Abstract
The mushroom of the genus Pleurotus in western China, called Bailinggu, is a precious edible fungus with high economic value. However, its taxonomical position is unclear. Some researchers regard it as a variety of P. eryngii, namely P. eryngii var. tuoliensis, whereas others consider it to be a subspecies of P. eryngii, viz. P. eryngii subsp. tuoliensis. A total of 51 samples representing seven genetic groups of the genus Pleurotus were subjected to a phylogenetic analysis of partial sequences of the translation elongation factor 1 alpha gene (ef1a), the RNA polymerase II largest subunit gene (rpb1), the RNA polymerase II second largest subunit gene (rpb2) and nuc rDNA internal transcribed spacers (ITS). Our data indicate that the mushroom Bailinggu is a lineage independent of P. eryngii and should be lifted as its own species, namely P. tuoliensis. In addition, its known distribution range consists of both western China and Iran.
The mushroom from the genus Pleurotus (Fr.) P. Kumm., which is found in western China and commercially called Bailinggu, is a precious edible fungus with a white fruiting body and crisp texture. Wild Bailinggu is usually associated with plants of the genus Ferula L. of the family Umbelliferae1. Therefore, its geographical distribution is closely related to that of the plants of the genus Ferula and is restricted to Yumin, Tuoli, Qinghe, Mulei and Shihezi of the Xinjiang Autonomous Region in western China at an altitude of 790–1400 m2. The first collection of Bailinggu was made by Mou on the roots of Ferula krylovii Korov. in Tuoli2, and then it was collected on F. ferulaeoides (Steud) Korov. in Mulei3. Based on its morphological features, host and altitude, Bailinggu was described as a new variety of Pleurotus eryngii (DC. ex Fr.) Quél., namely, P. eryngii var. tuoliensis Mou3.
The early studies on the taxonomy of Bailinggu were mainly based on morphological characteristics, leaving many open questions and controversies. Three different Latin names were successively used to name the wild mushroom Bailinggu. Huang4 considered Bailinggu to be a variety of the P. eryngii species complex and named it P. eryngii var. nebrodensis (Inzenga) Sacc. Mao5 regarded Bailinggu as another independent Pleurotus species P. nebrodensis (Inzenga) Quél., which was originally described from the Italian island Sicily. Moreover, some other mycologists regarded Bailinggu as P. eryngii var. ferulae (Lanzi) Sacc.
With the development of molecular techniques, mycologists began to investigate the taxonomic status of Bailinggu using molecular methods. The results obtained from ITS sequence and IGS-RFLP analyses indicated that Bailinggu from China was a different species from P. eryngii var. ferulae, which resulted in it being erroneously regarded as P. nebrodensis by Zhang et al.6. However, P. nebrodensis, which weakly parasitize Prangos ferulacea (L.) Lindl., is uniquely associated with Prangos Lindl. plants7. In contrast, Bailinggu from western China is associated with plants of the genus Ferula there. Furthermore, Kawai et al.8 found that Chinese Bailinggu was distinct from Sicilian P. nebrodensis based on ITS and IGS1 analyses, and the results indicated that Chinese Bailinggu evolved independently in China. The study conducted by Kawai et al.8 suggested the scientific name of P. eryngii var. tuoliensis. The phylogeny of the P. eryngii species complex based on the results of ITS and ef1α analyses supports the viewpoint of Kawai and his colleagues9,10. Based on mating experiments and ITS and IGS1 sequence analyses, Zervakis et al.11 upgraded its taxonomic status to subspecies, and treated Bailinggu as P. eryngii subsp. tuoliensis (C.J. Mou) Zervakis & Venturella.
Many studies have shown that single-copy protein encoding regions are more suited for revealing the relationships of closely related species12. Based on an analysis of ef1a and rpb2 sequence data, Rodriguez Estrada et al.13 treated P. eryngii var. nebrodensis as an independent species, which is consistent with the viewpoint of Venturella7. The present study include a phylogenetic analysis of several genetic groups in the Pleurotus genus that was implemented using four nuclear DNA fragments (ef1a, rpb1, rpb2 and ITS) to infer the taxonomic status of Bailinggu from western China and its relationships with other related species. The phylogenetic species were then delimited in this study according to the genealogical concordance phylogenetic species recognition (GCPSR) criterion14.
Results
Morphology
Pileus cochleariform to flabelliform, margin inrolled, convex; surface white, with cream-colored spots, with cracks and indistinct scales; flesh white, thick. Gills white, crowded, decurrent, 1–2 mm in width. Stipe lateral, solid, white, attenuate downwards (Fig. 1). Spore (9) 10–14 × (4.2) 5–6 μm, Q = 2.0–2.5 (Q = 2.2 ± 0.21), oblong-elliptic to elliptic, colorless and hyaline (Fig. 2A). Basidia 30–45 (50) × 7–9 μm, clavate, hyaline, thin-walled, four-spored (Fig. 2B).
Figure 1. Basidiomata of Pleurotus tuoliensis (GDGM 27082).
Figure 2. Microscopic characters of Pleurotus tuoliensis (GDGM 27082).
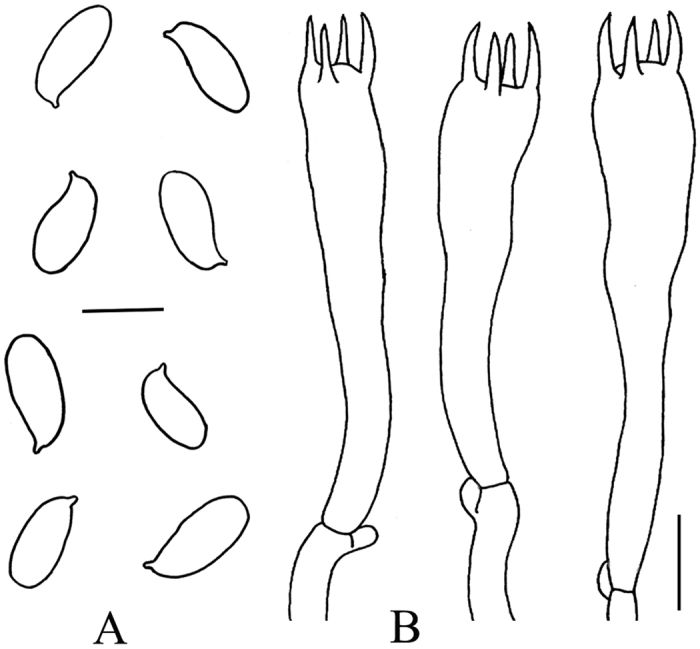
(A) Basidiospores; (B) Basidia. Bars: A and B = 10 μm.
Phylogenetic analysis and phylogenetic species recognition
Both ef1a (except CCMSSC 04235) and rpb2 (except CCMSSC 00929) were successfully amplified from 50 samples. A gene fragment of rpb1 was obtained from only 47 samples. After sequence alignment, editing and trimming, 525-bp, 1152-bp and 1093-bp segments, which contained 95, 307 and 102 parsimony informative sites, respectively, remained for phylogenetic analysis. The ITS dataset consisted of 50 sequences (with the exception of CCMSSC 00761) generated in this study and 48 related ITS sequences retrieved from GenBank (Table S1). The sequence alignment comprised 577 nucleotide positions in the ITS region used for the phylogenetic analysis.
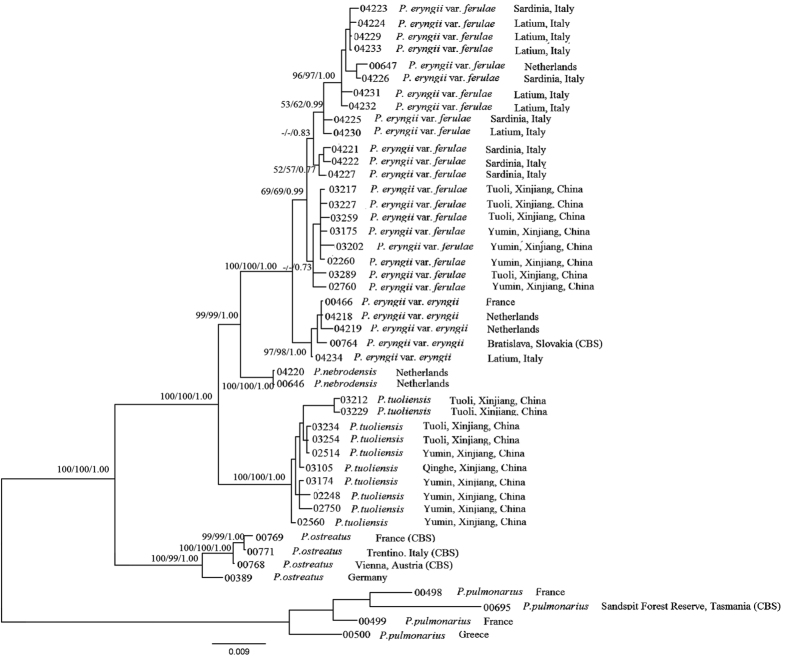
The phylogenetic trees that were reconstructed with three independent gene fragments (ef1a, rpb2 and rpb1) and inferred from a maximum likelihood (ML) analysis together with maximum likelihood bootstraps (LB), maximum parsimony bootstraps (PB) and Bayesian posterior probabilities (PP) are shown in Figs S1–S3, respectively. The three phylogenetic trees share the same topology. The phylogenetic tree obtained from the ML analysis with LB, PB and PP support based on the combined dataset (ef1a, rpb2 and rpb1) is shown in Fig. 3. Six major clades supported with high bootstrap values and posterior probabilities could be inferred, corresponding with samples of var. ferulae, var. eryngii, P. nebrodensis, P. tuoliensis (Bailinggu), P. ostreatus (Jacq.) P. Kumm. and P. pulmonarius (Fr.) Quél. Our results identified the mushroom Bailinggu as a monophyletic group supported by a bootstrap value of 100% and a posterior probability value of 1.00. According to the GCPSR criterion, the mushroom Bailinggu should be recognized as an independent phylogenetic species based on the fact that it is highly divergent from its sibling groups.
Figure 3. Phylogenetic tree of Pleurotus species inferred from maximum likelihood (ML) analysis based on the combined dataset (ef1a, rpb2, and rpb1).
Only maximum parsimony bootstraps (PB) and maximum likelihood bootstraps (LB) over 50% and Bayesian posterior probabilities (PP) over 0.70 are reported on the branches.
Phylogenetic relationships among Bailinggu and its related species
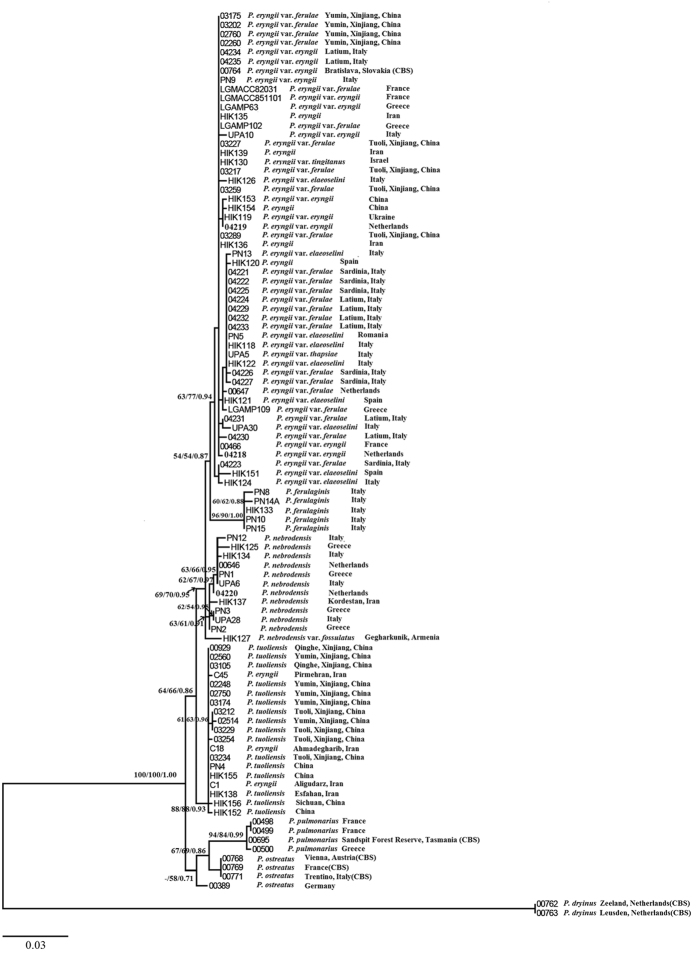
ML, maximum parsimony (MP) and Bayesian algorithm (BA) analyses based on the ITS dataset yielded similar tree topologies with some differences in bootstrap and posterior probability values. The tree inferred from the ML analysis is shown in Fig. 4. A phylogenetic reconstruction based on the ITS dataset clustered the P. eryngii species complex samples into four major clades, which are supported with moderate bootstrap and high posterior probability values. One clade consists of the varieties eryngii, ferulae, elaeoselini Venturella, Zervakis & La Rocca, thapsiae Venturella, Zervakis & Saitta, and tingitanus Lewinsohn. The other three clades correspond to P. ferulaginis Zervakis, Venturella & Cattarossi from Italy, P. nebrodensis from Europe and Asia, and P. tuoliensis (Bailinggu) from Asia. The samples of Bailinggu form a monophyletic group in the ITS tree, which exhibits the furthest genetic distance from the other groups of the P. eryngii species complex. These results are consistent with those obtained based on each single-copy protein-encoding gene. The phylogenetic relationships among P. eryngii var. eryngii, P. eryngii var. ferulae, P. eryngii var. elaeoselini, P. eryngii var. thapsiae, and P. eryngii var. tingitanus obtained using the ITS dataset remain resolved.
Figure 4. Phylogenetic tree of Pleurotus species inferred from maximum likelihood (ML) analysis based on ITS sequences.
Only maximum parsimony bootstraps (PB) and maximum likelihood bootstraps (LB) over 50% and Bayesian posterior probabilities (PP) over 0.70 are reported on the branches.
Discussion
Bailinggu is one of the most widely cultivated mushrooms in China. Recently, this species has been involved in the researches of genetic diversity evaluation15,16, temperature response mechanism17,18,19,20, fructification mechanism21, and bioactive substance exploitation22. However, the most essential information on the taxonomic status of Bailinggu and its phylogenetic relationships with its sibling species remain uncertain.
The mushrooms from Pleurotus genus that grow on the roots and stems of Umbelliferae plants belong to the P. eryngii species complex. The morphological characteristics of Bailinggu from western China conform to those of the P. eryngii species complex23. The morphological differences between Bailinggu and its related species are shown in Table 1. The pileus color of P. eryngii var. eryngii ranges from brown and beige-brown to light beige, whereas the pileus color of P. eryngii var. ferulae from Europe ranges from grey-brown to slate grey to beige brown. The macro-morphological characteristics of Bailinggu are similar to those of P. nebrodensis, but the basidiospores of Bailinggu are slightly smaller than those of P. nebrodensis. The pileus color of P. eryngii var. ferulae from China is brown to white, therefore, it is not possible to distinguish Bailinggu from P. eryngii var. ferulae from China based exclusively on their macroscopic and microscopic characteristics.
Table 1. The distinctive discriminating morphological characters for the Pleurotus eryngii species complex.
| Source | Species or variety | Spore size (μm) | Pileus color | Gill | Stipe position | |
|---|---|---|---|---|---|---|
| Length | Width | |||||
| Zervakis et al.25 | P. eryngii var. eryngii | 9.1–13.5 | 4.8–6.7 | Brown-red brown, warm brown, light beige to beige brown | Decurrent, cream to light beige, anastomoses | Central to eccentric |
| P. eryngii var. ferulae | 9.6–13.8 | 4.7–6.9 | Grey-brown to slate grey to beige brown | Decurrent, cream to light beige, anastomoses | Central to eccentric | |
| P. nebrodensis | 13.2–17.4 | 5.5–8.2 | Light ivory to cream | Deeply decurrent, whitish to pale yellow reticulum at stipe | Central to eccentric, radiating | |
| Kawai et al.8 | P. eryngii var. ferulae | 10–12 | 4.5–5.6 | Brown, pale brown | Decurrent, pale brown, pale yellow brown | Central, white |
| P. nebrodensis | 11–16 | 5–8 | White, centrally tinged pale brown | Decurrent, cream | Central, white | |
| Bailinggu | 9–14 | 4–6 | White | Decurrent, dull white, cream | Central, eccentric, white | |
| Zervakis et al.11 | Bailinggu | 8.7–14.3 | 4.5–6.3 | White to cream | × | × |
| P. ferulaginis | 11.0–16.0 | 4.0–5.5 | Whitish to ochraceous to beige to brown | White to cream to ivory | Decurrent | |
| Teng44 | P. eryngii var. ferulae | 12–14 | 5–6 | Brown, to white gradually | Decurrent, white, light yellow | Eccentric, white |
| Ying et al.45 | P. eryngii var. ferulae | 12–14 | 5–6 | Brown, to white gradually | Decurrent, white to light yellow | Eccentric, white |
| Mao5 | Bailinggu | 9–13.5 | 4.5–5.5 | White | Decurrent, white | Lateral, eccentric, white |
The crosses indicate that the trait has not been described.
The intersterility criterion is a derivative of the biological species criteria. Many cryptic species, such as the Armillaria mellea (Vahl) P. Kumm.24, have recently been recognized using this criterion. Previous mating compatibility tests of the P. eryngii species complex did not indicate any complete reproductive isolation among the genetic groups within the P. eryngii species complex. The mating rate between P. eryngii var. eryngii and P. eryngii var. ferulae was the highest, with a value of 98%8 or 93%10, but those between P. nebrodensis and P. eryngii var. eryngii and between P. nebrodensis and were significantly lower, with values of 6–18%25. Very few mating tests have been performed between Bailinggu and other genetic groups. According to the previous studies, Chinese Bailinggu showed much higher compatibility with P. eryngii var. eryngii (65%) and P. eryngii var. ferulae (82%) than with P. nebrodensis (15%) and P. ferulaginis (11%)8,11. This indicates that Bailinggu might be closer to P. eryngii var. eryngii and P. eryngii var. ferulae. However, some evidence that many fungi genetically isolated in nature retain the ancestral character of interbreeding14. Hilber26 found that P. eryngii var. eryngii and P. eryngii var. ferulae could mate with each other in the laboratory, but they appear to be reproductively isolated in the field and are associated with specific host plants.
The results of this study based on molecular data showed that Bailinggu is a separate phylogenetic species instead of a variety or subspecies of the P. eryngii complex according to the GCPSR criterion, although this mushroom retains high intercompatibility with P. eryngii var. eryngii and P. eryngii var. ferulae in the laboratory. A similar observation was found in a study of the P. ostreatus complex. Three intersterility groups or biological species (I, II, and VI) in the P. ostreatus complex were found to contain more than one phylogenetic species27. A phylogenetic reconstruction based on the ITS dataset and the combined dataset revealed that the genetic distance between Bailinggu and P. eryngii (var. eryngii, var. ferulae, var. elaeoselini, var. thapsiae, var. tingitanus) was greater than those between Bailinggu and P. ferulaginis and between Bailinggu and P. nebrodensis. This result is in conflict with the previous findings in the mating tests. Considering the geographical isolation of Bailinggu in nature, the results inferred from molecular data are more acceptable because DNA sequence divergence, be it allopatric or sympatric, might occur much earlier than the evolution of intersterility28,29.
Previous research using sequence analyses of ITS and IGS1 showed that Bailinggu is a phylogenetic sister group to P. eryngii11. However, our study indicates that P. ferulaginis is much more similar to P. eryngii in terms of not only morphology, distribution, and ecology but also DNA divergence. The phylogenetic analysis revealed that Bailinggu is a sister group to the eryngii-ferulaginis-nebrodensis clade and is not closely related to the other genetic groups of the P. eryngii species complex.
Reproductive isolation caused by host specialization is often observed in basidiomycetes, particularly plant pathogenic fungal species30. To the best of our knowledge, the P. eryngii species complex has developed a certain degree of host specificity. To detect whether the relationships among the genetic groups of the species complex correlate with those among their hosts, the phylogeny of the relevant hosts was reconstructed based on ITS1 and ITS2 sequences retrieved from GenBank (Fig. S4). The results showed that the eryngii, ferulae, elaeoselini, thapsiae, and tingitanus varieties are so closely related genetically that they could not be distinguished by ITS analysis, but the relationship among hosts of P. eryngii var. eryngii, P. ferulaginis, and P. nebrodensis is markedly closer. In contrast, the genetic relationships of Bailinggu with the ferulae, elaeoselini, thapsiae, and tingitanus varieties are distant, but the genetic relationships among their hosts are close, indicating that hosts might not be the main reason for the divergence of Bailinggu from other genetic groups. Its long geographical isolation might be the main reason for the distant genetic relationship among Bailinggu and other genetic groups.
Pleurotus eryngii, P. ferulaginis and P. nebrodensis are mainly distributed in the Mediterranean and surrounding areas, whereas recent studies found that P. eryngii and P. nebrodensis also occur in Asia11. The distributions of the two mushrooms are wide and continuous, but there is very limited information on the distribution of Bailinggu. The samples of Bailinggu used in the present study were mostly from western China, and partly from Iran11,31. The main distribution area of Bailinggu in China is located far from the distribution areas of other genetic groups with the exception of P. eryngii var. ferulae from China. There are no obvious differences in morphological characteristics or habitat between Bailinggu and P. eryngii var. ferulae from China. However, a sequence analysis showed a remarkable difference between them in terms of DNA sequence, which is consistent with previous results6. What efficient prezygotic barriers that maintain the separation of both gene pools will require further study. The pileus color of P. eryngii var. ferulae from China is different from that of P. eryngii var. ferulae from Europe. Moreover, the phylogenetic analysis showed that they cluster according to their geographical origins even though they still belong to the same genetic group. Geographical isolation and differences in biotope would likely lead to increasing divergence of an individual population to enhance differentiation32,33.
Conclusion
This study, which involved multiple or independent DNA gene fragment analyses in combination with a morphological analysis, showed that Bailinggu is highly divergent from its related groups at the DNA level but presents no significant differences in morphology or mating incompatibility. According to the GCPSR criterion, Bailinggu is an independent phylogenetic species in the P. eryngii complex, and based on its geographical isolation in nature, P. eryngii var. tuoliensis or P. eryngii subsp. tuoliensis should be upgraded to an independent species, and P. tuoliensis should be the scientific name for this mushroom. The taxonomic treatment is as follows:
Pleurotus tuoliensis (C.J. Mou) M.R. Zhao & J.X. Zhang, comb. nov. & stat. nov.
Fungal Name No.: FN570249.
Basionym: Pleurotus eryngii var. tuoliensis C.J. Mou, Acta Mycol. Sin. 6(3): 153 (1987) [MycoBank No.:133079]; Pleurotus eryngii subsp. tuoliensis (C.J. Mou) Zervakis & Venturella, Fungal Biology 118: 826 (2014) [MycoBank No.:807241].
Specimen examined: GDGM 27082 (Table 2).
Table 2. The information and GenBank accession numbers of the Pleurotus samples used in this study.
| Strain No. (CCMSSC) | Taxa | Geographic origin | ITS | ef1α | rpb2 | rpb1 |
|---|---|---|---|---|---|---|
| 03105 | P. tuoliensis | Qinghe, Xinjiang, China | KU612906 | KU612970 | KU612991 | KU612948 |
| 00929 | P. tuoliensis | Qinghe, Xinjiang, China | KU612907 | KU612971 | × | × |
| 03212 | P. tuoliensis | Tuoli, Xinjiang, China | KU612908 | KU612972 | KU612992 | KU612949 |
| 03229 | P. tuoliensis | Tuoli, Xinjiang, China | KU612909 | KU612973 | KU612993 | KU612950 |
| 03234 | P. tuoliensis | Tuoli, Xinjiang, China | KU612910 | KU612974 | KU612994 | KU612951 |
| 03254 | P. tuoliensis | Tuoli, Xinjiang, China | KU612911 | KU612975 | KU612995 | KU612952 |
| 03174 | P. tuoliensis | Yumin, Xinjiang, China | KU612912 | KU612976 | KU612996 | KU612953 |
| 02248 | P. tuoliensis | Yumin, Xinjiang, China | KU612913 | KU612977 | KU612997 | KU612954 |
| 02514* | P. tuoliensis | Yumin, Xinjiang, China | HM777041 | KU983512 | KU983514 | KU983513 |
| 02560 | P. tuoliensis | Yumin, Xinjiang, China | KU612914 | KU612978 | KU612998 | KU612955 |
| 02750 | P. tuoliensis | Yumin, Xinjiang, China | KU612915 | KU612979 | KU612999 | KU612956 |
| 03217 | P. eryngii var. ferulae | Tuoli, Xinjiang, China | KU612916 | KM000984 | KU613000 | KR493299 |
| 03227 | P. eryngii var. ferulae | Tuoli, Xinjiang, China | KU612917 | KM000995 | KU613001 | KR493310 |
| 03259 | P. eryngii var. ferulae | Tuoli, Xinjiang, China | KU612918 | KM000989 | KU613002 | KR493304 |
| 03289 | P. eryngii var. ferulae | Tuoli, Xinjiang, China | KU612919 | KM001005 | KU613003 | KR493320 |
| 03175 | P. eryngii var. ferulae | Yumin, Xinjiang, China | KU612920 | KM000979 | KU613004 | KR493294 |
| 03202 | P. eryngii var. ferulae | Yumin, Xinjiang, China | KU612921 | KM000954 | KU613005 | KR493269 |
| 02760 | P. eryngii var. ferulae | Yumin, Xinjiang, China | KU612922 | KM000962 | KU613006 | KR493277 |
| 02260 | P. eryngii var. ferulae | Yumin, Xinjiang, China | KU612923 | KM000935 | KU613007 | KR493250 |
| 00647 | P. eryngii var. ferulae | Netherlands | KU612924 | KU612980 | KU613008 | KU612957 |
| 04221 | P. eryngii var. ferulae | Sardinia, Italy | KU612925 | KR493212 | KU613009 | KR493322 |
| 04222 | P. eryngii var. ferulae | Sardinia, Italy | KU612926 | KR493213 | KU613010 | KR493323 |
| 04223 | P. eryngii var. ferulae | Sardinia, Italy | KU612927 | KR493214 | KU613011 | KR493324 |
| 04225 | P. eryngii var. ferulae | Sardinia, Italy | KU612928 | KR493215 | KU613012 | KR493325 |
| 04226 | P. eryngii var. ferulae | Sardinia, Italy | KU612929 | KR493216 | KU613013 | KR493326 |
| 04227 | P. eryngii var. ferulae | Sardinia, Italy | KU612930 | KR493217 | KU613014 | KR493327 |
| 04224 | P. eryngii var. ferulae | Latium, Italy | KU612931 | KR493218 | KU613015 | KR493328 |
| 04229 | P. eryngii var. ferulae | Latium, Italy | KU612932 | KR493219 | KU613016 | KR493329 |
| 04230 | P. eryngii var. ferulae | Latium, Italy | KU612933 | KU612981 | KU613017 | KU612958 |
| 04231 | P. eryngii var. ferulae | Latium, Italy | KU612934 | KR493220 | KU613018 | KR493330 |
| 04232 | P. eryngii var. ferulae | Latium, Italy | KU612935 | KR493221 | KU613019 | KR493331 |
| 04233 | P. eryngii var. ferulae | Latium, Italy | KU612936 | KR493222 | KU613020 | KR493332 |
| 04234 | P. eryngii var. eryngii | Latium, Italy | KU612937 | KU612982 | KU613021 | KU612959 |
| 04235 | P. eryngii var. eryngii | Latium, Italy | KU612938 | × | KU613022 | KU612960 |
| 00466 | P. eryngii var. eryngii | France | KU612939 | KU612983 | KU613023 | KU612961 |
| 00764 | P. eryngii var. eryngii | Bratislava, Slovakia (CBS 100.82) | EU424295 | KR493223 | KU613024 | KR493336 |
| 04219 | P. eryngii var. eryngii | Netherlands | KU612940 | KR493224 | KU613025 | KR493337 |
| 04218 | P. eryngii var. eryngii | Netherlands | KU612941 | KU612984 | KU613026 | KU612962 |
| 04220 | P. nebrodensis | Netherlands | KU612942 | KU612985 | KU613027 | KU612963 |
| 00646 | P. nebrodensis | Netherlands | KU612943 | KU612986 | KU613028 | KU612964 |
| 00768 | P. ostreatus | Vienna, Austria (CBS 102513) | KU612944 | KR493225 | KU613029 | KR493333 |
| 00769 | P. ostreatus | France (CBS 291.47) | EU424309 | KR493226 | KU613030 | KR493334 |
| 00771 | P. ostreatus | Trentino, Italy (CBS 375.51) | EU424310 | KR493227 | KU613031 | KR493335 |
| 00389 | P. ostreatus | Germany | KU612945 | KU612987 | KU613032 | KU612965 |
| 00498 | P. pulmonarius | France | KU612946 | KU612902 | KU613033 | KU612966 |
| 00499 | P. pulmonarius | France | EU424314 | KU612903 | KU613034 | KU612967 |
| 00500 | P. pulmonarius | Greece | KU612947 | KU612904 | KU613035 | KU612968 |
| 00695 | P. pulmonarius | Sandspit Forest Reserve, Tasmania, Australia (CBS 100130) | EU424311 | KU612905 | KU613036 | KU612969 |
| 00761 | P. dryinus | Harz, Germany (CBS 481.72) | × | KU612988 | KU613037 | × |
| 00762 | P. dryinus | Zeeland, Netherlands (CBS 724.83) | EU424293 | KU612989 | KU613038 | × |
| 00763 | P. dryinus | Leusden, Netherlands (CBS 804.85) | EU424294 | KU612990 | KU613039 | × |
*The fruiting body of this strain was collected by Qiang Chen at Zhuanchang (altitude 735 m) of Yumin county in Xinjiang Autonomous Region on April 22, 2009. And now it was deposited at the herbarium of Guangdong institute of Microbiology, the herbarium No. was GDGM 27082.
The crosses indicate that sequences were not available. CBS (Centraalbureau voor Schimmelcultures, the Netherlands). CBS numbers are presented in parentheses.
Materials and Methods
Taxon sampling
Fifty-one specimens representing seven different genetic groups of Pleurotus were used in this study (Table 2). These samples were obtained through field collection, donation, culture exchange and purchasing. Pure cultures of all samples were deposited at the China Center for Mushroom Spawn Standards and Control (CCMSSC).
Morphological observation
The morphological characteristics of fresh fruitbodies were observed and recorded in the field. The samples were dried at 40–50 °C, and microscopic features were observed with a light microscope. The size of the basidiospores was described in the form of (a)b–c(d), and 90% of the measurements were within the range of b and c; a and d (in the parentheses) are the minimum and maximum of the measurements, respectively, whereas the quotient (Q) of their dimensions was calculated as the ratio of the spore length (arithmetic average of all spores) to the spore width (arithmetic average of all spores).
DNA extraction, amplification and sequencing
Total DNA was extracted using a DP305-Plant Genome Extraction Kit (Tiangen, China). PCR amplifications were conducted using the following primer pairs: EF595F/EF116OR for the portion of the ef1a gene, fRPB2 5F/bRPB2 7.1R, b6.9F/b11R1 for the fragment of the rpb2 gene12, RPB1 2F (5′ ATTGCGGGCGACTAAAGG 3′) and RPB1 5R (5′ CTGCTCAAACTCGGAGATAA 3′) for the part of the rpb1 gene, and ITS1/ITS434 for the ITS region. Each amplification reaction system contained approximately 20 ng of DNA template, 0.2 mM dNTPs, 0.5 mM each primer, and 1 U of Ex Taq DNA polymerase (TaKaRa, Japan) in a final volume of 20 μL. The PCR was conducted using the following program: 94 °C for 4 min followed by 35 cycles of 94 °C for 50 s, 55 °C for 50 s, and 72 °C for 1 min. The reaction was completed by incubation at 72 °C for 10 min. The amplified products were separated by electrophoresis on 1.2% agarose gels and stained with ethidium bromide. Sequencing was performed by BGI Co., Ltd (Beijing, China). The PCR products from each sample that failed to yield direct sequencing results were cloned using a pGEM-T easy cloning kit (Promega, USA) and transformed into DH5α component Escherichia coli cells. Ten random transformed E. coli colonies were selected for sequencing, and the sequence data were deposited in GenBank (Table 2).
Sequence alignments
Each DNA sequence was assembled and edited manually if needed. Sequence alignments were performed using the MUSCLE algorithm in MEGA 5.035. Different alignments were performed for different analytical purposes. Multiple or independent DNA gene fragments (ef1a, rpb2 and rpb1) were used to reconstruct the phylogeny of mushrooms of the genus Pleurotus to infer the taxonomic status of the Chinese Bailinggu. For the Pleurotus genus, more ITS sequences than sequences of the other three genes were readily available in GenBank, and the relationships among Bailinggu and its sibling species were further investigated using the ITS dataset.
Phylogenetic analysis
Phylogenetic reconstructions using ef1a, rpb1, rpb2, the combined data set of the three genes, and ITS were performed using MP, ML and BA. The MP analyses were performed with PAUP* 4.0b1036. Heuristic searching with TBR branch swapping was implemented with 1000 random-addition sequence replicates. The bootstrap analysis was conducted with 1,000 replicates using the heuristic search37. ML analyses were conducted in PHYML3.038, and the bootstrap analysis was performed with 1000 replicates. BA analyses were run using MrBayes3.1.239. The Markov Chain Monte Carlo (MCMC) algorithm40 was utilized to calculate the Bayesian posterior probabilities. Four Markov chains were run for 5,000,000 generations with the trees sampled every 1000th generation. The average standard deviation of split frequencies was restricted to less than 0.01. The first 25% trees were discarded as burn-in. The optimum substitution model for each dataset was estimated by jModelTest41 according to the Corrected Akaike Information Criterion (AIC)42 for the ML analyses and the Bayesian information criterion (BIC)43 for the Bayesian analyses. For the ML analyses, the optimal substitution models for the four partitions determined using the AIC were as follows: TIM1 + G for ef1a and rpb1, SYM + G for rpb2, and TPM2uf + G for ITS. The Bayesian analyses were performed with the following selected substitution models: TrNef + G for ef1a and rpb2, TPM1 + G for rpb1, and TPM2uf + G for ITS. The samples without available sequences were not used in the phylogenetic reconstructions.
Phylogenetic species determination
The phylogenetic species were delimited in this study according to the genealogical concordance phylogenetic species recognition (GCPSR) criterion. Using this method, phylogenetic species were recognized as genealogically exclusive under GCPSR if they were concordantly supported by multiple independent loci14.
Additional Information
How to cite this article: Zhao, M. et al. The famous cultivated mushroom Bailinggu is a separate species of the Pleurotus eryngii species complex. Sci. Rep. 6, 33066; doi: 10.1038/srep33066 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Z. L. Yang at the Kunming Institute of Botany, Chinese Academy of Sciences and Dr. T. H. Li at Guangdong Institute of Microbiology for the critical reading and helpful discussion of the manuscript. Dr. M. Zhang from Guangdong Institute of Microbiology is acknowledged for helping to draw the illustration. The research was funded by the National Basic Research Program of China (2014CB138305) and China Agriculture Research System (CARS24). We thank Prof. Massimo Reverberi of La Sapienza University of Rome and Prof. Anton SM Sonnenberg of Wageningen University for providing us with samples.
Footnotes
Author Contributions M.Z., J.Z. and C.H. designed the experiments. Q.C. and W.G. collected the strains. M.Z. and X.W. performed the experiments. W.D. drawed the illustration. M.Z. and C.H. wrote the manuscript. All authors reviewed the manuscript.
References
- Chen Z. C. The study of Pleurotus eryngii (DC. ex Fr.) Quél. Arid Zone Res 8, 94–95 (1991). [Google Scholar]
- Cao Y. Q., Mou C. J., Chen Z. C. & Wang X. Y. The preliminary study on the biological characteristics of Pleurotus eryngii (DC. ex Fr.) Quél. Microbiol China 12, 97–101 (1985). [Google Scholar]
- Mou C. J., Cao Y. Q. & Ma J. L. A new variety of Pleurotus eryngii and its cultural characters. Acta Mycol Sin 6, 153–156 (1987). [Google Scholar]
- Huang N. L. Pleurotus eryngii var. nebrodensis in Cultivation of 18 species of rare and delicious mushroom (ed. Huang N. L.) 17–21. (Chinese Agriculture Press, 1996). [Google Scholar]
- Mao X. L. In The Macrofungi in China (ed. Mao X. L.) 64–66. (Henan Science and Technology Press, 2000). [Google Scholar]
- Zhang J. X., Huang C. Y., Ng T. B. & Wang H. X. Genetic polymorphism of ferula mushroom growing on Ferula sinkiangensis. Appl Microbiol Biotechnol 71, 304–309 (2006). [DOI] [PubMed] [Google Scholar]
- Venturella G. Typification of Pleurotus nebrodensis. Mycotaxon 75, 229–231 (2000). [Google Scholar]
- Kawai G., Babasaki K. & Neda H. Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu”: it belongs to Pleurotus eryngii (DC.: Fr.) Quél. and evolved independently in China. Mycoscience 49, 75–87 (2008). [Google Scholar]
- Mang S. M. & Figliuolo G. Species delimitation in Pleurotus eryngii species-complex inferred from ITS and EF-1α gene sequences. Mycology 1, 269–280 (2010). [Google Scholar]
- Huang C. Y. et al. Correction of scientific name for cultivated Bai-Ling-Gu in China. J Plant Genet Resour 12, 825–827, 832 (2011). [Google Scholar]
- Zervakis G. I. et al. A reappraisal of the Pleurotus eryngii complex – New species and taxonomic combinations based on the application of a polyphasic approach, and an identification key to Pleurotus taxa associated with Apiaceae plants. Fungal Biol 118, 814–834 (2014). [DOI] [PubMed] [Google Scholar]
- Xu J. P. Fudamental of fungal molecular population genetic analyses. Curr Issues in Mol Biol 8, 75–90 (2006). [PubMed] [Google Scholar]
- Rodriguez Estrada A. E., Jimenez-Gasco M. d. M. & Royse D. J. Pleurotus eryngii species complex: sequence analysis and phylogeny based on partial EF1alpha and RPB2 genes. Fungal Biol 114, 421–8 (2010). [DOI] [PubMed] [Google Scholar]
- Taylor J. W. et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31, 21–32 (2000). [DOI] [PubMed] [Google Scholar]
- Zhao M. R. et al. Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 8, e83253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. J. et al. Analysis of the mating type factors in natural populations of Pleurotus eryngii var. tuoliensis in China. Mycosystema 32, 248–252 (2013). [Google Scholar]
- Kong W. W., Huang C. Y., Chen Q., Zou Y. J. & Zhang J. X. Nitric oxide alleviates heat stress-induced oxidative damage in Pleurotus eryngii var. tuoliensis. Fungal Genet Biol 49, 15–20 (2012). [DOI] [PubMed] [Google Scholar]
- Kong W. W. et al. Nitric oxide is involved in the regulation of trehalose accumulation under heat stress in Pleurotus eryngii var. tuoliensis. Biotechnol Lett 34, 1915–1919 (2012). [DOI] [PubMed] [Google Scholar]
- Liu X. M., Zheng S. Y., Bau T., Huang C. Y. & Zhang J. X. Effect of growth temperature on the activities of amtioxidative enzymes in mycelial extracts of Pleurotus nebrodensis. Acta Edulis Fungi 17, 60–62 (2010). [Google Scholar]
- Meng L. J. et al. Biochemical pathway analysis of exogenous NO improving heat‐tolerance of Pleurotus eryngii var. tuoliensis. Mycosystema 34, 632–639 (2015). [Google Scholar]
- Chen M. M. et al. Screening and analysis of gene fragments related to fructification of Pleurotus eryngii var. tuoliensis. Biotechnology 23, 4–8 (2013). [Google Scholar]
- Lv H. et al. Nebrodeolysin, a novel hemolytic protein from mushroom Pleurotus nebrodensis with apoptosis-inducing and anti-HIV-1 effects. Phytomedicine 16, 198–205 (2009). [DOI] [PubMed] [Google Scholar]
- Zervakis G. & Balis C. A pluralistic approach in the study of Pleurotus species with emphasis on compatibility and physiology of the European morphotaxa. Mycol Res 100, 717–731 (1996). [Google Scholar]
- Anderson J. B. & Ullrich R. C. Biological species of Armillaria mellea in North America. Mycologia 71, 402–414 (1979). [Google Scholar]
- Zervakis G. I., Venturella G. & Papadopoulou K. Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species-complex as determined by RAPD analysis, isozyme profiles and ecomorphological characters. Microbiology 147, 3183–3194 (2001). [DOI] [PubMed] [Google Scholar]
- Hilber O. In Bibliotheca Mycologica Band 87 (ed. Hilber O.) 25–448 (Cramer, J. 1982). [Google Scholar]
- Vilgalys R. & Sun B. L. Ancient and recent patterns of geographic speciation in the Oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. P Natl Acad Sci USA 91, 4599–4603 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gac M. L. & Giraud T. Existence of a pattern of reproductive character displacement in Homobasidiomycota but not in Ascomycota. J Evolution Biol 21, 761–772 (2008). [DOI] [PubMed] [Google Scholar]
- Giraud T., Gladieux P. & Gavrilets S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol 25, 387–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T., Villaréal L. M. M. A., Austerlitz F., Gac M. L. & Lavigne C. Importance of the life cycle in sympatric host race formation and speciation of pathogens. Phytopathology 96, 280–287 (2006). [DOI] [PubMed] [Google Scholar]
- Ravash R. et al. Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Prog 9, 181–194 (2010). [Google Scholar]
- Giraud T., Refregier G., Gac M. L., Vienne D. M. d. & Hood M. E. Speciation in fungi. Fungal Genet Biol 45, 791–802 (2008). [DOI] [PubMed] [Google Scholar]
- Pildain M. B. et al. Molecular phylogeny of Armillaria from the Patagonian Andes. Mycol Prog 8, 181–194 (2009). [Google Scholar]
- White T. J., Bruns T., Lee S. & Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies In PCR protocols: a guide to methods and applications (eds Innis M. A., Gelfand D. H., Sninsky J. J. & White T. J.) 315–322 (Academic Press, 1990). [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. In PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4.0. (Sinauer Associates, 2002) [Google Scholar]
- Felsenstein J. Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML3.0. Systematic Biol 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Larget B. & Simon D. L. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol 16, 750–759 (1999). [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE T Automat Contr 19, 716–723 (1974). [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann Stat 6, 461–464 (1978). [Google Scholar]
- Teng S. C. In Fungi in China (ed. Teng S. C.) 580 (Science Press, 1963). [Google Scholar]
- Ying J. Z., Mao X. L., Ma Q. M., Zong Y. C. & Wen H. A. Icones of Medicinal Fungi from China (eds Ying J. Z., Mao X. L., Ma Q. M., Zong Y. C., Wen H. A.) 277 (Science Press, 1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.