Abstract
The Pediatric HIV/AIDS Cohort Study (PHACS), the largest ongoing longitudinal study of perinatal HIV-infected (PHIV) and HIV-exposed, uninfected (PHEU) children in the United States, comprises the Surveillance Monitoring of Antiretroviral Therapy [ART] Toxicities (SMARTT) Study in PHEU children and the Adolescent Master Protocol (AMP) that includes PHIV and PHEU children ≥7 years. Although race/ethnicity is often used to assess health outcomes, this approach remains controversial and may fail to accurately reflect the backgrounds of ancestry-diverse populations as represented in the PHACS participants.
In this study, we compared genetically determined ancestry (GDA) and self-reported race/ethnicity (SRR) in the PHACS cohort. GDA was estimated using a highly discriminative panel of 41 single nucleotide polymorphisms and compared to SRR. Because SRR was similar between the PHIV and PHEU, and between the AMP and SMARTT cohorts, data for all unique 1958 participants were combined.
According to SRR, 63% of study participants identified as Black/African-American, 27% White, and 34% Hispanic. Using the highest percentage of ancestry/ethnicity to identify GDA, 9.5% of subjects were placed in the incorrect superpopulation based on SRR. When ≥50% or ≥75% GDA of a given superpopulation was required, 12% and 25%, respectively, of subjects were placed in the incorrect superpopulation based on SRR, and the percent of subjects classified as multiracial increased. Of 126 participants with unidentified SRR, 71% were genetically identified as Eurasian.
GDA provides a more robust assessment of race/ethnicity when compared to self-report, and study participants with unidentified SRR could be assigned GDA using genetic markers. In addition, identification of continental ancestry removes the taxonomic identification of race as a variable when identifying risk for clinical outcomes.
Keywords: ancestry informative markers, genetically determined ancestry, HIV exposed uninfected, HIV-infected children, Pediatric HIV/AIDS Cohort Study, self-reported race/ethnicity
1. Introduction
An abundance of data supports an important role for racial and ethnic ancestry in health outcomes.[1–6] Typically, such ancestry information is obtained through self-report which can be incorrect due to inaccurate or incomplete knowledge of one's ancestral background, especially in admixed populations. Moreover, information regarding ancestry may be unavailable from data collected. Recent advances in genetics have enabled the accurate identification of continental ancestry. Genetically determined ancestry (GDA) allows for the identification of admixtures that may sufficiently alter the genetic background of an individual to affect risk for disease or rate of disease progression.[7–12] In addition, the use of race/ethnicity to define individual background is without generally agreed-upon definitions and have been called flawed surrogates for genetic factors in disease causation.[13]
The United States has frequently been referred to as a melting pot where people from many diverse backgrounds and continents have come together leading to population admixture. The genetic diversity of the US population can make it difficult to identify a person's race/ethnicity. However, a recent study from the U.S. National Children's Study (NCS) found 601 (98.8%) of 608 subjects to be in agreement when self-reported race/ethnicity (SRR) ancestry was compared to GDA. They concluded that GDA added little additional information and was unnecessary when longitudinally comparing subsequent health outcomes.[14] However, the NCS included predominantly subjects of European ancestry, with 72% reporting to be non-Hispanic Whites, a population group known to show little admixture. In addition, in the assessment of ancestry in the NCS, the highest percentage ancestry/ethnicity as determined by GDA was used to assign ancestry. Thus, in the NCS, a subject was identified as a specific ancestry even if that ancestral background accounted for <50% of the continental ancestry identified.
The Pediatric HIV/AIDS Cohort Study (PHACS) comprises 2 longitudinal studies: the Surveillance Monitoring of Antiretroviral Therapy [ART] Toxicities (SMARTT) Study in perinatal HIV-exposed, uninfected (PHEU) children, and the Adolescent Master Protocol (AMP) that includes children aged ≥7 years who are perinatal HIV-infected (PHIV) or PHEU.[15,16] The self-reported ancestry of the PHACS subjects differs substantially from the NCS participants. Although the NCS specifically attempted to enroll children representative of the US population, children participating in PHACS are disproportionately from minority groups that are over-represented in those with human immunodeficiency virus (HIV)/AIDS.[14] The objective of the present study was to determine if self-identified race and ethnicity accurately reflects the genetic ancestry of participants in PHACS. Our findings indicate that SRR does not fully capture the genetic diversity of PHACS participants. In addition, PHACS subjects with unidentified SRR could be assigned ancestry using genetic markers. Thus, GDA was found to provide a more robust assessment of ancestry when compared to self-report.
2. Methods
2.1. Study population
GDA was determined for 1958 children participating in either AMP or SMARTT. Among these 1958, 1324 children were enrolled only in SMARTT, 523 were enrolled only in AMP, and 111 were enrolled in both SMARTT and AMP. There were 427 PHIV children, all of whom were enrolled in AMP. The AMP study is designed to assess the long-term physical and mental health of children with PHIV and their transition to adulthood (AMP up). The SMARTT study is designed to evaluate PHEU infants and children for the short- and long-term safety of in utero and neonatal exposure of antiretrovirals (ARVs) and to explore the mechanisms of adverse events, including mitochondrial dysfunction and other potential adverse effects of intrauterine ARV exposure. This study followed the human experimentation guidelines of the U.S. Department of Health and Human Services and was approved by the University of California, San Diego institutional review board.
2.2. Identification of genetic ancestry
Genetic ancestry was determined from DNA extracted from saliva or peripheral blood mononuclear cells using a highly discriminative panel of ancestry informative markers (AIMs) consisting of 41 single nucleotide polymorphisms (SNPs) previously shown to discern accurately global ancestry.[17] This panel estimates continental origin by comparing each child's genotypes to allele frequencies found in a set of 3517 reference individuals originating from 107 populations around the world. Reference populations were grouped into the 7 world regions Europe, Africa, America, central/south Asia, south/west Asia, east Asia, and Oceania. Population structure and ancestry estimates were obtained in a trained clustering analysis using STRUCTURE v2.3.2.1 (Clinical Data Interchange Standards Consortium, Inc).[18,19] Five independent runs were performed for each analysis. Allele frequencies were updated using only individuals with population information at a migration prior of 0.05. Uniform priors were used for the degree of admixture (“infer α” option) and for the allele frequency (λ = 1 option). All other parameters were set at default. Continental ancestry calling was performed by assigning the predominant continental origin to each subject depending on the definition to be used. Each SNP within the panel was discerned by real-time PCR. Among the 118 children coenrolled in both AMP and SMARTT, and among 2 that were only enrolled in SMARTT, duplicate assessments were conducted and indicated high reproducibility. Of the 120 children, 90% had a maximum continental ancestry difference less than 10%, while 98% had a maximum continental ancestry difference less than 50%. The assessments from AMP samples were included for these 111 coenrolled subjects, and the first available was included for those only enrolled in SMARTT. Seven DNA samples yielded inconsistent PCR products, and these subjects were eliminated from the analyses. To graphically display the percent multiracial, subjects were categorized as multiracial if the maximum was below a sliding threshold. SAS 9.4 and R version 2.15.1 (SAS Integration Technologies, Alkes Price Developed Program) were used for the analyses. Population structure was further analyzed using principal component analysis (PCA) implemented in the EIGENSTRAT software.[20]
3. Results
3.1. Demographics
Since race and ethnicity frequencies were similar between the AMP and SMARTT cohorts, the 2 cohorts were combined in the analyses. Of the 1958 subjects, 1832 self-identified race/ethnicity; 63% self-identified as Black or African-American, 27% as White, 6% unknown, 2.1% multiracial, and 1% other; 34% self-identified as Hispanic (Table 1). Figure 1 shows the PCA for the AMP and SMARTT cohorts combined based on the genotype data of the 41 AIMs for the reference set of 3517 divided into 5 superpopulations similar to the NCS analysis.
Table 1.
Selected demographics of the combined PHACS AMP and SMARTT cohorts.
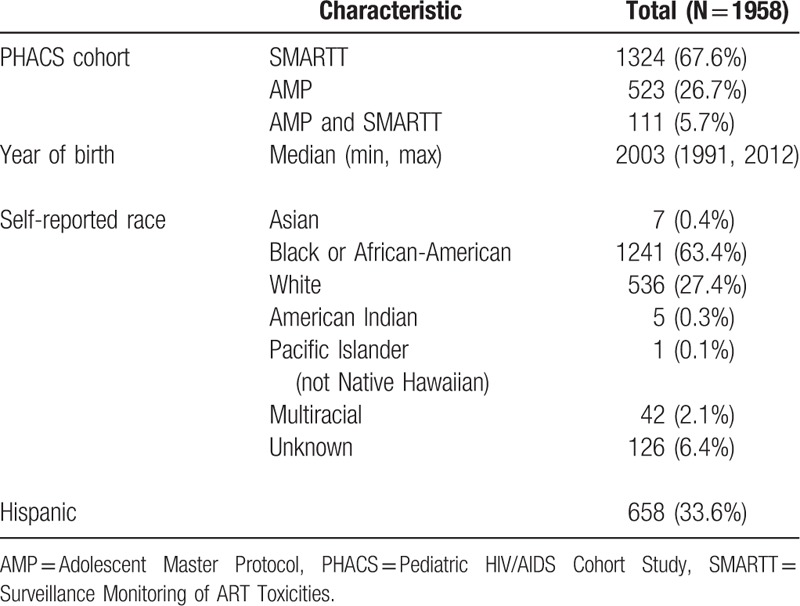
Figure 1.
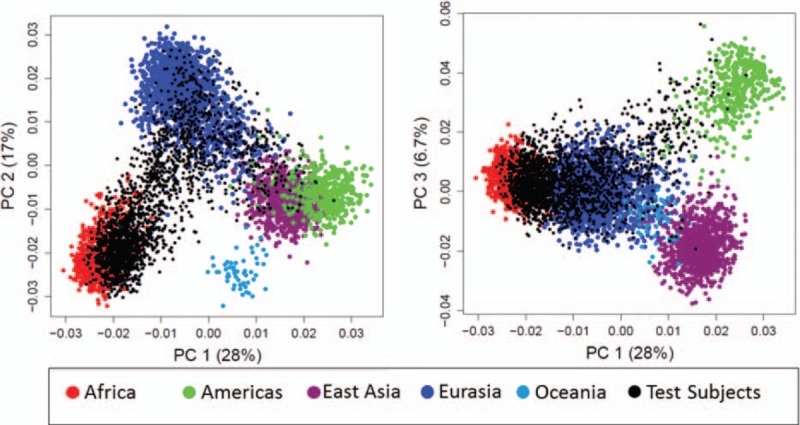
Principal component analysis for the Adolescent Master Protocol and Surveillance Monitoring of Antiretroviral Therapy Toxicities cohorts combined based on the genotype data of the 41 ancestry informative markers for the reference set of 3517 separated into 5 superpopulations.
3.2. Self-reported race/ethnicity compared to genetically determined ancestry
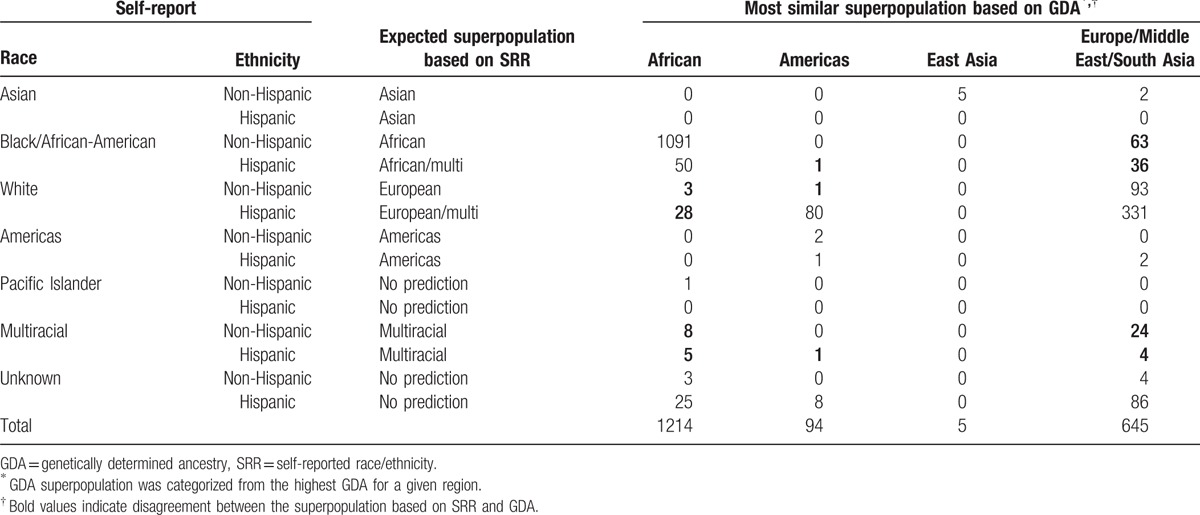
Using a similar analysis as that performed for the NCS (where the predominant ancestry was used even if it represented <50% of total identified ancestry) compared to self-report, 174 (9.5%) of 1832 PHACS participants with an identified ancestry are misidentified compared to ∼1.2% in the NCS (Table 2). Of those that misidentified, the greatest number were among study participants who self-reported as African-American non-Hispanic (n = 63) or African-American Hispanic (n = 36) and by GDA were found to be predominantly of Eurasian continental ancestry. Another 28 participants who by SRR identified as White Hispanic by GDA were found to be of predominantly of African ancestry. In total, of the 42 PHACS participants who self-identified as multiracial, 28 (66.6%) were predicted to be predominantly of Eurasian, 13 (30.9%) as African, and 1 (2.4%) as Americas ancestry.
Table 2.
Superpopulation classification based on the most representative GDA by self-reported race and ethnicity.
3.3. Self-reported ancestry/ethnicity compared to genetically determined ancestry with increasing cutoff for dominant GDA
Although using the predominant ancestry to define a single GDA provides valuable information regarding an individual's genetic background, it fails to account for a large amount of genetic diversity that could affect risk factors for disease. For this reason, we examined SRR compared to GDA at cutoffs of ≥50% and ≥75%. As can be seen in Tables 3 and 4, PHACS subjects with an SRR were misidentified with the ≥50% cutoff in 12% of cases (223/1832) and with the ≥75% cutoff in 26% of cases (478/1832) when compared to GDA. With each increase in the stringency required for the percent of a single GDA, there is a concurrent increase in the percent of subjects classified as multiracial (Fig. 2). A cutoff for the maximum GDA of <50% and <75% results in 8.5% and 49% classified as multiracial, respectively.
Table 3.
Superpopulation classification based on the most representative GDA after categorizing those with less than a maximum GDA of 50% as multiracial by self-reported race and ethnicity.
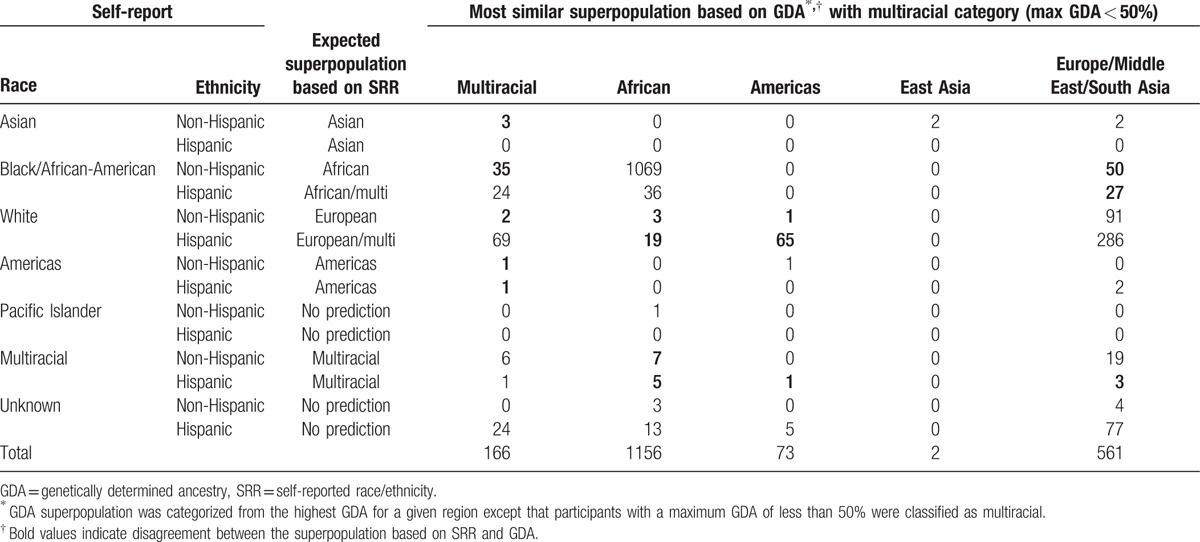
Table 4.
Superpopulation classification based on the most representative GDA after categorizing those with less than a maximum GDA of 75% as multiracial by self-reported race and ethnicity.
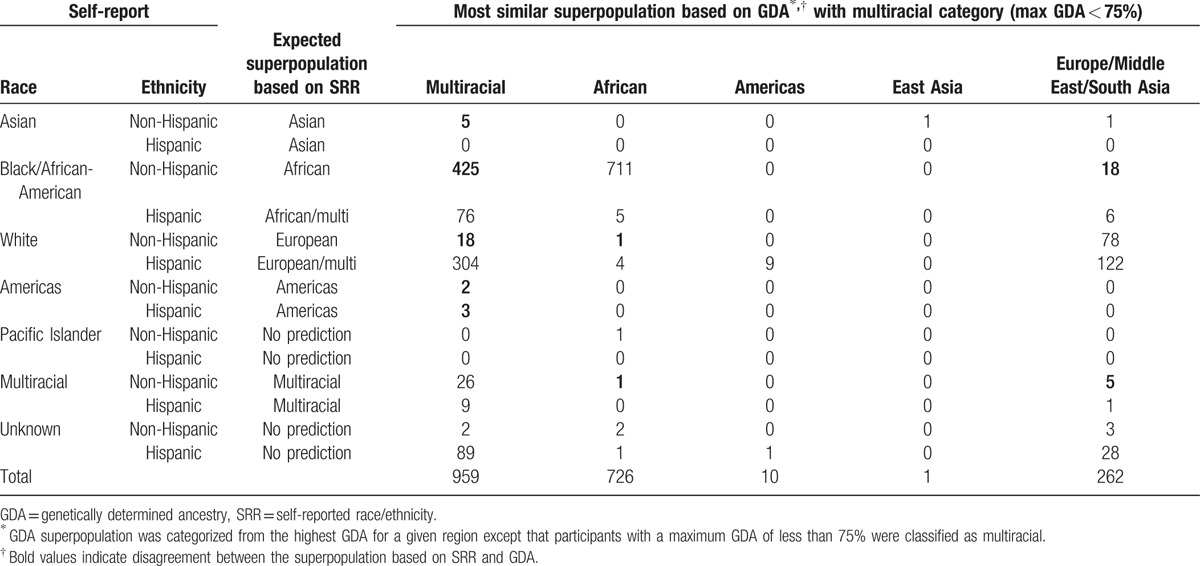
Figure 2.
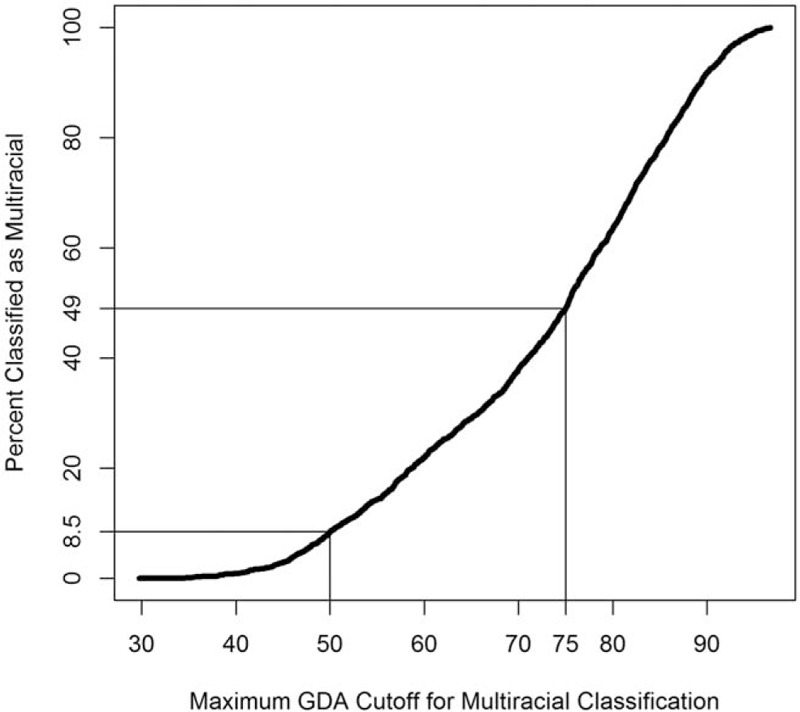
Percent classified as multiracial as a function of the maximum genetically determined ancestry required to classify a participant as multiracial.
3.4. Genetically determined ancestry can identify ancestry in PHACS subjects with unknown/unavailable self-reported race/ethnicity
Of PHACS participants, 126 subjects had no identified SRR either because of unknown background or no reported SRR. Most of these subjects were from Puerto Rico as reflected by 94% identified as having Hispanic ethnicity. As shown in Table 4, using the GDA cutoff of ≥75%, 72% were identified as multiracial, 2% African, 1% Americas, and 25% Eurasian.
3.5. Genetically determined ancestry and self-identified Hispanic
Using the approach employed in the NCS genetic ancestry study, self-identified Hispanic White subjects were most often identified by GDA to be of Eurasian ancestry (predominantly European) (331/439 [75%]) with 18% (n = 80) Americas (Table 2). In contrast, when Hispanic White subjects are classified as multiracial with a GDA below 75% (Table 4), only 122/439 (27.7%) are found to be of Eurasian ancestry and 2% from America.
4. Discussion
A major goal of the PHACS is to determine the long-term outcomes of in utero exposure to ARVs of infants born to HIV-infected mothers. Because race and ethnicity are associated with the clinical consequences of environmental exposures, it is important to assign accurately the correct racial and ethnic background of PHACS participants. Our findings highlight the diverse GDA backgrounds of children who are HIV-infected and -exposed in the United States, and reflect the over-representation of minorities of color among HIV-infected persons. The substantially higher number of minority children in the PHACS compared to those within the NCS cohort is likely important in explaining why when performing similar analyses, 9.5% of PHACS participants with an identified SRR were misidentified compared to only 1.2% of the NCS cohort.
A recent study performed by Bryc et al[21] examined genetic ancestry among African-Americans, Latinos, and European Americans in the United States. They found in their population of persons seeking genetic testing that African-Americans show an average proportion of 73.2% African ancestry, 24% European ancestry, and 0.8% Native Americas ancestry. Latinos constituted a highly diverse population with the mean ancestry of Hispanics in the United States having 18% Native American ancestry, 65.1% European ancestry, and 6.2% African ancestry. Among those with SRR as European, the percent African ancestry was greatly dependent on the region of the country. They estimated that at least 1.4% of those persons in the United States who by SRR identified as European carry at least 2% African ancestry. Of interest, persons with below 28% African ancestry self-reported as European ancestry. Our findings are consistent with those of Bryc et al and demonstrate a high proportion of genetic heterogeneity among children HIV-infected and -exposed in the United States. As would be expected, the discrepancy between SRR and GDA rises as the stringency for identification of a single race/ethnicity increases. The percentage of children who are misidentified by SRR increases to 12% when a cutoff of ≥50% is required and to 26% when a cutoff of ≥75% is used. Thus, a high proportion of PHACS participants could be considered multiracial depending on the cutoff used.
The identification of accurate ancestry is of particular importance to the interpretation of data obtained in PHACS. Multiple host genetic factors have been identified to play an important role in the rate of HIV disease progression, most notably human leukocyte antigen that is closely linked to continental ancestry. In addition, a major objective of PHACS is to determine the impact of in utero exposure to ARVs on clinical outcomes where continental ancestry is likely to affect the risk for an observed abnormality. In addition, progressive HIV infection is associated with the increased risk of opportunistic infections, many of which are modified by polymorphisms that alter the host immune response.
A potential limitation of our study is the use of only 41 AIMs to assess continental ancestry. The major constraint of this panel was found for subjects from Eurasia, where low fixation index (a genetic distance measure for interpopulation differentiation compared to intrapopulation variation) values of 0.06 to 0.09 among Europe, the Middle East, and central/south Asia indicate little genetic diversity. However, this AIMs panel has been validated on using a large reference set including over 4000 subjects collected from 120 global populations and offers a feasible and highly accurate estimation of continental ancestry when genome-wide genotyping or full genome sequencing is not available.[17]
Cardiovascular disease is known to be strongly associated with genetic risk factors. Recently, the identification of GDA in the Multicenter AIDS Cohort Study (MACS) was found to be useful in assigning risk for hyperlipidemia and cardiovascular disease.[22] In the MACS cohort of HIV-infected men, GDA more accurately identified risk for abnormal lipid profiles and other metabolic complications associated with ART than SRR. Moreover, these differences persisted after controlling for covariates most often associated with lipid abnormalities found with HIV and ARVs. They suggest that unidentified genetic variants could affect lipid metabolism, HIV infection, drug metabolism, inflammation, or the immune response and conclude that risk for HIV-related cardiovascular disease is closely linked with GDA. Asthma is a complex pulmonary disease that has strong associations with host genetics and environmental factors.[23–25] Using GDA, children and adults with African ancestry have been found to be at increased risk for asthma and in some studies at higher risk for severe exacerbations.[26,27] The PHACS is specifically designed to identify the presence of any long-term complications in HIV-infected and HIV-uninfected but exposed children in the US. Studies performed, to date, evaluating the PHACS participants have identified an increased risk for asthma among HIV-infected children[28] and potential indicators of early cardiovascular disease.[29–32] Thus, considering the role of genetic ancestry in assigning risk for phenotypic conditions identified to be increased among HIV-infected and -exposed children is important especially in this highly admixed cohort.
With the recent advances in genome biology, many experts have called for an end to the use of race as a variable in genetic and clinical research.[13,33] As noted by Yudell et al,[33] race historically has been used as a taxonomic categorization based on common phenotypic features such as skin color to determine the relationship between genes and ancestry. However, ancestry reflects an individual's relationship with persons within their genealogical history. Assigning of race without appreciation of an individual's genetic diversity has led to underdiagnosing or miscategorization of diseases. For example, sickle cell disease is underdiagnosed in Whites since it is often considered a disease of Blacks, while cystic fibrosis is underdiagnosed in Blacks because it is considered a disease of Whites. As suggested by Yudell et al, “Scientific journals and professional societies should encourage use of terms like ‘ancestry’ or ‘population’ to describe human groupings ….” Through the use of the highly discriminative panel of 41 SNPs described here, the continental ancestry of PHIV and PHEU children has been identified and can eliminate the use of the taxonomic and often inaccurate identification of race. This is of particular importance in the heterogeneous population of HIV-infected persons within the United States.
In summary, to our knowledge, this is the largest study to apply AIMs to identify GDA in a cohort of HIV-infected and -exposed children. Our findings demonstrate that consistent with the demographics of HIV/AIDS in the United States, minority groups are over-represented in PHACS. However, although SRR misidentified only 1% of participants in the NCS, 9.5% of PHACS participants are misidentified. If ≥75% of a given superpopulation is required to be classified as a single race, ∼50% of subjects would be classified as multiracial. This implies that self-reported race does not fully capture the genetic diversity of PHACS subjects. In addition, PHACS subjects with unidentified SRR can be assigned ancestry using genetic markers. Thus, GDA provides a more robust assessment of race/ethnicity when compared to self-report. Moreover, identification of continental ancestry removes the taxonomic identification of race as a variable when identifying risk for clinical outcomes.
Acknowledgments
We thank Rodney Trout for his technical assistance. We thank the children and families for their participation in PHACS, and the individuals and institutions involved in the conduct of PHACS. The following institutions, clinical site investigators, and staff participated in conducting PHACS AMP in 2014 in alphabetical order: Ann & Robert H. Lurie Children's Hospital of Chicago: Ram Yogev, Margaret Ann Sanders, Kathleen Malee, Scott Hunter; Baylor College of Medicine: William Shearer, Mary Paul, Norma Cooper, Lynnette Harris; Bronx Lebanon Hospital Center: Murli Purswani, MahboobullahBaig, Anna Cintron; Children's Diagnostic & Treatment Center: Ana Puga, Sandra Navarro, Patricia Garvie, James Blood; Children's Hospital, Boston: Sandra Burchett, Nancy Karthas, Betsy Kammerer; Jacobi Medical Center: Andrew Wiznia, Marlene Burey, Molly Nozyce; Rutgers—New Jersey Medical School: ArryDieudonne, Linda Bettica, Susan Adubato; St. Christopher's Hospital for Children: Janet Chen, Maria Garcia Bulkley, Latreaca Ivey, Mitzie Grant; St. Jude Children's ResearchHospital: Katherine Knapp, Kim Allison, Megan Wilkins; San Juan Hospital/Department of Pediatrics: Midnela Acevedo-Flores, Heida Rios, Vivian Olivera; Tulane University Health Sciences Center: Margarita Silio, Medea Jones, Patricia Sirois; University of California, San Diego: Stephen Spector, Kim Norris, Sharon Nichols; University of Colorado Denver Health Sciences Center: Elizabeth McFarland, Alisa Katai, Jennifer Dunn, Suzanne Paul; University12 of Miami: Gwendolyn Scott, Patricia Bryan, Elizabeth Willen.
Footnotes
Abbreviations: AIDS = Acquired Immunodeficiency Syndrome, AIMs = ancestry informative markers, AMP Study = Adolescent Master Protocol Study, ART = antiretroviral therapy, ARVs = antiretrovirals, DNA = deoxyribonucleic acid, GDA = genetically determined ancestry, HIV = human immunodeficiency virus, NCS = National Children's Study, PCA = principal component analysis, PCR = polymerase chain reaction, PHACS = Pediatric HIV/AIDS Cohort Study, PHEU = perinatal HIV-exposed, uninfected, PHIV = perinatal HIV-infected, SMARTT Study = Surveillance Monitoring of ART Toxicities Study, SNPs = single nucleotide polymorphisms, SRR = self-reported race/ethnicity.
This research was supported in part by 1R01NS077874 (SAS). The Pediatric HIV/AIDS Cohort Study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development with cofunding from the National Institute on Drug Abuse, the National Institute of Allergy and Infectious Diseases, the Office of AIDS Research, the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, the National Institute on Deafness and Other Communication Disorders, the National Heart Lung and Blood Institute, the National Institute of Dental and Craniofacial Research, and the National Institute on Alcohol Abuse and Alcoholism, through cooperative agreements with the Harvard T.H. Chan School of Public Health (HD052102; Principal Investigator: George Seage; Project Director: Julie Alperen) and the Tulane University School of Medicine (HD052104; Principal Investigator: Russell Van Dyke; Co-Principal Investigator: Kenneth Rich); Data management services were provided by Frontier Science and Technology Research Foundation (Principal Investigator: Suzanne Siminski), and regulatory services and logistical support were provided by Westat, Inc (Principal Investigator: Julie Davidson).
Authors’ contributions: SAS contributed to the conception and design, acquisition of data, analysis and interpretation of data, and wrote the manuscript.
SSB contributed to the conception and design, acquisition of data, analysis and interpretation of data, and critically edited the manuscript.
CMN, AXM, MUP, PLW, RH, RVD, and GRS contributed to the analysis and interpretation of data, and critically edited the manuscript. KKS contributed to the acquisition of data, analysis and interpretation of data, and critically edited the manuscript.
Data presented in this manuscript are available upon request at: https://phacsstudy.org/Our-Research/Data-Request-Form.
The authors have no conflicts of interest to disclose.
References
- 1.Ntzani EE, Liberopoulos G, Manolio TA, et al. Consistency of genome-wide associations across major ancestral groups. Hum Genet 2012; 131:1057–1071. [DOI] [PubMed] [Google Scholar]
- 2.Balfour PC, Jr, Rodriguez CJ, Ferdinand KC. The role of hypertension in race–ethnic disparities in cardiovascular disease. Curr Cardiovasc Risk Rep 2015; 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gijsberts CM, Groenewegen KA, Hoefer IE, et al. Race/ethnic differences in the associations of the Framingham risk factors with carotid IMT and cardiovascular events. PLoS One 2015; 10:e0132321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham CK, Balasubramanian R, Delke I, et al. The impact of race/ethnicity on mother-to-child HIV transmission in the United States in Pediatric AIDS Clinical Trials Group Protocol 316. J Acquir Immune Defic Syndr 2004; 36:800–807. [DOI] [PubMed] [Google Scholar]
- 5.Abrahao R, Lichtensztajn DY, Ribeiro RC, et al. Racial/ethnic and socioeconomic disparities in survival among children with acute lymphoblastic leukemia in California, 1988–2011: a population-based observational study. Pediatr Blood Cancer 2015; 62:1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pu J, Zhao B, Wang EJ, et al. Racial/ethnic differences in gestational diabetes prevalence and contribution of common risk factors. Paediatr Perinat Epidemiol 2015; 29:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halder I, Shriver M, Thomas M, et al. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat 2008; 29:648–658. [DOI] [PubMed] [Google Scholar]
- 8.Cheng CY, Reich D, Haiman CA, et al. African ancestry and its correlation to type 2 diabetes in African Americans: a genetic admixture analysis in three U.S. population cohorts. PLoS One 2012; 7:e32840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dries DL. Genetic ancestry, population admixture, and the genetic epidemiology of complex disease. Circ Cardiovasc Genet 2009; 2:540–543. [DOI] [PubMed] [Google Scholar]
- 10.Kidd JM, Gravel S, Byrnes J, et al. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet 2012; 91:660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libiger O, Schork NJ. A method for inferring an individual's genetic ancestry and degree of admixture associated with six major continental populations. Front Genet 2012; 3:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brummel SS, Singh KK, Maihofer AX, et al. Associations of genetically determined continental ancestry with CD4+ count and plasma HIV-1 RNA beyond self-reported race and ethnicity. J Acquir Immune Defic Syndr 2016; 71:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins FS. What we do and don’t know about ‘race’, ‘ethnicity’, genetics and health at the dawn of the genome era. Nat Genet 2004; 36:S13–S15. [DOI] [PubMed] [Google Scholar]
- 14.Smith EN, Jepsen K, Arias AD, et al. Genetic ancestry of participants in the National Children's Study. Genome Biol 2014; 15:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Dyke RB, Patel K, Siberry GK, et al. Antiretroviral treatment of US children with perinatally acquired HIV infection: temporal changes in therapy between 1991 and 2009 and predictors of immunologic and virologic outcomes. J Acquir Immune Defic Syndr 2011; 57:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams PL, Seage GR, 3rd, Van Dyke RB, et al. A trigger-based design for evaluating the safety of in utero antiretroviral exposure in uninfected children of human immunodeficiency virus-infected mothers. Am J Epidemiol 2012; 175:950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nievergelt CM, Maihofer AX, Shekhtman T, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig Genet 2013; 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 21.Bryc K, Durand EY, Macpherson JM, et al. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet 2015; 96:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholaou MJ, Martinson JJ, Abraham AG, et al. HAART-associated dyslipidemia varies by biogeographical ancestry in the multicenter AIDS cohort study. AIDS Res Hum Retroviruses 2013; 29:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portelli MA, Hodge E, Sayers I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy 2015; 45:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijk FN, de Jongste JC, Postma DS, et al. Genetics of onset of asthma. Curr Opin Allergy Clin Immunol 2013; 13:193–202. [DOI] [PubMed] [Google Scholar]
- 25.Custovic A, Marinho S, Simpson A. Gene-environment interactions in the development of asthma and atopy. Expert Rev Respir Med 2012; 6:301–308. [DOI] [PubMed] [Google Scholar]
- 26.Rumpel JA, Ahmedani BK, Peterson EL, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol 2012; 130:1302–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flores C, Ma SF, Pino-Yanes M, et al. African ancestry is associated with asthma risk in African Americans. PLoS One 2012; 7:e26807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siberry GK, Leister E, Jacobson DL, et al. Increased risk of asthma and atopic dermatitis in perinatally HIV-infected children and adolescents. Clin Immunol 2012; 142:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipshultz SE, Miller TL, Wilkinson JD, et al. Cardiac effects in perinatally HIV-infected and HIV-exposed but uninfected children and adolescents: a view from the United States of America. J Int AIDS Soc 2013; 16:18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipshultz SE, Shearer WT, Thompson B, et al. Cardiac effects of antiretroviral therapy in HIV-negative infants born to HIV-positive mothers: NHLBI CHAART-1 (National Heart, Lung, and Blood Institute Cardiovascular Status of HAART Therapy in HIV-Exposed Infants and Children cohort study). J Am Coll Cardiol 2011; 57:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipshultz SE, Williams PL, Wilkinson JD, et al. Cardiac status of children infected with human immunodeficiency virus who are receiving long-term combination antiretroviral therapy: results from the Adolescent Master Protocol of the Multicenter Pediatric HIV/AIDS Cohort Study. JAMA Pediatr 2013; 167:520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipshultz SE, Williams PL, Zeldow B, et al. Cardiac effects of in-utero exposure to antiretroviral therapy in HIV-uninfected children born to HIV-infected mothers. AIDS 2015; 29:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yudell M, Roberts D, DeSalle R, et al. Science and Society. Taking race out of human genetics. Science 2016; 351:564–565. [DOI] [PubMed] [Google Scholar]


