Abstract
Background:
Global DNA methylation has been reported to be associated with urothelial cell carcinoma (UCC) by studies using blood samples collected at diagnosis. Using the Illumina HumanMethylation450 assay, we derived genome-wide measures of blood DNA methylation and assessed them for their prospective association with UCC risk.
Methods:
We used 439 case–control pairs from the Melbourne Collaborative Cohort Study matched on age, sex, country of birth, DNA sample type, and collection period. Conditional logistic regression was used to compute odds ratios (OR) of UCC risk per s.d. of each genome-wide measure of DNA methylation and 95% confidence intervals (CIs), adjusted for potential confounders. We also investigated associations by disease subtype, sex, smoking, and time since blood collection.
Results:
The risk of superficial UCC was decreased for individuals with higher levels of our genome-wide DNA methylation measure (OR=0.71, 95% CI: 0.54–0.94; P=0.02). This association was particularly strong for current smokers at sample collection (OR=0.47, 95% CI: 0.27–0.83). Intermediate levels of our genome-wide measure were associated with decreased risk of invasive UCC. Some variation was observed between UCC subtypes and the location and regulatory function of the CpGs included in the genome-wide measures of methylation.
Conclusions:
Higher levels of our genome-wide DNA methylation measure were associated with decreased risk of superficial UCC and intermediate levels were associated with reduced risk of invasive disease. These findings require replication by other prospective studies.
Keywords: urothelial cell carcinoma, bladder cancer, DNA methylation, peripheral blood, biomarker, EWAS
Urothelial cell carcinoma (UCC) includes tumours of the transitional epithelium of the renal pelvis, the ureter, proximal urethra, and, predominantly, the urinary bladder. Urothelial cell carcinoma rarely occurs before the age of 40 years and is more common in men than women (Shariat et al, 2010). Smoking and occupation are the most clearly established environmental risk factors (Burger et al, 2013). Epigenetic changes such as DNA methylation are thought to play a major role in tumourigenesis through their influence on gene expression and genomic stability. Thus, epigenetic variations may serve as biomarkers of UCC risk (Marsit et al, 2010). DNA methylation is dependent on the one-carbon metabolism pathway and consists of the addition of methyl groups (CH3) to cytosines in CpG dinucleotides, forming 5-methyl cytosines (5-mC) via DNA methyltransferases (Brennan and Flanagan, 2012a). Ageing and environmental factors associated with the risk of UCC, such as smoking and occupational exposure to carcinogens, are reported to reduce global DNA methylation levels (Cho et al, 2007). DNA hypomethylation can potentially activate oncogenes and cause genetic instability if affecting repetitive genomic DNA elements that may lead to the initiation of carcinogenesis (Besaratinia et al, 2013). Global hypomethylation of DNA from peripheral blood collected years before diagnosis has been associated with the risk of several common cancers when measured with bisulphite sequencing assessment of % 5-mC content or surrogate measures of global DNA methylation (Woo and Kim, 2012; Brennan and Flanagan, 2012b; Mendoza-Perez et al, 2015). Recently, genome-wide measures of DNA methylation derived from the Illumina HumanMethylation450 assay (Illumina Inc., San Diego, CA, USA) have been prospectively associated with the risk of several cancers, including breast cancer (Severi et al, 2014; van Veldhoven et al, 2015) and B-cell lymphoma (Wong Doo et al, 2016). Although the Illumina HumanMethylation450 assay has limited genome coverage, it is fully annotated by genomic function, region, and CpG density, allowing interpretation of the functions of DNA methylation according to CpG location, and not exclusively as the total DNA 5-mC content (Jones, 2012).
The few earlier studies that have investigated associations between genome-wide DNA methylation and UCC have used different assessment methods, such as measuring repetitive sequences, predominantly long interspersed nucleotide elements (LINE-1), that act as surrogate markers for the whole genome. These studies (Moore et al, 2008; Marsit et al, 2010; Wilhelm et al, 2010; Cash et al, 2012; Ji et al, 2013; Andreotti et al, 2014; Tajuddin et al, 2014) are heterogeneous in terms of methodology, and all but one (Andreotti et al, 2014) are retrospective. Findings have been equivocal, with two Chinese studies (Cash et al, 2012; Ji et al, 2013) and an early Spanish hospital-based case–control study (Moore et al, 2008) reporting that global hypomethylation (measured with whole-genome bisulphite sequencing) in blood leukocyte DNA was potentially associated with increased UCC risk. A follow-up investigation of the Spanish case–control study (Tajuddin et al, 2014) reported that both low and high levels of LINE-1 methylation were associated with increased risk. Conversely, an analysis of pooled data from two cohort studies (Andreotti et al, 2014) suggested that high levels of global DNA methylation (LINE-1) are associated with increased risk of UCC, particularly for male smokers. Although a diverse range of other studies have investigated methylation status at specific CpG sites (Bilgrami et al, 2014; Li et al, 2014), in terms of environmental exposures (Salas et al, 2014; Rager et al, 2015) and prognosis (Kitchen et al, 2015; Lin et al, 2015), the relationship between different levels of methylation and risk of UCC, particularly in the context of the global levels of 5-mC across the genome, remains unclear.
Thus, our aim was to build on the limited existing evidence, and investigate prospectively the potential association between genome-wide DNA methylation and the risk of developing UCC using peripheral blood collected from participants in the Melbourne Collaborative Cohort Study (MCCS). Given the complex and diverse nature of UCC, our secondary aims were to investigate associations according to disease subtype, and to assess whether any relationship was modified by sex or lifestyle factors such as smoking and diet.
Materials and methods
Study sample
Study participants were selected from the MCCS, a prospective cohort study of 41 514 healthy adult volunteers (24 469 women) aged between 27 and 76 years (99.3% aged 40–69) when recruited between 1990 and 1994 (Giles and English, 2002). Peripheral blood was drawn at recruitment (1990–1994) or at subsequent follow-up (2003–2007). These samples were collected as dried blood spots (DBS) on Guthrie cards or as mononuclear cells or as buffy coats. Cases of UCC were identified by record linkage with the Victorian Cancer Registry that receives mandatory notification of all new cancer cases in Victoria, Australia. Incident UCC cases were identified up to 31 December 2012, using ICD-0–3 morphology codes 8120, 8122, 8130, or 8131. Diagnostic pathology reports were reviewed and classified according to the International Classification of Disease (ICD-O-3 WHO classification). Disease subtypes were defined according to behaviour, with invasive UCC including any tumour that had penetrated or invaded the basement membrane. Superficial UCC included papillary transitional/urothelial cell neoplasm of low malignant potential (PUNLMP) or carcinoma in situ (CIS) that was completely confined within the epithelium. Cases with uncertain behaviour type, including PUNLMPs (N=5), and with a topography code corresponding to vagina (C529), were excluded from the analyses. Subjects with any history of UCC before blood collection were excluded. Controls were individually matched to cases by sex, year of birth, country of birth, DNA source (DBS, mononuclear cells, buffy coats), and DNA collection period (baseline or follow-up). Each control had to have reached the age at diagnosis of their matched case without having developed UCC (incidence density sampling).
Ethics
Study participants provided informed consent in accordance with the Declaration of Helsinki. The study was approved by Cancer Council Victoria's Human Research Ethics Committee and performed in accordance with the institution's ethical guidelines.
DNA extraction and bisulphite conversion
The DNA was extracted from lymphocytes and buffy coat specimens, stored at −80 °C, using QIAamp mini spin columns (Qiagen, Hilden, Germany), and from dried blood spots collected onto Guthrie Card Diagnostic Cellulose filter paper (Whatman, Kent, UK) and stored in air-tight containers at room temperature using a previously reported method (Joo et al, 2013). Briefly, 21 blood spots of 3.2 mm diameter were punched from the Guthrie card and lysed in phosphate-buffered saline using TissueLyser (Qiagen). The resulting supernatant was processed using Qiagen mini spin columns according to the manufacturer's protocol. The DNA was quantified using the Quant-iT Picogreen dsDNA assay measured on the Qubit Fluorometer (Life Technologies, Grand Island, NY, USA), with a minimum of 0.75 μg DNA considered acceptable for methylation analysis. Bisulphite conversion was performed using EZ DNA Methylation-Gold single-tube kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. Post-conversion quality control was performed using SYBR Green-based quantitative PCR, an in-house assay, designed to determine the success of bisulphite conversion by comparing amplification efficiency of the test sample with unconverted negative high-quality DNA control. Test samples that amplified five or more quantitative cycles earlier than the negative control were assayed on the Infinium HumanMethylation450 BeadChip array. For all case–control pairs, the DNA was extracted at a similar point in time.
DNA methylation assay
Samples were processed in batches of 96 samples (8 Infinium HumanMethylation450 BeadChips per batch). In order to minimise potential plate and chip effects, samples from each matched case–control pair were plated to adjacent wells on the same BeadChip, with plate, chip, and position assigned randomly (Harper et al, 2013). The Infinium HumanMethylation450 BeadChip analysis was performed according to the manufacturer's instructions. A total of 200 ng of bisulphite converted DNA was whole genome amplified and hybridised onto the BeadChips. The TECAN automated liquid handler (Tecan Group Ltd, Mannedord, Switzerland) was used for the single-base extension and staining steps.
Data processing
Initial methylation data normalisation was performed in R programming software (R Core Team, 2015) using the minfi Bioconductor package (Gentleman et al, 2004). Subset-quantile within array normalisation was then used to correct the type I/type II probe bias (Maksimovic et al, 2012). Normalisation procedures were performed using the functions preprocessIllumina and preprocessSWAN in minfi (Aryee et al, 2014).
Samples were excluded if >5% CpG sites (CpGs) had a detection P-value of >0.01, regarded as missing values, whereas CpGs were excluded from further analysis if >20% of samples had missing values. As several technical replicate samples were included as part of quality control procedures, only the sample with the best overall detection P-value was kept in the analysis. After initial quality checks, the exclusion of 10 case–control pairs left 439 available for analysis. The DNA was obtained from DBS, mononuclear cells, and buffy coats for 178, 98, and 163 case–control pairs, respectively.
Genome-wide measures of DNA methylation
We excluded from the measures of genome-wide DNA methylation CpGs likely to be measured inaccurately, as described by Naeem et al (2014) based on a comparison with measures obtained using whole-genome bisulphite sequencing (Ziller et al, 2015). Thus, we excluded probes mapping to multiple genomic locations, probes containing single-nucleotide polymorphisms, and probes from repetitive elements (the latter being tested in a separate analysis as a surrogate measure of global DNA methylation (Brennan and Flanagan, 2012b)). We further restricted the analyses to the most reliable probes, defined as those with an intraclass correlation coefficient above 0.3, based on 129 technical replicate pairs from Guthrie cards or lymphocytes included in this and other MCCS nested case–control studies (Dugué et al, 2015). Genome-wide DNA methylation measures were computed across reliable CpGs of the Infinium HumanMethylation450 BeadChip (Supplementary Table 1).
Statistical analysis
Methylation β- and M-values were calculated using the library minfi in R (Aryee et al, 2014). M-values are defined as log2 (meth/unmeth), where meth and unmeth are the intensities of the methylated and unmethylated probes, respectively. We defined the genome-wide measure of DNA methylation for each individual as the median M-value across all included CpGs (Du et al, 2010). As the rank is conserved when converting β-values to M-values, our findings have similar interpretation as for the corresponding global β-value measure. Methylation β-values by patient characteristics are provided in Supplementary Table 2. Associations between genome-wide measures of DNA methylation and risk of UCC were assessed by fitting conditional logistic regression models and estimating odds ratios (OR) per s.d. increase of the genome-wide measure. All models were adjusted for other confounding variables (potentially associated with both UCC and methylation levels) such as smoking, socioeconomic status, alcohol consumption, body mass index, time since blood draw, folate intake, and vitamin B12 intake. These variables were defined at the time of blood draw using either baseline or follow-up questionnaires. Missing data (<0.5% in any of the confounders) were imputed with the median or mode of observed values of the corresponding variable.
Using the annotation file provided by Illumina, CpGs were classified according to their distribution across the genome, that is, their location in CpG islands, shores, shelves, or other, and location with regard to promoter regions (Price et al, 2013). Promoter regions were defined as loci spanning 1500 bp upstream of transcription start sites, within enhancer-associated regions or within the 5′ untranslated region. Promoter regions were further divided according to their CpG content and ratio, known to influence methylation profile and gene expression (Weber et al, 2007), and analysed according to promoter CpG density (high-CpG promoters (HC), intermediate-CpG promoters (IC), and low-CpG promoters (LC)).
We performed subgroup analyses, stratifying by sex, DNA source, period of blood sample collection, and aggressiveness of the tumour. The effect of time since blood collection (⩽5, 5–10 and, ⩾10 years) on associations with the genome-wide measure of DNA methylation including all CpGs was also assessed. Effect modification by smoking, sex, time since blood collection, and other variables was examined by testing the significance of their interaction with the genome-wide DNA methylation variable. The shape of the relation between DNA methylation and risk of UCC was examined by plotting ORs for quintiles of the more global measure of methylation.
Lastly, given the strong association of smoking with UCC risk and potentially with genome-wide measures of DNA methylation, we conducted sensitivity analyses according to the smoking status variable: first, by using a finer categorisation of smoking status and adding to the models other elements of dose (such as for current smokers less or more than 20 cigarettes per day; for former smokers having quit less or more than 15 years ago; and the age at starting smoking for ever smokers); second, by restricting the analysis to case–control pairs with same smoking status.
All analyses were carried out using R version 3.2.1 (Vienna, Austria).
Results
Altogether, 439 UCC cases were included in the analysis, including 193 (43.9%) invasive and 246 (56.1%) superficial cases. The median follow-up time was 6.3 years, interquartile range (IQR): 3.5 to 10.5 years. The UCC cases were more likely than controls to be current or former smokers at the time of blood collection (Table 1). Other potential confounders such as alcohol consumption, body mass index, folate and vitamin B12 intake, and socioeconomic status were not significantly associated with the risk of UCC (Table 1). After the removal of potentially less reliable probes, a total of 196 260 CpGs were included in the analysis. The overall proportions of probes within each genomic region were conserved (Supplementary Table 1).
Table 1. Characteristics of study participants and estimated ORs and 95% CIs for UCC associated with risk factors.
| Matching variables | Controls N=439 | Cases N=439 | |
|---|---|---|---|
| Age at blood draw | Median=65.6 | Median=65.3 | |
| IQR=60.3–69.2 | IQR=60.2–69.6 | ||
| DNA source | |||
| Dried blood spot | 178 (41%) | 178 (41%) | |
| PBMC | 98 (22%) | 98 (22%) | |
| Buffy coat | 163 (37%) | 163 (37%) | |
| Sex | |||
| Male | 335 (76%) | 335 (76%) | |
| Country of birth | |||
| Aus/NZ/UK | 302 (69%) | 302 (69%) | |
| Italy | 79 (18%) | 79 (18%) | |
| Greece | 58 (13%) | 58 (13%) | |
| Other risk factors (at blood draw) | Controls N=439 | Cases N=439 | Adjusted ORa (95% CI) |
| Smoking | |||
| Never | 185 (42%) | 128 (29%) | Ref. |
| Current | 56 (13%) | 78 (18%) | 2.1 (1.3–3.3) |
| Former | 198 (45%) | 233 (53%) | 1.9 (1.4–2.7) |
| Alcohol consumptionb | |||
| None | 102 (23%) | 112 (26%) | Ref. |
| Low | 273 (62%) | 253 (58%) | 0.8 (0.5–1.1) |
| Moderate | 39 (9%) | 41 (9%) | 0.9 (0.5–1.5) |
| High | 25 (6%) | 33 (7%) | 1.0 (0.5–2.8) |
| BMI | |||
| <25 kg m−2 | 135 (30%) | 113 (26%) | Ref. |
| 25–30 kg m−2 | 216 (51%) | 224 (51%) | 1.1 (0.8–1.5) |
| ⩾30 kg m−2 | 88 (19%) | 102 (23%) | 1.2 (0.8–1.8) |
| SES indicatorc | |||
| Quintile 1 | 61 (14%) | 78 (18%) | Ref. |
| Quintile 2 | 96 (22%) | 96 (22%) | 0.9 (0.6–1.3) |
| Quintile 3 | 83 (19%) | 84 (19%) | 0.9 (0.5–1.3) |
| Quintile 4 | 92 (21%) | 82 (19%) | 0.8 (0.5–1.2) |
| Quintile 5 | 107 (24%) | 99 (23%) | 0.8 (0.5–1.3) |
| Folate intaked | |||
| Quintile 1 | 85 (19%) | 89 (20%) | Ref. |
| Quintile 2 | 85 (19%) | 91 (21%) | 1.2 (0.7–1.9) |
| Quintile 3 | 88 (20%) | 89 (20%) | 1.0 (0.6–1.6) |
| Quintile 4 | 84 (19%) | 91 (21%) | 1.2 (0.7–2.0) |
| Quintile 5 | 97 (22%) | 79 (18%) | 0.9 (0.5–1.6) |
| Vitamin B12d | |||
| Quintile 1 | 89 (20%) | 85 (19%) | Ref. |
| Quintile 2 | 84 (19%) | 92 (21%) | 1.1 (0.7–1.7) |
| Quintile 3 | 93 (21%) | 84 (19%) | 1.0 (0.6–1.6) |
| Quintile 4 | 78 (18%) | 97 (22%) | 1.4 (0.9–2.2) |
| Quintile 5 | 95 (22%) | 81 (18%) | 0.9 (0.6–1.5) |
| Clinical variables | Cases N=439 | Adjusted ORa (95% CI) | |
| Time between blood draw and diagnosis | Median=6.3 | Per year increase | |
| IQR=3.5–10.4 | 1.1 (0.9–1.3) | ||
| Age at diagnosis | Median=73.3 | ||
| IQR=67.1–77.9 | |||
| Tumour invasiveness | |||
| Invasive | 193 (44%) | ||
| Superficial | 246 (56%) | ||
| Tumour grade | |||
| Grade 1 | 108 (25%) | ||
| Grade 2 | 104 (24%) | ||
| Grade 3 | 147 (33%) | ||
| Unknown | 80 (18%) |
Abbreviations: Aus=Australia; BMI=body mass index; CI=confidence interval; IQR=interquartile range; NZ=New Zealand; OR=odds ratio; PBMC=peripheral blood mononuclear cell; Ref=reference; SES=socioeconomic status; UCC=urothelial cell carcinoma.
Mutually adjusted ORs using a conditional logistic regression model with matching on age, DNA source, sex, and country of birth, and included covariates: smoking, alcohol consumption, body mass index, socioeconomic status, folate and vitamin B12 intakes, and time between blood draw and diagnosis.
Alcohol consumption was defined according to the Australian National Health and Medical Research Council categories as follows: None=0 g per day (males and females); Low=1–39 g per day (males) and 1–19 g per day (females); Moderate=40–59 g per day (males) and 20–39 g per day (females); High=60+ g per day (males) and 40+ g per day (females).
Index of Relative Socioeconomic Disadvantage score from the Socio Economic Indexes for Area defined by the Australian Bureau of Statistics, divided into quintiles (quintile 1 is the most disadvantaged).
Folate and vitamin intakes were computed using the Melbourne Collaborative Cohort Study (MCCS) 121-item Food Frequency Questionnaire.
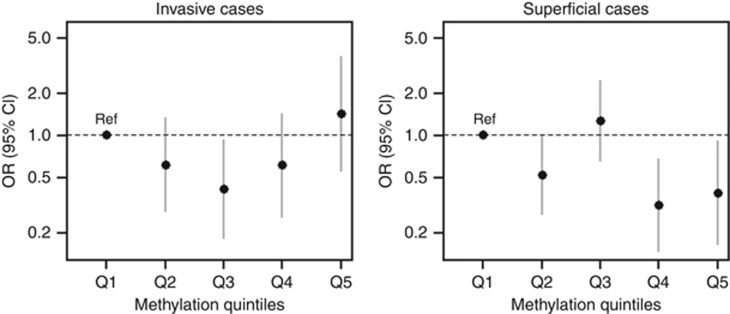
Although our genome-wide measure of DNA methylation based on all CpGs was not associated with the risk of UCC overall (Table 2), the risk of superficial UCC was significantly decreased for individuals with higher levels of DNA methylation (OR=0.71, 95% CI: 0.54–0.94; P=0.02). Lower ORs for superficial disease were consistently observed for genome-wide measures including CpGs of more regulatory regions: OR=0.82, 95% CI: 0.63–1.07 for gene promoters, with OR=0.75, 95% CI: 0.57–0.98 for intermediate CpG density promoters, OR=0.80, 95% CI: 0.63–1.02 for other regulatory gene regions (mostly enhancers), and OR=0.73, 95% CI: 0.56–0.95 for CpG shores, Table 3. The estimated relative risk of invasive UCC did not follow a linear trend with our genome-wide measure of methylation (OR=1.06; 95% CI: 0.79–1.43; P=0.70; Table 2), but rather intermediate levels of DNA methylation were associated with a significantly lower risk (Figure 1). On the contrary, genome-wide measures of DNA methylation for CpGs in non-regulatory regions tended to be associated with a decreased risk of invasive UCC, although the observed trends were not significant (non-regulatory regions: OR=0.76, 95% CI: 0.56–1.04; gene bodies: OR=0.82, 95% CI: 0.62–1.10; CpG shelves: OR=0.79, 95% CI: 0.59–1.06; Table 3). Measures at repetitive elements (filtered by the Naeem procedure) were highly correlated with measures at gene bodies (Spearman's ρ=0.95) and findings were virtually the same (invasive UCC: OR=0.84, 95% CI: 0.63–1.14; superficial UCC: OR=1.04, 95% CI: 0.80–1.34; data not shown).
Table 2. ORs for UCC and genome-wide measure of DNA methylation by disease subtypes and potential modifiers.
| ORa | 95% CI | P-value | P-value for heterogeneity | |
|---|---|---|---|---|
| Overall | 0.88 | 0.72–1.06 | 0.17 | |
| Invasiveness | ||||
| Invasive | 1.06 | 0.79–1.43 | 0.70 | |
| Superficial | 0.71 | 0.54–0.94 | 0.02 | 0.09 |
| Time since blood draw | ||||
| <5 Years | 0.77 | 0.54–1.08 | 0.13 | |
| 5–10 Years | 1.10 | 0.78–1.57 | 0.58 | |
| >10 Years | 0.72 | 0.43–1.20 | 0.20 | 0.26 |
| DNA source | ||||
| PBMC | 0.69 | 0.42–1.15 | 0.22 | |
| DBS | 0.93 | 0.61–1.42 | 0.74 | |
| Buffy coats | 0.81 | 0.61–1.08 | 0.16 | 0.67 |
| Blood collection period | ||||
| Baseline (1990–1994) | 0.89 | 0.68–1.16 | 0.38 | |
| Follow-up (2003–2007) | 0.78 | 0.57–1.07 | 0.13 | 0.29 |
| Sex | ||||
| Male | 0.89 | 0.72–1.11 | 0.31 | |
| Female | 0.66 | 0.36–1.22 | 0.18 | 0.50 |
| Smoking status at blood collection | ||||
| Never | 0.94 | 0.71–1.25 | 0.67 | |
| Current | 0.67 | 0.46–0.99 | 0.04 | |
| Former | 0.92 | 0.72–1.17 | 0.48 | 0.27 |
Abbreviations: CI=confidence interval; DBS=dried blood spot; OR=odds ratio; PBMC=peripheral blood mononuclear cell; UCC=urothelial cell carcinoma.
ORs per s.d. in median M-value were calculated using conditional logistic regression models, with matching on age, sex, ethnicity, type of sample, plate, and chip, and adjusting for smoking status, alcohol intake, body mass index (BMI), time since blood draw, folate intake, vitamin B12 intake, and socioeconomic status.
Table 3. OR for UCC and genome-wide measures of DNA methylation by CpG subgroup and disease subtype.
|
All cases |
Invasive cases |
Superficial cases |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N CpGs | ORa | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| CpG region | ||||||||||
| Islands | 71 728 | 0.97 | 0.81–1.15 | 0.72 | 1.04 | 0.79–1.36 | 0.77 | 0.89 | 0.69–1.15 | 0.39 |
| Shores | 51 833 | 0.87 | 0.73–1.05 | 0.15 | 1.02 | 0.77–1.36 | 0.86 | 0.73 | 0.56–0.95 | 0.02 |
| Shelves | 14 261 | 0.94 | 0.79–1.12 | 0.50 | 0.79 | 0.59–1.06 | 0.11 | 1.07 | 0.84–1.37 | 0.58 |
| None | 58 438 | 0.99 | 0.83–1.18 | 0.90 | 0.83 | 0.63–1.10 | 0.20 | 1.13 | 0.89–1.43 | 0.33 |
| Regulatory regions | ||||||||||
| Promoters | 94 695 | 0.93 | 0.77–1.11 | 0.41 | 1.04 | 0.78–1.38 | 0.79 | 0.82 | 0.63–1.07 | 0.13 |
| Other regulatory | 46 154 | 0.92 | 0.78–1.09 | 0.32 | 1.02 | 0.78–1.33 | 0.88 | 0.80 | 0.63–1.02 | 0.07 |
| Not regulatory | 55 411 | 0.92 | 0.77–1.11 | 0.39 | 0.76 | 0.56–1.04 | 0.08 | 1.05 | 0.81–1.35 | 0.71 |
| Within promoters | ||||||||||
| HCP | 51 208 | 1.01 | 0.84–1.22 | 0.90 | 1.03 | 0.76–1.39 | 0.85 | 0.99 | 0.76–1.28 | 0.93 |
| ICP | 21 983 | 0.89 | 0.74–1.08 | 0.24 | 1.06 | 0.79–1.41 | 0.71 | 0.75 | 0.57–0.98 | 0.03 |
| LCP | 21 504 | 0.97 | 0.82–1.15 | 0.73 | 0.84 | 0.64–1.11 | 0.22 | 1.06 | 0.84–1.35 | 0.60 |
Abbreviations: CI=confidence interval; HCP=high-density CpG promoter; ICP=intermediate-density CpG promoter; LCP=low-density CpG promoter; OR=odds ratio; UCC=urothelial cell carcinoma.
ORs per s.d. in median M-value were calculated using conditional logistic regression models, with matching on age, sex, ethnicity, type of sample, plate, and chip, and adjusting for smoking status, alcohol intake, body mass index (BMI), folate intake, vitamin B12 intake, and socioeconomic status.
Figure 1.
Urothelial cell carcinoma (UCC) risk according to the genome-wide measure of DNA methylation quintiles. Ref=lowest quintile of the genome-wide measure of DNA methylation.
The results of the assessment of effect modification by smoking, time between blood collection and cancer diagnosis, and sex are presented in Table 4. Although smoking did not seem to modify the association between genome-wide DNA methylation and UCC risk overall (P for heterogeneity, Phet=0.30), there was a significantly stronger association between our genome-wide measure of DNA methylation and risk of superficial UCC according to smoking status (current smokers: OR=0.47, 95% CI: 0.27–0.83; former smokers: OR=0.65, 95% CI: 0.44–0.94; never smokers: OR=0.99, 95% CI: 0.66–1.47; Phet=0.03). The OR estimates for superficial UCC varied by time since blood collection (Phet=0.07), but there was no consistent trend with time (OR=0.66, 95% CI: 0.44–0.98; OR=1.04, 95% CI: 0.65–1.65; and OR=0.51, 95% CI: 0.28–0.94 for <5 years, 5–10 years, and ⩾10 years, respectively). We did not find evidence of other interactions with our genome-wide measure of DNA methylation for either invasive or superficial disease; stronger associations were estimated for women (UCC overall: OR=0.66, 95% CI: 0.36–1.22; superficial UCC: OR=0.59, 95% CI: 0.24–0.96), but these were not significantly different to those for men. Because of small numbers, CIs widened considerably when UCC cases were further divided according to tumour aggressiveness (Supplementary Table 4).
Table 4. Genome-wide measure of DNA methylation and UCC risk by disease subtype: effect modification by smoking, time since blood collection, and sex.
|
All cases |
Invasive cases |
Superficial cases |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ORa | 95% CI | P | ORb | 95% CI | P | ORb | 95% CI | P | |
| Smoking status at blood collection | |||||||||
| Never | 0.94 | 0.71–1.25 | 0.65 | 0.90 | 0.59–1.37 | 0.63 | 0.99 | 0.66–1.47 | 0.95 |
| Current | 0.68 | 0.46–0.99 | 0.05 | 1.06 | 0.61–1.86 | 0.83 | 0.47 | 0.27–0.83 | 0.01 |
| Former | 0.91 | 0.72–1.17 | 0.47 | 1.21 | 0.85–1.73 | 0.30 | 0.65 | 0.44–0.94 | 0.02 |
| Heterogeneity test |
P=0.30 |
P=0.51 |
P=0.03 |
||||||
| Time since blood collection | |||||||||
| <5 Years | 0.77 | 0.55–1.09 | 0.14 | 1.00 | 0.64–1.60 | 0.97 | 0.66 | 0.44–0.98 | 0.04 |
| 5–10 Years | 1.10 | 0.78–1.57 | 0.58 | 0.93 | 0.59–1.47 | 0.62 | 1.04 | 0.65–1.65 | 0.89 |
| >10 Years | 0.72 | 0.43–1.20 | 0.21 | 1.72 | 0.87–3.40 | 0.12 | 0.51 | 0.28–0.94 | 0.03 |
| Heterogeneity test |
P=0.34 |
P=0.22 |
P=0.07 |
||||||
| Sex | |||||||||
| Male | 0.89 | 0.72–1.11 | 0.31 | 1.09 | 0.80–1.50 | 0.56 | 0.82 | 0.62–1.08 | 0.16 |
| Female | 0.66 | 0.36–1.22 | 0.18 | 1.43 | 0.66–3.09 | 0.36 | 0.59 | 0.24–0.96 | 0.05 |
| Heterogeneity test | P=0.50 | P=0.20 | P=0.19 | ||||||
Abbreviations: CI=confidence interval; OR=odds ratio; UCC=urothelial cell carcinoma.
ORs per s.d. in median M-value were calculated using conditional logistic regression models, with matching on age, sex, ethnicity, type of sample, plate, and chip, and adjusting for smoking status, alcohol intake, time since blood draw, body mass index (BMI), folate intake, vitamin B12 intake, and socioeconomic status.
Adjusting for the matching variables, and smoking status and time since blood draw only, because of relatively small numbers of observed cases.
Sensitivity analyses using a finer categorisation of the smoking variable or restricting the analysis to case–control pairs with the same smoking status did not materially change our results, either for our genome-wide measure of DNA methylation or for the analyses by CpG content and location relative to gene (not shown).
Discussion
Although our genome-wide measure of DNA methylation was not associated with risk of UCC overall, the risk of superficial UCC was significantly decreased for individuals with higher methylation levels (OR=0.71, 95% CI: 0.54–0.94). This association was significant after adjustment for several risk factors including smoking and was stronger for current smokers (OR=0.47, 95% CI: 0.27–0.83) and former smokers (OR=0.65, 95% CI: 0.44–0.94).
Although variable, there was no apparent trend in the association between hypomethylation and the risk of superficial UCC by time since blood draw. This may indicate that hypomethylation plays a causal role in superficial UCC carcinogenesis rather than being a marker of an already present malignancy, or potential circulating cell-free tumour DNA. We observed a lower risk of invasive UCC for individuals with intermediate levels of our genome-wide measure of DNA methylation. Such nonlinear associations are difficult to interpret but have sometimes been reported in the context of cancer risk (Chuang et al, 2011; Skinner et al, 2012), in particular in a study of LINE-1 methylation and bladder cancer risk (Tajuddin et al, 2014). Although the associations between DNA methylation levels and risk of UCC did not appear to vary substantially by genomic region, there was a trend of decreasing risk of invasive UCC in non-regulatory regions, and a decreasing trend of superficial UCC in regulatory regions.
Comparison with other studies
Our results are consistent with previous reports from studies investigating different tumour streams using the same methodology (prospective design) and assay (Illumina Infinium HumanMethylation450 BeadChip array) in that genome-wide measures of methylation may be associated with early stages of carcinogenesis (Severi et al, 2014; Wong Doo et al, 2016). It is more difficult to make a direct comparison across the existing literature relating specifically to risk of UCC because of the heterogeneous study designs and measures. Most previous studies on UCC risk have used PCR-based methylation detection at repetitive elements, mainly LINE-1. Our results were similar to findings from a US case–control study of 285 cases and 465 controls (Wilhelm et al, 2010) that reported that lower levels of LINE-1 methylation in peripheral blood were associated with higher risk of UCC, in particular for non-invasive disease (OR=1.94; 95% CI: 1.17–3.22) and for current smokers (OR=2.43; 95% CI: 1.46–4.03). They observed that the risk was higher for females that we also observed, but our estimates were not statistically significant (Phet=0.50 for the risk of UCC overall, and Phet=0.17 for superficial cases). One of the earliest studies on DNA methylation and bladder cancer, a large Spanish hospital-based, case–control study of 775 cases and 397 controls (Moore et al, 2008), also reported that genomic DNA hypomethylation as measured by cytosine methylation (% 5-mC) in leukocyte DNA was associated with an increased risk of bladder cancer. This was consistent with the results from two Chinese case–control studies (Cash et al, 2012; Ji et al, 2013) that reported an increased risk of UCC with lower LINE-1 levels using lymphocyte DNA from 510 cases and 528 controls, and with global hypomethylation measured in BLCA-4 repeat regions using blood leukocyte DNA from 312 cases and 361 controls, respectively. A more recent report from the large Spanish case–control study (Tajuddin et al, 2014) of 952 cases and 892 controls reported a nonlinear association with LINE-1 methylation, suggesting that both low and high levels of global DNA methylation were associated with risk of bladder cancer. Further stratified analyses of LINE-1 methylation levels according to disease aggressiveness (low- and high-grade superficial and muscle invasive bladder cancer) found similar results with no heterogeneity between phenotypes. As far as can be determined, there were no analyses according to disease subtype for the only other prospective study investigating the association between global DNA methylation and risk of bladder cancer (Andreotti et al, 2014). This combined study measured LINE-1 methylation using prediagnostic blood samples from two cohort studies (Prostate, Lung, Colorectal, and Ovarian cancer screening trial (PLCO) and Alpha-Tocopherol and Beta-carotene prevention study (ATBC)). The pooled analysis of these cohorts (Andreotti et al, 2014) found that higher levels of global DNA methylation were associated with increased risk of bladder cancer.
Study sample differences need to be taken into account when interpreting conflicting reports from the literature. It should be noted that the study by Andreotti et al (2014) comprised two different cohorts: one an all-male Finnish cohort restricted to ever-smokers (ATBC 391 cases/778 controls), and the other a cohort of both sexes including smokers and non-smokers (PLCO 299 cases/676 controls). This may help to explain the reported differences in DNA methylation levels between their two study samples and our results. Variation in DNA methylation levels between populations have been reported elsewhere and may reflect differences in lifestyle factors such as smoking and diet (Cash et al, 2012).
Effect modification by smoking status on the association between DNA methylation levels and risk of UCC has been consistently observed across multiple studies. We observed the strongest effects for risk of superficial UCC for current and former smokers. Similarly, the first published Spanish case–control study (Moore et al, 2008) reported current smokers in the lowest methylation quartile to be at the highest risk of bladder cancer. Andreotti et al (2014) also reported in their pooled analysis study that the effect was more pronounced for male smokers (highest vs lowest quartile, OR=2.03, 95% CI: 1.52–2.72). In contrast, a Chinese case–control study found that the association between hypomethylation and UCC risk was particularly strong for never smokers (lowest tertile OR=1.91; 95% CI: 1.17–3.13) (Cash et al, 2012).
Interpretation of the findings
Traditionally, global DNA methylation refers to the level of 5-mC content in a sample relative to total cytosine (unmethylated+5-mC) and has been assessed with various techniques over time (Kuo et al, 1980; Wagner and Capesius, 1981; Gama-Sosa et al, 1983; Antequera et al, 1984; Bestor et al, 1984; Fraga et al, 2002; Friso et al, 2002). These techniques provide accurate measures of global 5-mC, but are labour intensive and require large amounts of DNA. Given the limitations of these traditional approaches to measuring global 5-mC, several surrogate measures have been developed. The most popular method involves measuring DNA methylation following PCR amplification of repetitive DNA segments, including LINE (long interspersed numerical elements; mainly LINE-1) and SINE (short interspersed numerical elements; mainly Alu) (Yang et al, 2004) that together comprise upwards of 30% of human genomic DNA (Cordaux and Batzer, 2009). An increasingly popular surrogate measure of global DNA methylation is that calculated using data obtained from genome-wide DNA methylation profiling. This usually represents the mean/average or median DNA methylation value from many thousands, to several million, primarily unique CpG sites throughout the genome. The widely used Illumina Infinium HumanMethylation platform is enriched for gene-associated CpG sites, particularly those surrounding CpG-rich islands (Price et al, 2013).
There are several important caveats to using any surrogate markers of global 5-mC. Most genomic DNA methylation is found in repetitive elements, such as transposons and endogenous retroviruses (Schulz et al, 2006), but commonly used PCR-based repeat measures generally only assess methylation at a subset of desired LINE-1 or Alu elements because of the presence of a range of subfamilies of varying frequency and the large amount of sequence degeneration in each family over time (Lander et al, 2001). Although LINE-1 and Alu sequences account for ∼17% and 11% of the human genome (Lander et al, 2001), representing ∼12% and 25% of all CpG dinucleotides respectively (Schmid, 1996), only a subset of each can be interrogated by any given technique. Finally, the mechanism of regulation of DNA methylation at different classes of unique and repetitive DNAs vary and, therefore, measuring one ‘type' of methylation site is unlikely to be representative of global methylation levels – for example, LINE-1 methylation varies in some prostate cancers in the absence of any measurable change in overall genomic methyl-cytosine content (Schmid, 1996).
Although simplified approaches for global 5-mC DNA methylation estimation are now widely used as surrogates for total genomic DNA methylation, there is uncertainty about their comparability and the extent to which they reflect measurements of total methyl cytosine content of DNA. Numerous studies have tested the relevance of such measures to global 5-mC as measured by HPLC with varying results according to tissue and disease state of interest. The emerging picture is that no surrogate assay can accurately detect biologically important differences in global genomic DNA methylation in all instances, with this needing to be ascertained on a case-by-case basis (Weisenberger et al, 2005; Cho et al, 2007; Choi et al, 2007; Price et al, 2012), particularly in the context of human malignancy (Brennan and Flanagan, 2012b).
In our study, we used various genome-wide measures of DNA methylation derived from the HM450K assay and did not assess the correlation of our measures with global 5-mC measured, for example, with whole-genome bisulphite sequencing. Because the structure of the HM450K assay is skewed towards genes, we examined various genomic regions separately to obtain more specific genome-wide DNA methylation measures (Price et al, 2013). Measures including CpGs of more regulatory regions were associated with decreased risk of superficial UCC (in CpG shores: OR=0.73, in promoter regions: OR=0.82, in other regulatory regions: OR=0.80). Measures including CpGs of less regulatory regions, that is, in regions where DNA methylation is thought to help maintain genomic stability (Jones, 2012), were associated with a nonsignificant decrease of invasive UCC (CpG shelves: OR=0.79; not regulatory regions: OR=0.76; gene bodies: OR=0.82). We also computed genome-wide measures of DNA methylation at repetitive elements (24 847 CpG sites localising entirely within repetitive DNA sequences of the genome). This genome-wide measure was highly correlated with that measured in gene bodies and similar associations were observed (OR=0.84 for invasive UCC and OR=1.04 for superficial UCC). These gene body and repetitive elements measures, thought to be essential for maintaining genomic stability (Jones, 2012), were the closest to what is commonly referred to as ‘global DNA methylation' that we could obtain with the Illumina 450K assay.
Strengths and limitations
One of the major strengths of our study was its prospective design. Using blood samples collected before diagnosis allowed us to examine genome-wide measures of DNA methylation as potential biomarkers of risk. Measures of DNA methylation in retrospective studies may reflect molecular changes due to carcinogenesis, including treatment. An additional strength was the high CpG coverage of the Illumina HumanMethylation450 array that was not available at the time most previous studies were conducted. Our analysis was restricted to the most reliable CpGs, that is, those for which highest correlations with gold-standard methylation measurement methods are observed (Naeem et al, 2014), and with highest technical reproducibility (Bose et al, 2014; Dugué et al, 2015; Shvetsov et al, 2015). Other selection thresholds (e.g., ICC >0.5 and ICC >0.1) for the reliability of the probes included in our analysis did not meaningfully change the OR estimates (Supplementary Table 3).
We also had detailed information available on participants' characteristics collected at blood collection. Our design involved careful matching on age, DNA source, and ethnicity, and adjustment for various potential risk factors for UCC was made. In addition, and importantly, potential batch effects were corrected for by placing matched cases and controls next to each other on the same chip of the assay, with pairs at a random position, resulting in minimal technical bias (Harper et al, 2013).
There were also some limitations of our study, including the heterogeneity of the DNA source, although case–control pairs were matched on DNA source. We tested the feasibility of using these different sources of DNA in epigenetic studies and found them to be highly correlated and suitable for this purpose (Joo et al, 2013). Furthermore, we found no evidence that associations between methylation and risk of UCC differed by DNA source (Phet=0.69). Potential imbalances by imperfect representation of ethnicity when matching for country of birth may also have existed in our design, as we did not have information on ethnicity or genetic ancestry. In the MCCS, virtually all participants were of white European origin, born in Australia, the UK, New Zealand, Italy and Greece, between 1920 and 1955, minimising the possibility of influence by population stratification. In addition, country of birth was not associated with our genome-wide measures of DNA methylation (P=0.76, Supplementary Table 2).
Blood cell composition has been shown to vary substantially by age and may influence the measured level of DNA methylation, and hence other authors have considered the correction of epigenetic analyses for cell composition content to be warranted (Houseman et al, 2012; Jaffe and Irizarry, 2014). In our study, individuals were matched on age at diagnosis, and other factors that may be related to leukocyte composition. We also adjusted the results for smoking status, and age at blood collection, and hence confounding by blood cell composition is unlikely to have occurred with our study design. Although CIs widened after adjustment for cell composition, the point estimates remained very similar (Supplementary Table 4), and this may be explained by the points mentioned above as well as by the relatively homogeneous age at baseline in our cohort, age groups for which most cell types seem to display a similar cell composition (Jaffe and Irizarry, 2014). This is further illustrated in Supplementary Figure 1 that shows that the relationship between age and blood cell composition was virtually identical for cases and controls.
Because of small numbers we had inadequate statistical power to estimate with precision any associations between UCC and methylation for tumours diagnosed close to blood collection (69 invasive and 99 superficial tumours diagnosed <5 years after blood collection) or by aggressiveness of disease.
Finally, although studies have shown relatively good agreement between HM450K methylation measures and those obtained with more accurate and costly techniques such as whole-genome bisulphite sequencing, this independent validation was not made in our study. It should be noted, however, that we used the Naeem procedure to discard the possibility of systematic errors in the HM450K assay, and that the reliability of the genome-wide measures of DNA methylation defined in our study was high (ICC=0.8) based on a large number of technical replicate pairs (Dugué et al, 2016).
Future directions
The findings of our study confirm that UCC is not a single disease, but rather a heterogeneous group of ‘divergent clinical and pathological phenotypes' (Marsit et al, 2010) and our study points to epigenetic differences between superficial and invasive UCC. Further examination of DNA methylation in the context of detoxification processes, the one-carbon metabolism pathway, and gene–environment interactions may help to elucidate the mechanisms underlying differential DNA methylation between subtypes of disease, and individuals at risk of UCC (Aine et al, 2015a, 2015b). Because of the exploratory nature of our analyses, the translational possibilities of our findings are at present limited by lack of (1) independent validation of our measures by gold-standard methylation measurement with whole-genome bisulphite sequencing, and (2) replication of our results in other studies using a similar design. Our study may, nevertheless, generate more research focussed on region-specific hypomethylation and UCC risk.
Conclusion
Our study identified associations between a genome-wide measure of DNA methylation in peripheral blood collected several years before diagnosis and subsequent risk of superficial UCC. This association was strongest for smokers. For invasive UCC, the risk appeared to be lowest for individuals with intermediate DNA methylation levels. These findings need to be replicated by other studies of similar prospective design, and future investigations should focus on the underlying mechanisms that explain the differences in DNA methylation patterns for disease subtypes.
Acknowledgments
This study was made possible by the contribution of many people, including the original investigators and the diligent team who recruited the participants and who continue working on follow-up. We also express our gratitude to the many thousands of Melbourne residents who took part in the study and provided blood samples. This work was supported by the National Health and Medical Research Council project grant number 1043616.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Aine M, Eriksson P, Liedberg F, Hoglund M, Sjodahl G (2015a) On molecular classification of bladder cancer: out of one, many. Eur Urol 68(6): 921–923. [DOI] [PubMed] [Google Scholar]
- Aine M, Eriksson P, Liedberg F, Sjodahl G, Hoglund M (2015b) Biological determinants of bladder cancer gene expression subtypes. Sci Rep 5: 10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti G, Karami S, Pfeiffer RM, Hurwitz L, Liao LM, Weinstein SJ, Albanes D, Virtamo J, Silverman DT, Rothman N, Moore LE (2014) LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics 9(3): 404–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Tamame M, Villanueva JR, Santos T (1984) DNA methylation in the fungi. J Biol Chem 259(13): 8033–8036. [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10): 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A, Cockburn M, Tommasi S (2013) Alterations of DNA methylome in human bladder cancer. Epigenetics 8(10): 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Hellewell SB, Ingram VM (1984) Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol 4(9): 1800–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgrami SM, Qureshi SA, Pervez S, Abbas F (2014) Promoter hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. SpringerPlus 3: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose M, Wu C, Pankow JS, Demerath EW, Bressler J, Fornage M, Grove ML, Mosley TH, Hicks C, North K, Kao WH, Zhang Y, Boerwinkle E, Guan W (2014) Evaluation of microarray-based DNA methylation measurement using technical replicates: the Atherosclerosis Risk In Communities (ARIC) Study. BMC Bioinformatics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan K, Flanagan JM (2012a) Epigenetic epidemiology for cancer risk: harnessing germline epigenetic variation. Methods Mol Biol 863: 439–465. [DOI] [PubMed] [Google Scholar]
- Brennan K, Flanagan JM (2012b) Is there a link between genome-wide hypomethylation in blood and cancer risk? Cancer Prev Res (Phila) 5(12): 1345–1357. [DOI] [PubMed] [Google Scholar]
- Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, Lotan Y (2013) Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 63(2): 234–241. [DOI] [PubMed] [Google Scholar]
- Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, Gao YT, Nelson HH, Kelsey KT (2012) LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int J Cancer 130(5): 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH (2007) Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 211(3): 269–277. [DOI] [PubMed] [Google Scholar]
- Choi IS, Estecio MR, Nagano Y, Kim DH, White JA, Yao JC, Issa JP, Rashid A (2007) Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors). Mod Pathol 20(7): 802–810. [DOI] [PubMed] [Google Scholar]
- Chuang SC, Stolzenberg-Solomon R, Ueland PM, Vollset SE, Midttun O, Olsen A, Tjonneland A, Overvad K, Boutron-Ruault MC, Morois S, Clavel-Chapelon F, Teucher B, Kaaks R, Weikert C, Boeing H, Trichopoulou A, Benetou V, Naska A, Jenab M, Slimani N, Romieu I, Michaud DS, Palli D, Sieri S, Panico S, Sacerdote C, Tumino R, Skeie G, Duell EJ, Rodriguez L, Molina-Montes E, Huerta JM, Larranaga N, Gurrea AB, Johansen D, Manjer J, Ye W, Sund M, Peeters PH, Jeurnink S, Wareham N, Khaw KT, Crowe F, Riboli E, Bueno-de-Mesquita B, Vineis P (2011) A U-shaped relationship between plasma folate and pancreatic cancer risk in the European Prospective Investigation into Cancer and Nutrition. Eur J Cancer 47(12): 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10(10): 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM (2010) Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugué PA, English DR, MacInnis RJ, Joo JE, Jung CH, Milne RL (2015) The repeatability of DNA methylation measures may also affect the power of epigenome-wide association studies. Int J Epidemiol 44(4): 1460–1461. [DOI] [PubMed] [Google Scholar]
- Dugué PA, English DR, MacInnis RJ, Jung CH, Bassett JK, Fitzgerald LM, Wong EM, Joo JE, Hopper JL, Southey MC, Giles GG, Milne RL (2016) Reliability of DNA methylation measures from dried blood spots and mononuclear cells using the HumanMethylation450k BeadArray Sci Rep 26(6): 30317. [DOI] [PMC free article] [PubMed]
- Fraga MF, Uriol E, Borja Diego L, Berdasco M, Esteller M, Canal MJ, Rodriguez R (2002) High-performance capillary electrophoretic method for the quantification of 5-methyl 2'-deoxycytidine in genomic DNA: application to plant, animal and human cancer tissues. Electrophoresis 23(11): 1677–1681. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J (2002) A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA 99(8): 5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M (1983) The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 11(19): 6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10): R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles GG, English DR (2002) The Melbourne Collaborative Cohort Study. IARC Sci Publ 156: 69–70. [PubMed] [Google Scholar]
- Harper KN, Peters BA, Gamble MV (2013) Batch effects and pathway analysis: two potential perils in cancer studies involving DNA methylation array analysis. Cancer Epidemiol Biomarkers Prev 22(6): 1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 15(2): R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji HX, Zhao Q, Pan JH, Shen WH, Chen ZW, Zhou ZS (2013) Association of BLCA-4 hypomethylation in blood leukocyte DNA and the risk of bladder cancer in a Chinese population. Pathol Oncol Res 19(2): 205–210. [DOI] [PubMed] [Google Scholar]
- Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7): 484–492. [DOI] [PubMed] [Google Scholar]
- Joo JE, Wong EM, Baglietto L, Jung CH, Tsimiklis H, Park DJ, Wong NC, English DR, Hopper JL, Severi G, Giles GG, Southey MC (2013) The use of DNA from archival dried blood spots with the Infinium HumanMethylation450 array. BMC Biotechnol 13: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen MO, Bryan RT, Haworth KE, Emes RD, Luscombe C, Gommersall L, Cheng KK, Zeegers MP, James ND, Devall AJ, Fryer AA, Farrell WE (2015) Methylation of HOXA9 and ISL1 predicts patient outcome in high-grade non-invasive bladder cancer. PLoS One 10(9): e0137003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo KC, McCune RA, Gehrke CW, Midgett R, Ehrlich M (1980) Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res 8(20): 4763–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ (2001) Initial sequencing and analysis of the human genome. Nature 409(6822): 860–921. [DOI] [PubMed] [Google Scholar]
- Li G, Liu Y, Yin H, Zhang X, Mo X, Tang J, Chen W (2014) E-cadherin gene promoter hypermethylation may contribute to the risk of bladder cancer among Asian populations. Gene 534(1): 48–53. [PubMed] [Google Scholar]
- Lin YL, Gui SL, Ma JG (2015) Aberrant methylation of CDH11 predicts a poor outcome for patients with bladder cancer. Oncol Lett 10(2): 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic J, Gordon L, Oshlack A (2012) SWAN: subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol 13(6): R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Houseman EA, Christensen BC, Gagne L, Wrensch MR, Nelson HH, Wiemels J, Zheng S, Wiencke JK, Andrew AS, Schned AR, Karagas MR, Kelsey KT (2010) Identification of methylated genes associated with aggressive bladder cancer. PLoS One 5(8): e12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Perez J, Gu J, Herrera LA, Tannir NM, Matin SF, Karam JA, Huang M, Chang DW, Wood CG, Wu X (2015) Genomic DNA hypomethylation and risk of renal cell carcinoma: a case-control study. Clin Cancer Res 22(8): 2074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C, Carrato A, Dosemeci M, Garcia-Closas M, Esteller M, Fraga M, Rothman N, Malats N (2008) Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol 9(4): 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H, Wong NC, Chatterton Z, Hong MK, Pedersen JS, Corcoran NM, Hovens CM, Macintyre G (2014) Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics 15: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, Cotton AM, Penaherrera MS, McFadden DE, Kobor MS, Robinson W (2012) Different measures of ‘genome-wide' DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics 7(6): 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, Robinson WP, Kobor MS (2013) Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin 6(1): 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/.
- Rager JE, Tilley SK, Tulenko SE, Smeester L, Ray PD, Yosim A, Currier JM, Ishida MC, Gonzalez-Horta Mdel C, Sanchez-Ramirez B, Ballinas-Casarrubias L, Gutierrez-Torres DS, Drobna Z, Del Razo LM, Garcia-Vargas GG, Kim WY, Zhou YH, Wright FA, Styblo M, Fry RC (2015) Identification of novel gene targets and putative regulators of arsenic-associated DNA methylation in human urothelial cells and bladder cancer. Chem Res Toxicol 28(6): 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas LA, Villanueva CM, Tajuddin SM, Amaral AF, Fernandez AF, Moore LE, Carrato A, Tardon A, Serra C, Garcia-Closas R, Basagana X, Rothman N, Silverman DT, Cantor KP, Kogevinas M, Real FX, Fraga MF, Malats N (2014) LINE-1 methylation in granulocyte DNA and trihalomethane exposure is associated with bladder cancer risk. Epigenetics 9(11): 1532–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CW (1996) Alu: structure, origin, evolution, significance and function of one-tenth of human DNA. Prog Nucleic Acids Res Mol Biol 53: 283–319. [DOI] [PubMed] [Google Scholar]
- Schulz WA, Steinhoff C, Florl AR (2006) Methylation of endogenous human retroelements in health and disease. Current Topics Microbiol Immunol 310: 211–250. [DOI] [PubMed] [Google Scholar]
- Severi G, Southey MC, English DR, Jung CH, Lonie A, McLean C, Tsimiklis H, Hopper JL, Giles GG, Baglietto L (2014) Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat 148(3): 665–673. [DOI] [PubMed] [Google Scholar]
- Shariat SF, Sfakianos JP, Droller MJ, Karakiewicz PI, Meryn S, Bochner BH (2010) The effect of age and gender on bladder cancer: a critical review of the literature. BJU Int 105(3): 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvetsov YB, Song MA, Cai Q, Tiirikainen M, Xiang YB, Shu XO, Yu H (2015) Intraindividual variation and short-term temporal trend in DNA methylation of human blood. Cancer Epidemiol Biomarkers Prev 24(3): 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner HG, Gangnon RE, Litzelman K, Johnson RA, Chari ST, Petersen GM, Boardman LA (2012) Telomere length and pancreatic cancer: a case-control study. Cancer Epidemiol Biomarkers Prev 21(11): 2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajuddin S, Amaral A, Fernández A, Chanock S, Silverman D, Tardón A, Carrato A, García-Closas M, Jackson B, Toraño E (2014) LINE-1 methylation in leukocyte DNA, interaction with phosphatidylethanolamine N-methyltransferase variants and bladder cancer risk. Br J Cancer 110(8): 2123–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veldhoven K, Polidoro S, Baglietto L, Severi G, Sacerdote C, Panico S, Mattiello A, Palli D, Masala G, Krogh V, Agnoli C, Tumino R, Frasca G, Flower K, Curry E, Orr N, Tomczyk K, Jones ME, Ashworth A, Swerdlow A, Chadeau-Hyam M, Lund E, Garcia-Closas M, Sandanger TM, Flanagan JM, Vineis P (2015) Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin Epigenetics 7(1): 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, Capesius I (1981) Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochim Biophys Acta 654(1): 52–56. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39(4): 457–466. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW (2005) Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 33(21): 6823–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, Karagas MR, Marsit CJ (2010) Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res 16(5): 1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Doo N, Makalic E, Joo JE, Vajdic CM, Schmidt DF, Wong EM, Jung CH, Severi G, Park DJ, Chung J, Baglietto L, Prince HM, Seymour JF, Tam C, Hopper JL, English DR, Milne RL, Harrison SJ, Southey MC, Giles GG (2016) Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature B-cell neoplasms. Epigenomics 8(1): 55–66. [DOI] [PubMed] [Google Scholar]
- Woo HD, Kim J (2012) Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One 7(4): e34615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32(3): e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Hansen KD, Meissner A, Aryee MJ (2015) Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods 12(3): 230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.