salmonella typhimurium. Science. 1985 May 24;228(4702):999–1002. doi: 10.1126/science.2408335. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz M., Gold H., Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989 Apr;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Becker D. M., Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Haas E. S., James B. D., Hunt D. A., Liu J. S., Pace N. R. Phylogenetic analysis and evolution of RNase P RNA in proteobacteria. J Bacteriol. 1991 Jun;173(12):3855–3863. doi: 10.1128/jb.173.12.3855-3863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S. C., Pace B., Pace N. R. Characterization of ribonuclease P from the archaebacterium Sulfolobus solfataricus. J Biol Chem. 1990 Aug 5;265(22):12927–12932. [PubMed] [Google Scholar]
- Davis T. N., Thorner J. Isolation of the yeast calmodulin gene using synthetic oligonucleotide probes. Methods Enzymol. 1987;139:248–262. doi: 10.1016/0076-6879(87)39090-1. [DOI] [PubMed] [Google Scholar]
- Dihanich M. E., Najarian D., Clark R., Gillman E. C., Martin N. C., Hopper A. K. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doersen C. J., Guerrier-Takada C., Altman S., Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985 May 25;260(10):5942–5949. [PubMed] [Google Scholar]
- Doria M., Carrara G., Calandra P., Tocchini-Valentini G. P. An RNA molecule copurifies with RNase P activity from Xenopus laevis oocytes. Nucleic Acids Res. 1991 May 11;19(9):2315–2320. doi: 10.1093/nar/19.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R., Hopper A. K., Martin N. C. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5172–5176. doi: 10.1073/pnas.84.15.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Micrococcus luteus: conservation and variations among eubacteria. Gene. 1990 Sep 1;93(1):73–78. doi: 10.1016/0378-1119(90)90138-h. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Pace N. R. RNase P of Bacillus subtilis has a RNA component. J Biol Chem. 1980 Aug 25;255(16):7507–7509. [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Hanic-Joyce P. J., Gray M. W. Processing of transfer RNA precursors in a wheat mitochondrial extract. J Biol Chem. 1990 Aug 15;265(23):13782–13791. [PubMed] [Google Scholar]
- Hansen F. G., Hansen E. B., Atlung T. Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in Escherichia coli. Gene. 1985;38(1-3):85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- Hemmings B. A., Zubenko G. S., Hasilik A., Jones E. W. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Jan;78(1):435–439. doi: 10.1073/pnas.78.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Koerner T. J., Myers A. M., Lee S., Tzagoloff A. Isolation and characterization of the yeast gene coding for the alpha subunit of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 1987 Mar 15;262(8):3690–3696. [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Söll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Jul;5(7):1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N., Wesolowski D., Gold H., Bartkiewicz M., Guerrier-Takada C., McClain W. H., Altman S. Characteristics of ribonuclease P from various organisms. Cold Spring Harb Symp Quant Biol. 1987;52:233–238. doi: 10.1101/sqb.1987.052.01.028. [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Engelke D. R. Partial characterization of an RNA component that copurifies with Saccharomyces cerevisiae RNase P. Mol Cell Biol. 1989 Jun;9(6):2536–2543. doi: 10.1128/mcb.9.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamula M. J., Baer M., Craft J., Altman S. An immunological determinant of RNase P protein is conserved between Escherichia coli and humans. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8717–8721. doi: 10.1073/pnas.86.22.8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. C., Miller D. L., Underbrink K., Ming X. Structure of a precursor to the yeast mitochondrial tRNAMetf. Implications for the function of the tRNA synthesis locus. J Biol Chem. 1985 Feb 10;260(3):1479–1483. [PubMed] [Google Scholar]
- Martin N. C., Underbrink-Lyon K. A mitochondrial locus is necessary for the synthesis of mitochondrial tRNA in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4743–4747. doi: 10.1073/pnas.78.8.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983 Oct;34(3):911–917. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Morales M. J., Wise C. A., Hollingsworth M. J., Martin N. C. Characterization of yeast mitochondrial RNase P: an intact RNA subunit is not essential for activity in vitro. Nucleic Acids Res. 1989 Sep 12;17(17):6865–6881. doi: 10.1093/nar/17.17.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D. P., Schmidt F. J. Sequences encoding the protein and RNA components of ribonuclease P from Streptomyces bikiniensis var. zorbonensis. Gene. 1992 Aug 1;117(1):61–66. doi: 10.1016/0378-1119(92)90490-g. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt D. T., Haas E. S., Daniels C. J. The RNA component of RNase P from the archaebacterium Haloferax volcanii. J Biol Chem. 1991 Mar 25;266(9):5689–5695. [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 1985 Dec 1;4(12):3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H. H., Wise C. A., Clark-Walker G. D., Martin N. C. A gene required for RNase P activity in Candida (Torulopsis) glabrata mitochondria codes for a 227-nucleotide RNA with homology to bacterial RNase P RNA. Mol Cell Biol. 1991 Mar;11(3):1662–1667. doi: 10.1128/mcb.11.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O. Nucleotide sequence of a Proteus mirabilis DNA fragment homologous to the 60K-rnpA-rpmH-dnaA-dnaN-recF-gyrB region of Escherichia coli. Gene. 1990 Sep 1;93(1):27–34. doi: 10.1016/0378-1119(90)90131-a. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Stark B. C., Kole R., Bowman E. J., Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbrink-Lyon K., Miller D. L., Ross N. A., Fukuhara H., Martin N. C. Characterization of a yeast mitochondrial locus necessary for tRNA biosynthesis. Deletion mapping and restriction mapping studies. Mol Gen Genet. 1983;191(3):512–518. doi: 10.1007/BF00425771. [DOI] [PubMed] [Google Scholar]
- Wessel D., Flügge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984 Apr;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wise C. A., Martin N. C. Dramatic size variation of yeast mitochondrial RNAs suggests that RNase P RNAs can be quite small. J Biol Chem. 1991 Oct 15;266(29):19154–19157. [PubMed] [Google Scholar]
- Wise C., Martin N. C. Sequence analysis of Saccharomyces exiguus mitochondrial DNA reveals an RNase P RNA gene flanked by two tRNA genes. Nucleic Acids Res. 1991 Sep 11;19(17):4773–4773. doi: 10.1093/nar/19.17.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S., Gamulin V., Burkard U., Söll D. The RNA component of RNase P in Schizosaccharomyces species. FEBS Lett. 1990 Oct 1;271(1-2):189–193. doi: 10.1016/0014-5793(90)80403-6. [DOI] [PubMed] [Google Scholar]