Summary
N eisseria meningitidis, a major cause of bacterial meningitis and septicaemia, secretes multiple virulence factors, including the adhesion and penetration protein (App) and meningococcal serine protease A (MspA). Both are conserved, immunogenic, type Va autotransporters harbouring S6‐family serine endopeptidase domains. Previous work suggested that both could mediate adherence to human cells, but their precise contribution to meningococcal pathogenesis was unclear. Here, we confirm that App and MspA are in vivo virulence factors since human CD46‐expressing transgenic mice infected with meningococcal mutants lacking App, MspA or both had improved survival rates compared with mice infected with wild type. Confocal imaging showed that App and MspA were internalized by human cells and trafficked to the nucleus. Cross‐linking and enzyme‐linked immuno assay (ELISA) confirmed that mannose receptor (MR), transferrin receptor 1 (TfR1) and histones interact with MspA and App. Dendritic cell (DC) uptake could be blocked using mannan and transferrin, the specific physiological ligands for MR and TfR1, whereas in vitro clipping assays confirmed the ability of both proteins to proteolytically cleave the core histone H3. Finally, we show that App and MspA induce a dose‐dependent increase in DC death via caspase‐dependent apoptosis. Our data provide novel insights into the roles of App and MspA in meningococcal infection.
Introduction
Autotransporters are the largest class of secreted protein virulence factors among Gram‐negative pathogens (Henderson et al., 2004). Since the prototype autotransporter IgA1 protease of Neisseria gonorrhoeae was first described (Koomey et al., 1982), over 1500 putative autotransporters have been identified (Celik et al., 2012). Of the three main subtypes, the most common is type Va (or classical); these are characterized by common structural components, including an N‐terminal signal peptide, a central functional passenger domain, and a C‐terminal translocator or β‐domain (Leyton et al., 2012). The signal peptide mediates inner‐membrane translocation, whereas the β‐domain is essential for translocation of the passenger domain to the bacterial surface. Once there, the passenger domain may be cleaved and secreted, either auto‐catalytically or via a secondary protease, cleaved but remain non‐covalently bound to the translocator domain, or remain uncleaved and displayed at the cell surface (Henderson et al., 2004; Leyton et al., 2012). The functions of autotransporter passenger domains are diverse but typically relate to virulence (Benz and Schmidt, 2011).
Neisseria meningitidis is normally a harmless commensal that inhabits the human nasopharynx; it is also capable of causing devastating invasive disease, including meningitis and septicaemia (Stephens, 2009). The organism elaborates numerous cell surface and secreted virulence factors, a number of which are autotransporter proteins, which allow them to colonize and infect the host (Virji, 2009). Eight autotransporters have been identified in meningococci: IgA1 protease; MspA (meningococcal serine protease A); App (adhesion and penetration protein); NhhA; AutA; AutB; NadA and NalP, the majority of which were first discovered in our laboratory (Ait‐Tahar et al., 2000; Hadi et al., 2001; Turner et al., 2002; Turner et al., 2006). IgA1 protease, App and MspA have been assigned, based upon their likely common ancestry and other shared features including the nature, order and relative positions of their catalytic residues, to the S6‐peptidase family in the MEROPS database (Rawlings et al., 2012). This family of secreted proteins also encompasses homologues of IgA1 protease and App expressed by Haemophilus influenzae (Poulsen et al., 1989; St Geme et al., 1994), proteins belonging to the serine protease autotransporter of Enterobacteriaceae (SPATE) group (Ruiz‐Perez and Nataro, 2014) and other, as yet uncharacterized, proteins (Rawlings et al., 2012). Typical functions of characterized S6‐peptidase family autotransporters relate to adherence or cleavage of extracellular or intracellular host proteins resulting in toxicity and/or immune modulation (Henderson et al., 2004; Ruiz‐Perez and Nataro, 2014).
More specifically, previous work has shown that App is expressed during meningococcal infection and carriage, stimulates B‐ and T‐cells, and can induce a bactericidal antibody response (Hadi et al., 2001; van Ulsen et al., 2001). Importantly, App is an adhesin required for optimal meningococcal adhesion to epithelial cells (Serruto et al., 2003). After export to the cell surface, App is (at least partially) auto‐cleaved and released from the cell surface (Hadi et al., 2001), a mechanism suggested to assist in meningococcal detachment and dispersal (Serruto et al., 2003). App auto‐cleavage releases a ca. 100 kDa fragment harbouring the active peptidase domain, but in the presence of NalP (a phase‐variably expressed meningococcal surface protease), a 140 kDa secreted App fragment, containing the peptidase domain and an additional α‐peptide region, predominates (van Ulsen et al., 2003). The secretion of two forms, the smaller resulting from auto‐proteolysis and the larger due to NalP processing, has also been described for IgA1 protease and MspA (van Ulsen et al., 2003).
MspA (also known as AusI) is a 157 kDa protein that shows ca. 33% and ca. 36% overall amino acid identity to App and IgA1 protease respectively (Turner et al., 2006). Like NalP, MspA expression is phase‐variable (van Ulsen et al., 2006; Oldfield et al., 2013). Similar to App and IgA1 protease, MspA is expressed in the host and can induce a bactericidal antibody response (Turner et al., 2006). Escherichia coli expressing recombinant MspA were shown to have increased adherence to some, but not all, human cell types tested, suggesting a possible role in adherence (Turner et al., 2006).
Here, we confirm that App and MspA are meningococcal virulence factors in vivo using a human CD46 transgenic mouse model. In addition, specific roles for the secreted fragments of App and MspA have yet to be established. Here, we show that recombinant proteins encompassing the secreted fragments of both autotransporters are trafficked to the host cell nucleus, proteolytically cleave histone and trigger caspase‐dependent apoptosis.
Results
In vivo confirmation that App and MspA are meningococcal virulence factors
In order to confirm a role for App and MspA in in vivo infection, the serogroup B meningococcal strain MC58 and a series of mutants in which either or both genes were deleted (MC58Δapp, MC58ΔmspA and MC58ΔappΔmspA) were used to infect groups of 10 hCD46Ge transgenic mice. These mice have previously been shown to express CD46 in a human‐like pattern (Mrkic et al., 1998) and develop meningococcal disease (Johansson et al., 2003, Johansson et al., 2005, Sjölinder and Jonsson, 2007). A further group of mice received suspension fluid alone (negative control).
Survival rate of 100% was observed in all groups prior to 48 h post‐infection; the survival rate of the negative control group remained at 100% at all subsequent time points (not shown). Survival rates for the MC58‐infected group of mice were 60% and 0% at 48 and 72 h respectively (Fig. 1A). Higher survival rates were apparent in groups infected with mutant strains (Fig. 1A). For the MC58ΔmspA group, survival rates of 80%, 20% and 20% were observed at 48, 72 and 96 h post‐infection, respectively; for MC58Δapp, survival rates of 60%, 40% and 20% were observed at 48, 72 and 96 h, respectively (P < 0.05, Fisher's exact test, MC58 compared with MC58Δapp‐infected groups at 72 h). Highest survival rates were observed for the group infected with MC58ΔappΔmspA (90%, 40% and 30% at 48, 72 and 96 h, respectively) (P < 0.05, wild type compared with MC58ΔappΔmspA at 72 h). Blood samples were also collected from the tail vein for the measurement of bacteraemia levels. Significantly fewer bacteria were found in the blood of animals infected with MC58ΔmspA at 6 h post‐infection and in animals infected with MC58ΔappΔmspA at 6 and 48 h post‐infection (Fig. 1B). Taken together, our in vivo data provide confirmation that App and MspA are in vivo meningococcal virulence factors.
Figure 1.
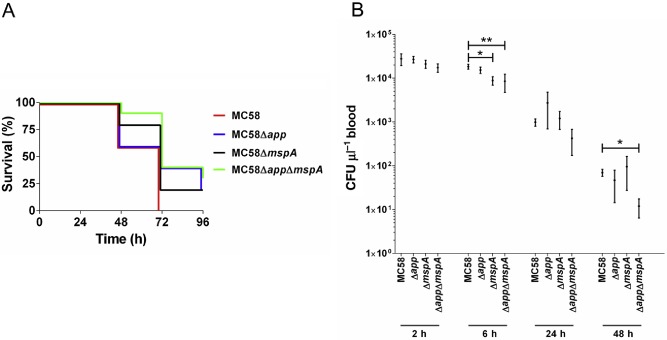
Infection of hCD46Ge transgenic mice with MC58 and its Δapp, ΔmspA and ΔappΔmspA mutant derivatives. Groups of 10 mice were infected intraperitoneally with 1.2 × 109 CFU of bacteria.
A. Survival rates tended to be higher in groups infected with Δapp, Δmsp A or ΔappΔmsp A (P < 0.05, Fisher's exact test, MC58 compared with MC58 Δapp‐infected groups or MC58ΔappΔmsp A strain at 72 h). A negative control group (n = 7), which received re‐suspension fluid alone, showed 100% survival at all time points (not shown).
B. CFU counts were determined after plating out serial dilutions of blood. Significantly fewer bacteria were found in the blood of animals infected with MC58Δmsp A and MC58ΔappΔmsp A at 6 h post‐challenge, and in animals infected with MC58ΔappΔmsp A at 48 h post‐infection. Mean CFU (± SEM) from surviving animals shown. *P < 0.05; **P < 0.01; Kruskal–Wallis and Dunn's test.
App and MspA are internalized into human cells and translocate to the nucleus
To determine whether the extracellular forms of App and MspA interact directly with host cells, the passenger domain of each protein was expressed under non‐denaturing conditions as a fusion with the E. coli chaperone trigger factor (TF), yielding rpdApp and rpdMspA respectively. FITC‐labelled rpdApp and rpdMspA were then co‐incubated with human monocyte‐derived dendritic cells (DCs). Examination by confocal microscopy clearly demonstrated that both rpdApp and rpdMspA were internalized and at least partially localized in the nucleus (Fig. 2). Clear pattern differences were observed between the two proteins; App formed crescent‐like accumulations at the edge of the nucleus, whereas MspA staining was more diffuse inside both the cytoplasm and the nucleus (Fig. 2A and B). Recombinant TF alone, purified in the same way as the recombinant autotransporters, did not bind DCs (Fig. 2C and D). In separate experiments, recombinant App and MspA passenger domains tagged with 6 × histidine tags and purified under denaturing conditions were also taken up by DCs, confirming that the observed internalization was mediated by the autotransporter passenger domains and not the TF tag (data not shown). In addition, rpdApp and rpdMspA (but not TF) bound were internalized and at least partially localized to the nucleus in human brain microvascular endothelial cells (HBMECs; Supporting Information Fig. S1).
Figure 2.
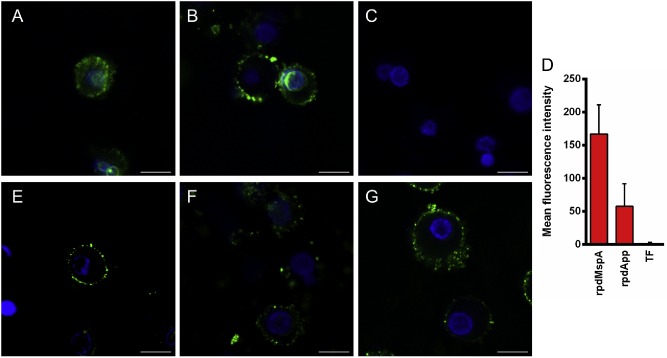
Uptake of rpdMspA and rpdApp into dendritic cells (DCs). Monocyte‐derived human DCs were incubated with FITC‐labelled rpdMspA (A), rpdApp (B) or TF (C). Cell nuclei were stained with Hoechst 33258. Co‐localization of labelled recombinant protein and nuclei (D) was determined using Zeiss LSM image examiner software. Mean fluorescence intensities were derived from ≥ 13 different fields from three independent experiments. Error bars: mean of values + SD. For inhibition, DCs were pre‐treated with mannan (1 mg ml−1; E), human transferrin (1 mg ml−1; F) or both inhibitors (G) for 30 min at 37°C before being incubated with FITC‐labelled rpdMspA. Cells were washed and fixed before analysis by confocal laser scanning microscopy. All images were scanned at a resolution of 1024 × 1024 pixels using the same laser and gains settings. The cells are representative of cells observed from three independent experiments. Scale bars = 10 μm.
App and MspA enter DCs via the mannose and transferrin receptors
Specific interaction between the autotransporters and host cells suggested that App and MspA bind to, and exploit, specific host molecules. To identify potential interacting partners, we used a re‐tagging approach, in conjunction with Matrix‐assisted laser desorption/ionization time of flight (MALDI‐ToF), as previously described (Ilver et al., 1998). DCs were incubated with recombinant proteins conjugated to the light‐activated cross‐linker Sulfo‐SBED. Following photoactivation and subsequent exposure to reducing conditions to allow transfer of the reactive biotin moiety to molecules in close proximity to the autotransporters, the DCs were washed, lysed, subjected to Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and probed with streptavidin. Reactive bands were excised and identified by MALDI‐ToF. For both autotransporters, two host cell surface molecules were identified: the mannose receptor (MR; gi|55665998; mass: 169 599; score: 1423 and 829 for rpdMspA and rpdApp, respectively) and transferrin receptor 1 (TfR1; gi|4507457; mass: 85 416; score: 1022 and 1,663 for rpdMspA and rpdApp respectively).
To confirm these interactions, a series of experiments using independent approaches were conducted. Firstly, the specific physiological ligands for MR and TfR1 (mannan and transferrin, respectively) were used to block internalization into DCs. Both substrates blocked the internalization of both autotransporters, which remained associated with the host cell surface (MspA is shown in Fig. 2E and F). Interestingly, the observed effects of the individual ligands were virtually identical to those of the agents used in combination (Fig. 2G). Secondly, in enzyme‐linked immuno assay (ELISA) experiments, rpdApp and rpdMspA were both shown to bind specifically to TfR1; binding was highly specific when compared to bovine serum albumin (BSA), which was used as a negative control (Fig. 3).
Figure 3.
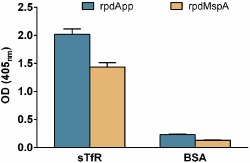
rpdApp and rpdMspA bind transferrin receptor 1. ELISA plates were coated with sTfR (2 μg ml−1) or 1% BSA. Following overnight incubation, wells were washed and rpdApp or rpdMspA (5 μg ml−1) added. Wells were probed with guinea pig anti‐rpdApp or rpdMspA, respectively, followed by anti‐guinea pig IgG alkaline phosphatase conjugate. Each test was performed in triplicate wells and on a least three independent occasions. Values represent the mean + SEM from three experiments.
Mannose receptor is a multi‐domain protein comprising an N‐terminal cysteine‐rich (CR) domain, a fibronectin type II domain (FNII) and eight tandemly arranged C‐type lectin‐like domains (CTLD1‐8) (Martinez‐Pomares, 2012). To confirm binding of the two autotransporters to MR, and to localize the MR region responsible for binding, we utilized two human Fc‐tagged recombinant MR fragments in ELISAs. The first comprised CR‐FNII‐CTLD1‐3‐Fc, whereas the second comprised CTLD4‐7‐Fc (Martinez‐Pomares et al., 2006) (Fig. 4, inset). CTLD4‐7‐Fc showed strong binding to both rpdApp and rpdMspA in a calcium‐dependent manner; CR‐FNII‐CTLD1‐3‐Fc bound neither protein, irrespective of the presence or absence of Ca2+ (Fig. 4). Together, our data confirm that App and MspA show calcium‐specific binding to the CTLD4‐7 region of MR.
Figure 4.
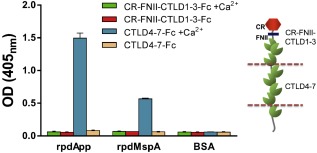
Calcium‐specific binding of rpdApp and rpdMspA to mannose receptor. ELISA plates were coated with rpdApp, rpdMspA (5 μg ml−1) or 1% BSA. Following incubation, wells were washed and MR fragments CR‐FNII‐CTLD1‐3‐Fc or CTLD4‐7‐Fc dissolved in lectin buffer supplemented with or without 10 mM Ca2+ were added. Bound MR was detected with a mouse anti‐human immunoglobulin Fc‐specific antibody coupled to alkaline phosphatase. Each test was performed in triplicate wells and on a least three independent occasions. Values represent the mean + SD from three experiments. The domain structure of MR is indicated in the schematic (inset). CR = cysteine‐rich domain (red); FNII = fibronectin type II domain (blue); CTLDs = C‐type lectin‐like domains (green).
App and MspA interaction with histones
In addition to MR and TfR1, our cross‐linking experiments also identified several histone proteins as potential rpdApp and rpdMspA‐interacting molecules [for rpdApp, Histone H2B (score 557), H4 (score 489) and H2A (score 623) and for rpdMspA, histone H2B (score 1625), H3.1 (score 1759) and H4 (score 378)]. Histones are major structural components of chromatin and play a role in chromatin condensation and gene regulation (Khorasanizadeh, 2004). Five major histone families have been described: H1/H5; H2A; H2B; H3; and H4. We examined the binding of rpdApp and rpdMspA to a panel of representative recombinant human histones (Fig. 5A). Both bound to each of the histones tested, with rpdApp showing greater binding to all histones compared with rpdMspA. Both proteins bound the core histones (H2A, H2B, H3.1 and H4) at higher levels than to the linker histone (H1) (Fig. 5A).
Figure 5.
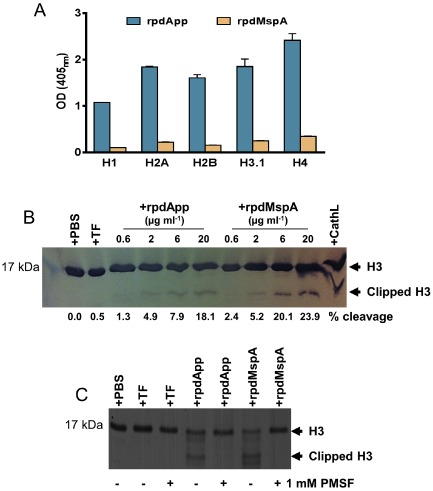
rpdApp or rpdMspA interaction with histones.
A. ELISA plates were coated with recombinant human histones (5 μg ml−1). Following washing, rpdApp and rpdMspA (5 μg ml−1) were added; bound autotransporters were detected using rabbit anti‐TF, followed by anti‐rabbit IgG alkaline phosphatase conjugate. Specific binding was calculated by subtracting the BSA (negative control) values from the histone‐autotransporter absorbance. Tests were performed in triplicate wells and on two independent occasions. Values represent the mean + SD from two experiments.
B. Dose‐dependent cleavage of recombinant histone H3.1 by rpdMspA and rpdApp. Histones were incubated with rpdMspA or rpdApp at 37°C for 15 h. Cathepsin L (CathL; 6 μg ml−1) and TF (20 μg ml−1) were used as positive and negative controls for cleavage, respectively. Reactions were stopped by the addition of SDS‐PAGE sample buffer and cleavage products subjected to SDS‐PAGE. Proteins were strained with Simply Blue Safe stain (Invitrogen). The percentage of clipped H3.1 shown for each lane was determined by densitometry using ImageJ software for data acquisition.
C. Cleavage of histone H3.1 by rpdMspA and rpdApp could be inhibited by 1 mM PMSF, confirming that the serine endopeptidase activity of rpdApp and rpdMspA was responsible for cleavage.
Core histones are heavily post‐translationally modified in order to provide functional variation (Rothbart and Strahl, 2014). One major modification of histone H3 is proteolytic cleavage of the N‐terminal tail region (Duncan et al., 2008; Santos‐Rosa et al., 2009). Given that MspA and App harbour serine endopeptidase domains, we examined the ability of rpdApp and rpdMspA to proteolytically cleave H3. Our data showed that rpdApp and rpdMspA could cleave H3 in a dose‐dependent manner, with the latter exhibiting higher protease activity (Fig. 5B). Inhibition of autotransporter‐mediated clipping of H3 using the serine protease inhibitor phenylmethanesulfonyl fluoride (PMSF) confirmed that cleavage was due to the serine endopeptidase activity of rpdApp and rpdMspA (Fig. 5C).
App and MspA induce cell death by caspase‐dependent apoptosis
Histone post‐translational modifications are involved in numerous processes, including gene expression, DNA replication and repair, and apoptosis (Zhang and Pradhan, 2014). To further understand the effect of exposing DCs to rpdApp and rpdMspA, we investigated the ability of both proteins to induce cell death. After exposure, DCs were collected, stained with propidium iodide (PI), and the proportion of PI‐stained (dead) and unstained (live) cells was determined by flow cytometry. Our data showed that rpdApp and rpdMspA (but importantly not TF alone) could induce a dose‐dependent increase in cell death (Fig. 6). As expected, treatment with staurosporine or nocodazole (known apoptosis‐inducing agents) also led to a significant increase in DC cell death. Importantly, addition of heat‐inactivated or proteinase K‐treated rpdApp and rpdMspA did not result in a significant increase in DC cell death, eliminating the possibility that trace amounts of contaminating LPS might account for the effect (Fig. 6). Further experiments showed that Z‐VAD‐FMK (a pan‐caspase inhibitor) (Van Noorden, 2001) could significantly inhibit the rpdApp‐ and rpdMspA‐mediated effects, confirming that these proteins induce cell death by a caspase‐dependent pathway (Fig. 7).
Figure 6.
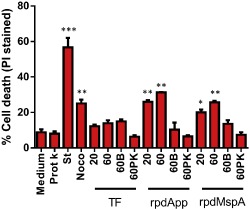
rpdApp and rpdMspA induce a dose‐dependent increase in human dendritic cell death. 1 × 105 human monocyte‐derived dendritic cells were stimulated with TF, rpdApp or rpdMspA at 20 or 60 μg ml−1. To eliminate the effect of contaminating LPS in the protein preparations, 60 μg ml−1 heat‐inactivated (60B) or proteinase K‐treated (60PK) proteins were used as negative controls. Staurosporine (St; 2 μM) and nocodazole (Noco; 60 μg ml−1) were used as positive controls for cell death respectively. After exposure, cells were collected and stained with propidium iodide (PI) followed by flow cytometric analysis, and PI‐positive cells (dead) were compared with unstained cells (live) for the determination of cell death. Statistically significant differences, compared with medium alone, were assessed by one‐way ANOVA and Student's t‐test: *P < 0.05; **P < 0.01; ***P < 0.001. Values shown are the mean + SD from five independent donors.
Figure 7.
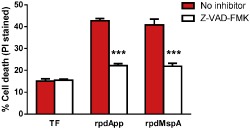
Inhibition of rpdApp‐ and rpdMspA‐mediated cell death using Z‐VAD‐FMK. Human monocyte‐derived dendritic cells were stimulated with TF, rpdApp or rpdMspA at 60 μg ml−1 in the presence or absence of 10 μM Z‐VAD‐FMK. After exposure, the cells were collected and stained with propidium iodide (PI) followed by flow cytometric analysis, and PI‐positive cells (dead) were compared with unstained cells (live) for the determination of cell death. Statistically significant differences, between cell death in the presence or absence of inhibition, were assessed by one‐way ANOVA and Student's t‐test: ***P < 0.001. Values shown are the mean + SD from three independent donors.
Discussion
Previously, we identified and partially characterized App and MspA: two meningococcal proteins that share a number of common features; both are immunogenic, type Va autotransporters, which mediate bacterial–host cell adhesion (Hadi et al., 2001; Serruto et al., 2003; Turner et al., 2006). Additionally, their functional passenger domains, harbouring serine protease motifs, are released from the cell surface via auto‐cleavage or by the action of NalP (van Ulsen et al., 2003). Here, we provide further evidence of commonalities between App and MspA, while additional molecule‐specific functions cannot be excluded, especially since App and MspA only exhibit ca. 34% overall amino acid identity.
The significance of App and MspA in disease was confirmed using a human CD46‐expressing murine infection model. Mutation of app or mspA did not dramatically suppress disease; unsurprising given that meningococcal pathogenesis is a multifaceted process involving complex meningococcal–host interactions (Virji, 2009). While the relatively small group size of 10 mice per group (for reasons of cost and availability) may have precluded some differences in survival rates between groups being statistically significant; nevertheless, statistically significant differences were observed in virulence and in vivo growth/survival at some time points. Highest survival rates were observed for the mutant lacking both autotransporters, suggestive of (at least partial) functional redundancy between the two proteins. Previous work suggesting a role for both proteins in adhesion (Hadi et al., 2001; Serruto et al., 2003; Turner et al., 2006), and subsequent experiments in this study showing that both proteins traffic towards the host cell nucleus, clip histone H3 and induce apoptosis, support this hypothesis. Redundancy is a recurring feature of meningococcal pathogenesis, enabling maintenance of key phenotypes (e.g. adhesion or iron acquisition), while also allowing for evasion of host defences via the modulation of surface epitopes (Hill and Virji, 2012). App is constitutively expressed across all meningococcal serogroups (Hadi et al., 2001). In contrast, not all strains harbour mspA, some strains contain a mspA gene with a single nucleotide mutation, which places a premature stop codon in frame, and MspA expression is phase‐variable due to the presence of a poly‐cytosine tract within the coding sequence that is presumed to be susceptible to slipped‐strand mispairing during replication (van Ulsen et al., 2006; Oldfield et al., 2013).
Our data show that both autotransporters specifically bind to both MR and TfR1. These receptors mediate endocytosis via separate mechanisms and recognize different types of substrates. TfR1 is a transmembrane homo‐dimeric glycoprotein, expressed by virtually all cells, which is required for internalization of iron‐loaded transferrin via endosomes through clathrin‐dependent receptor‐mediated endocytosis (Hayes et al., 1997; Aisen, 2004; Syed et al., 2006). It is also involved in the cellular entry of several viruses, including hepatitis C virus (Martin and Uprichard, 2013), New World haemorrhagic fever arenaviruses (Demogines et al., 2013), mouse mammary tumour virus (Wang et al., 2008), and canine and feline parvoviruses (Parker et al., 2001). To our knowledge, this is the first study showing TfR1 involvement in the uptake of a bacterial ligand.
Mannose receptor is a member of the C‐type (calcium‐dependent) lectin receptor superfamily, which is involved in innate immune responses as well as mediating selective endocytosis (Sallusto et al., 1995; Linehan et al., 1999). MR is expressed mainly by DCs and macrophages and contains an N‐terminal CR domain, an FNII, and a CTLD1‐8 (Martinez‐Pomares, 2012). Through these various domains, MR can bind a variety of endogenous and exogenous ligands, with the CTLD domains (implicated here in App and MspA binding) commonly binding allergens and microbial products (Taylor et al., 1992). MR has been implicated in the cellular uptake of several bacteria, including Enterococcus faecalis, Listeria monocytogenes, E. coli, Bacillus subtilis and Streptococcus pneumoniae (Zamze et al., 2002; Macedo‐Ramos et al., 2011; Hashino et al., 2012; Tsuruta et al., 2013). Clearly, MR and TfR1 have little in common; nevertheless, both their substrates (when used alone) blocked the internalization, but not the binding, of rpdApp and rpdMspA into DCs, perhaps implying that autotransporter internalization requires the cooperation of both receptors. Interestingly, we observed that maximal nuclear localization of rpdApp and rpdMspA occurred later in HBMECs than DCs (8 h and 45 min, respectively). We hypothesize that this results from differential receptor expression, but this requires experimental confirmation. Additionally, further work is required to define the regions of both receptors and their autotransporter ligands, which are involved in uptake.
There is an emerging recognition that secretion of proteins that target the host cell nucleus, subsequently altering host cell biology, is an important pathogenic mechanism of Gram‐negative bacteria (Bhavsar et al., 2007). Examples of such secreted proteins include cytolethal distending toxins (McSweeney and Dreyfus, 2004; Matangkasombut et al., 2010), Anaplasma phagocytophilum AnkA (Park et al., 2004), Salmonella enterica SspH1 (Haraga and Miller, 2003) and Yersinia YopM (Benabdillah et al., 2004); our data suggest that App and MspA are also members of this group. Once in the eukaryotic cytoplasm, proteins < 40 kDa can access the nucleus by passive diffusion; larger proteins are commonly transported by nuclear transporter receptors following recognition of nuclear localization signals (NLSs) in the cargo protein (Gerace, 1992; Lange et al., 2007; Marfori et al., 2011). NLSs are short motifs rich in basic amino acids, such as arginine and lysine (Marfori et al., 2011). Importantly, some secreted fragments of Neisserial IgA1 protease contain NLS motifs; these have been shown to promote the accumulation of reporter proteins in the eukaryotic cell nucleus (Pohlner et al., 1995). App also contains at least one putative NLS (933RRRSRRSR940 in MC58 App) (Hadi et al., 2001), whereas NLS sequences are not evident in MspA (Turner et al., 2006). Whether the putative NLSs in App are functional, and precisely how App and MspA are trafficked to the nucleus will require additional experimentation. Intriguingly, the NLSs present in both IgA1 protease and App are present in their respective α‐peptide regions. These are present in the larger secreted fragments of IgA1 protease and App (160 and 140 kDa, respectively), but absent from the smaller, auto‐proteolytically released, forms (110 and 100 kDa respectively) (van Ulsen et al., 2003). The biological significance of releasing the different forms of these autotransporters is currently unknown, but fragments containing or lacking the α‐peptide may differ in their ability to access the nucleus and may therefore elicit different cellular responses. The recombinant proteins used in this study correspond closely to the larger, α‐peptide‐containing, fragments secreted by N. meningitidis. These were used because the low abundance of both App and MspA in culture supernatants has impeded the purification of experimentally useful amounts of autotransporter fragments direct from the complex meningococcal secretome and consequently hampered the definition of the sites of auto‐proteolytic and NalP‐mediated cleavage.
Eukaryotic chromosomal DNA is packaged into nucleosomes; these contain two copies each of the core histones H2A, H2B, H3 and H4 (Luger et al., 1997). Histone modifications, including methylation, acetylation and phosphorylation, play important roles in numerous cellular processes, including DNA replication and gene expression (Strahl and Allis, 2000; Khorasanizadeh, 2004). Histone H3 is the target of many modifications; one is the proteolytic cleavage of the N‐terminal tail region. For example, cathepsin L cleavage of the H3 N‐terminus is associated with gene activity during mouse cell development (Duncan et al., 2008), and in Saccharomyces cerevisiae, H3 clipping has been associated with gene activation during stationary phase (Santos‐Rosa et al., 2009). Our data show that rpdApp and rpdMspA can bind core histone proteins and can proteolytically cleave H3 in a dose‐dependent manner. Our results also suggest that rpdMspA binds histones less efficiently than rpdApp, yet cleaves H3 more efficiently. The generation of rpdApp and rpdMspA derivatives harbouring substitutions at their respective catalytic serine residues will allow a more accurate determination of their relative histone‐binding efficiencies in the absence of cleavage. Interestingly, based on their mobility on SDS‐PAGE gels, the clipped H3 fragments generated by both autotransporters appeared similar to those generated by cathepsin L, suggesting cleavage at, or near, the same region (A21‐K27) (Duncan et al., 2008), but this requires additional experimental confirmation.
Since histone H3 tail clipping can regulate gene expression and histone modifications can influence additional cellular processes including apoptosis (Füllgrabe et al., 2010), we examined the effect of App and MspA on DCs. This showed that both proteins induce cell death by a caspase‐dependent pathway. In the study by Robinson et al., meningococcal secreted proteins (MSPs) induced complex changes in the gene expression profile of human meningothelial cells, leading to an increased resistance to apoptosis (Robinson et al., 2004). Thus, effects mediated by specific proteins (i.e. App and MspA) have to be considered in the context of a complex multifaceted interaction involving numerous other ligand–receptor and cell signalling events.
In summary, our data provide novel and important insights into the roles of App and MspA in meningococcal infection and notably provide further evidence of commonalities between them, namely they both (i) bind mannose receptor and transferrin receptor, (ii) translocate to the nucleus, (iii) bind and proteolytically leave histone, and (iv) induce cell death.
Experimental procedures
Bacterial strains and culture conditions
Escherichia coli strain JM109 was used for plasmid propagation, and either XL10‐Gold or BL21‐Gold (DE3) was used for recombinant protein expression (Table 1). Strains were grown at 37°C in lysogeny broth (LB) or on LB agar supplemented, where appropriate, with ampicillin (100 μg ml−1), streptomycin and spectinomycin (100 μg ml−1) or kanamycin (25 μg ml−1). N. meningitidis strains were grown at 37°C, in an atmosphere of air plus 5% CO2, on Columbia agar with chocolated horse blood (Oxoid) or in brain–heart infusion (BHI) broth (Oxoid) supplemented, where appropriate, with streptomycin and spectinomycin (100 μg ml−1) and/or kanamycin (50 μg ml−1) (Table 1 ).
Table 1.
Characteristics of bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| JM109 | Plasmid propagation strain | Promega |
| XL10‐Gold | Expression strain for pQE70‐based plasmids | Stratagene |
| BL21‐Gold (DE3) | Expression strain for pCold TF‐based plasmids | Stratagene |
| N. meningitidis | ||
| MC58 | Wild‐type serogroup B strain | ATCC |
| MC58Δapp | app replaced with Ω cassette | Hadi et al. (2001) |
| MC58ΔmspA | mspA replaced with kanamycin cassette | This study |
| MC58ΔappΔmspA | mspA mutant derivative of MC58Δapp | This study |
| Plasmids | ||
| pTOPOMspAflΔΩ | mspA mutagenesis plasmid containing Ω cassette | Turner et al. (2006) |
| pJMK30 | Source of kanamycin resistance cassette | van Vliet et al. (1998) |
| pTOPOMspAflΔKan | mspA mutagenesis plasmid containing kanamycin cassette | This study |
| pGEM‐T Easy | PCR cloning vector | Promega |
| pGEMTEpdapp | Region of MC58 app encoding 43G–1179N in pGEM‐T Easy | This study |
| pGEMTEpdmspA | Region of MC58 mspA encoding 27S–1152A in pGEM‐T Easy | This study |
| pQE70 | Expression vector | Qiagen |
| pQE70pdapp | Expression vector encoding 6 × histidine‐tagged 43G–1179N fragment of MC58 App | This study |
| pQE70pdmspA | Expression vector encoding 6 × histidine‐tagged 27S–1152A fragment of MC58 MspA | This study |
| pCold TF | Protein expression vector | Takara Bio |
| pCold TF‐App | Expression vector encoding 6 × histidine‐tagged 43G–1179N of MC58 App fused to trigger factor | GenScript |
| pCold TF‐MspA | Expression vector encoding 6 × histidine‐tagged 27S–1152A of MC58 MspA fused to trigger factor | This study |
Construction of app, mspA and app / mspA double mutants
We previously described a streptomycin/spectinomycin‐resistant app mutant of strain MC58 (Hadi et al., 2001). To construct an mspA mutant with a different selectable marker, the streptomycin/spectinomycin cassette was removed from pTOPOMspAflΔΩ (Turner et al., 2006) (Table 1) by KpnI digestion and replaced with a 1.5 kb kanamycin resistance cassette extracted from pJMK30 after digestion with KpnI. The resulting plasmid, pTOPOMspAflΔKan, was confirmed by DNA sequencing and subsequently used to transform MC58 and MC58Δapp by natural transformation and allelic exchange to yield strains MC58ΔmspA and MC58ΔappΔmspA respectively (Table 1).
Murine infection experiments
These utilized an hCD46Ge transgenic mouse line (CD46+/+) (Mrkic et al., 1998). Animals were age‐ (6–8 weeks) and sex‐matched and verified for the CD46+/+ genotype by polymerase chain reaction (PCR). Meningococcal strains were grown for 18 h at 37°C, in an atmosphere of air plus 5% CO2, on Gonococcus (GC) agar base (Difco) with Kellogg's supplement. Bacteria were resuspended in GC broth, and each mouse challenged intraperitoneally with 1.2 × 109 colony‐forming units (CFU) in 100 μl of GC broth. Bacterial inoculums were verified by retrospective viable counts. Ten mice were inoculated per group, except for the negative control group (n = 7), which were sham‐challenged with 100 μl of GC broth. All investigations were carried out blind. The health status of all mice was closely monitored for 96 h. At the indicated time points, whole blood samples were collected from the tail vein. Bacteraemia levels were determined by serial dilution of blood samples, plating aliquots onto agar and subsequently determining numbers of CFU. All mouse experiments were performed at the animal facility of the Wenner‐Gren Institute, Stockholm University. Animal care and experiments were conducted according to the institution's guidelines for animal husbandry and all protocols were approved by the Swedish Ethical Committee on Animal Experiments (Approval ID: N380/08).
Construction of plasmids encoding recombinant proteins
Primers app‐QE70‐For_nos/app‐QE70‐Rev_noB and mspa‐QE70‐For_nos/mspa‐QE70‐Rev_noB (Table 2) were used to amplify fragments of ca. 3.4 kb encoding amino acids 43G to 1179N and 27S to 1152A, from N. meningitidis MC58 app and mspA respectively. PCR products were cloned into pGEM‐T Easy, used according to the manufacturer's instructions, to obtain pGEMTEpdapp and pGEMTEpdmspA respectively (Table 1). The cloned inserts were subsequently removed by digestion with BglII and SphI and ligated into pQE70, digested with the same enzymes, to yield plasmids pQE70pdapp and pQE70pdmspA respectively.
Table 2.
Primers used in this study
| Primer | Sequence | Restriction site |
|---|---|---|
| app‐QE70‐For_nos | CGCGCATGCATGGACACACTTATTTCGGCATC | SphI |
| app‐QE70‐Rev_noB | GCGAGATCTATTGGCATAACGGCTGATC | BglII |
| mspa‐QE70‐For_nos | CGCGCATGCAATCCATTGTCCGCAACGATG | SphI |
| mspa‐QE70‐Rev_noB | GCGAGATCTGGCCGACCGGCTGAT | BglII |
| pColdTF‐MspA(F) | GGCTGGTACCATGCATTCCATTGTCCGCAAC | KpnI |
| pColdTF‐MspA(R) | GCGTAAGCTTTTATTAAGATCTGGCCGACCGGCTG | HindIII |
pCold TF‐App, encoding App 43G to 1179N with N‐terminal TF and 6 × histidine tags, was constructed by GenScript Corporation utilizing the cold shock expression vector pCold TF. pCold TF‐MspA encoding MspA 27S to 1152A with N‐terminal TF and 6 × histidine tags was constructed by amplifying the relevant mspA fragment from MC58 using primers pColdTF‐MspA(F) and pColdTF‐MspA(R) (Table 2). After digestion with KpnI and HindIII, the PCR product was ligated to KpnI‐ and HindIII‐digested pCold TF to yield pCold TF‐MspA.
Protein expression and purification
rpdApp and rpdMspA were expressed in E. coli XL10‐Gold harbouring plasmids pQE70pdapp or pQE70pdmspA, respectively, purified under denaturing conditions and re‐natured by dialysis. Briefly, cultures were grown to an OD600 of 0.6 and induced with 1 mM IPTG for 3 h. Cells were harvested by centrifugation at 3,000 × g for 10 min, resuspended in 5 ml of denaturing wash buffer (200 mM Tris–HCl, pH 7.5, 100 mM EDTA, 10% Triton X‐100) per gram (wet weight) of cells. About 1 mg ml−1 lysozyme (Sigma) was added and the suspension was incubated at room temperature for 30–60 min. Cell suspensions were subjected to sonication (10 cycles of 10 s on, 20 s off) using an MSE Soniprep 150, frozen at −80°C, thawed and subjected to repeat sonication. The samples were then centrifuged at 5000 × g for 30 s, and the supernatant was collected and centrifuged at 10 000 × g for 30 min at 4°C. Pellets were re‐suspended in denaturing wash buffer, incubated at room temperature for 15 min and centrifuged at 10 000 × g for 30 min at 4°C. Pellets were solubilized in 1 × SDS‐PAGE sample buffer [0.125 M Tris–HCl (pH 6.8), 1% SDS, 5% glycerol, 0.0004% bromophenol blue] and proteins separated by SDS‐PAGE (7.5% gels; Protean–II, Bio‐Rad). Gels were stained briefly with Simply Blue Safe stain (Invitrogen) and the recombinant protein excised, transferred to Midi D‐Tube dialysers [molecular weight cut‐off (MWCO) 6–8 kDa, Novagen) and electro‐eluted according to the recommendations of the manufacturer. Proteins were then dialysed in a fresh D‐Tube against phosphate‐buffered saline (PBS) for 48 h and concentrated using Microcon centrifugal filter units (30 kDa MWCO).
For expression and purification of recombinant proteins under non‐denaturing conditions, E. coli BL21‐Gold (DE3) cells harbouring pCold TF‐App, pCold TF‐MspA or pCold TF were grown at 37°C with shaking to an OD600 of 0.6, after which the culture was cooled to 15°C for 30 min without shaking. IPTG was added to 0.5 mM and the culture incubated with shaking at 15°C for 24 h. Following incubation, cells were harvested by centrifugation at 5000 × g at 4°C for 10 min. The cell pellet was resuspended in 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0. Cells were lysed by intermittent sonication for 5 min in an ice‐water bath, and the bacterial lysate was centrifuged at 11 000 × g at 4°C for 30 min. The supernatant was removed, filtered through a 0.22 μm sterile syringe filter (Sartorius) and recombinant protein purified using HisPur cobalt resin (Thermo Scientific) using a gravity flow column (Sigma). Briefly, the supernatant–resin mixture was washed with 10 column volumes of 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0. After washing, recombinant protein was eluted in 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0, concentrated using 30 kDa MWCO centrifugal concentrators (Millipore) and dialysed against sterile PBS.
Generation of antisera and immunoblotting
Dunkin‐Hartley female guinea pigs were immunized subcutaneously four times at 3 week intervals with 50 μg of natively purified rpdApp or rpdMspA emulsified in Freud's complete (first immunization only) or incomplete adjuvant. Guinea pig anti‐App or anti‐MspA antisera were used in immunoblotting experiments 1:4000 diluted in blocking buffer (1% BSA in PBS/0.05% Tween 20) and incubated for 2 h. After washing three times in PBS‐T, membranes were incubated for 2 h with anti‐guinea pig immunoglobulin G (IgG)‐alkaline phosphatase conjugate (CovalAb) (1:4,000 diluted in blocking buffer). After washing with PBS‐T, the blots were developed using BCIP/NBT‐Blue liquid substrate (Sigma). Alternatively, detection of 6 × histidine‐tagged recombinant proteins was performed using mouse anti‐penta histidine monoclonal antibody (Qiagen; 1:10,000 diluted), followed by anti‐mouse IgG alkaline phosphatase conjugate (Sigma; 1:30 000 diluted).
Generation of DCs and cell culture
Heparinized venous blood was obtained from healthy human volunteers and peripheral blood mononuclear cells (PBMCs) isolated by density gradient centrifugation over Ficoll‐Histopaque (Sigma). Isolated PBMCs were washed three times in Hank's balanced salt solution (Gibco) and resuspended in RPMI 1640 medium (Gibco). Purified PBMCs were then incubated at 4°C for 30 min with mouse anti‐human CD14‐coated micro beads (Miltenyi Biotec), washed with RPMI 1640 and separated magnetically using a magnetic activated cell sorting (MACS) separator (Miltenyi Biotec). CD14 negative cells were washed off the column and retained (CD14‐positive) cells eluted after removal of the column from the magnetic separator. Eluted CD14‐positive cells (monocytes) were washed twice with RPMI 1640 and seeded into 6‐well flat‐bottomed culture plates at approximately 1 × 106 cells per well in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 2 mM L‐glutamine. To stimulate the differentiation of monocytes into DCs, 50 ng ml−1 granulocyte‐macrophage colony‐stimulating factor (GM‐CSF; Perotech) and 250 IU ml−1 IL‐4 (R&D Systems) were added to the media. DCs were harvested after incubation for 6 days at 37°C in an atmosphere of 5% CO2.
Human brain microvascular endothelial cells (HBMECs; ATCC) were cultured on human fibronectin‐coated flasks (BD Biosciences) in endothelial cell medium (ECM; ScienCell‐USA) supplemented with 5% FBS, endothelial cell growth supplement (ECGS; ScienCell‐USA), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. Cells were maintained at 37°C in an atmosphere of 5% CO2.
Cell internalization assays
Proteins were FITC (DCs) or Cy5 (HBMECs) labelled using the Lightning‐Link conjugation kit (Innova Biosciences) according to the manufacturer's instructions. DC uptake assays were carried out in RPMI 1640 media supplemented with 10% FBS, at 37°C in an atmosphere of air plus 5% CO2. DCs (1 × 106 cells) were plated on poly‐L‐lysine‐coated 35 mm glass base dishes (Iwaki) and incubated for 1 h. DCs were then incubated with labelled rpdApp, rpdMspA or TF (30 μg ml−1) for 45 min. Following incubation, cell nuclei were stained with 10 μg ml−1 Hoechst 33258 (Sigma) in PBS for 10 min at room temperature and rinsed thrice with PBS. The cells were fixed in 1% paraformaldehyde in PBS for 10 min and then washed thrice before analysis by confocal microscopy. Imaging was performed using a Zeiss LSM 710 confocal laser scanning microscope, equipped with lasers at 488 and 364 nm for excitation of FITC and Hoechst 33258 respectively. For blocking experiments, DCs were incubated for 30 min with human transferrin (Sigma; 1 mg ml−1) or S. cerevisiae mannan (Sigma; 1 mg ml−1) for 30 min at 37°C prior to the addition of labelled proteins.
For HMBECs, confluent monolayers were harvested and seeded onto fibronectin‐coated coverslips in 24‐well plates at a seeding density of 5 × 105 cells per well and incubated for 16 h. Cells were then treated with labelled rpdApp, rpdMspA or TF (30 μg ml−1) for 8 h. After treatment, cells were washed thrice and fixed in 1% paraformaldehyde for 10 min at room temperature and rinsed thrice with PBS. Cells were mounted using proLong Gold anti‐fade with DAPI (Life Technologies) to stain cell nuclei. Confocal imaging was performed using a Zeiss LSM 710 equipped with lasers at 633 and 405 nm for excitation of Cy5 and DAPI respectively. All images were scanned at a resolution of 1024 × 1024 pixels using the same laser and gains settings. The images collected in the two channels were later merged and analysed using Zeiss LSM software. Co‐localization for DCs was determined using Zeiss LSM image examiner software using the threshold method; a value of 25% for both FITC and Hoechst (Blue, nucleus staining) channels was applied. For HMBECs, co‐localization was determined using Velocity 6.3.1 software (PerkinElmer).
Re‐tagging
Dendritic cells were incubated for 2 h with rpdApp or rpdMspA to which the Sulfo‐SBED cross‐linker (Pierce) had been conjugated according to the manufacturer's recommendations (5 μg of cross‐linker tagged protein to 106 cells). After incubation, cells were harvested by centrifugation at 250 × g for 5 min, resuspended in PBS‐T and exposed to UV light (365 nm) for 5 min. Cells were washed, resuspended in 10 mM dithiothreitol (DTT) (Sigma) as a reducing agent and incubated for 10 min. Following three washes in PBS, cells were boiled and proteins resolved by SDS‐PAGE. Biotin‐tagged proteins were identified by probing with streptavidin. Reactive proteins bands were excised, digested with sequencing‐grade trypsin (Promega) and analysed using a Micromass Tof‐Spec E (Micromass).
ELISA
Soluble human transferrin receptor (sTfR1; BioVendor) was diluted in PBS to 2 μg ml−1 and 100 μl aliquots added to 96‐well microtiter plates (PolySorp, amino surface immobilizer; Nunc). PBS + 1% BSA was used as a negative control. Plates were incubated overnight at 4°C, washed in PBS‐T, and blocked in PBS + 1% BSA for 1 h. 100 μl of aliquots of recombinant protein (5 μg ml−1 in PBS) were added and incubated overnight at 4°C. Plates were washed and incubated with guinea pig anti‐App or anti‐MspA (1:4,000 diluted) overnight at 4°C. After washing, plates were incubated with alkaline phosphatase‐conjugated anti‐guinea pig IgG (CovalAb; diluted 1:4000) for 1 h. Following washing, bound antibody was detected using p‐nitrophenyl phosphate substrate (pNPP; Sigma) diluted in buffer (100 mM Tris–HCl, 100 mM NaCl, 1 mM MgCl2, pH 9.5) to a final concentration of 1 mg ml−1. The absorbance at 405 nm was measured using a BioTek EL800 plate reader.
To detect binding of MR fragments to rpdApp or rpdMspA, CR‐FNII‐CTLD1‐3‐Fc and CTLD4‐7‐Fc were purified as previously reported (Martinez‐Pomares et al., 2006), and ELISAs undertaken as above, with modifications. Wells were coated with rpdApp or rpdMspA (5 μg ml−1) or 1% BSA for 5 h at 4°C. Plates were washed with 10 mM Tris–HCl (pH 7.5), 10 mM Ca2+, 154 mM NaCl and 0.05% Tween 20 (lectin buffer). Following blocking, wells were incubated with MR fragments (2 μg ml−1) for 2 h. Wells were subsequently washed and probed with alkaline phosphatase‐conjugated anti‐human IgG (Fc‐specific) antibody (Sigma) diluted 1:5,000. When assays were performed to examine binding in the presence or absence of Ca2+, lectin buffer and calcium‐free lectin buffer were used respectively.
Non‐post‐translationally modified, non‐tagged, recombinant human histones (New England Biolabs) were used to assess binding with rpdApp and rpdMspA. Briefly, wells were coated overnight with histones (5 μg ml−1) or PBS + 1% BSA. Following PBS‐T washes, wells were blocked with PBS + 1% BSA for 1 h, then recombinant protein (5 μg ml−1) was added for 5 h at 4°C. Wells were washed and rabbit anti‐TF antibody (GenScript) (1:1000 diluted) was added. Following overnight incubation, wells were washed and anti‐rabbit IgG alkaline phosphatase conjugate (Sigma; 1:30 000 in 1% BSA) was added for 1 h before bound antibody is detected using pNPP substrate.
Histone clipping
Recombinant histone H3.1 (New England Biolabs; 200 μg ml−1 final concentration) and natively purified recombinant protein (0.6–20 μg ml−1 final concentration) were mixed in a total volume of 40 μl of PBS and incubated at 37°C for 15 h. Mouse cathepsin L (MP Biomedical; 6 μg ml−1 final concentration) was used as a positive control for cleavage. Reactions were stopped by the addition of 5 × sample buffer [0.62 M Tris–HCl (pH 6.8), 5% SDS, 25% glycerol, 12.5% β‐mercaptoethanol, 0.25 M DTT, 0.002% bromophenol blue], followed by immediate boiling for 5 min. Aliquots of cleavage products were subjected to SDS‐PAGE, and gels were strained with Simply Blue Safe stain (Invitrogen). Densitometry was performed on a 16 bit greyscale TIFF file of the scanned gel image using ImageJ software for data acquisition. PMSF (1 mM; Sigma) was added to assays for inhibition experiments.
Apoptosis assays
About 1 × 105 DCs were stimulated with recombinant proteins (20 or 60 μg ml−1) for 24 h in RPMI 1640 media. To eliminate the effect of contaminating LPS in the protein preparations, heat‐inactivated (100°C for 15 min) or proteinase K‐treated (100 μg ml−1 Sigma 56°C for 60 min) proteins were used as negative controls. Exposure for 4 h to staurosporine (2 μM; Sigma) and nocodazole (60 μg ml−1; Sigma) was used as a positive control for cell death. Inhibition experiments were performed as above using 60 μg ml−1 recombinant proteins and in the presence or absence of 10 μM Z‐VAD‐FMK (Z‐Val‐Ala‐Asp‐fluoromethylketone; Sigma). After exposure, cells were washed and resuspended in PBS containing 1 μg ml−1 PI (Sigma), followed by flow cytometric analysis using a BD FACSAria flow cytometer. Data were analysed using the FACSDiva and WinMDI flow cytometry software, and PI‐positive cells (dead) were compared with unstained cells (live) for the determination of cell death.
Supporting information
Fig. S1. Uptake of rpdMspA and rpdApp into human brain microvascular endothelial cells (HBMECs). HBMECs were incubated with Cy5‐labelled rpdMspA (A), rpdApp (B) or TF (C). Cell nuclei were stained with DAPI. Co‐localisation of labelled recombinant protein and nuclei (D) was determined using Velocity 6.3.1 software (Perkin Elmer). Mean fluorescence intensities were derived from ≥ 3 different fields. Error bars: mean of values + SD. All images were scanned at a resolution of 1024 × 1024 pixels, using the same laser and gains settings. The cells are representative of cells observed from multiple experiments. Scale bars = 10 μm.
Acknowledgements
We thank Dr J. Stoof (University of Nottingham, UK) for the technical assistance during the construction of plasmid pTOPOMspAflΔKan.
References
- Aisen, P. (2004) Transferrin receptor 1. Int J Biochem Cell Biol 36: 2137–2143. [DOI] [PubMed] [Google Scholar]
- Ait‐Tahar, K. , Wooldridge, K.G. , Turner, D.P.J. , Atta, M. , Todd, I. , and Ala'Aldeen, D.A.A. (2000) Auto‐transporter A protein of Neisseria meningitidis: a potent CD4+ T‐cell and B‐cell stimulating antigen detected by expression cloning. Mol Microbiol 37: 1094–1105. [DOI] [PubMed] [Google Scholar]
- Benabdillah, R. , Mota, L.J. , Lutzelschwab, S. , Demoinet, E. , and Cornelis, G.R. (2004) Identification of a nuclear targeting signal in YopM from Yersinia spp. Microb Pathog 36: 247–261. [DOI] [PubMed] [Google Scholar]
- Benz, I. , and Schmidt, M.A. (2011) Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol 301: 461–468. [DOI] [PubMed] [Google Scholar]
- Bhavsar, A.P. , Guttman, J.A. , and Finlay, B.B. (2007) Manipulation of host‐cell pathways by bacterial pathogens. Nature 449: 827–834. [DOI] [PubMed] [Google Scholar]
- Celik, N. , Webb, C.T. , Leyton, D.L. , Holt, K.E. , Heinz, E. , Gorrell, R. , et al (2012) A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One 7: e43245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines, A. , Abraham, J. , Choe, H. , Farzan, M. , and Sawyer, S.L. (2013) Dual host‐virus arms races shape an essential housekeeping protein. PLoS Biol 11: e1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, E.M. , Muratore‐Schroeder, T.L. , Cook, R.G. , Garcia, B.A. , Shabanowitz, J. , Hunt, D.F. , and Allis, C.D. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135: 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe, J. , Hajji, N. , and Joseph, B. (2010) Cracking the death code: apoptosis‐related histone modifications. Cell Death Differ 17: 1238–1243. [DOI] [PubMed] [Google Scholar]
- Gerace, L. (1992) Molecular trafficking across the nuclear pore complex. Curr Opin Cell Biol 4: 637–645. [DOI] [PubMed] [Google Scholar]
- Hadi, H.A. , Wooldridge, K.G. , Robinson, K. , and Ala'Aldeen, D.A.A. (2001) Identification and characterization of App: an immunogenic autotransporter protein of Neisseria meningitidis . Mol Microbiol 41: 611–623. [DOI] [PubMed] [Google Scholar]
- Haraga, A. , and Miller, S.I. (2003) A Salmonella enterica serovar typhimurium translocated leucine‐rich repeat effector protein inhibits NF‐kappa B‐dependent gene expression. Infect Immun 71: 4052–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashino, M. , Tachibana, M. , Shimizu, T. , and Watarai, M. (2012) Mannose receptor, C type 1 contributes to bacterial uptake by placental trophoblast giant cells. FEMS Immunol Med Microbiol 66: 427–435. [DOI] [PubMed] [Google Scholar]
- Hayes, G.R. , Williams, A.M. , Lucas, J.J. , and Enns, C.A. (1997) Structure of human transferrin receptor oligosaccharides: conservation of site‐specific processing. Biochemistry 36: 5276–5284. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R. , Navarro‐Garcia, F. , Desvaux, M. , Fernandez, R.C. , and Ala'Aldeen, D.A.A. (2004) Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68: 692–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D.J. , and Virji, M. (2012) Meningococcal ligands and molecular targets of the host. Methods Mol Biol 799: 143–152. [DOI] [PubMed] [Google Scholar]
- Ilver, D. , Arnqvist, A. , Ogren, J. , Frick, I.M. , Kersulyte, D. , Incecik, E.T. , et al (1998) Helicobacter pylori adhesin binding fucosylated histo‐blood group antigens revealed by retagging. Science 279: 373–377. [DOI] [PubMed] [Google Scholar]
- Johansson, L. , Rytkönen, A. , Bergman, P. , Albiger, B. , Källström, H. , Hökfelt, T. , et al (2003) CD46 in meningococcal disease. Science 301: 373–375. [DOI] [PubMed] [Google Scholar]
- Johansson, L. , Rytkönen, A. , Wan, H. , Bergman, P. , Plant, L. , Agerberth, B. , et al (2005) Human‐like immune responses in CD46 transgenic mice. J Immunol 175: 433–440. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh, S. (2004) The nucleosome: from genomic organization to genomic regulation. Cell 116: 259–272. [DOI] [PubMed] [Google Scholar]
- Koomey, J.M. , Gill, R.E. , and Falkow, S. (1982) Genetic and biochemical analysis of gonococcal IgA1 protease: cloning in Escherichia coli and construction of mutants of gonococci that fail to produce the activity. Proc Natl Acad Sci U S A 79: 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, A. , Mills, R.E. , Lange, C.J. , Stewart, M. , Devine, S.E. , and Corbett, A.H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282: 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton, D.L. , Rossiter, A.E. , and Henderson, I.R. (2012) From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 10: 213–225. [DOI] [PubMed] [Google Scholar]
- Linehan, S.A. , Martinez‐Pomares, L. , Stahl, P.D. , and Gordon, S. (1999) Mannose receptor and its putative ligands in normal murine lymphoid and nonlymphoid organs: in situ expression of mannose receptor by selected macrophages, endothelial cells, perivascular microglia, and mesangial cells, but not dendritic cells. J Exp Med 189: 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K. , Mader, A.W. , Richmond, R.K. , Sargent, D.F. , and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Macedo‐Ramos, H. , Campos, F.S. , Carvalho, L.A. , Ramos, I.B. , Teixeira, L.M. , De Souza, W. , et al (2011) Olfactory ensheathing cells as putative host cells for Streptococcus pneumoniae: evidence of bacterial invasion via mannose receptor‐mediated endocytosis. Neurosci Res 69: 308–313. [DOI] [PubMed] [Google Scholar]
- McSweeney, L.A. , and Dreyfus, L.A. (2004) Nuclear localization of the Escherichia coli cytolethal distending toxin CdtB subunit. Cell Microbiol 6: 447–458. [DOI] [PubMed] [Google Scholar]
- Marfori, M. , Mynott, A. , Ellis, J.J. , Mehdi, A.M. , Saunders, N.F.W. , Curmi, P.M. , et al (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813: 1562–1577. [DOI] [PubMed] [Google Scholar]
- Martin, D.N. , and Uprichard, S.L. (2013) Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A 110: 10777–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Pomares, L. (2012) The mannose receptor. J Leukoc Biol 92: 1177–1186. [DOI] [PubMed] [Google Scholar]
- Martinez‐Pomares, L. , Wienke, D. , Stillion, R. , McKenzie, E.J. , Arnold, J.N. , Harris, J. , et al (2006) Carbohydrate‐independent recognition of collagens by the macrophage mannose receptor. Eur J Immunol 36: 1074–1082. [DOI] [PubMed] [Google Scholar]
- Matangkasombut, O. , Wattanawaraporn, R. , Tsuruda, K. , Ohara, M. , Sugai, M. , and Mongkolsuk, S. (2010) Cytolethal distending toxin from Aggregatibacter actinomycetemcomitans induces DNA damage, S/G2 cell cycle arrest, and caspase‐independent death in a Saccharomyces cerevisiae model. Infect Immun 78: 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrkic, B. , Pavlovic, J. , Rulicke, T. , Volpe, P. , Buchholz, C.J. , Hourcade, D. , et al (1998) Measles virus spread and pathogenesis in genetically modified mice. J Virol 72: 7420–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield, N.J. , Matar, S. , Bidmos, F.A. , Alamro, M. , Neal, K.R. , Turner, D.P.J. , et al (2013) Prevalence and phase variable expression status of two autotransporters, NalP and MspA, in carriage and disease isolates of Neisseria meningitidis . PLoS One 8: e69746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kim, K.J. , Choi, K.S. , Grab, D.J. , and Dumler, J.S. (2004) Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol 6: 743–751. [DOI] [PubMed] [Google Scholar]
- Parker, J.S. , Murphy, W.J. , Wang, D. , O'Brien, S.J. , and Parrish, C.R. (2001) Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol 75: 3896–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner, J. , Langenberg, U. , Wolk, U. , Beck, S.C. , and Meyer, T.F. (1995) Uptake and nuclear transport of Neisseria IgA1 protease‐associated alpha‐proteins in human cells. Mol Microbiol 17: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Poulsen, K. , Brandt, J. , Hjorth, J.P. , Thogersen, H.C. , and Kilian, M. (1989) Cloning and sequencing of the immunoglobulin A1 protease gene (iga) of Haemophilus influenzae serotype b. Infect Immun 57: 3097–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N.D. , Barrett, A.J. , and Bateman, A. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40: D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, K. , Taraktsoglou, M. , Rowe, K.S. , Wooldridge, K.G. , and Ala'Aldeen, D.A.A. (2004) Secreted proteins from Neisseria meningitidis mediate differential human gene expression and immune activation. Cell Microbiol 6: 927–938. [DOI] [PubMed] [Google Scholar]
- Rothbart, S.B. , and Strahl, B.D. (2014) Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 1839: 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Perez, F. , and Nataro, J.P. (2014) Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol Life Sci 71: 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Geme, J.W., 3rd , de la Morena, M.L. , and Falkow, S. (1994) A Haemophilus influenzae IgA protease‐like protein promotes intimate interaction with human epithelial cells. Mol Microbiol 14: 217–233. [DOI] [PubMed] [Google Scholar]
- Sallusto, F. , Cella, M. , Danieli, C. , and Lanzavecchia, A. (1995) Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J Exp Med 182: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos‐Rosa, H. , Kirmizis, A. , Nelson, C. , Bartke, T. , Saksouk, N. , Cote, J. , and Kouzarides, T. (2009) Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol 16: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto, D. , Adu‐Bobie, J. , Scarselli, M. , Veggi, D. , Pizza, M. , Rappuoli, R. , and Aricò, B. (2003) Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol Microbiol 48: 323–334. [DOI] [PubMed] [Google Scholar]
- Sjölinder, H. , and Jonsson, A.B. (2007) Imaging of disease dynamics during meningococcal sepsis. PLoS One 2: e241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, D.S. (2009) Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis . Vaccine 27: B71–B77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D. , and Allis, C.D. (2000) The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Syed, B.A. , Sargent, P.J. , Farnaud, S. , and Evans, R.W. (2006) An overview of molecular aspects of iron metabolism. Hemoglobin 30: 69–80. [DOI] [PubMed] [Google Scholar]
- Taylor, M.E. , Bezouska, K. , and Drickamer, K. (1992) Contribution to ligand binding by multiple carbohydrate‐recognition domains in the macrophage mannose receptor. J Biol Chem 267: 1719–1726. [PubMed] [Google Scholar]
- Tsuruta, T. , Inoue, R. , Nagino, T. , Nishibayashi, R. , Makioka, Y. , and Ushida, K. (2013) Role of the mannose receptor in phagocytosis of Enterococcus faecalis strain EC‐12 by antigen‐presenting cells. Microbiologyopen 2: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, D.P.J. , Wooldridge, K.G. , and Ala'Aldeen, D.A.A. (2002) Autotransported serine protease A of Neisseria meningitidis: an immunogenic, surface‐exposed outer membrane, and secreted protein. Infect Immun 70: 4447–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, D.P.J. , Marietou, A.G. , Johnston, L. , Ho, K.K. , Rogers, A.J. , Wooldridge, K.G. , and Ala'Aldeen, D.A.A. (2006) Characterization of MspA, an immunogenic autotransporter protein that mediates adhesion to epithelial and endothelial cells in Neisseria meningitidis . Infect Immun 74: 2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ulsen, P. , van Alphen, L. , Hopman, C.T. , van der Ende, A. , and Tommassen, J. (2001) In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol Med Microbiol 32: 53–64. [DOI] [PubMed] [Google Scholar]
- van Ulsen, P. , van Alphen, L. , ten Hove, J. , Fransen, F. , van der Ley, P. , and Tommassen, J. (2003) A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol 50: 1017–1030. [DOI] [PubMed] [Google Scholar]
- van Ulsen, P. , Adler, B. , Fassler, P. , Gilbert, M. , van Schilfgaarde, M. , van der Ley, P. , et al (2006) A novel phase‐variable autotransporter serine protease, AusI, of Neisseria meningitidis . Microbes Infect 8: 2088–2097. [DOI] [PubMed] [Google Scholar]
- Van Noorden, C.J. (2001) The history of Z‐VAD‐FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem 103: 241–251. [DOI] [PubMed] [Google Scholar]
- Virji, M. (2009) Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nature 7: 274–286. [DOI] [PubMed] [Google Scholar]
- van Vliet, A.H. , Wooldridge, K.G. , and Ketley, J.M. (1998) Iron‐responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol 180: 5291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E. , Obeng‐Adjei, N. , Ying, Q. , Meertens, L. , Dragic, T. , Davey, R.A. , and Ross, S.R. (2008) Mouse mammary tumor virus uses mouse but not human transferrin receptor 1 to reach a low pH compartment and infect cells. Virology 381: 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamze, S. , Martinez‐Pomares, L. , Jones, H. , Taylor, P.R. , Stillion, R.J. , Gordon, S. , and Wong, S.Y. (2002) Recognition of bacterial capsular polysaccharides and lipopolysaccharides by the macrophage mannose receptor. J Biol Chem 277: 41613–41623. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , and Pradhan, S. (2014) Mammalian epigenetic mechanisms. IUBMB Life 66: 240–256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Uptake of rpdMspA and rpdApp into human brain microvascular endothelial cells (HBMECs). HBMECs were incubated with Cy5‐labelled rpdMspA (A), rpdApp (B) or TF (C). Cell nuclei were stained with DAPI. Co‐localisation of labelled recombinant protein and nuclei (D) was determined using Velocity 6.3.1 software (Perkin Elmer). Mean fluorescence intensities were derived from ≥ 3 different fields. Error bars: mean of values + SD. All images were scanned at a resolution of 1024 × 1024 pixels, using the same laser and gains settings. The cells are representative of cells observed from multiple experiments. Scale bars = 10 μm.


