Abstract
Helicobacter pylori is the dominant species of the human gastric microbiota and is present in the stomach of more than half of the human population worldwide. Colonization by H. pylori causes persistent inflammatory response and H. pylori-induced gastritis is the strongest singular risk factor for the development of gastric adenocarcinoma. However, only a small proportion of infected individuals develop malignancy. Besides H. pylori, other microbial species have also been shown to be related to gastritis. We previously reported that interspecies microbial interaction between H. pylori and S. mitis resulted in alteration of their metabolite profiles. In this study, we followed up by analyzing the changing protein profiles of H. pylori and S. mitis by LC/Q-TOF mass spectrometry to understand the different response of the two bacterial species in a multi-species micro-environment. Differentially-expressed proteins in mono- and co-cultures could be mapped into 18 biological pathways. The number of proteins involve in RNA degradation, nucleotide excision repair, mismatch repair, and lipopolysaccharide (LPS) biosynthesis were increased in co-cultured H. pylori. On the other hand, fewer proteins involve in citrate cycle, glycolysis/ gluconeogenesis, aminoacyl-tRNA biosynthesis, translation, metabolism, and cell signaling were detected in co-cultured H. pylori. This is consistent with our previous observation that in the presence of S. mitis, H. pylori was transformed to coccoid. Interestingly, phosphoglycerate kinase (PGK), a major enzyme used in glycolysis, was found in abundance in co-cultured S. mitis and this may have enhanced the survival of S. mitis in the multi-species microenvironment. On the other hand, thioredoxin (TrxA) and other redox-regulating enzymes of H. pylori were less abundant in co-culture possibly suggesting reduced oxidative stress. Oxidative stress plays an important role in tissue damage and carcinogenesis. Using the in vitro co-culture model, this study emphasized the possibility that pathogen-microbiota interaction may have a protective effect against H. pylori-associated carcinogenesis.
Keywords: Helicobacter pylori, Streptococcus mitis, LC/Q-TOF mass spectrometry, phosphoglycerate kinase (PGK), thioredoxin (TrxA)
Introduction
The human stomach was considered to be microbiologically sterile before the successful culturing of Helicobacter pylori from gastric biopsy tissue (Marshall and Warren, 1984). It was shown that gastritis and stomach ulcers in humans are caused by the Gram-negative, urease producing bacterium (Marshall and Warren, 1984). Later, it also became clear that this bacterium is a major risk factor in the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma (Kusters et al., 2006). In developing countries, 70–90% of the population is infected with H. pylori; while in developed countries, the prevalence of H. pylori is 25–50% (Solnick et al., 2003; Obiageli and Ivan, 2016).
Besides H. pylori, the human stomach can also contain transient oral, esophageal or intestinal bacteria and is highly dominated by Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (Dicksved et al., 2007). These microorganisms may either be permanent members of the gastric microbiota but not picked up due to limitation of conventional microbiological culturing methods or may be in transit in the stomach (e.g., together with food intake). However, a change of the physiological conditions of the stomach, as occurs during acid-reducing drug therapy, corpus atrophy or gastric cancer, provides an opportunity for foreign microbes to enter and colonize the stomach (Dicksved et al., 2007).
Streptococci are members of the normal intestinal flora of healthy individuals which exert antagonistic activities against many intestinal pathogens (Heczko et al., 2006). A strong correlation was found between the presence of Streptococcus salivarius and H. pylori where 83% of the S. salivarius positive biopsies also harbored H. pylori. S. salivarius is known to have urease activity which creates a less acidic environment and could further enhance the survival and incidence of H. pylori (Ryan et al., 2008). Hence, streptococci may potentially survive and develop in an acidic gastric environment as an indigenous microbiota of the gastric mucosa, which may in turn inhibit the colonization by H. pylori (Adolfsson et al., 2004; Johnson-Henry et al., 2004; Uziel et al., 2004). In our previous studies, we have shown that Streptococcus mitis can be isolated from human gastric tissue biopsies (Khosravi et al., 2014a) and co-culturing S. mitis and H. pylori released metabolites that induced H. pylori to transform into viable but non-culturable (VBNC) coccoidal form in vitro (Khosravi et al., 2014b). On the other hand, culturability of S. mitis in the co-culture was enhanced. In this current paper, we completed our analysis by analyzing the changing protein profiles of H. pylori and S. mitis to understand the different response of the two bacterial species in a multi-species micro-environment. While it is not surprising that H. pylori changes to coccoid in a multi-species micro-environment, the enhancement of S. mitis survival capability deserve further investigation to determine any potential pathogenic role of this bacterium in the human gastric environment in the presence of H. pylori.
Materials and methods
Bacterial strains
S. mitis ATCC 6249 and H. pylori NCTC 11637 (ATCC 43504) obtained from the American Type Culture Collection (ATCC, USA) were selected as model microorganisms to simulate interaction in a multispecies micro-environment. Culturing of both organisms was performed on chocolate agar plates supplemented with 7% horse blood and was incubated at 37°C in a humidified incubator with 10% CO2 for 3 days (Khosravi et al., 2014b).
Co-culture experiment
A bacterial co-culturing system was setup for this study in 12-well plates with a cell culture insert of 0.4 μm polyethylene terephathalate (PET) membrane (BD Biosciences, USA) that physically separate the two bacteria only allowing secreted compounds to penetrate as previously described (Khosravi et al., 2014b). Briefly, for the co-culture assay, 3 days old H. pylori and 1 day old S. mitis from chocolate-agar plates were used to make a suspension of OD600 ~0.02 (106–107 cfu/ml) and OD600 ~0.008 (105–106 cfu/ml) respectively in an enrichment medium of Brain heart infusion broth (BHI) supplemented with 0.4% yeast extract and 1% β-cyclodextrin. An aliquot of 2 ml suspension of H. pylori was distributed in each well of the 12-well plates. An aliquot of 0.5 ml suspension of S. mitis was added to the insert. The cultures were incubated at 37°C in a humidified incubator with 10% CO2 for 1–4 days. Experiments were carried out as independent biological triplicates.
Protein extraction
The ProteoSpin detergent-free total protein isolation kit (Norgen Biotek, Canada) with the Halt protease and phosphatase inhibitors cocktail (Thermo Scientific, USA) was used for the isolation and purification of total protein from bacteria pellet according to the manufacturer's instructions. The lysates were subsequently treated with 10 mM dithiothreitol (DTT; Bio-Rad, USA) at 37°C for 10 min and alkylated with 55 mM iodoacetamide (IAA; Bio-Rad) for 30 min at room temperature. The proteins in the sample were digested with 1:50 (trypsin: protein) of MS-grade Pierce trypsin protease (Thermo Scientific, USA) at 37°C overnight. The samples were desalted using a Pierce C-18 spin column (Thermo Scientific, USA) and dried to completeness in a refrigerated CentriVap centrifugal vacuum concentrator (Labconco, USA) before mass spectrometry analysis.
Protein profiling by LC/Q-TOF MS system
Tryptic peptides were analyzed on the 1260 Infinity HPLC-Chip System coupled with the 6540 UHD Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) LC/MS systems (Agilent, USA). For analysis, the injection volume was 2 μl of tryptic digest (200 ng/μl). The HPLC-Chip was the Large Capacity C18 Chip (G4240-6210), which comprised a 160 nL enrichment column and a 150 mm analytical column. HPLC-grade water with 0.1% formic acid and acetonitrile with 0.1% formic acid were used as mobile phases A and B respectively. HPLC-grade acetonitrile and formic acid were procured from Friendemann Schmidt (Australia) and Sigma (USA), respectively. Instrument settings were as described in Chan et al. (2015).
Data analysis
Mass spectrometric data were processed and analyzed using the Peaks software, version 7.5 (Bioinformatics Solutions Inc., Canada) for MS/MS-based identification and de novo sequencing. De novo sequencing was carried out with the default parameters, except that: (i) parent mass error tolerance was 1.5 Da, (ii) fragment mass error tolerance was 0.5 Da, (iii) trypsin as digestion enzyme, (iv) carbamidomethylation (+57.02 Da, C) as fixed modification, (v) oxidation (+15.99 Da, M) as variable modification, (vi) maximum variable post-translation modification allowed per peptide was three and (vii) H. pylori (strain NCTC 11637/ATCC 43504; 1633 proteins) and S. mitis (ATCC 6249; 1793 proteins) UniProtKB reference proteomes databases were used for identifications. Peptides were identified with PEAKS DB and filtered at 1% false discover rate (FDR). Proteins were filtered at 1 minimum unique peptide. Label-free quantification of protein abundances were estimated in each sample by correlation the average of the feature intensities of the three most highly responding peptides per protein.
Statistical analysis
Statistical analyses were performed using the IBM SPSS version 21.0 software. One-way ANOVA and Two-tailed student's t-test were performed. P-value of <0.05 was considered significant.
Results and discussion
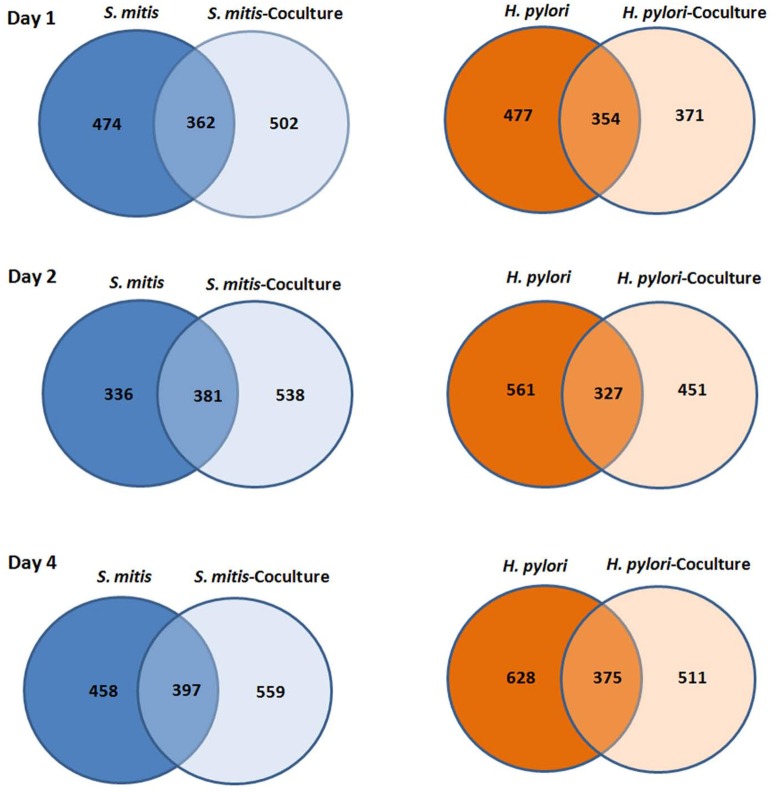
A total of 1514 H. pylori and 1414 S. mitis proteins - identified based on −logP ≥20, ≥1 unique peptide(s) (FDR <1%) and were detected in ≥2 of the triplicates - in both mono- and co-cultures (day 1, 2, and 4) are presented as Venn diagrams (Figure 1). Complete list of proteins identified can be found in the Supplementary Materials with confidence of identification and peptide data.
Figure 1.
Venn diagram of number of proteins H. pylori and S. mitis found in mono and co-cultures at day 1, 2, and 4.
H. pylori proteins that were found to be significantly different between mono-cultured and co-cultured H. pylori were mapped to 12 and six biological pathways respectively (Table 1). Proteins involve in RNA degradation, nucleotide excision repair, mismatch repair, and lipopolysaccharide (LPS) biosynthesis were relative more abundant in co-cultured H. pylori. On the other hand, proteins involve in citrate cycle, glycolysis/ gluconeogenesis, aminoacyl-tRNA biosynthesis, translation, metabolism, and cell signaling were less abundant in co-cultured H. pylori. This is consistent with the observation that in the presence of S. mitis, H. pylori was transformed to coccoid (Khosravi et al., 2014b). H. pylori coccoid has been demonstrated to have low metabolic enzymes (FBA, EDD, AcnB, FumC, OorA, and ICD as shown in Table 2B) but proteins involved in DNA biosynthesis remained high (Loke et al., 2016). Despite that H. pylori coccoid cannot be cultured in vitro, it has been reported that the coccoid had a stronger inhibitory effect on proliferation and weaker apoptotic effect than its spiral counterpart (Li et al., 2013), which suggest that the coccoid may be an important factor in gastric cancer progression. However, contradictory to the earlier report (Loke et al., 2016), proteins involve in epithelial cell signaling during H. pylori infection were reduced and those involved in LPS biosynthesis (Table 1) were increased in H. pylori coccoid induced by co-culturing with S. mitis. These differences may highlight differences between H. pylori coccoids induced by various means (prolonged culturing vs. co-culturing with S. mitis) and age of coccoids (3 months old vs. 4 days old). Furthermore, the role of H. pylori coccoids in a multi-species environment and its impact on gastric pathogenesis has not been fully assessed.
Table 1.
KEGG pathway and GO enrichment analysis of H. pylori proteins in mono- and co-cultures using functional annotation tool of DAVID.
| Sample group | Pathway | Count | Percentage | Proteins | Total | Pop Hits | Pop Total | Fold Enrichment | P-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bonferroni | Benjamini | FDR | |||||||||
| HP | hpp00020:Citrate cycle (TCA cycle) | 9 | 6.02 | AcnB, PorD, OorB, GltA, OorA, OorC, PorB, FumC, ICD | 67 | 15 | 2109 | 16.79 | 2.22E−05 | 2.22E−05 | 1.41E−04 |
| HP | hpj00970:Aminoacyl-tRNA biosynthesis | 7 | 5.26 | LeuS, ArgS, AspS, MetG, GatA, GLTX1, AlaS | 67 | 25 | 2109 | 8.81 | 0.02 | <0.01 | 0.10 |
| HP | hpj00010:Glycolysis / Gluconeogenesis | 6 | 4.51 | PorD, FBA, PorB, GAP_2, ENO, JHP_1030 | 67 | 16 | 2109 | 11.80 | 0.02 | <0.01 | 0.11 |
| HP | hpj00230:Purine metabolism | 6 | 3.76 | JHP_1168, ADK, UreA, UreB, GuaB, NDK | 67 | 34 | 2109 | 4.63 | 0.98 | 0.36 | 22.35 |
| HP | hpj05120:Epithelial cell signaling in Helicobacter pylori infection | 6 | 3.76 | CagY, UreA, UreB, VacA, CagE, ORF15 | 67 | 34 | 2109 | 4.63 | 0.98 | 0.36 | 22.35 |
| HP | hpp03010:Ribosome | 5 | 3.76 | RplI, RplN, RpsA, RplD, RplL | 67 | 49 | 2109 | 3.21 | 0.10 | 0.67 | 56.63 |
| HP | hpj00240:Pyrimidine metabolism | 4 | 3.01 | JHP_1168, NDK, TRXB_2, PyrF | 67 | 35 | 2109 | 3.60 | 1 | 0.70 | 70.95 |
| HP | hpj00030:Pentose phosphate pathway | 3 | 2.26 | FBA, EDD, TktA | 67 | 13 | 2109 | 7.26 | 0.10 | 0.67 | 54.023 |
| HP | hpj00250:Alanine, aspartate and glutamate metabolism | 3 | 2.26 | GlmS, AspB, AspA | 67 | 13 | 2109 | 7.26 | 0.10 | 0.67 | 54.023 |
| HP | hpj00480:Glutathione metabolism | 3 | 2.26 | PepA, GGT, ICD | 67 | 6 | 2109 | 15.74 | 0.93 | 0.28 | 15.46 |
| HP | hpj00620:Pyruvate metabolism | 3 | 2.26 | PorD, PpsA, PorB | 67 | 12 | 2109 | 7.87 | 0.10 | 0.65 | 48.72 |
| HPc | hpj00240:Pyrimidine metabolism | 6 | 13.95 | PyrG, DnaN, TRXB_1, DnaX, RpoB, PNP | 24 | 35 | 2109 | 15.06 | <0.01 | <0.01 | 0.03 |
| HPc | hpj00230:Purine metabolism | 5 | 11.63 | GppA, DnaN, DnaX, RpoB, PNP | 24 | 34 | 2109 | 12.92 | 0.03 | <0.01 | 0.41 |
| HPc | hpj03018:RNA degradation | 4 | 9.30 | RNJ, PPK, RHO, PNP | 24 | 10 | 2109 | 35.15 | <0.01 | <0.01 | 0.13 |
| HPc | hpj03430:Mismatch repair | 3 | 6.98 | JHP_0847, DnaN, DnaX | 24 | 15 | 2109 | 17.58 | 0.51 | 0.11 | 10.59 |
| HPc | hpp00540:Lipopolysaccharide biosynthesis | 3 | 6.98 | KdsB, LpxA, LpxD | 24 | 19 | 2109 | 13.88 | 0.67 | 0.15 | 16.32 |
| HPc | hpj03420:Nucleotide excision repair | 2 | 4.65 | JHP_0847, UvrB | 24 | 9 | 2109 | 19.53 | 0.10 | 0.55 | 63.38 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; HP, H. pylori mono-culture; HPc, H. pylori and S. mitis co-culture. Count, the number of genes associated with this gene set; percentage, calculated by “gene associated with this gene set “/” total number of query genes;” total, the number of genes in query list mapped to any gene set in this ontology; pop hits, the number of genes annotated to this gene set on the background list; pop total, the number of genes on the background list mapped to any gene set in this ontology; fold enrichment, the ratio of the proportions on query genes and the background information which are associated with the gene set; Bonferroni, Bonferroni adjusted p-value; Benjamini, Benjamini adjusted p-value; FDR, FDR adjusted p-value.
Among proteins identified, 27 proteins satisfied the criteria to be selected for label-free quantification analysis using the Peaks software. In contrast to 23 proteins that were significantly different in expression level between mono- and co-cultured H. pylori (Table 2B), only 4 proteins were found to be significantly different between mono- and co-cultured S. mitis (Table 2A). This suggests that multi-species environment may have a greater impact on H. pylori than S. mitis.
Table 2A.
List of S. mitis proteins with significant expression difference in mono- and co-cultures.
| Protein | KEGG Pathway | Unique peptide | Avg. Mass | S. mitis monoculture | S. mitis co-culture | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 4 | Day 1 | Day 2 | Day 4 | Day 1 | Day 2 | Day 4 | ||||
| 50S ribosomal protein L13 (RplM) | smb03010:Ribosome | 2 | 16143 | 0 | 0 | 0 | 0 | 0 | 6.82E+03 | >0.05 | >0.05 | 2.62E−08 |
| UPF0297 protein RN80_02805 | – | 3 | 10227 | 5.15E+02 | 2.78E+02 | 0 | 1.73E+03 | 1.77E+03 | 8.30E+02 | 0.015 | 7.51E−04 | >0.05 |
| Phosphocarrier protein HPr (PtsH) | – | 7 | 8939 | 3.50E+04 | 9.07E+03 | 1.25E+05 | 4.52E+03 | 3.76E+04 | 1.91E+03 | >0.05 | 0.029 | 0.029 |
| Phosphoglycerate kinase (Pgk) | smb00010:Glycolysis/Gluconeogenesis | 3 | 41978 | 0 | 0 | 0 | 1.01E+03 | 8.57E+02 | 7.03E+01 | 0.028 | >0.05 | >0.05 |
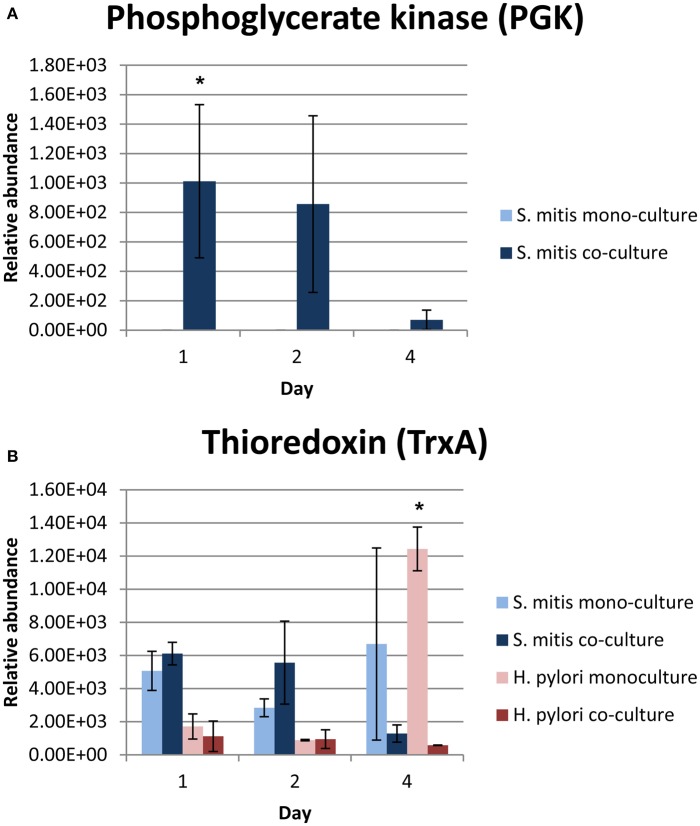
Among the differentially expressed proteins, phosphoglycerate kinase (PGK), which is required for ATP generation in both the glycolytic pathway of aerobes and the fermentation process of anaerobes (Yoshida and Tani, 1983), was only expressed by co-cultured S. mitis (Table 2A and Figure 2A). In addition, PGK is one of the predominant surface-associated proteins of streptococci, such as S. oralis (Wilkins et al., 2003) and group B streptococci (Hughes et al., 2002). Interestingly, sera directed against PGK was shown to protect neonatal animals from S. agalactiae infection suggesting that this protein may be essential for multiplication or adhesion of streptococci in vivo (Hughes et al., 2002). The expression of S. mitis PGK in the presence of H. pylori might have contributed to the enhanced survival of S. mitis, which was demonstrated earlier by our group (Khosravi et al., 2014b).
Figure 2.
Relative abundance of (A) PGK and (B) TrxA proteins in mono- and co-cultures. *Denote statistical significant differences with p-value = 0.028 (PGK) and <0.001 (TrxA) compared between mono- and co-cultures by 2-tailed one-way ANOVA.
Consistent with the reduction in abundance of enzymes involve in citrate cycle detected in the co-cultured H. pylori (Table 1), the expression of citrate cycle enzymes (AcnB, FumC, OorA, and ICD) were also found to be lower in co-cultured H. pylori (Table 2B), The citrate cycle is most sensitive to reactive oxygen species (ROS; Janero and Hreniuk, 1996). Thus, reduced level of expression of citrate cycle enzyme may indicate reduced oxidative stress response of H. pylori in the presence of S. mitis. This viewpoint is further supported by reduced expression of glutathione metabolism enzymes (ICD and PepA), thioredoxin (TrxA), flavodoxin (FldA), and thiol peroxidases (TPX and TsaA) in co-cultured H. pylori.
Table 2B.
List of H. pylori proteins with significant expression difference in mono- and co-cultures.
| Protein | KEGG Pathway | Unique peptide | Avg. Mass | H. pylori monoculture | H. pylori co-culture | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 4 | Day 1 | Day 2 | Day 4 | Day 1 | Day 2 | Day 4 | ||||
| JHP_0156 | – | 2 | 27461 | 2.88E+02 | 6.69E+02 | 1.74E+02 | 1.21E+01 | 3.00E+01 | 3.67E+00 | 3.16E−06 | 8.92E−05 | >0.05 |
| Fructose-bisphosphate aldolase (FBA) | hpj00010:Glycolysis / Gluconeogenesis; hpj00030:Pentose phosphate pathway |
12 | 33798 | 3.43E+02 | 6.43E+01 | 1.23E+04 | 9.77E+01 | 3.07E+00 | 4.73E+00 | >0.05 | >0.05 | 0.004 |
| Phosphogluconate dehydratase (EDD) | hpj00030:Pentose phosphate pathway | 4 | 66603 | 6.80E+02 | 8.46E+01 | 3.02E+03 | 1.93E+01 | 8.37E+00 | 0 | 6.19E−05 | >0.05 | 0.008 |
| Aconitate hydratase (AcnB) | hpj00020:Citrate cycle (TCA cycle) | 11 | 92742 | 1.05E+03 | 2.02E+03 | 4.26E+03 | 5.37E+01 | 7.87E+01 | 0 | 0.046 | 8.61E−04 | 1.72E−04 |
| Fumarase (FumC) | hpp00020:Citrate cycle (TCA cycle) | 2 | 50920 | 0 | 0 | 1.98E+03 | 0 | 0 | 0 | >0.05 | >0.05 | 0.026 |
| 2-Oxoglutarate oxidoreductase subunit A (OorA) | hpp00020:Citrate cycle (TCA cycle) | 4 | 41573 | 5.16E+02 | 5.96E+01 | 2.94E+03 | 0 | 0 | 0 | >0.05 | >0.05 | 0.025 |
| Isocitrate dehydrogenase (ICD) | hpj00480:Glutathione metabolism; hpp00020:Citrate cycle (TCA cycle) |
7 | 47462 | 6.92E+02 | 7.67E+02 | 1.76E+03 | 9.23E+01 | 3.06E+00 | 0.00E+00 | >0.05 | 9.20E−04 | >0.05 |
| Aminopeptidase (PepA) | hpj00480:Glutathione metabolism | 4 | 54612 | 1.40E+02 | 2.56E+01 | 1.63E+03 | 3.04E+01 | 2.71E+01 | 3.33E+01 | >0.05 | >0.05 | 0.002 |
| Thiol peroxidase (TPX) | K11065 thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | 6 | 18262 | 1.77E+03 | 5.40E+02 | 2.90E+03 | 1.26E+02 | 1.61E+02 | 0 | >0.05 | >0.05 | 0.003 |
| Probable peroxiredoxin (TsaA) | K03386 peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15]; Exosome [BR:hpj04147] | 5 | 22259 | 1.11E+03 | 6.15E+02 | 3.51E+03 | 0 | 0 | 0 | 0.007 | >0.05 | 0.036 |
| Thioredoxin (TrxA) | K03671 thioredoxin 1; Chaperones and folding catalysts [BR:hpj03110] | 9 | 11855 | 1.71E+03 | 8.88E+02 | 1.24E+04 | 1.12E+03 | 9.47E+02 | 5.78E+02 | >0.05 | >0.05 | 9.98E−05 |
| Flavodoxin (FldA) | K03839 flavodoxin I | 9 | 17473 | 8.38E+03 | 4.88E+03 | 3.17E+04 | 3.25E+03 | 1.66E+03 | 6.83E+01 | 0.003 | 0.028 | 2.25E−05 |
| JHP_0216 | K03981 thiol:disulfide interchange protein DsbC [EC:5.3.4.1]; Chaperones and folding catalysts [BR:hpj03110]; Secretion system [BR:hpj02044] | 3 | 29490 | 2.82E+02 | 2.11E+02 | 5.16E+03 | 6.35E+01 | 1.31E+02 | 4.10E+01 | >0.05 | >0.05 | 0.009 |
| 70kDa chaperone (DnaK) | hpj03018:RNA degradation | 9 | 67122 | 3.03E+03 | 3.70E+03 | 4.75E+03 | 3.18E+02 | 0 | 0 | 2.15E−04 | 0.032 | 0.019 |
| JHP_0301 | K07226 heme iron utilization protein | 3 | 28584 | 5.35E+01 | 2.01E+01 | 1.44E+03 | 4.83E+00 | 0 | 0 | >0.05 | >0.05 | 0.037 |
| Response regulator (CheY) | hpj02020:Two-component system; hpj02030:Bacterial chemotaxis |
5 | 13926 | 5.01E+02 | 1.09E+03 | 2.79E+03 | 0 | 0 | 0 | >0.05 | >0.05 | 0.020 |
| Urease subunit B (UreB) | hpj05120:Epithelial cell signaling in Helicobacter pylori infection; hpj00230:Purine metabolism; hpj00220:Arginine biosynthesis |
8 | 61684 | 4.21E+03 | 1.30E+03 | 7.23E+03 | 1.04E+02 | 0 | 0 | 6.49E−04 | >0.05 | 2.39E−04 |
| Urease subunit A (UreA) | hpj05120:Epithelial cell signaling in Helicobacter pylori infection; hpj00230:Purine metabolism; hpj00791:Atrazine degradation |
7 | 26568 | 2.34E+03 | 4.23E+03 | 5.42E+03 | 1.79E+02 | 7.40E+01 | 7.83E+00 | 0.018 | >0.05 | 0.025 |
| 50S Ribosomal protein L7/L12 (RplL) | hpp03010:Ribosome | 2 | 13314 | 3.11E+03 | 1.19E+03 | 3.05E+03 | 0 | 0 | 0 | 1.90E−04 | 0.036 | 0.003 |
| Elongation factor Ts (TSF) | Translation factors [BR:hpj03012] | 2 | 39859 | 0 | 1.44E+02 | 3.55E+02 | 0 | 0 | 0 | >0.05 | >0.05 | 6.01E−04 |
| Elongation factor G (FusA) | Translation factors [BR:hpj03012] | 10 | 77127 | 3.02E+02 | 0 | 2.20E+03 | 3.07E+02 | 4.50E+01 | 9.61E+01 | >0.05 | 0.027 | 0.002 |
| Elongation factor Tu (TUF) | Translation factors [BR:hpj03012]; Exosome [BR:hpj04147] | 10 | 43730 | 4.51E+03 | 1.19E+03 | 9.60E+03 | 0 | 8.83E+00 | 0 | 0.002 | 0.036 | 0.014 |
| GTP-binding protein TypA/BipA homolog (TypA) | K06207 GTP-binding protein | 3 | 66649 | 9.77E+01 | 2.93E+01 | 2.45E+03 | 0 | 6.23E+00 | 0 | >0.05 | 0.026 | 0.023 |
The expression of thioredoxin (TrxA), a small redox-regulating protein that is involved in maintaining the thiol/ disulfide balance in both prokaryotes and eukaryotes (Holmgren, 1985), was significantly reduced in 4 days old co-cultured H. pylori (Figure 2B). This protein is essential for protecting bacteria, such as Bacillus subtilis (Scharf et al., 1998; Uziel et al., 2004), Bacteroides fragilis (Tally et al., 1975; Rolfe et al., 1997) and Salmonella species (Bjur et al., 2006), against oxidative stress for survival and replication. Interestingly, TrxA is also highly expressed in many cancers, including lung (Kim et al., 2003), cervix (Hedley et al., 2004), pancreatic (Han et al., 2002), colorectal (Raffel et al., 2003), hepatocellular carcinomas (Choi et al., 2002), gastric carcinomas (Grogan et al., 2000) and breast cancer (Cha et al., 2009). TrxA has been postulated to contribute toward cancer progression by playing crucial roles in maintaining cellular redox homeostasis and cell survival (Trachootham et al., 2008). Up-regulation of TrxA and related proteins has been postulated to present a dynamic redox change to drive proliferation and malignant progression of tumors (Karlenius and Tonissen, 2010). In the presence of S. mitis, the expression of H. pylori TrxA was reduced suggesting that S. mitis may potentially reduce the risk of H. pylori-associated gastric cancer development and/ or progression in the human stomach.
Alkylhydroperoxide reductase of H. pylori, which protects the bacterium from a hyperoxidative environment by reduction of toxic organic hydroperoxides, has been shown to function as a molecular chaperone for prevention of protein misfolding under oxidative stress (Chuang et al., 2006). This study highlights the importance of translation (elongation factors) and protein folding (chaperones) to H. pylori in response to oxidative stress. Thus, low level of expression of chaperones, such as TrxA, JHP_0216 and DnaK (aka 70 kDa chaperone; Table 2B), implies relatively low oxidative stress level in co-culture H. pylori. It has been shown that both bacterial factors and host inflammatory response causes oxidative stress on the gastric epithelium during H. pylori infection that may lead to apoptosis and tissue damage (Ding et al., 2007). Thus, low oxidative stress confers by H. pylori in a multi-species environment can be expected to be less pathogenic.
In conclusion, using S. mitis and H. pylori as model organism, data from this in vitro study suggest that in a multi-species setting, S. mitis may be able to benefit from cross-talking with H. pylori to enhancing its survival in the adverse gastric environment. Simultaneously, S. mitis may protect H. pylori from excessive oxidative stress. This in vitro co-culture model emphasizes the possibility that inter-species interaction may protect the host against bacterial-associated pathogenesis and carcinogenesis. However, this study is preliminary and the human gastric environment is highly complex and dynamic. Therefore, more evidences are required in order to fully understand the implication of H. pylori-gastric microbiota crosstalk and its impact on the development of gastroduodenal diseases in human.
Author contributions
Conceived and designed the experiments: YK, MFL, KLG, JV. Performed the experiments: YK. Analyzed the data: YK, MFL.
Funding
University of Malaya-Ministry of Higher Education (UM-MOHE) High Impact Research (HIR) grant (reference UM.C/625/1/HIR/MOHE/CHAN-02; account no. A000002-50001, “Molecular Genetics”).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01462
References
- Adolfsson O., Meydani S. N., Russell R. M. (2004). Yogurt and gut function. Am. J. Clin. Nutr. 80, 245–256. [DOI] [PubMed] [Google Scholar]
- Bjur E., Eriksson-Ygberg S., Åslund F., Rhen M. (2006). Thioredoxin promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 74, 5140–5151. 10.1128/IAI.00449-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha M. K., Suh K. H., Kim I. H. (2009). Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J. Exp. Clin. Cancer Res. 28:93. 10.1186/1756-9966-28-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. G., Loke M. F., Ong B. L., Wong Y. L., Hong K. W., Tan K. H., et al. (2015). Multiphasic strain differentiation of atypical mycobacteria from elephant trunk wash. Peer J. 3:e1367. 10.7717/peerj.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. H., Kim T. N., Kim S., Baek S. H., Kim J. H., Lee S. R., et al. (2002). Overexpression of mitochondrial thioredoxin reductase and peroxiredoxin III in hepatocellular carcinomas. Anticancer Res. 22, 3331–3335. [PubMed] [Google Scholar]
- Chuang M. H., Wu M. S., Lo W. L., Lin J. T., Wong C. H., Chiou S. H. (2006). The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. U.S.A. 103, 2552–2557. 10.1073/pnas.0510770103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J., Flöoistrup H., Bergström A., Rosenquist M., Pershagen G., Scheynius A., et al. (2007). Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl. Environ. Microbiol. 73, 2284–2289. 10.1128/AEM.02223-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. Z., Minohara Y., Fan X. J., Wang J., Reyes V. E., Patel J., et al. (2007). Helicobacter pylori infection induces oxidative stress and programmed cell death in human gastric epithelial cells. Infect. Immun. 75, 4030–4039. 10.1128/IAI.00172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan T. M., Fenoglio-Prieser C., Zeheb R., Bellamy W., Frutiger Y., Vela E., et al. (2000). Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum. Pathol. 31, 475–481. 10.1053/hp.2000.6546 [DOI] [PubMed] [Google Scholar]
- Han H., Bearss D. J., Browne L. W., Calaluce R., Nagle R. B., Von Hoff D. D. (2002). Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 62, 2890–2896. [PubMed] [Google Scholar]
- Heczko P. B., Strus M., Kochan P. (2006). Critical evaluation of probiotic activity of lactic acid bacteria and their effects. J. Physiol. Pharmacol. 9, 5–12. [PubMed] [Google Scholar]
- Hedley D., Pintilie M., Woo J., Nicklee T., Morrison A., Birle D., et al. (2004). Up-regulation of the redox mediators thioredoxin and apurinic/ apyrimidinic excision (APE)/Ref-1 in hypoxic microregions of invasive cervical carcinomas, mapped using multispectral, wide-field fluorescence image analysis. Am. J. Pathol. 164, 557–565. 10.1016/s0002-9440(10)63145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1985). Thioredoxin. Annu. Rev. Biochem. 54, 237–271. 10.1146/annurev.bi.54.070185.001321 [DOI] [PubMed] [Google Scholar]
- Hughes M. J., Moore J. C., Lane J. D., Wilson R., Pribul P. K., Younes Z. N., et al. (2002). Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 70, 1254–1259. 10.1128/IAI.70.3.1254-1259.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janero D. R., Hreniuk D. (1996). Suppression of TCA cycle activity in the cardiac muscle cell by hydroperoxide-induced oxidant stress. Am. J. Physiol. 270, C1735–C1742. [DOI] [PubMed] [Google Scholar]
- Johnson-Henry K. C., Mitchell D. J., Avitzur Y., Galindo-Mata E., Jones N. L., Sherman P. M. (2004). Probiotics reduce bacterial colonization and gastric inflammation in H. pylori-infected mice. Dig. Dis. Sci. 49, 1095–1102. 10.1023/B:DDAS.0000037794.02040.c2 [DOI] [PubMed] [Google Scholar]
- Karlenius T. C., Tonissen K. F. (2010). Thioredoxin and cancer: a role for thioredoxin in all states of tumor oxygenation. Cancers (Basel) 2, 209–232. 10.3390/cancers2020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi Y., Dieye Y., Loke M. F., Goh K. L., Vadivelu J. (2014b). Streptococcus mitis induces conversion of Helicobacter pylori to coccoid cells during co-culture in vitro. PLoS ONE 11:e112214. 10.1371/journal.pone.0112214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi Y., Dieye Y., Poh B. H., Ng C. G., Loke M. F., Goh K. L., et al. (2014a). Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. ScientificWorldJournal 2014:610421. 10.1155/2014/610421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Chae H. Z., Kim Y. J., Kim Y. H., Hwangs T. S., Park E. M., et al. (2003). Preferential elevation of Prx, I., and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell. Biol. Toxicol. 19, 285–298. 10.1023/B:CBTO.0000004952.07979.3d [DOI] [PubMed] [Google Scholar]
- Kusters J. G., van Vliet A. H. M., Kuipers E. J. (2006). Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19, 449–490. 10.1128/CMR.00054-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Han L., Chen J., Lin X., Chen H., She F. (2013). Proliferative and apoptotic effects of gastric epithelial cells induced by coccoid Helicobacter pylori. J. Basic Microbiol. 53, 147–155. 10.1002/jobm.201100370 [DOI] [PubMed] [Google Scholar]
- Loke M. F., Ng C. G., Vilashni Y., Lim J., Ho B. (2016). Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci. Rep. 6:26784. 10.1038/srep26784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. J., Warren J. R. (1984). Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1, 1311–1315. 10.1016/S0140-6736(84)91816-6 [DOI] [PubMed] [Google Scholar]
- Obiageli E. R., Ivan E. C. (2016). Prevalence of Helicobacter pylori and its associated peptic ulcer infection among adult residents of Aba, Southeastern Nigeria. Int. J. Curr. Microbiol. App. Sci. 5, 16–21. 10.20546/ijcmas.2016.506.002 [DOI] [Google Scholar]
- Raffel J., Bhattacharyya A. K., Gallegos A., Cui H., Einspahr J. G., Alberts D. S., et al. (2003). Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J. Lab. Clin. Med. 142, 46–51. 10.1016/S0022-2143(03)00068-4 [DOI] [PubMed] [Google Scholar]
- Rolfe R. D., Hentges G. J., Barrett J. T., Campbell B. J. (1997). Oxygen tolerance of human intestinal anaerobes. Am. J. Clin. Nutr. 30, 1762–1769. [DOI] [PubMed] [Google Scholar]
- Ryan K. A., Daly P., Li Y., Hooton C., O'Toole P. W. (2008). Strain-specific inhibition of Helicobacter pylori by Lactobacillus salivarius and other lactobacilli. J. Antimicrob. Chemother. 61, 831–834. 10.1093/jac/dkn040 [DOI] [PubMed] [Google Scholar]
- Scharf C., Riethdorf S., Ernst H., Engelmann S., Volker U., Hecker M. (1998). Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180, 1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick J. V., Chang K., Canfield D. R., Parsonnet J. (2003). Natural acquisition of Helicobacter pylori infection in newborn rhesus macaques. J. Clin. Microbiol. 41, 5511–5517. 10.1128/JCM.41.12.5511-5516.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Stewart P. R., Sutter V. L., Rosenblatt J. E. (1975). Oxygen tolerance of fresh clinical anaerobic bacteria. J. Clin. Microbiol. 1, 161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D., Lu W., Ogasawara M. A., Nilsa R. D., Huang P. (2008). Redox regulation of cell survival. Antioxid. Redox Signal. 10, 1343–1374. 10.1089/ars.2007.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel O., Borovok I., Schreiber R., Cohen G., Aharonowitz Y. (2004). Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186, 326–334. 10.1128/JB.186.2.326-334.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins J. C., Beighton D., Homer K. A. (2003). Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69, 5290–5296. 10.1128/AEM.69.9.5290-5296.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Tani K. (1983). Phosphoglycerate kinase abnormalities: functional, structural and genomic aspects. Biomed. Biochim. Acta 42, S263–S267. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.