SUMMARY
Exome sequencing analysis of over 2,000 children with complex malformations of cortical development identified 5 independent homozygous deleterious mutations in KATNB1, encoding the regulatory subunit of the microtubule severing enzyme katanin. Mitotic spindle formation is defective in patient-derived fibroblasts, a consequence of disrupted interactions of mutant KATNB1 with KATNA1, the catalytic subunit of katanin, and other microtubule associated proteins. Loss of KATNB1 orthologs in zebrafish (katnb1) and flies (kat80) results in microcephaly, recapitulating the human phenotype. In the developing Drosophila optic lobe, kat80 loss specifically affects the asymmetrically dividing neuroblasts, which display supernumerary centrosomes and spindle abnormalities during mitosis, leading to cell cycle progression delays and reduced cell numbers. Furthermore, kat80 depletion results in dendritic arborization defects in sensory and motor neurons, affecting neural architecture. Taken together, we provide insight into the mechanisms by which KATNB1 mutations cause human cerebral cortical malformations, demonstrating its fundamental role during brain development.
INTRODUCTION
A mechanistic understanding of human brain development has only recently begun to be elaborated at the gene level, with the discovery of disease causing mutations in monogenic forms of malformations of cerebral cortical development (MCD). MCD syndromes have traditionally been classified on the basis of imaging findings that correlate with disturbances at distinct phases of cortical development, including proliferation of neural progenitors (e.g. leading to genetic forms of microcephaly), neuronal migration (e.g. pachygyria, lissencephaly, subcortical and periventricular heterotopias), and postmigratorial development and organization (e.g. schizencephaly, polymicrogyria) (Barkovich et al., 2012). Phenotypic overlap between these MCD disorders is commonly observed, with a single gene mutation leading to multiple cortical abnormalities, suggesting that diverse cerebral malformations can have a unified underlying causation (Bilguvar et al., 2010).
Genetic studies have also highlighted significant heterogeneity in the molecular pathways underlying MCD, with the possible exception of autosomal recessive primary microcephaly (MCPH) which is associated with a plethora of genes (e.g. ASPM, CDK5RAP2, CASC5, CENPJ, CEP63, CEP135, CEP152, STIL and WDR62) that encode proteins involved in cytoskeletal control of the mitotic apparatus, including centrosomes and mitotic spindle poles (Bettencourt-Dias et al., 2011; Bilguvar et al., 2010; Kaindl et al., 2010; Thornton and Woods, 2009).
Despite dramatic differences in brain size and complexity, animal models have proven invaluable in elucidating the biology of MCD, for example by confirming the importance of centrosome in microcephaly (Kaindl et al., 2010). In Drosophila, homozygous loss of either asp (abnormal spindle, ortholog of human ASPM, mutated in MCPH5, OMIM#608716) or cnn (centrosomin, ortholog of human CDK5RAP2, mutated in MCPH3, OMIM#604804), affects asymmetric cell division during development (Bond et al., 2005; Wakefield et al., 2001). Similarly, both mouse and zebrafish models of human SCL/TAL1-interrupting locus gene (STIL), mutated in MCPH7 (OMIM#612703), have shown that STIL plays a role in centrosome duplication and function and mitotic spindle organization and signaling (Izraeli et al., 1999; Pfaff et al., 2007).
The centrosome functions as the primary microtubule-organizing center of the cell and in humans, mutations in microtubule-associated proteins (DCX, LIS1, NDE1)(Alkuraya et al., 2011; Bakircioglu et al., 2011; Gleeson et al., 1998; Reiner et al., 1993) or tubulin isoforms (TUBA1A, TUBA8, TUBB2B and TUBB3) (Abdollahi et al., 2009; Jaglin et al., 2009; Kumar et al., 2010; Tischfield et al., 2010) also underlie defects in cellular proliferation, neuronal migration and cortical organization. Proper functioning of microtubules is in turn dependent on the tight control of their length, number, as well as cargo movement (Shu et al., 2004; Tanaka et al., 2004).
A concerted action of polymerizing and severing enzymes regulates microtubule length. Indeed, mutations in SPAST, encoding the microtubule severing enzyme spastin, result in progressive axonal degeneration and autosomal dominant spastic paraplegia (SPG4, OMIM# 182601), thus linking microtubule remodeling to neurodegeneration (Hazan et al., 1999). Katanin, the only other well-characterized microtubule severing enzyme, composed of a catalytic, p60 (KATNA1), and a regulatory, p80 (KATNB1), subunit, acts by disrupting contacts within the polymer lattice (McNally and Vale, 1993). In developing neurons, katanin localizes to microtubules and centrosomes and is essential for microtubule shortening and release (Ahmad et al., 1999). Katanin functions in cell division (McNally et al., 2006; Zhang et al., 2007), neuronal morphogenesis (Karabay et al., 2004; Yu et al., 2008) and assembly and disassembly of cilia and flagella (Casanova et al., 2009; Sharma et al., 2007).
p60/KATNA1 is a member of the AAA (ATPases Associated with diverse cellular Activities) domain containing protein family, whereas p80/KATNB1 binds to p60 and targets it to subcellular structures including the centrosome, further mediating its interactions with Dynein, LIS1 and NDEL1 (Hartman et al., 1998; McNally et al., 2000). A missense mutation in the highly conserved WD40 domain of Katnb1 has been shown to cause azoospermia and male sterility in mice (O’Donnell et al., 2012).
Here, by studying patients with MCD, we identify homozygous mutations in KATNB1 that result in a spectrum of MCD disorders, including microcephaly co-occurring with lissencencephaly or less severe neuronal migration abnormalities such as periventricular or subcortical heterotopias. Knockdown of KATNB1 orthologs in zebrafish (Danio rerio; katnb1) and Drosophila (kat80) results in a small brain, recapitulating the human phenotype. Further, in Drosophila, kat80 is essential for the formation of the mitotic spindle and its loss results in supernumerary centrosomes and delayed anaphase onset, preferentially affecting asymmetrically dividing neuroblasts in vivo. Lastly, KATNB1 predominantly regulates neuronal dendritic arborization. Taken together, these findings demonstrate a fundamental role of KATNB1 in human cerebral cortical development and pathology.
RESULTS
Whole-exome sequencing identifies recessive mutations in KATNB1 in patients with malformations of cortical development (MCD)
We performed whole-exome capture and next generation sequencing of germline DNA of over 2,000 children, who were mainly products of consanguineous unions. In Family 1 (NG-961), the 2 affected siblings (kinship coefficient 0.23, Table S1) displayed cognitive delay and seizures (Table S1). Physical exam revealed microcephaly, with magnetic resonance imaging (MRI) demonstrating subcortical heterotopia (Fig. 1A) (Table S1). Exome-sequencing (Table S1) identified 2 homozygous predicted deleterious missense variants, one was previously reported and affected the glucosaminyl (N-acetyl) transferase 2 gene (encoding a blood group II antigen) (p.Glu298Lys, rs139794913) resulting in cataracts, a phenotype not seen in our patients. The other, a p.Ser535Leu mutation, was novel and affected the KATNB1 gene (Table S1).
Figure 1. KATNB1 mutations in MCD patients.
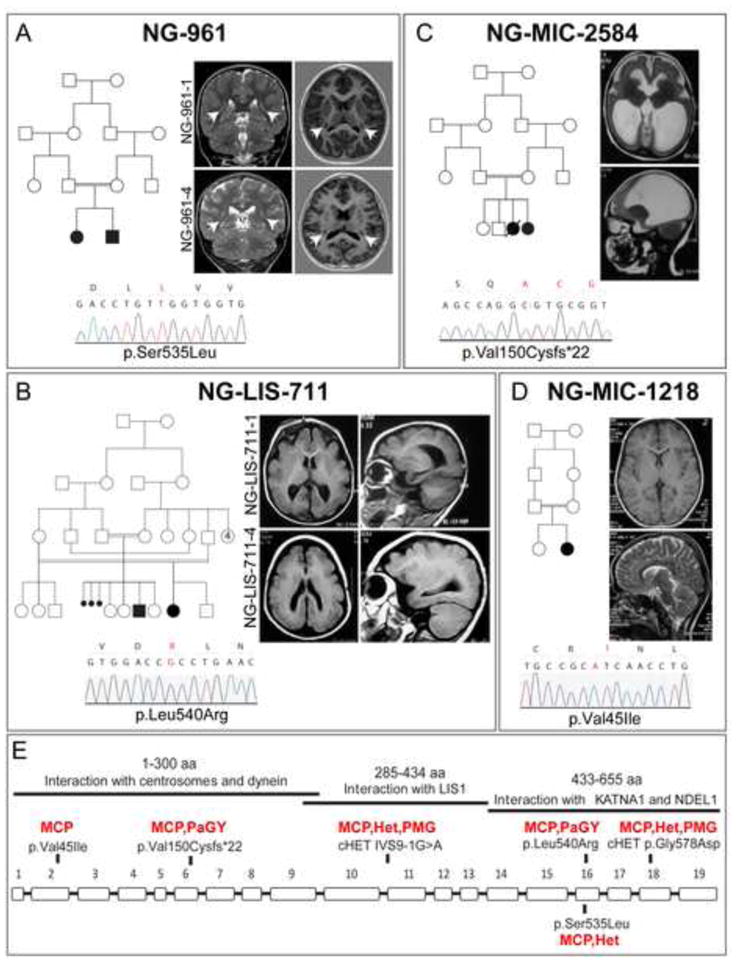
(A) Kindred NG-961. Pedigree structure depicting a 1st cousin consanguineous union is shown on the left. Coronal T2-weighted images (left) and axial T1-weighted images (right) show symmetric nodular grey matter heterotopia in the bilateral corona radiata, indicated with white arrows, in both affected siblings. Sanger sequencing confirmation of the p.Ser535Leu mutation in KATNB1 is shown at the bottom. (B) Kindred NG-LIS-711. Complex pedigree structure is shown at left. Axial (left) and sagittal (right) T1-weighted images show diffuse pachygyria. A chromatogram of Sanger sequencing result, which confirmed the KATNB1 p.Leu540Arg homozygous mutation in both affected children, is shown at the bottom. (C) Kindred NG-MIC-2584. Pedigree structure demonstrating a 1st cousin consanguineous union with two affected children (one deceased) is shown on the left. Axial (upper) and sagittal (lower) T2 -weighted images reveal a microlissencephalic brain with grossly dilated ventricles. Sanger sequencing confirmation of the p.Val150Cysfs*22 homozygous mutation is shown at the bottom. (D) Kindred NG-MIC-1218. Pedigree structure (top left) and axial T1- (upper) and sagittal T2- (lower) weighted images revealing a microcephalic brain with grossly normal architecture. The patient is homozygous for the p.Val45Ile mutation (bottom). (E) Exon-intron structure of KATNB1 is shown. Solid bars on top indicate the functional interaction domains and their localization to the KATNB1 protein. The location of each mutation and the associated phenotype are noted. MCP: microcephaly; Het: heterotopia; PaGY: pachygyria; PMG: polymicrogyria. (See also Figure S1 and Table S1).
Family 2 (NG-LIS-711), with a history of 3 early childhood deaths, also had 2 affected members (kinship coefficient of 0.12, Table S1) that exhibited severe cognitive delay and autistic features (Table S1). MRI scans confirmed microcephaly and revealed severe simplified gyral pattern and corpus callosum abnormality (Fig. 1B) (Table S1). Exome sequencing (Table S1) detected only 2 homozygous variants, one affected the metallothionein-4 gene (p.TyrTrp300CysArg) while the other was a KATNB1 predicted deleterious missense mutation (p.Leu540Arg) (Table S1).
In Family 3, a 21 month-old female (NG-MIC-2584) (who also had a sister that died 25 days after birth) presented to medical attention with jaundice, respiratory distress and severe delay in motor and mental development and was found to be microcephalic (Table S1). MRI demonstrated microlissencephaly and grossly dilated ventricles (Fig. 1C). Exome sequencing revealed a homozygous frameshift mutation (p.Val150Cysfs*22) in KATNB1.
In Family 4, the patient (NG-MIC-1218-4) exhibited mild cognitive delay (Table S1). Her MRI was mainly remarkable for microcephaly (Fig. 1D). Exome sequencing of the patient and her unaffected sibling (kinship coefficient of 0.23) (Table S1), identified 21 homozygous variants observed only in the patient. Twelve of these variants were novel, including a predicted deleterious missense mutation (p.Val45Ile) in KATNB1.
Prompted by the identification of multiple independent homozygous mutations in KATNB1, we then searched our exome sequencing databases for any potential compound heterozygous patients and identified a fifth patient (NG-PNH 226). This was a single affected offspring of a non-consanguineous union whose clinical and radiologic findings have previously been reported (Wieck et al., 2005). She displayed severe cognitive and motor developmental delay and advanced microcephaly, MRI confirmed the microcephaly and revealed partial genesis of corpus callosum, polymicrogyria and posterior predominant periventricular nodular heterotopia (Table S1). Analysis of her exome sequencing results did not identify any homozygous variants in the known MCD genes but revealed 2 heterozygous KATNB1 mutations: a splice acceptor (IVS9-1G>A; g.57787300G>A) variant and a predicted deleterious missense mutation (p.Gly578Asp; g.57790282G>A) (Fig. 1E, Table S1). Sanger sequencing proved the patient to be compound heterozygous for the KATNB1 mutations with each of the variants being inherited from one of the parents (Fig. S1).
To summarize, brain imaging of KATNB1 mutant patients demonstrates severe microcephaly with diffuse frontal predominant undersulcation (only the frontal lobe in NG-MIC-1218) with variable gyral size, mildly thick cortex with subtle irregularity of the cortical-white matter border but no clear microgyri, variable subcortical or periventricular heterotopia, variable hypogenesis of the corpus callosum, and relatively preserved brainstem and cerebellum. The cortical malformation thus differs from both classic pachygyria (lissencephaly) and polymicrogyria, but resembles the cortical malformation seen in other severe congenital microcephaly syndromes, especially NDE1 (Alkuraya et al., 2011; Bakircioglu et al., 2011). At the genetic level, exome sequencing of these patients revealed all of them harbor rare homozygous or compound heterozygous LOF, splice site or predicted deleterious missense KATNB1 mutations, including p.Val45Ileu, p.Val150Cysfs*22, p.Ser535Leu, p.Leu540Arg and IVS9-1G>A;p.Gly578Asp (Fig. 1 and Table S1). All mutations were confirmed by Sanger sequencing and segregated as expected, with both parents of the affected children being heterozygous for the respective mutations. None of the mutations were observed in the public databases or, among the 3,000 exomes sequenced at Yale including 1,460 ethnically matched control chromosomes. These findings provide conclusive evidence that mutations in KATNB1 are the underlying genetic cause of the observed MCD phenotypes.
Mutations in KATNB1 disrupt mitotic spindle architecture
To study the functional effects of the KATNB1 mutations at a cellular level, we used dermal fibroblast cultures established from skin biopsies of two patients (siblings NG-961-1 and NG-961-4, carrying the p.Ser535Leu mutation) and an unaffected heterozygous parent (NG-961-2). Western analysis revealed reduced KATNB1 (but not of its interactors) in patient-derived cells (Fig. S2A) without an obvious impact on the microtubule network as there were no apparent differences in either the intensity of KATNB1 immunostaining or the microtubule cytoskeleton in interphase cells (Fig. 2A–C). However, in dividing cells, the mutations affected the mitotic spindle with patient-derived cells displaying disorganized microtubules (Fig. 2D–F′) and reduced KATNB1 immunostaining (Fig. 2G–I′). Since KATNB1 facilitates the recruitment of KATNA1 and its interactors, such as NDEL1, to target locations, we next investigated NDEL1 and KATNA1 localization in mitotic cells and found both to be reduced in the mitotic spindle (Fig. 2J–L′, M–O′, respectively). Also, patient derived fibroblasts displayed aberrant number of centrosomes (Fig. 2P–R′). Next, we expressed wild type or mutant KATNB1 (p.Ser535Leu) in HeLa cells and found that tubulin staining of the mitotic spindle and localization of the mutant forms to the centrosomes were reduced, suggesting that the defect in KATNB1 is sufficient to cause mitotic spindle perturbation (Fig. 2S–T″, Fig. S2). Moreover, immunoprecipitation of HeLa cell lysates transfected with WT or mutant forms of KATNB1 along with KATNA1 or NDEL1 showed reduced interaction of the mutant forms of KATNB1 with both KATNA1 and NDEL1 (Fig. 2U–V). This supports an essential role for the interaction of these proteins for their targeting to the mitotic spindle.
Figure 2. C-terminal mutant forms of KATNB1 disrupt the mitotic spindle and display reduced interaction with NDEL1 and KATNA1.
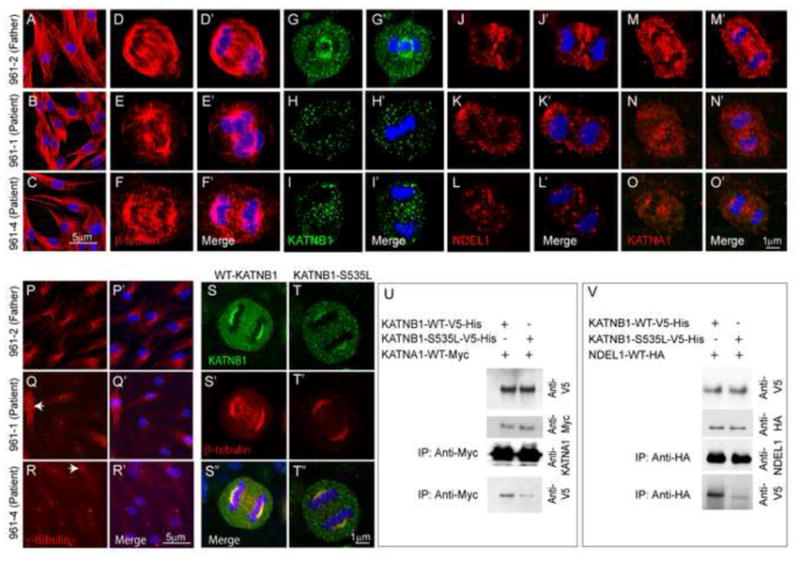
(A–C) As evidenced by beta-tubulin staining, microtubule architecture of the interphase dermal fibroblasts, derived from patients and their parents, is intact. However, the mitotic spindle is significantly disrupted and malformed in patient-derived cells in anaphase (D–F). Patient fibroblasts also show reduced localization of KATNB1 (G–I), NDEL1 (J–L) and KATNA1 (M–O) to the mitotic spindle and increased number of centrosomes (arrow) as seen by staining for γ-tubulin (P–R). Panels marked with a prime (′) show merged images of primary antigen and DAPI (blue) staining (D′–F′, G′–I′, J′–L′, M′–O′, P′–R′). Consistent with the observations in patient fibroblasts, transfection of HeLa cells with wild type and mutant forms of KATNB1 results in reduced localization of mutant form of KATNB1 (green) to centrosomes and abnormal spindle formation (tubulin staining, red) in anaphase cells (S–T″). The specific KATNB1 mutation assayed/investigated is indicated at the top of the panel. Co-immunoprecipitation of wild type and mutant forms of KATNB1 with KATNA1 (U) and NDEL1 (V) shows reduced interaction of mutant KATNB1 with both proteins. Scale bar: 5 μm (A–C, P–R) 1μm (D–O, S–T). All confocal images were captured using identical settings. (See also Figure S2)
We also assessed the effect of N-terminal domain mutation in dermal fibroblast cultures from the index case of family NG-2584 (carrying the p.Val150Cysfs*22 mutation) and an unaffected parent. As with the p.Ser535Leu mutant fibroblasts, we failed to detect an impact of the mutation on microtubule architecture in interphase cells; but mitotic cells displayed spindle morphology defects (Fig. 3A–D′) and supernumerary centrosomes (Fig. 3C–D′). In mitotic cells, spindle pole localization of KATNB1 was strikingly affected (Fig. 3G–H′) while that of LIS1 was slightly reduced (Fig. 3I–J′). Also, dynein levels at the spindle poles and the spindle proper were dramatically reduced (Fig. 3K–N′).
Figure 3. N-terminal mutant forms of KATNB1 display reduced interaction with dynein and disrupted mitotic spindle.
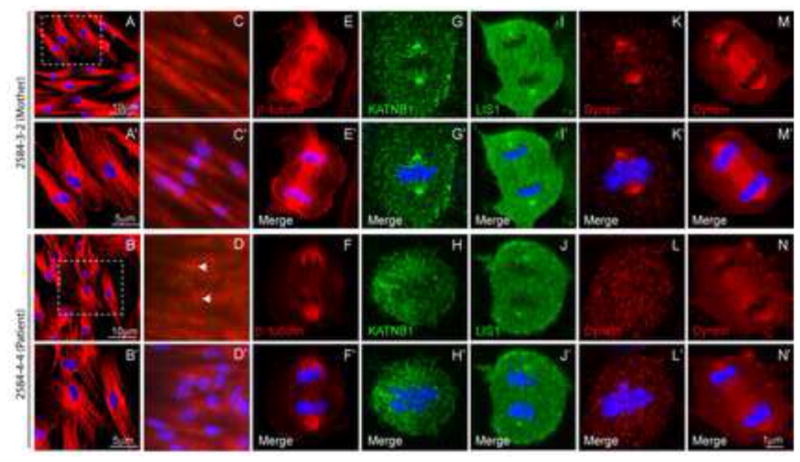
β-tubulin staining shows microtubule architecture to be intact in interphase dermal fibroblasts derived from patients and their parents (A–B). However, patient-derived cells display increased number of centrosomes (arrow) as seen by staining for γ-tubulin (C–D) and significantly disrupted and malformed mitotic spindle in anaphase (E–F). Patient fibroblasts also show reduced localization of KATNB1 (G–H), LIS1 (I–J) and dynein (K–N) to the mitotic spindle and spindle poles. Panels marked with a prime (′) show merged images of primary antibody and DAPI (blue) staining (A′–N′). All confocal images were captured using identical settings.
KATNB1 expression in the developing brain
The above findings demonstrate that KATNB1 mutations impact the overall spindle dynamics and integrity by affecting the assembly of the NDEL1/KATNA1/Dynein/LIS1 complex at the spindle poles and microtubules. To understand how these mutations lead to severe cortical abnormalities in humans, we next investigated KATNB1 expression in the developing human brain by interrogating the Human Brain Transcriptome Database (Kang et al., 2011) and found it to be stably expressed throughout fetal development, starting shortly after conception (Fig. 4A). Expression levels remained elevated into infancy particularly in neocortex, hippocampus and striatum, with high levels still detected in the adult brain. The high levels of expression and localization in neural progenitor cells and postmitotic neurons during early development suggest a continuing role of KATNB1 in neuronal proliferation, migration, and laminar organization of the human cortex.
Figure 4. KATNB1 is highly expressed in the developing brain.
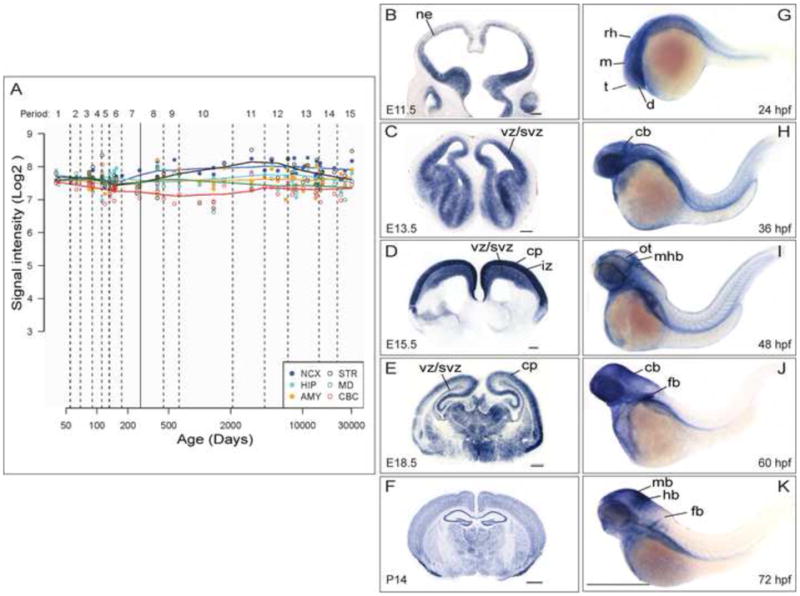
(A) KATNB1 is expressed across all regions and developmental periods in the human brain. KATNB1 exon array signal intensity. NCX, neocortex; STR, striatum; HIP, hippocampus; MD, mediodorsal nucleus of the thalamus; AMY, amygdala; CBC, cerebellar cortex. In developing mouse brain, Katnb1 is expressed in neural progenitors until mid-neurogenesis (E11.5, E13.5) (B, C), and then is also expressed in postmitotic neurons in the cortical plate (E15.5, E18.5) (D, E), and widespread expression was observed in postnatal brain (P14) (F). ne: neuropithelium; vz: ventricular zone; svz: subventricular zone; iz: intermediate zone; cp: cortical plate. Scale bar: G–I: 200 μm, J–K: 500 μm. Similarly, katnb1 is expressed in the brain in the developing zebrafish embryo (G–K): Lateral views of whole-mount in situ hybridization of the brain and torso of zebrafish embryos reveal the expression pattern of katnb1 at 24 hours post fertilization (hpf) (G), 36 hpf (H), 48 hpf (I), 60 hpf (J) and 72 hpf (K). During early developmental stages (G–H), katnb1 mRNA expression is ubiquitous throughout the embryo, including the cephalic region. As the embryos develop further (I–K), katnb1 mRNA expression becomes restricted to neural tissue. Black lines point to various anatomical structures. d, diencephalon; t, telencephalon; m, mesencephalon; rh, rhombomeres; cb, cerebellum; ot, optic tectum; mhb, midbrain hindbrain boundary; mb, midbrain; hb, hindbrain; fb, fin bud. Scale bar: 500 μm.
We then investigated KATNB1 expression in the developing mouse and zebrafish brains. In mouse, Katnb1 was initially expressed in neural progenitors until mid-neurogenesis, and subsequently in postmitotic neurons in the cortical plate; in postnatal brain, Katnb1 was expressed widely (Fig. 4B–F). Similarly, katnb1 was also highly expressed in the developing zebrafish brain. During early developmental stages, katnb1 mRNA expression was ubiquitous throughout the embryo, including the cephalic region. As the embryo develops further, katnb1 mRNA expression profile became more restricted (Fig. 4G–K).
Loss of KATNB1 orthologs in zebrafish and Drosophila results in microcephaly
Based on the finding of diffuse katnb1 expression in the developing zebrafish brain, we initially used this model organism to study KATNB1 function. Knocking down katnb1, the single ortholog, by morpholino injection resulted in a significant reduction of the midbrain size (P = 9.16×10e−7) (Fig. 5A–D, E), recapitulating the major phenotypic finding in humans.
Figure 5. Knockdown of KATNB1 orthologs in zebrafish and Drosophila results in small brain phenotype.
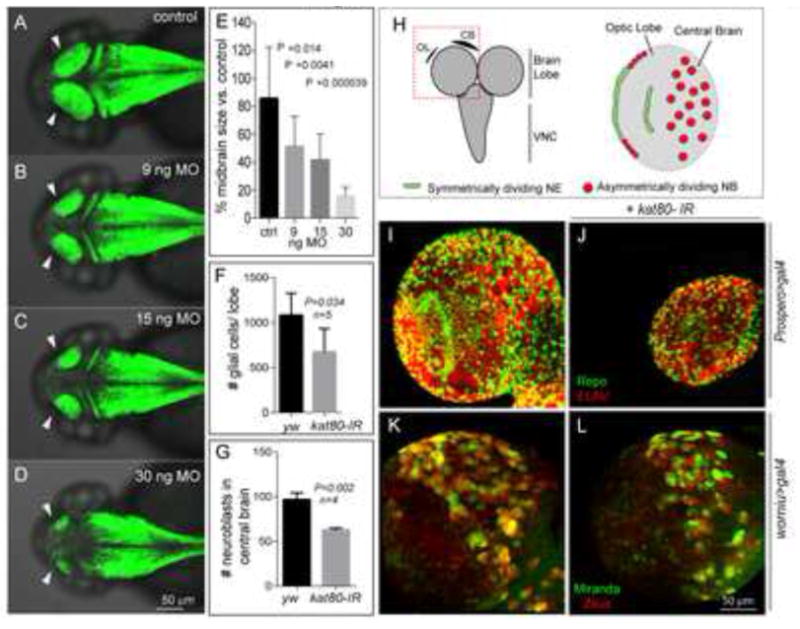
katnb1 morpholino reduces zebrafish midbrain size: Confocal microscopy shows that the katnb1 morphants at (B) 9 nanogram (ng), (C) 15 ng, and (D) 30 ng have smaller midbrains (arrows) as compared with (A) control at 2 days post fertilization (dpf). The reduction in brain size is statistically significant (E). Zebrafish brain is labeled with green fluorescence by Tg(HuC:Kaede). (H) Left panel: Schematic of the Drosophila brain; box indicates brain lobe imaged. Right panel: Schematic of a single brain lobe marks the location of symmetrically dividing neuroepithelium (NE, green) and asymmetrically dividing neuroblasts (NBs, red). (I, J) Expression of kat80-IR with prospero-Gal4 results in a dramatically reduced brain size in 3rd instar larvae. There is an overall reduction in the number of neurons and glia generated as seen by ELAV (red) and Repo (green) staining, respectively. (K, L) kat80-IR expressed under worniu-Gal4, UAS-mir::GFP, UAS-zeus::mCherry results in a significant reduction in NB number in central brain. Images in F–I are 3D projection of identical Z-sections. (F) Quantification of glial cell counts seen in panel F–G. There is significantly reduced number of glial cells in kat80-IR larvae (Error bars indicate SD, yw: 1080±110; kat80-IR: 673±116; two-tailed test, P=0.034). (G) GFP- and RFP-positive cells were quantified using 3D projections of identical Z-stacks from worniu>gal4 and worniu>kat80-IR brains, which reveal a significant reduction in central brain NBs per brain lobe (yw: 96.5±7.9; kat80-IR: 62.5±2.3; P = 0.002). (See also Figure S3)
Next, to gain a detailed mechanistic insight into the biology of KATNB1, we extended our studies to Drosophila, a model that has been successfully implemented to study human MCD-associated genes. kat80 [the single fly ortholog of human KATNB1 (Goldstein and Gunawardena, 2000)], has been shown to be ubiquitously expressed in both embryonic and larval stages (Chintapalli et al., 2007; Frise et al., 2010). To examine the potential role of kat80 in regulating brain size, we employed the GAL4/UAS system (Xu and Rubin, 1993). We used Prospero-GAL4 to drive expression of kat80 RNAi (kat80-IR) in neural progenitor cells (neuroblasts) and their newly born progeny (ganglion mother cells), which constitute the majority of cells in the developing larval brain (Fig. 5H) (Isshiki et al., 2001). kat80-IR resulted in markedly reduced brain size (microcephaly) of 3rd instar larvae as compared with controls (Fig. 5I–J) with a concomitant reduction in the number of differentiated cells (Fig. 5F) (1,080 ± 110 versus 673 ± 116 in wild-type (yw) and kat80-IR flies, respectively; P=0.034). Approximately 30% of the kat80-IR brains were reduced to 1/10th of normal size (Fig. 5J).
kat80 loss in Drosophila results in mitotic spindle abnormalities, delay in anaphase onset and mitotic failure
The above findings demonstrate that KATNB1 regulates neurogenesis in both vertebrates and invertebrates but do not reveal the underlying molecular mechanism. As mutations affecting neuroblast (NB) numbers are also known to impact brain size (Lee et al., 2006), we next examined whether kat80-IR expressing larval brains had fewer cells. Larval central brain NBs are specified during embryogenesis, then enter quiescence, progressively exiting during larval life to reach a total number of ~100 per brain lobe at 3rd instar. NBs can be identified by their large size and expression of molecular markers, including the cell polarity protein Miranda (Mir) and transcription factor Worniu (Wor) (Ashraf et al., 2004; Lai et al., 2012). We used Wor-GAL4 to express kat80-IR and GFP::Miranda to identify NBs and scored the number of Wor/Mir-positive cells. We found that 3rd instar kat80-IR larval brains had on average ~70 NBs per lobe as compared to ~100 in wild-type brains (Fig. 5K–L, G; P < 0.002), indicating that kat80 regulated brain size at least partly by controlling the NB number, which could be due to either excessive cell death or reduced cell proliferation. TUNEL staining showed no apparent ectopic cell death (Fig. S3A–D″) in kat80-IR clones, indicating that, at least in the 3rd instar larvae, kat80 knockdown impacts brain size independent of cell death.
Given the impact of KATNB1 loss on mitotic spindle (Fig. 2D–F′, 3C–D′), we postulated that in kat80-IR brains the remaining ~70 NBs could have cell cycle progression defects, further impacting the production of NB progeny and leading to microcephaly. Hence, we examined cell cycle progression in NBs using time-lapse imaging, and scored the time elapsed between nuclear envelope breakdown (NEBD, determined by the initial detection of microtubules in the center of the cell) and anaphase onset (AO, defined by the first sign of separation of sister chromatids), as previously described (Siller et al., 2005) (Fig. 6A–C, J). We made use of Worniu-Gal4 to drive GFP::Miranda in order to mark the NBs and the microtubule-binding protein Zeus::mCherry to label the mitotic spindle. The NEBD-AO interval was significantly elongated in kat80-IR NBs as compared with controls (17.86 min ± 3.59; versus 10.33 min± 0.82, respectively; P = 0.0075), with AO extending over 2 hours in ~13% (N=30) of kat80-IR NBs, indicating that kat80 knockdown significantly delayed NB cell cycle progression.
Figure 6. kat80-IR delays anaphase onset in Drosophila central brain neuroblasts, causing a reduction in their numbers.
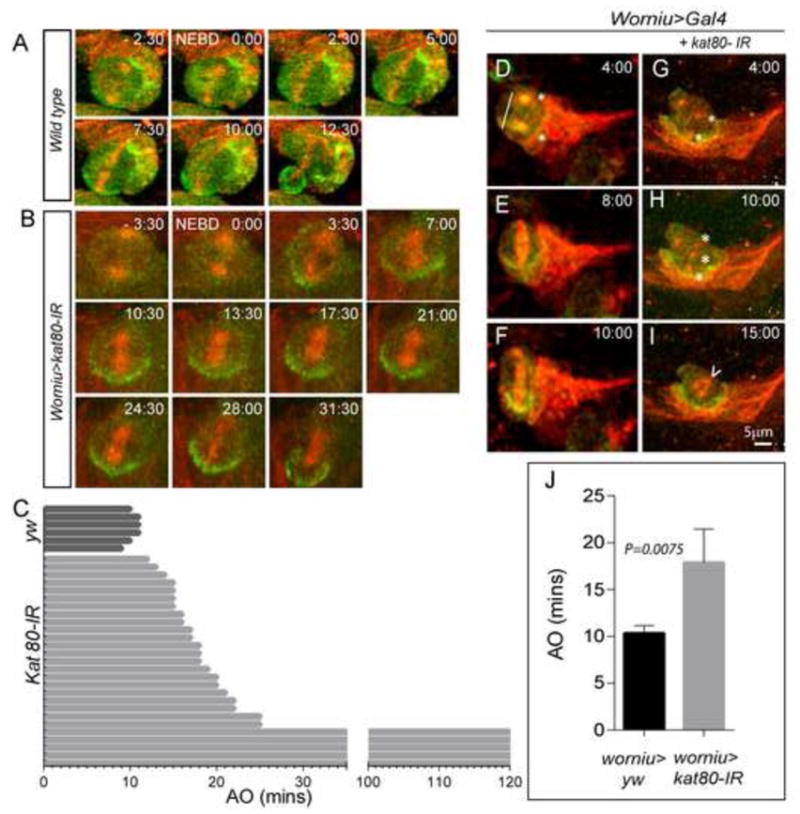
(A–C) kat80-IR was expressed under worniu-Gal4, UAS-mir::GFP, UAS-zeus::mCherry. Thirty NBs from worniu>kat80-IR and 6 NBs from worniu>gal4 3rd instar larval brains were used for time- lapse imaging. Wild type NBs exhibit anaphase onset at ~ 10.33 ± 0.82 minutes after nuclear envelope breakdown. kat80-IR leads to increase in anaphase onset time with an average of about 17.9 ± 3.59 minutes (error bars indicate standard deviation; two-tailed test, P=0.008). In addition, 4 NBs failed to display anaphase onset even after 2 hours of imaging (C). Snapshots of live imaging of 3rd instar larval brains expressing kat80-IR under worniu>Gal4, mir-GFP, zeus-mcherry. A wild type (D–F) and a kat80-IR (G–I) NB undergoing division are shown. kat80-IR expression results in multiple centrosomes (asterisks in G–H), and multipolar and barrel-shaped spindles (arrowhead in I). (J) Quantification of time to anaphase onset of 30 kat80-IR NBs compared with wild type (yw) cells reveals significant delay in mutant NBs.
Since patient derived fibroblasts showed spindle and centrosome defects (Fig 2D–F, P–R; 3C–D, M–N), we also examined kat80-IR NBs for similar abnormalities. We observed supernumerary centrosomes and multipolar and/or barrel shaped spindles (Fig. 6D–I), suggesting that kat80 regulates Drosophila brain size in vivo by controlling both number as well as cell cycle progression of NBs.
Differential effects of kat80 loss in the optic lobe
Unlike the embryonically derived central brain NBs, those in the optic lobe are specified from neural epithelium (NE), which expands during larval life by undergoing symmetric divisions in the epithelial plane (Fig. 5H). In contrast, optic lobe NB specification is accompanied by a coordinated switch from symmetric to asymmetric division (Fig. 5E), a 90-degree rotation of the division axis, and expression of specific molecular markers (Egger et al., 2007; Egger et al., 2011; Homem and Knoblich, 2012). Thus, the optic lobe is ideal to study symmetrically versus asymmetrically dividing cells. We used GH146-Gal4 to drive kat80-IR expression in the outer and inner proliferation centers of the optic lobe NE (Berdnik et al., 2008). Overall, no defects in NE morphology or spindle orientation were detected (Fig. 7A–E′), indicating that kat80 does not regulate spindle integrity or cell cycle progression of symmetrically dividing NE cells. In sharp contrast, but similar to our observations in the central brain, the number of mitotic NBs (Mir-positive) was significantly reduced in the kat80-IR expressing larval brains compared with controls (Fig. 7F–G′, J–L). In addition, kat80-IR brains showed supernumerary centrosomes (average number/cell=3) and twice as many cells in metaphase (57±7% versus 26.9±1.5% in kat80-IR versus wild-type (yw), respectively) (Fig. 7H–I′, M). These observations strongly suggested that kat80 loss specifically affects asymmetrically dividing neural progenitor cells, at least in the Drosophila optic lobe.
Figure 7. kat80-IR results in centrosomal defects and reduced neuroblasts in Drosophila optic lobe.
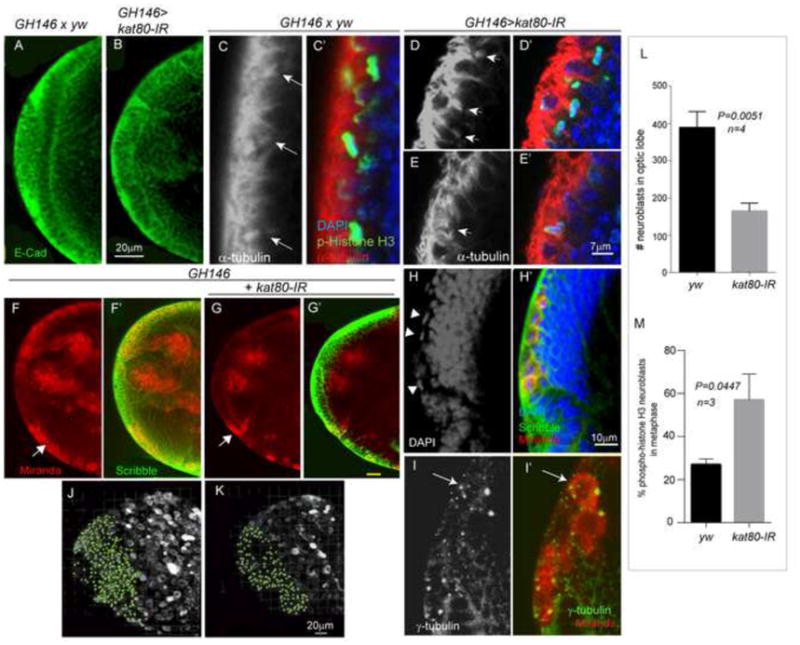
Expression of kat80-IR with GH146-gal4 does not affect the morphology of the neuroepithelium (NE, marked by E-cadherin staining (green) in A, B) or spindle orientation (C–E′). In C–E′, arrows mark the mitotic spindles and staining with alpha tubulin (red), phospho-histone H3 (pH3, marking the metaphase plate in the neuroepithelial cells, green) and DAPI (marking the nuclei, blue) are shown. Panels C, D, E show α-tubulin staining only (gray scale) for easier visualization of the mitotic spindle. (F–G′) Expression of kat80-IR in the optic lobe results in significantly reduced number of NBs (arrow). Miranda (marking NBs, red) and Scribble (marking NE cells) staining is shown in wild-type (yw, F-F′) versus kat80-IR (G-G′) larval brains. H and H′ are high magnification images of the kat80-IR brain in G, indicating that the NBs in kat80-IR brains are mainly in metaphase (arrowheads). (I-I′) GH146>kat80-IR brains also show increased number of centrosomes in NBs as seen by gamma-tubulin staining (green) in miranda-positive NBs (red). (J–K) 3D projections of identical Z-sections of GH146>kat80-IR and wildtype (yw) brains showing reduced number of miranda positive NBs in the optic lobe of 3rd instar larval brains. (L) Quantification of the miranda positive cells in the optic lobe shows significantly reduced NBs in the kat80-IR brains (yw: 389±24.8; kat80-IR:165 ±10.5; two-tailed t-test P = 0.005). (M) Quantification of phosphor-histone H3 (pH3) positive NBs in kat80 depleted brains shows an increase in the number of pH3 positive NBs in metaphase (also visible in panel H) (yw: 26.9±1.5; kat80-IR: 57±7; two-tailed t-test, P =0.04) suggesting delayed anaphase onset. (See also Figure S4)
Kat80 loss impacts on dendritic arborization
Finally, previous reports demonstrated changes in neuronal architecture and dendritic arborization not only in MCD patients (Barak et al., 2011; Kaindl et al., 2010), but also in other intellectual disability and related syndromes, including the Rett syndrome (Armstrong, 2005). Therefore, we studied potential neuronal structural abnormalities of the differentiated neurons in the kat80-IR Drosophila larvae. We examined the dendritic arborization (da) sensory neurons, which innervate the overlying larval epidermis and fall into 4 categories based on dendritic branching pattern and complexity (Grueber et al., 2007). Due to their accessibility and stereotyped morphology, da neurons serve as a model for dendritic growth, maintenance, and tiling (Jan and Jan, 2010). We used pickpocket-Gal4 (ppk-Gal4) (Grueber et al., 2007) to drive kat80-IR expression in class IV da neurons, which are characterized by highly branched dendritic trees. In kat80-IR Drosophila larvae, dendritic arborization was dramatically reduced (Fig. 8A–B) and the total number of dendritic termini was significantly diminished (Fig. 8E, P <0.01), suggesting that dendritic extension and number in peripheral neurons were compromised by kat80 loss.
Figure 8. kat80-IR results in reduced dendritic arborization in central and peripheral nervous system.
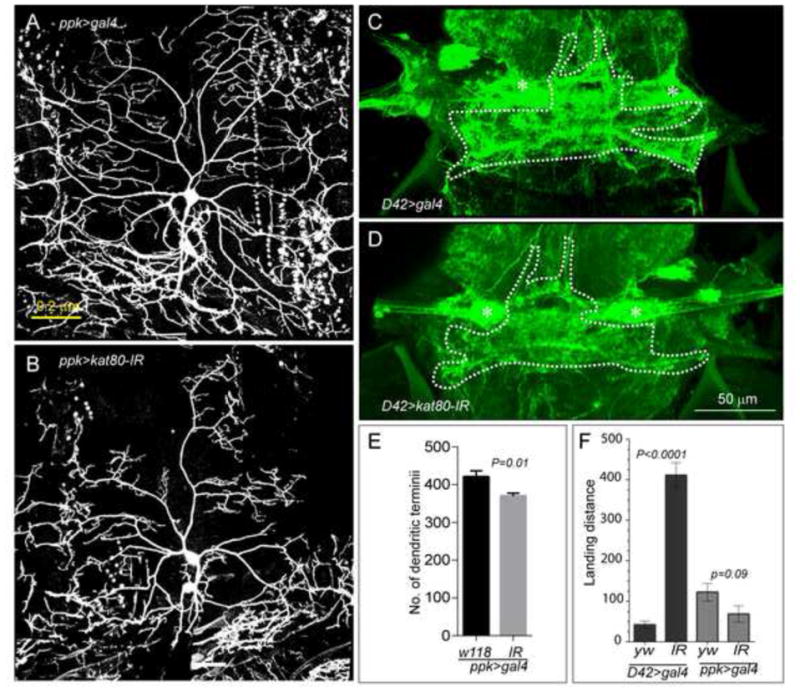
(CNS and PNS, respectively) (A–B) Larval class IV sensory neurons in the PNS were visualized using UAS>CD8-GFP expressed under the control of the PPK-GAL4 driver. Morphological analysis of dendrites of class IV dendrites, which display distinct morphology, was only performed in segments A3 and A4. We observed a significant reduction in dendrite extension in kat80-IR larvae (B) as compared to the wild-type, shown in A. (C–D) kat80-IR reduced dendritic arbor of motoneuron 5 in the CNS (D) as compared to wild-type flies (C). Dendrites of adult flight motoneurons in the CNS were visualized by expressing UAS>CD8-GFP under the control of the D42-GAL4 or C380-GAL4 driver. Asterisk marks the motoneuron 5 cell body. (E) The total number of terminal dendrites is statistically significantly reduced in kat80-IR versus wild-type larvae as counted manually on z-projections: (mean± SEM: WT = 420 ± 15.67; kat80-IR = 369.6 ± 7.8, P = 0.01 (N= 10 cells, 7 larvae for WT and N= 10 cells, 5 larvae for kat80-IR larvae)). (F) The effect of reduced dendritic arborization of flight motoneurons was assessed in a flight assay. Expression of kat80-IR using the D42 driver (expressed in adult motoneurons controlling wing muscles) resulted in severely impaired flight response, as assessed by the landing distance in the cylinder (left 2 columns in black, landing distance in millimeters: mean ± SEM: D42 driver: WT (yw)= 42.4 ± 8.7; kat80-IR = 411.7 ± 30.7 (P=0.0001)). In contrast, kat80-IR expression in PNS sensory neurons under the ppk driver did not affect the flight response, as expected (right 2 columns in grey, landing distance in millimeters ppk driver: mean ± SEM: WT = 122.6 ± 21.8; kat80-IR = 69 ± 19.9). N= 20 adult males for each genotype. (See also Figure S5)
We next extended our observations to the Drosophila CNS, by examining dendritic arborization of adult flight motoneurons (MN1–5) that innervate the dorsal longitudinal flight muscle (Consoulas et al., 2002; Ikeda and Koenig, 1988). Among all singly identifiable flight motoneurons, MN5 serves as a paradigm of dendritic architecture (Consoulas et al., 2002; Vonhoff and Duch, 2010). Using D42-Gal4 to express kat80-IR primarily in adult flight motoneurons (Vonhoff and Duch, 2010) resulted in reduced dendritic arborization of MN5 (Fig. 8C–D). Because dendritic defects in flight motoneurons alter flight performance (Vonhoff et al., 2012), we tested flight ability and found a significantly reduced flight response in adult D42>kat80-IR versus wild type flies (Fig. 8F). Innervation of flight muscles was normal, as confirmed by imaging of MN axons and their targeting into the neuromuscular junction (NMJ) (Fig. S5A–B). Further, although the number of boutons was not altered in the NMJ of D42>katp80-IR larvae, their diameter was significantly larger (Fig. S5C–H), suggesting defective axonal transport. Finally, we did not detect any significant changes in axonal structure (data not shown). Taken together, these observations suggested that kat80 regulates dendritic arborization of sensory and motor neurons in Drosophila.
DISCUSSION
Here, we report the identification of homozygous deleterious mutations in KATNB1 in multiple independent patients with pleomorphic cerebral cortical phenotypes of varying severity, consisting primarily of microlissencephalies, in which microcephaly co-occurred with neuronal migration abnormalities, ranging from white matter nodular heterotopia to lobar or global pachygyria, as well as cortical organization problems, including polymicrogyria.
KATNB1 encodes the p80 regulatory subunit of the microtubule severing enzyme katanin. A subset of the identified mutations (p.Leu540Arg, p.Ser535Leu and p.Gly578Asp) localize to the C-terminal region of KATNB1, which is known to interact with the p60/KATNA1 catalytic subunit as well as NDEL1; while 2 mutations (p.Val45Ileu, p.Val150Cysfs*22) map to the N-terminal region, which interacts with the molecular motor protein dynein and LIS1 (O’Donnell et al., 2012; Toyo-Oka et al., 2005). Patient-derived dermal fibroblasts displayed disorganized mitotic spindles and expressed lower amounts of KATNB1, similar to findings in the Taily mouse, which carries a hypomorphic allele of p80 (O’Donnell et al., 2012). The disease-causing mutations affected the interaction of KATNB1 with NDEL1 and KATNA1, disrupting their efficient localization to the centrosome and to the mitotic spindle during division. This is consistent with previous findings demonstrating that NDEL1 is required for Katanin localization to the centrosome during cell division (Toyo-Oka et al., 2005), suggesting that KATNB1, KATNA1 and NDEL1 are inter-dependent for their respective localization to the centrosomes. Similarly, fibroblasts harboring an N-terminal mutation in KATNB1, display spindle defects and a significant reduction in the amount of dynein localizing to the spindle and centrosomes. Thus, KATNB1 mutations result not only in decreased KATNB1 protein levels but also reduced localization of the katanin complex and other effector molecules to target areas, causing mitotic spindle defects.
To gain mechanistic understanding into KATNB1 function, we used Drosophila, a model system that has provided invaluable insight into the mechanism of action of genes involved in human MCD syndromes (Gonzalez et al., 1990; Liu et al., 2000; Rujano et al., 2013; Saunders et al., 1997; Siller and Doe, 2008; Yamamoto et al., 2014). Loss of the KATNB1 ortholog resulted in microcephaly in both Drosophila and zebrafish, recapitulating the human phenotype. In addition, in the accompanying paper by Hu, et al., Katnb1 knockout in mice leads to severe cortical abnormalities by affecting centriole and cilia biogenesis during development (Hu et al., 2014).
In Drosophila, loss of kat80 results in fewer NBs, which display cell cycle progression delay, ectopic supernumerary centrosomes and aberrant mitotic spindles. The reduced NB numbers in central brain is most likely a result of their failing to exit quiescence. During mitosis, anaphase chromatid-to-pole motion is tightly linked to de-polymerization of the opposite ends of chromosome-associated microtubules. Synchronized microtubule dynamics result from the highly regulated, concerted action of several proteins, including kat60 (Ghosh et al., 2012; Zhang et al., 2007). Since kat80 is important for targeting the kat60 catalytic subunit, our observation of delayed onset of anaphase in kat80-IR NBs demonstrates the central role of katanin in anaphase.
Unlike central brain NBs, which are derived from embryonic neural stem cells, the Drosophila optic lobe NBs originate from the neuroepithelium, in a process that resembles the development of the vertebrate cerebral cortex where progression from symmetric to asymmetric neurogenic divisions occur at early larval stages (Egger et al., 2007). Hence, Drosophila presents a unique system to dissect the molecular impact of any gene mutation on asymmetric versus symmetric cell division. For example, while asp loss leads to spindle defects and prometaphase arrest in central brain NBs, it causes chromosome segregation defects resulting in aneuploidy and apoptosis in optic lobe neuroepithelial cells (Gonzalez et al., 1990; Rujano et al., 2013; Saunders et al., 1997).
Surprisingly, kat80 loss did not significantly impact symmetrically dividing progenitors in optic lobe NE, yet resulted in severe reduction of asymmetrically dividing NBs, clearly demonstrating the differential effects of KATNB1 mutations on asymmetrically versus symmetrically dividing cells. The reduction in NB numbers could be due to deregulation of signaling mechanisms that control NB specification e.g., the Notch and JAK/STAT pathways. However, we found no effect of kat80 loss on either pathway in the optic lobe (Fig. S4), suggesting that kat80 regulates NB numbers independent of these cues. Indeed, it is known that in the optic lobe, intracellular signaling events, and not spindle orientation, regulate NB specification and hence cell fate (Egger et al., 2007). Thus, in kat80-IR larvae, the deficit in optic lobe NBs is also likely due to a cell fate specification defect, a notion also supported by our observation that, at some frequency, we do indeed find kat80-IR larvae with normal size brains containing supernumerary NBs (KM-G, unpublished observations). This would suggest a novel role for katanin in cell fate specification, which was previously shown to be independent of spindle orientation. Therefore, in the Drosophila larval brain, kat80 plays a dual role such that its loss not only compromises the initial pool of cells per se (exit from quiescence or cell fate specification), but also their proliferative capability (delayed anaphase onset, spindle and centrosomal defects), resulting in severe microcephaly.
Finally, microtubule severing and transport are known to play a central role in neuritogenesis (Franker and Hoogenraad, 2013). We observed a striking reduction of dendritic arborization of both central and peripheral neurons in kat80-IR flies, a finding that is consistent with the reported significant reduction of the dendritic field area and the number of the dendritic termini in kat-60L1 mutants (Stewart et al., 2012) and the role of Kat60 in dendritic elaboration (Mao et al., 2014). Dendrite pruning involves a noticeable degree of microtubule severing, especially in sensory neurons, and indeed MCDs have been associated previously with defective neuritogenesis (Gleeson et al., 1998; Shu et al., 2004). Furthermore, dendritic anomalies have been shown to be associated with intellectual disability (also seen in the patients reported here) and related syndromes caused by chromosomal aberrations (e.g., Down and Williams syndromes) and single gene mutations (e.g., Rett, Fragile-X and Rubinstein-Taybi disorders) (Armstrong, 2005; Kaufmann and Moser, 2000).
Our findings demonstrate the fundamental importance of microtubule dynamics in brain development across species. Perturbation of this evolutionarily conserved cellular process leads to complex cerebral cortical malformations caused by abnormalities in microtubule severing. The successful identification of new genes, such as KATNB1, implicated in cerebral cortical development provides unique insights into how the human brain develops normally and how this process may be derailed. This clearly has a significant impact not only in the area of basic neuroscience, as it promises to reveal key players in the fundamental mechanisms that govern the development of the human brain, but also has implications for understanding the pathophysiology of common neurodevelopmental disorders.
EXPERIMENTAL PROCEDURES
Human subjects
The study protocol was approved by the Yale Human Investigation Committee. Upon fully informed consent, genomic DNA was extracted from peripheral blood samples of patients, their parents, and unaffected siblings when available. DNA samples from affected individuals were subjected to whole-exome capture and sequencing, and to genome-wide genotyping for selected individuals. Identified mutations were confirmed and tested for segregation in the respective pedigrees by Sanger sequencing.
Fly Genetics
Oregon R or yw flies were used as wild-type controls. Other fly strains used include: w;worniu-GAL4, UAS-Miranda::GFP, UAS-Zeus::mCHERRY/Cyo;Dr/TM6b (Chris Doe); w;UAS-CD8-GFP;D42-gal4,chagal80; c380-Gal4, UAS-CD8-GFP;;cha-gal80; hsFLP;Act-FRT-CD2-FRTgal4,UAS-CD8(n); y[1] w[1118]; P{w[+m*]=GawB}GH146 (Bloomington Stock Center); kat80 RNAi lines were obtained from VDRC. Three independent kat80RNAi lines showed similar results.
Zebrafish experiments
katnb1 morpholino was injected into 1-cell stage wild type and dorsal view images of 72 hpf control and morphant embryos were taken by a Leica M205 FA dissecting microscope with LAS AF software. Statistical analysis was carried out in Microsoft Excel.
All other experimental methods are described in Supplemental Methods.
Supplementary Material
Supplemental Figure S1. Electropherograms obtained via Sanger sequencing analysis of patient NG-PNH-226 and her parents. Whole-exome sequencing identified two compound heterozygous mutations in this patient (marked in red, wild-type (WT) marked in green). (A) The G>A transition at the splice acceptor site variant; (B) shows the G>A transition leading to the missense p.Gly578Asp variant. Co-segregation analysis revealed that the patient inherited the splice acceptor site variant from her mother and the missense variant from her father. (Related to Figure 1)
Supplemental Figure S2. Reduced protein levels in patient-derived dermal fibroblasts and possible aneuploidy upon expression of mutant forms of KATNB1. (A) Western analysis indicates reduced levels of KATNB1 in patient- as compared with parent-derived fibroblasts. (B–C″) HeLa cells transfected with WT and mutant forms of KATNB1. Reduced localization to the kinetochores and lagging chromosomes during metaphase (arrow) upon expression of mutant KATNB1. (Related to Figure 2)
Supplemental Figure S3. kat80-IR does not lead to increased apoptosis. TUNEL staining of Drosophila 3rd instar larval brains with kat80-IR clones does not show increased apoptosis within the clones. (Related to Figure 5)
Supplemental Figure S4. kat80-IR does not affect Notch or JAK/STAT signaling in the optic lobe. The GH146 driver was used to express kat80-IR in Drosophila larval brains. 3rd instar larval brains were stained for Notch and STAT. Images are 3D projections of equivalent Z stacks and were captured using identical confocal settings. (Related to Figure 7)
Supplemental Figure S5. kat80-IR does not affect muscle innervation. (A, B). Adult flies expressing kat80-IR under C380-GAL4 driver, which directs expression in adult flight motoneurons (MN1–MN5), show normal innervation of the dorsal longitudinal flight muscle (DLM) via the PDMN nerve. (C–F). Bouton size but not numbers were affected by kat80-IR: NMJ at muscle 4 (C, D) and muscle 13 (E, F) were imaged and quantified for size (G) and number (H). Bouton size was significantly increased in kat80-IR 3rd instar larvae: muscle 4: yw: 2.71±0.098; kat80-IR:3.276 ±0.118; P = 0.0023; muscle 13: yw: 2.755±0.0853; kat80-IR:3.306 ±0.088; P = 0.0004. (Related to Figure 8)
Clinical Characteristics of patients with KATNB1 mutations, Additional physical exam findings of patients, Radiological features of patients, Calculated relationships, Overlapping HBD segments between relatives, HBD segments of patients, KATNB1 mutations identified by whole-exome sequencing and HBD variants of patients. (Related to Figure 1)
Acknowledgments
We thank the patients and families who contributed to this study. This work was supported by the Yale Program on Neurogenetics, National Institutes of Health (NIH) Grants U54HG006504 (Yale Center for Mendelian Disorders, to RPL, MG, M. Gerstein, and SM), U01MH081896 (to NS) and R01MH103616 (to MG and KB). This work was also supported by R01NS041537 and P01HD070494 (to JGG). RPL and JGG are Investigators of the Howard Hughes Medical Institute. We are grateful to the Gregory M. Kiez and Mehmet Kutman Foundation for its continuing support.
Footnotes
AUTHOR CONTRIBUTIONS
KMG designed, performed and analyzed in vitro and in vivo (Drosophila) experiments to characterize the KATNB1 mutations and wrote the manuscript. AOC and AES performed genetic analysis, identified KATNB1 mutations and summarized the genetic, clinical and radiological findings. CC helped design Drosophila experiments and write the manuscript and CC & TX helped analyze the data. OH generated and validated all constructs used in the study. FV worked on the dendritic arborization and bouton analysis studies and FV & HK analyzed the data. GTA experimentally identified and verified human mutations. SN and WH performed expression analyses in mouse and human tissue and SN, WH, AL and NS analyzed the data. ST performed the zebrafish experiments and ST& NCC analyzed the data. AOC, CD, MSZ, HAAH, JBR, HG, HK, EGS, ROR, OG, HP, SK & WBD ascertained and recruited patients; diagnosed and clinically evaluated patients and collected samples. JS performed genetic investigation in patients. BB, CC, CC, DD & JB assisted in experimental work. FJM performed radiological analysis. EZEO and KY performed bioinformatic analysis. SM & RPL oversaw exome sequencing of the patient samples. AL wrote the manuscript. KB analyzed the genetic data. JGG analyzed the genetic data and led the research. MG analyzed the genetic data, wrote the manuscript and led the research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdollahi MR, Morrison E, Sirey T, Molnar Z, Hayward BE, Carr IM, Springell K, Woods CG, Ahmed M, Hattingh L, et al. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. American journal of human genetics. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad FJ, Yu W, McNally FJ, Baas PW. An essential role for katanin in severing microtubules in the neuron. The Journal of cell biology. 1999;145:305–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected] American journal of human genetics. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong DD. Neuropathology of Rett syndrome. Journal of child neurology. 2005;20:747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, Ganguly A, Roote J, Ip YT. Worniu, a Snail family zinc-finger protein, is required for brain development in Drosophila. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;231:379–386. doi: 10.1002/dvdy.20130. [DOI] [PubMed] [Google Scholar]
- Bakircioglu M, Carvalho OP, Khurshid M, Cox JJ, Tuysuz B, Barak T, Yilmaz S, Caglayan O, Dincer A, Nicholas AK, et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. American journal of human genetics. 2011;88:523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak T, Kwan KY, Louvi A, Demirbilek V, Saygi S, Tuysuz B, Choi M, Boyaci H, Doerschner K, Zhu Y, et al. Recessive LAMC3 mutations cause malformations of occipital cortical development. Nature genetics. 2011;43:590–594. doi: 10.1038/ng.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain: a journal of neurology. 2012;135:1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Current biology: CB. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends in genetics: TIG. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nature genetics. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Casanova M, Crobu L, Blaineau C, Bourgeois N, Bastien P, Pages M. Microtubule-severing proteins are involved in flagellar length control and mitosis in Trypanosomatids. Molecular microbiology. 2009;71:1353–1370. doi: 10.1111/j.1365-2958.2009.06594.x. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature genetics. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Consoulas C, Restifo LL, Levine RB. Dendritic remodeling and growth of motoneurons during metamorphosis of Drosophila melanogaster. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22:4906–4917. doi: 10.1523/JNEUROSCI.22-12-04906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural development. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly. 2011;5:237–241. doi: 10.4161/fly.5.3.15640. [DOI] [PubMed] [Google Scholar]
- Franker MA, Hoogenraad CC. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- Frise E, Hammonds AS, Celniker SE. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Molecular systems biology. 2010;6:345. doi: 10.1038/msb.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh DK, Dasgupta D, Guha A. Models, Regulations, and Functions of Microtubule Severing by Katanin. ISRN Molecular Biology. 2012;2012:14. doi: 10.5402/2012/596289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, Walsh CA. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Gunawardena S. Flying through the drosophila cytoskeletal genome. The Journal of cell biology. 2000;150:F63–68. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, Glover DM. Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci. 1990;96(Pt 4):605–616. doi: 10.1242/jcs.96.4.605. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Durr A, Wincker P, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nature genetics. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Hu WF, Pomp O, Kodani A, Henke K, Ben-Omran T, Mochida GH, Yu TW, Woodworth MB, Bonnard C, Shboul M, et al. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 2014 doi: 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Koenig JH. Morphological identification of the motor neurons innervating the dorsal longitudinal flight muscle of Drosophila melanogaster. The Journal of comparative neurology. 1988;273:436–444. doi: 10.1002/cne.902730312. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, Kuehn MR. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature. 1999;399:691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nature genetics. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nature reviews Neuroscience. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, Gerard B, Verloes A, Mani S, Gressens P. Many roads lead to primary autosomal recessive microcephaly. Progress in neurobiology. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabay A, Yu W, Solowska JM, Baird DH, Baas PW. Axonal growth is sensitive to the levels of katanin, a protein that severs microtubules. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:5778–5788. doi: 10.1523/JNEUROSCI.1382-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cerebral cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Pilz DT, Babatz TD, Cushion TD, Harvey K, Topf M, Yates L, Robb S, Uyanik G, Mancini GM, et al. TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Human molecular genetics. 2010;19:2817–2827. doi: 10.1093/hmg/ddq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Miller MR, Robinson KJ, Doe CQ. The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Developmental cell. 2012;23:849–857. doi: 10.1016/j.devcel.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nature cell biology. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- Mao CX, Xiong Y, Xiong Z, Wang Q, Zhang YQ, Jin S. Microtubule-severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development. 2014;141:1064–1074. doi: 10.1242/dev.097774. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. The Journal of cell biology. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, Bazirgan OA, McNally FJ. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci. 2000;113(Pt 9):1623–1633. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- O’Donnell L, Rhodes D, Smith SJ, Merriner DJ, Clark BJ, Borg C, Whittle B, O’Connor AE, Smith LB, McNally FJ, et al. An essential role for katanin p80 and microtubule severing in male gamete production. PLoS genetics. 2012;8:e1002698. doi: 10.1371/journal.pgen.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Molecular and cellular biology. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Sanchez-Pulido L, Pennetier C, le Dez G, Basto R. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nature cell biology. 2013;15:1294–1306. doi: 10.1038/ncb2858. [DOI] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. The Journal of cell biology. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. The Journal of cell biology. 2007;178:1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Developmental biology. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Molecular biology of the cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Tsubouchi A, Rolls MM, Tracey WD, Sherwood NT. Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:11631–11642. doi: 10.1523/JNEUROSCI.0729-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. The Journal of cell biology. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends in genetics: TIG. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyo-Oka K, Sasaki S, Yano Y, Mori D, Kobayashi T, Toyoshima YY, Tokuoka SM, Ishii S, Shimizu T, Muramatsu M, et al. Recruitment of katanin p60 by phosphorylated NDEL1, an LIS1 interacting protein, is essential for mitotic cell division and neuronal migration. Human molecular genetics. 2005;14:3113–3128. doi: 10.1093/hmg/ddi339. [DOI] [PubMed] [Google Scholar]
- Vonhoff F, Duch C. Tiling among stereotyped dendritic branches in an identified Drosophila motoneuron. The Journal of comparative neurology. 2010;518:2169–2185. doi: 10.1002/cne.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, Williams A, Ryglewski S, Duch C. Drosophila as a model for MECP2 gain of function in neurons. PloS one. 2012;7:e31835. doi: 10.1371/journal.pone.0031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield JG, Bonaccorsi S, Gatti M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. The Journal of cell biology. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieck G, Leventer RJ, Squier WM, Jansen A, Andermann E, Dubeau F, Ramazzotti A, Guerrini R, Dobyns WB. Periventricular nodular heterotopia with overlying polymicrogyria. Brain: a journal of neurology. 2005;128:2811–2821. doi: 10.1093/brain/awh658. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Molecular biology of the cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. The Journal of cell biology. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Electropherograms obtained via Sanger sequencing analysis of patient NG-PNH-226 and her parents. Whole-exome sequencing identified two compound heterozygous mutations in this patient (marked in red, wild-type (WT) marked in green). (A) The G>A transition at the splice acceptor site variant; (B) shows the G>A transition leading to the missense p.Gly578Asp variant. Co-segregation analysis revealed that the patient inherited the splice acceptor site variant from her mother and the missense variant from her father. (Related to Figure 1)
Supplemental Figure S2. Reduced protein levels in patient-derived dermal fibroblasts and possible aneuploidy upon expression of mutant forms of KATNB1. (A) Western analysis indicates reduced levels of KATNB1 in patient- as compared with parent-derived fibroblasts. (B–C″) HeLa cells transfected with WT and mutant forms of KATNB1. Reduced localization to the kinetochores and lagging chromosomes during metaphase (arrow) upon expression of mutant KATNB1. (Related to Figure 2)
Supplemental Figure S3. kat80-IR does not lead to increased apoptosis. TUNEL staining of Drosophila 3rd instar larval brains with kat80-IR clones does not show increased apoptosis within the clones. (Related to Figure 5)
Supplemental Figure S4. kat80-IR does not affect Notch or JAK/STAT signaling in the optic lobe. The GH146 driver was used to express kat80-IR in Drosophila larval brains. 3rd instar larval brains were stained for Notch and STAT. Images are 3D projections of equivalent Z stacks and were captured using identical confocal settings. (Related to Figure 7)
Supplemental Figure S5. kat80-IR does not affect muscle innervation. (A, B). Adult flies expressing kat80-IR under C380-GAL4 driver, which directs expression in adult flight motoneurons (MN1–MN5), show normal innervation of the dorsal longitudinal flight muscle (DLM) via the PDMN nerve. (C–F). Bouton size but not numbers were affected by kat80-IR: NMJ at muscle 4 (C, D) and muscle 13 (E, F) were imaged and quantified for size (G) and number (H). Bouton size was significantly increased in kat80-IR 3rd instar larvae: muscle 4: yw: 2.71±0.098; kat80-IR:3.276 ±0.118; P = 0.0023; muscle 13: yw: 2.755±0.0853; kat80-IR:3.306 ±0.088; P = 0.0004. (Related to Figure 8)
Clinical Characteristics of patients with KATNB1 mutations, Additional physical exam findings of patients, Radiological features of patients, Calculated relationships, Overlapping HBD segments between relatives, HBD segments of patients, KATNB1 mutations identified by whole-exome sequencing and HBD variants of patients. (Related to Figure 1)


