Introduction
Microorganisms remain as one of the most important source of pharmaceutically important drugs (Berdy, 2005; Debbab et al., 2010; Waditee-Sirisattha et al., 2016). Among the bacteria domain, Streptomyces genus has received considerable attention by the Scientific community for its seemingly unmatched capability of producing useful bioactive metabolites. The Streptomyces genus was initially proposed by Waksman and Henrici (1943); as the largest genus of Actinobacteria, it is comprised of over 780 species with validly published names (http://www.bacterio.cict.fr/). These gram-positive bacteria have contributed remarkably in natural product discovery as they synthesize compounds with diverse chemical structures and biological activities such as anticancer, antibacterial, antioxidant, antifungal, and immunosuppresants activities (Berdy, 2005; Gallagher et al., 2010; Manivasagan et al., 2013; Ser et al., 2015a, 2016a).
Over the years, drug screening programs have been focusing on identification of terrestrial microorganisms and investigation of their bioactive potential (Burg et al., 1979; Marcus et al., 1999). However, these efforts have resulted in rediscovery of the known bioactive compounds. Thus, researchers begun to venture into new and/or underexplored areas in the hope of discovering novel, potent bioactive metabolites. The mangrove ecosystem represents one of the world's most dynamic environments that produces commercial forest products, supports coastal fisheries and protects coastlines (Alongi, 2008). Factors such as fluctuations in salinity and tidal gradient are believed to be the driving force for metabolic adaptations, which could in turn lead to production of valuable metabolites. In fact, the growing interest in bioactive potentials of mangrove-derived Streptomyces has been demonstrated by the isolation of Streptomyces pluripotens (Lee et al., 2014a), S. fradiae (Prakash et al., 2015), S. cheonanensis (Mangamuri et al., 2016), and S. malaysiense (Ser et al., 2016b).
For this study, Streptomyces sp. MUSC 125 was initially isolated from mangrove soil in the east coast of Peninsular Malaysia (Lee et al., 2014b). As an attempt to explore the antioxidant capacity of MUSC 125, metal-chelating assay was performed and discovered its potential in quenching ferrous ions with activity ranging from 10.02 to 51.14% (unpublished data). Thus, the strain was selected for whole genome sequencing to obtain further understanding on its genomic potential.
Materials and methods
Isolation and culture of strain MUSC 125
Streptomyces sp. MUSC 125 was isolated from Tanjung Lumpur mangrove forest located in the city of Kuantan, State of Pahang, in December of the year 2012 (Lee et al., 2014b). Purified cultures were maintained on ISP medium 2 slants at room temperature for short-term storage and as glycerol suspensions (20%, v/v) at −80°C for long-term storage.
Genome sequencing and bioinformatics analysis of MUSC 125
The genomic DNA of MUSC 125 was extracted with Masterpure™ DNA purification kit (Epicentre, Illumina Inc., Madison, WI, USA) followed by RNase (Qiagen, USA) treatment (Ser et al., 2015b). Quality of the extracted DNA was examined using NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit version 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Subsequently, DNA library was prepared using Nextera™ DNA Sample Preparation kit (Nextera, USA), while the library quality was validated by Bioanalyzer 2100 high sensitivity DNA kit (Agilent Technologies, Palo Alto, CA) prior to sequencing. The genome of strain MUSC 125 was sequenced on MiSeq platform with MiSeq Reagent Kit 2 (2 × 250 bp; Illumina Inc., Madison, WI, USA). Following that, the paired-end reads were trimmed and de novo assembled with CLC Genomics Workbench version 5.1 (CLC bio, Denmark). Gene prediction was performed using Prodigal version 2.6, whereas rRNA and tRNA were predicted using RNAmmer and tRNAscan SE version 1.21 (Lowe and Eddy, 1997; Lagesen et al., 2007; Hyatt et al., 2010). The assembly was uploaded for annotation to Rapid Annotation using Subsystem Technology (RAST) (Aziz et al., 2008).
Results
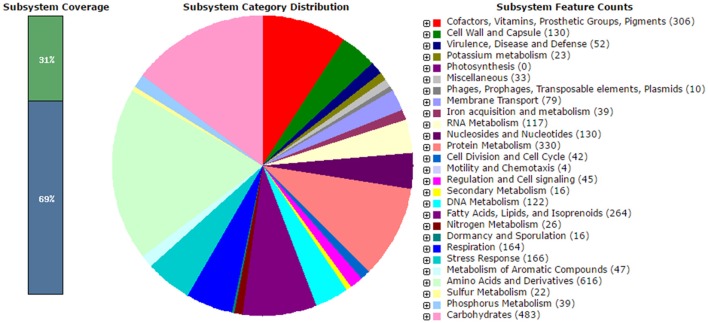
Sequencing of MUSC 125 genome using Illumina technology generated a total of 4,518,422 reads. After adapter trimming, the reads were de novo assembled into 164 contigs using CLC Genomics workbench. The genome size of MUSC 125 is 7,656,461 bp with G + C content of 70.00% (Table 1). The whole genome project of MUSC 125 was deposited at DDBJ/EMBL/GenBank under accession number JUIG00000000 and the version described in this paper is the first version (JUIG01000000). The analyses of the draft genome identified 5991 open reading frames (ORFs), 68 tRNAs, and 4 rRNA (5S, 16S, 23S rRNA). The RAST annotation has assigned these genes into 419 subsystems, with maximum number of genes associated with amino acids and derivatives metabolism (8.84%), followed by carbohydrates (6.93 %) and protein metabolism subsystems (4.73%) (Figure 1).
Table 1.
General features of Streptomyces sp. MUSC 125 draft genome.
| Streptomyces sp. MUSC 125 | |
|---|---|
| Genome size (bp) | 7,656,461 |
| G+C content (%) | 70.00 |
| Genome coverage | 94.0x |
| Contigs | 164 |
| Contigs N50 (bp) | 139,282 |
| Number of ORFs | 5991 |
| tRNA genes | 68 |
| rRNA genes (5S, 16S, 23S) | 4 |
| Bioproject ID | PRJNA261099 |
| Biosample ID | SAMN03070123 |
| Genome accession | JUIG00000000 |
Figure 1.
Annotation of MUSC 125 genome using Rapid Annotation using Subsystem Technology (RAST).
Direct link to deposited data and information to users
The genome sequence of Streptomyces sp. MUSC 125 (Biosample ID: SAMN03070123) can be accessed at NCBI using the accession number JUIG00000000. The genome project data are also available at GenBank under the genome Bioproject ID PRJNA261099. Users can download and use the data freely for research purpose only with acknowledgement to us and citing this paper as reference to the data.
Author contributions
The experiments, data analysis and manuscript writing were performed by H-LS and W-ST, while N-SA, W-FY, K-GC, B-HG, and L-HL. provided vital guidance and technical support. L-HL founded the research project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by PVC Award Grant (Project No. PVC-ECR-2016), External Industry Grant (Biotek Abadi Vote No. GBA-808813), MOSTI eScience funds (Project No. 06-02-10-SF0300) awarded to L-HL and MOSTI eScience funds (Project No. 02-02-10-SF0215) awarded to B-HG, and a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H- 50001-A000027 and No. A000001-50001) and PPP Grant (PG090-2015B) awarded to K-GC.
References
- Alongi D. M. (2008). Mangrove forests: resilience, protection from tsunamis, and responses to global climate change. Estuar. Coast. Shelf Sci. 76, 1–13. 10.1016/j.ecss.2007.08.024 [DOI] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdy J. (2005). Bioactive microbial metabolites. J. Antibiot. 58, 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., Hartman R., et al. (1979). Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367. 10.1128/AAC.15.3.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbab A., Aly A. H., Lin W. H., Proksch P. (2010). Bioactive compounds from marine bacteria and fungi. Microb. Biotech. 3, 544–563. 10.1111/j.1751-7915.2010.00179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K. A., Fenical W., Jensen P. R. (2010). Hybrid isoprenoid secondary metabolite production in terrestrial and marine actinomycetes. Curr. Opin. Biotechnol. 21, 794–800. 10.1016/j.copbio.2010.09.010 [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rodland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. H., Zainal N., Azman A. S., Eng S. K., Ab Mutalib N. S., Yin W. F., et al. (2014a). Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 64, 3297–3306. 10.1099/ijs.0.065045-0 [DOI] [PubMed] [Google Scholar]
- Lee L. H., Zainal N., Azman A. S., Eng S. K., Goh B. H., Yin W. F., et al. (2014b). Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 698178, 1–14. 10.1155/2014/698178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangamuri U., Muvva V., Poda S., Naragani K., Munaganti R. K., Chitturi B., et al. (2016). Bioactive metabolites produced by Streptomyces cheonanensis VUK-A from Coringa mangrove sediments: isolation, structure elucidation and bioactivity. 3 Biotech 6, 1–8. 10.1007/s13205-016-0398-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivasagan P., Venkatesan J., Sivakumar K., Kim S. K. (2013). Production, characterization and antioxidant potential of protease from Streptomyces sp. MAB18 using poultry wastes. BioMed Res. Int. 2013:496586. 10.1155/2013/496586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus N. H., Pomponi S., Rhines P., Tester P., Vena J. (1999). Marine derived pharmaceuticals and related bioactive compounds, in Monsoons to Microbes: Understanding the Oceanss Role in Human Health, eds Fiedler H. P., Brunter C., Riedlinger J., Bull A. T., Knutsen G., Goodfellow (Washington, DC: National Academies Press; ), 7186. [Google Scholar]
- Prakash S., Ramasubburayan R., Iyapparaj P., Arthi A. P. R., Ahila N. K., Ramkumar V. S., et al. (2015). Environmentally benign antifouling potentials of triterpene-glycosides from Streptomyces fradiae: a mangrove isolate. RSC Adv. 5, 29524–29534. 10.1039/C4RA15335A [DOI] [Google Scholar]
- Ser H. L., Palanisamy U. D., Yin W. F., Abd Malek S. N., Chan K. G., Goh B. H., et al. (2015a). Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4- dione, hexahydro- in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 6:854. 10.3389/fmicb.2015.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser H. L., Palanisamy U. D., Yin W. F., Chan K. G., Goh B. H., Lee L. H. (2016b). Streptomyces malaysiense sp. nov., a novel Malaysian mangrove soil actinobacterium with antioxidative activity and cytotoxic potential against human cancer cell lines. Sci. Rep. 6:24247. 10.1038/srep24247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser H. L., Tan L. T. H., Palanisamy U. D., Abd Malek S. N., Yin W. F., Chan K. G., et al. (2016a). Streptomyces antioxidans sp. nov., a novel mangrove soil actinobacterium with antioxidative and neuroprotective potentials. Front. Microbiol. 7:899. 10.3389/fmicb.2016.00899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser H. L., Tan W. S., Ab Mutalib N. S., Cheng H. J., Yin W. F., Chan K. G., et al. (2015b). Genome sequence of Streptomyces pluripotens MUSC 135T exhibiting antibacterial and antioxidant activity. Mar. Gen. 24, 281–283. 10.1016/j.margen.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Waditee-Sirisattha R., Kageyama H., Takabe T. (2016). Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiol. 2, 42–54. 10.3934/microbiol.2016.1.42 [DOI] [Google Scholar]
- Waksman S. A., Henrici A. T. (1943). The nomenclature and classification of the actinomycetes. J. Bacteriol. 46, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]