SUMMARY
The cell walls of the shoot apical meristem (SAM), containing the stem cell niche that gives rise to the above-ground tissues, are crucially involved in regulating differentiation. It is currently unknown how these walls are built and refined or their role, if any, in influencing meristem developmental dynamics. We have combined polysaccharide linkage analysis, immuno-labelling and transcriptome profiling of the SAM to provide a spatio-temporal plan of the walls of this dynamic structure. We find that meristematic cells express only a core subset of 152 genes encoding cell wall glycosyltransferases (GTs). Systemic localization of all these GT mRNAs by in situ hybridization reveals members with either enrichment in or specificity to apical subdomains such as emerging flower primordia, and a large class with high expression in dividing cells. The highly localized and coordinated expression of GTs in the SAM suggests distinct wall properties of meristematic cells, and specific differences between newly forming walls and their mature descendants. Functional analysis demonstrates that a subset of CSLD genes is essential for proper meristem maintenance, confirming the key role of walls in developmental pathways.
INTRODUCTION
Plant biomass, our only renewable bioresource, is largely composed of cell walls. The primary plant cell wall is a complex composite made up almost entirely of polysaccharides (90–95%) with some (glyco)proteins (5–10%) [1–4]. It serves to provide strength and mechanical support to plant tissues and provides resistance to the high turgor pressure inside each cell. Local reinforcement coupled with wall loosening, achieved by rapid remodeling, permits not only growth, but also the generation of a variety of cell shapes, ranging from the cylindrical cells of the epidermis and endodermis of the root to more complex shapes such as those of the leaf epidermal pavement cells [5]. All growing cells contain a primary wall and further specialization is observed in certain cell types during tissue differentiation [1, 6].
The shoot apical meristem (SAM) is a dome-shaped structure that contains the above-ground stem cell pool; slowly proliferating cells that are found at the top of the dome within a region termed the central zone (CZ). The progeny of these cells are gradually displaced to the peripheral zone (PZ) where cells grow and divide at a higher rate [7]. Auxin maxima within the PZ determine the sites of either flower or leaf primordial initiation, characterized by maximal rates of cell expansion [8, 9]. It has been proposed that the dome shape structure is maintained through a feedback loop whereby the stress patterns, dictated by tissue geometry, influence the organization of the cytoskeleton and reinforcements of the cell wall [10]. This loop affects auxin movements around the SAM via changes in the polarity of the PIN1 auxin transporter [11]. Differences in cell wall rigidity appear to demarcate the different functional domains [12] with the CZ being three-fold stiffer (having a higher elastic modulus) than faster-growing cells at the flanks of the SAM [13, 14]. Thus, local differences exist in walls and these result from, and contribute to, developmental dynamics.
The primary wall of most flowering plants consists typically of cellulose, non-cellulosic polysaccharides, pectins and glycoproteins/proteoglycans. While cellulose is an unsubstituted homo-polymer of glucose (Glc), most polysaccharides have backbone substitutions ranging from a small number of sugar residues (e.g. xyloglucan, XG) to very complex branching patterns observed in some pectic polysaccharides, for example arabinogalactan (AG), rhamnogalacturonan (RG), and proteoglycans (AGPs). Both glycan backbone chain elongation and substitutions are carried out by polysaccharide synthases/glycosyltransferase (GT) enzymes, classified into families and subfamilies based on phylogenetic similarities. Arabidopsis thaliana possesses in the order of 560 GTs, and many of these are expected to be involved in cell wall synthesis (CaZy, http://www.cazy.org/).
Despite the central role played by the cell wall in the SAM and subsequent development, very little information exists as to its composition and that of the early developing organs [14–18]. Here we have combined data from polysaccharide linkage analysis, steady-state mRNA quantification and localization, and polysaccharide immuno-labelling, to build up a comprehensive picture of the construction of the SAM primary walls and their cognate biosynthetic GTs. We find that meristematic cell walls are constructed by a reduced subset of GTs with spatially and temporally regulated expression patterns. The data suggest a distinction between new walls formed at cell division and pre-existing walls and suggests a key role by a GT of the CSLD gene family, required for proper growth and maintenance of cell number in the SAM.
RESULTS
Isolating shoot meristematic cells for polysaccharide and expression profiling
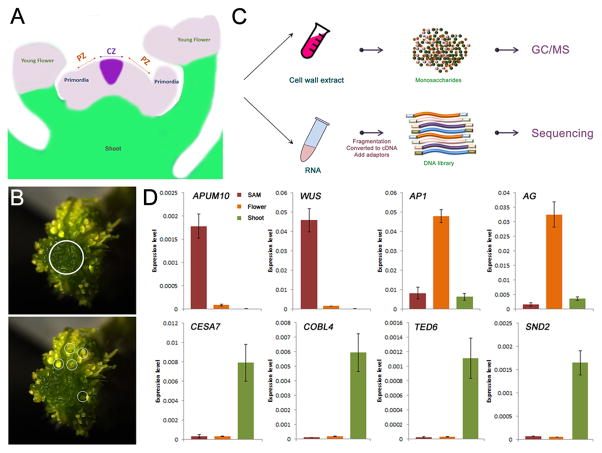
To reveal the structure and synthesis of stem cell walls, we tried to harvest pure meristematic cells from Arabidopsis shoot apex. Due to small meristem size, it would require around 105 dissected SAMs to generate the 4 mg fresh weight required for reliable polysaccharide linkage analysis. We thus looked to the enlarged, stem-cell enriched SAMs of the clavata3 (clv3) mutant [19] (Figure S1). Meristems were carefully dissected from 60 clv3-2 shoot apices, and young flowers were also included in our analysis to represent a mixed sample of meristematic, highly proliferative and developing tissues (Figures 1A–C) [20]. The samples were subjected to cell wall linkage analysis and RNA sequencing (Figure 1B). To evaluate the quality of these dissected samples (i.e. no contamination of developed shoot tissues in the meristematic cells), we performed quantitative RT-PCR to check genes with specific expression pattern in different tissues. Consistently, APUM10 and WUS, two SAM-enriched genes, showed high expression in the SAM sample, and floral organ specific genes, AP1 and AG, expressed highly in the flower sample. By contrast, the transcripts of genes involved in secondary cell wall biosynthesis and vascular formation, including CESA7, COBL4, TED6 and SND2, could be barely detected in the meristematic tissues (Figure 1D).
Figure 1. Dissection of Meristematic Cells for Wall Analysis.
(A) Schematic representation of the organization of Arabidopsis shoot apex. Meristematic cells (purple) in both shoot apical meristem (SAM) and flower primordia were collected for analysis. CZ, central zone; PZ, peripheral zone.
(B and C) Schematic flowchart displaying the strategy used in this study. The meristematic cells carefully dissected from clv3-2 SAM and flower primordia (before Stage 6 according to [20]) (C) were used for cell wall preparation and RNA extraction. Cell wall composition was determined by linkage analysis. Glycosyltransferase (GT) gene expression was identified by RNA sequencing (B).
(D) The expression of genes specific to SAM (APUM10 and WUS), flower (AP1 and AG) and shoot vasculature (CESA7, COBL4, TED6 and SND2) in dissected shoot apical meristem and young flower. The shoot sample was included for comparison. Arabidopsis UBIQUITIN10 gene was used as an internal control. Shown are mean values from three replicates, error bars represent standard deviation values.
Composition and Spatial Organization of Cell Wall Components
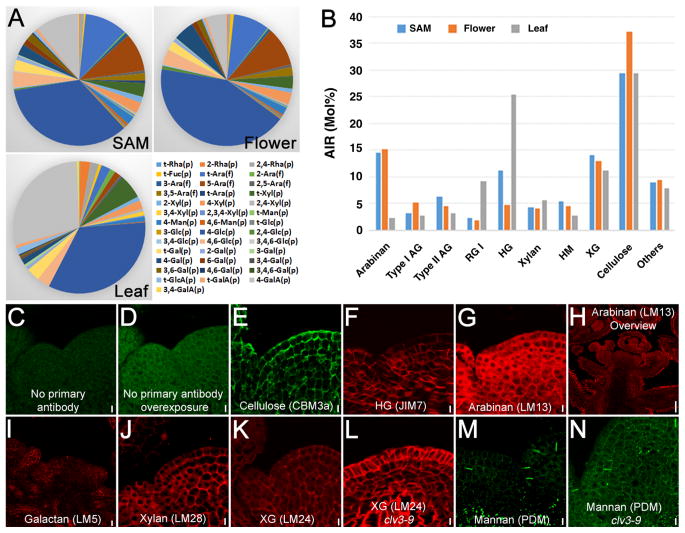
The alcohol insoluble residue (AIR) from samples were analyzed to determine their polysaccharide composition by monosaccharide linkage analyses based upon a priori knowledge of the relative proportions of these linkages in a particular polysaccharide [21]. Most of the monosaccharides identified previously from leaf could also be detected in both meristem and flower, although the relative composition was different (Figure 2A). Polysaccharide calculations show the walls of meristematic cells to be composed of, in order of decreasing abundance, cellulose, pectin and non-cellulosic polysaccharides (Figure 2B). A marked increase in the amount of homogalacturonan (HG), composed of linear chains of galacturonic acid (GalA) residues, was found in SAM compared to flowers (11.3 vs. 4.8%), while type I AG (3.3 vs 5.3%) and cellulose (29.4 vs 37.1%) exhibited decreases in the SAM. Compared to leaf, both the SAM and young floral tissue contained at least 3-fold less RG-I, a 2- to 6- fold reduction in HG and an approximate 6-fold increase in pectic arabinan. Total pectin content is reduced in SAM and flowers compared to the level in leaves. Cellulose levels in the leaf (29.5 %) were found to be nearly identical to the SAM (29.4 %). Overall the data show some marked differences in cell wall pectic and non-cellulosic composition between SAM and its derivatives, the flower and leaf.
Figure 2. Composition and Spatial Distribution of Wall Components in Shoot Apex.
(A) Monosaccharide linkage composition of AIR cell wall preparations.
(B) Calculation of polysaccharide composition based on monosaccharide linkage analysis shown in (A).
(C–N) Immunofluorescence labelling showing the spatial distribution of wall components. The longitudinal sections of Arabidopsis shoot apex were incubated with cell wall antibody probes. No primary antibody (anti-rabbit secondary) control is shown in (C) and overexposed control in (D). Scale bar = 25 μm. Scale bars in (C–G) and (J–N) = 5 μm, in (H) = 20 μm, and in (I) = 10 μm.
To reveal the spatial organization of cell wall structure in the SAM, we took advantage of immunohistochemistry using different antibodies or recombinant fluorescent proteins that recognize individual wall components. Consistent with cellulose being the major polysaccharide, CBM3a that binds crystalline cellulose, was found to label all walls within the SAM. Labeling of walls across the apex by CBM3a was not uniform, suggesting either differences in the organization of wall polymers (thereby affecting accessibility of the probe to cellulose), variation in the degrees of cellulose crystallinity or absolute abundance (Figure 2E). Similarly inhomogeneous labeling was observed for JIM7 (HG), which might reflect some variation in the degree of HG methyl-esterification (Figure 2F). Detection of pectic α-1,5-arabinan by LM13 resulted in a complex pattern of labeled walls in both SAM and floral tissues (Figure 2G), and a very intense signal was seen in the epidermal cells of older developing tissues (Figure 2H). Labeling of β-1,4-galactan by LM5 was found predominantly in primordia and the outer whorl of young developing flowers (Figure 2I), correlating closely with the double amount of 1,4-Gal in flowers compared to the SAM from linkage analysis (5.1 vs 2.7 %, Table S1). Both heteroxylan (HX) and xyloglucan (XG) existed in all walls of meristematic cells, as revealed by LM28 and LM24 labeling (Figures 2J and 2K). However, the signals could be observed only after pectolyase treatment, suggesting that these polysaccharides are masked by pectin. Although most of the epitopes exhibited similar organization patterns in clv3 compared to wild-type, XG seems to show distinct patterns, with a higher signal in the L1 layer cells (Figure 2L). This implies a secondary effect of an enlarged central zone upon either XG synthesis or organization. In addition, we found that PDM, a mannan antibody that recognizes heteromannans (glucomannans and galactomannans), only labelled certain regions of cells that oriented anticlinally in upper cell layers but were situated either anticlinally or periclinally in the lower cell layers (Figures 2M and 2N), reminiscent of the newly formed cross walls arising after cell division events.
Analysis of the transcriptome reveals differential expression of a subset of GT genes in the SAM
With this insight into the cell wall composition of the SAM and young flowers, we could then examine the specific differential expression patterns of GTs in these tissues, since these enzymes determine, to a large extent, which wall polysaccharides and glycoproteins are being made. We performed RNA sequencing using the highly purified SAM tissues and young flowers that were equal to those used for linkage analysis. A third sample consisting of pooled total RNA from several plant tissues was also analyzed for comparison. Comparison of the RNA levels from our purified meristematic cells with previously published cell-type expression profiling data revealed high spatial sensitivity to detect genes expressed within particular tissues (see Supplemental Experimental Procedures). For example, APUM10/PUMLIO10, a gene that is only expressed in the CLV3 domain, was detected exclusively in the SAM samples (Figure S2A). Gene Ontology (GO) analysis of differentially expressed genes revealed cell wall categories to be under-represented in the SAM and flower (Figure S2B). Further analysis found differences in GT transcript abundance between SAM, flower and the whole plant samples (Figure S2C), suggesting that meristematic cells employ only a subset of GT enzymes to build the wall. For example, in the GT8 subfamily of enzymes that give rise to GlcA substitution of xylan [22–24], only GUX3 expression could be detected in SAM and flower. Among the 10 FUT genes responsible for XG and AGI synthesis, only three exhibited considerable expression in meristematic tissues. This contraction also extends to the protein backbones of proteoglycans, including those that make up AGPs and extensins, where only a subset of mRNAs are present in the SAM (Table S2). A literature search allowed the assignment of more gene families or subfamilies that encode cell wall-related GTs to polysaccharide linkages. A total of 152 SAM-expressed GTs (Transcripts Per Million, TPM value >1) were mapped to their respective polysaccharides/glycoproteins (Table S2).
Consistent with cellulose being the major polysaccharide, the core primary wall cellulose synthase subunits of the GT2-CESA family, encoded by CESA1 and CESA3, are highly co-expressed (TPM values >100 for all samples), as observed in all primary wall tissues, and are found together with other CESA mRNAs (from CESA2, CESA5, CESA6) that encode proteins that provide the 3rd component of the active complex [25] with both CESA5 and CESA6 exhibiting higher mRNA levels in flowers.
The pectin backbone (HG and RGI) (Figure 2B), is synthesized by GalATs, encoded by the GT8-GAUT subfamily [26], the transcripts of which are widely represented in the SAM. The proposed HG core complex is encoded by GAUT7 and GAUT1 and these are both expressed (Table S2) consistent with their products forming a core biosynthetic complex [27]. Despite the low quantities of RGI in both SAM and flower compared to leaf, expression of some potential RGI-associated GTs (GATL3, 5, 6, 8, 9) were found at higher levels than for the pooled samples (Table S2). The absence of XGD1 mRNA in the floral sample suggests that flowers either do not contain xylogalacturonan or that it is made by the product of the related EMB175 gene.
XG biosynthesis has only one confirmed GT2 family member, namely CSLC4, responsible for backbone (1,4-Glc) synthesis [28]. Furthermore, all the genes encoding enzymes that make up the recently identified XG multi-protein complex; CSLC4, XXT2, XXT5, MUR3 [29] and XLT2 [30] are expressed in the SAM at levels comparable with the pooled sample (Table S2). CSLC6 displays the highest expression of all CSLC subfamily members in the SAM; however, a function in XG assembly has not yet been demonstrated. The GT34 family contains seven members, of which data supports five genes encoding XG xylosyltransferases (XXT1-5). There is no evidence that the remaining two, designated GTL6 and GTL7 encode proteins with this type of GT activity [31]. GTL6 is one of the most highly expressed GT genes in the SAM (Table S2, mean TPM = 210). The gene product exhibits similarity to galacto(gluco)mannan galactosyltransferases (GMGTs) in other plants [32]. Indeed, recent data demonstrate that the gene, renamed MUCI10, encodes a GMGT involved in seed mucilage formation [33]. We have therefore assigned these two GMGTs to mannan as the enzymes that add the terminal Gal residue onto the 4-Man backbone giving rise to the 1,4,6-Man residue that most likely represents substituted heteromannan. The backbone is synthesized by members of the CSLA family where CSLA2 transcripts are the most abundant (Table S2). CSLA7, essential for embryonic development [34], and CSLA10 exhibit higher expression in the flower compared to other tissues (Table S2).
For HX, a proportion detected in the floral linkage analysis will be expected to come from the vascular-derived secondary walls, however, it is not expected that there would be the same in the SAM preparations since mRNA transcripts from known genes involved in xylem and secondary wall formation have been shown either to be absent or at barely detectable levels (Figures 1D and S2D). While the key genes encoding the enzymes for 1,4-Xyl backbone synthesis have been shown to be IRX9, IRX10 and IRX14 [35, 36], transcriptome data suggests apical enrichment of the corresponding paralog in each case (IRX9L, IRX10L and IRX14L; Table S2). Of the secondary wall-associated members, only IRX10 and IRX14 are expressed in the SAM. Also, F8H rather than FRA8 is the likely GT involved in making the reducing-end sequence of the xylan chain. While the presence of a GT transcript is expected to correlate with its cognate polysaccharide, within the complex cell wall composite we might expect to see spatio-temporal correlation between different polysaccharides reflected in the expression of associated GTs. Using collections of publicly available microarray expression data, a pairwise correlation coefficient network [37] was constructed from SAM GT transcripts higher than 10 TPM (Figure S2D). At the center of the network are the CESA1, 3, 6 genes that have a pairwise Pearson coefficient r >0.9, consistent with the encoded proteins forming a primary wall cellulose synthase complex (CSC) found in diverse tissue types [25]. Figure S2D (boxed) shows a sub-network that comprises genes with transcripts that peak in the young floral samples and where the core is occupied by CSLC8. Together with CSLC5, this may represent construction of a XG although no function has yet been assigned to the encoded GTs at present. Of the other floral network genes; GATL5 is known to be involved in RGI formation [38], which may extend to GATL3; CSLD5 has been attributed to cellulose synthesis, although there is a report suggesting a role in mannan biosynthesis [39–41]; GSL12 is a callose synthase [42]; and GALT31a (and possibly AT5G41460) assemble AGPs [43].
Visualization of GT mRNAs at the wild type shoot apex reveals subdomain specific expression of cell wall GTs
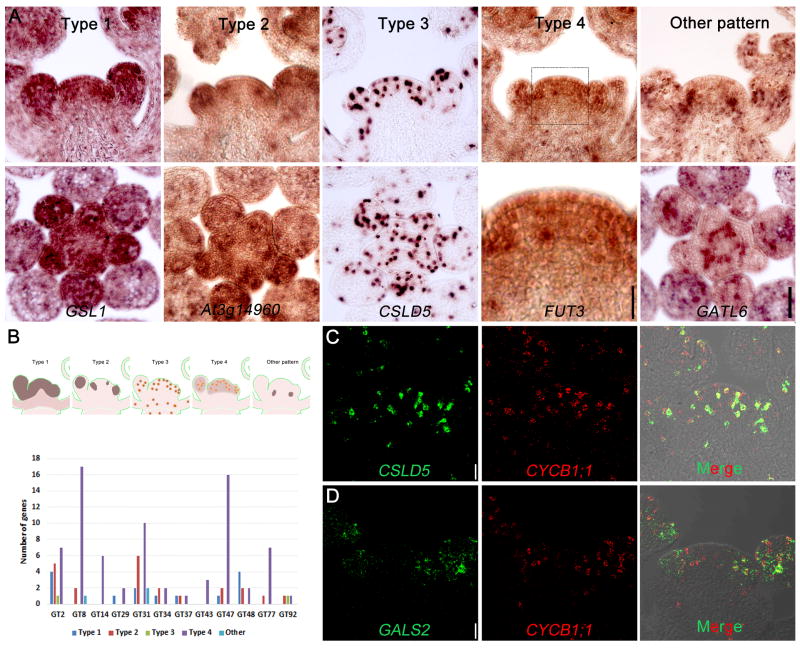
Co-expression of different GT family members suggests that the encoded enzymes, and their respective polysaccharides, may contribute to either specific types of walls or developmental stages. We therefore performed systematic RNA in situ hybridization assays (Figures 3A, Table S3) to examine the expression patterns of the SAM-expressed GTs at single-cell spatial resolution including those transcripts that either show relative increases, reductions or no change between SAM and flower samples. Transcript localization data confirmed that most of the candidate GT-encoded genes exhibit high expression in WT SAM as well as flower primordia. Patterns of expression were found to be similar between WT and clv3 (Figure S3). Based on the expression pattern, these genes were divided into five categories (Figure 3A): Type 1 represents a uniform distribution across the apex (e.g. GSL1); Type 2 represents apical patchy distribution, suggesting flower primordia-specific enrichment (e.g At3g14960); Type 3 have intense scattered spots (e.g. CSLD5); Type 4 represents both spotted and general apical enrichment (e.g. FUT3); and Type 5 consists of patterns not classified in the above. Within the Type 5 group, GATL6 mRNA gives a high signal in the approximate location of vascular initials and in developing outer whorls of the flower, further confirmed by visualizing the protein (Figure S4A–C). A summary of the classification of mRNA localization patterns for GT families, together with representative images of all SAM-expressed cell wall GT mRNA hybridizations, are shown in Figure 3B and Table S3. Some of these patterns are similar to those reported for non GT-encoding genes in the tomato SAM [44] with Arabidopsis Types 3 and 4 exhibiting similarities to the tomato pattern 5, represented by histone 2A and suggesting a degree of cell division-linked expression [45]. The majority (74/115) of the Arabidopsis GT mRNA patterns are classified as Type 4. There are 14 genes that have a Type 1 pattern. The expression of these genes could also be detected in other tissues (e.g. shoot tissue), suggesting that they might play fundamental roles in building the cell wall. Indeed, genes grouped into this type include CESA3 for primary wall cellulose and CSLC4, XXT1, and FUT1 that encode XG GTs. This broad expression of GTs is consistent with the broad labeling of the polysaccharides. In contrast to the Type 1 CESA3 pattern, RNAs for two of the subunits of the 3rd polypeptide of the CSC, encoded by either CESA5 or CESA6, gave a distribution pattern (Type 2) confirming up-regulation in floral primordia. For XG, other GTs include MUR3, At5g62220 (Type 4) and XXT2 (Type 2). The RNA for a putative GalT, encoded by GT15, while found at low levels across the SAM, appears to be confined to the inner whorls of the developing flower (Table S3).
Figure 3. Localisation of Glycosyltransferase (GT) Gene mRNAs in Shoot Apex by In Situ Hybridization.
(A) The expression patterns of glycosyltransferase genes expressed in shoot apex. The GTs were classified into five patterns according to their mRNA distribution. One representative gene for each type was shown in both longitudinal (top panel) and transverse (bottom panel) sections. Scale bar = 50 μm except for Type 4 bottom panel, which is 20 μm.
(B) A sketch showing the expression patterns of GTs and summary of gene number in each GT family that are classified into different expression patterns.
(C, D) Co-expression of GT genes with Cyclin B1;1 that marks dividing cells by dual labelling fluorescent in situ hybridization. Scale bar = 20 μm.
For acidic pectic polysaccharide synthesis, the majority of GAUT family transcripts (GAUTs 1,3,4,6,7,9,10,11,13,14,15) have similar expression patterns, in these cases a Type 4 distribution. In contrast, of the five genes that comprise the GT92 family encoding enzymes that specifically make β-1,4-galactan [46], the three shoot-apex expressed genes GALS1, GALS2 and GALS3 have divergent expression patterns, grouped into Type 2, 3 and 4, respectively (Tables S2, S3). The mRNA pattern of GALS2 and GALS3, concentrated at sites of primordial initiation and during floral development, were largely consistent with the signal observed with the corresponding epitope (Figure 2I).
Some members of the GT31 and GT29 families have been proposed to be involved in the assembly of the β-1,6-Gal residues that provide the core substitutions of the β-1,3-galactan backbone of type II AGs [43, 47]. Of particular interest is At1g32930 (GALT31A) since (i) its mRNA is florally enriched (Table S2), (ii) its patchy expression pattern matches that expected of early forming flower primordia (Table S3, Figure S4D) and (iii) insertion mutants have been found to arrest during embryo development [43]. To confirm whether the patchy expression is indeed confined to regions of organ initiation, we observed a fluorescent reporter of GALT31A promoter activity, together with a YFP plasma membrane reporter for observation of cell boundaries. A top view of a confocal z projection shows the foci of GALT31A expression coincide with floral primordia and at sites where primordia would be expected to initiate based on a spiral phyllotactic pattern (Figure S4E,F) and confirming that, at least for some genes, the patchy mRNA localization coincides with new flower development.
All transcripts encoding known xylan biosynthetic enzymes have a Type 4 distribution and for the heteromannan backbone synthesis, CSLA expression patterns were found to be either of Type 2 or 4 patterns. The Gal substitutions along the backbone appear to be formed through the action of MUCI10/GTL6. MUCI10 mRNA signal was very high in some cells (Table S3) consistent with the relatively high levels of mRNA found in the transcriptome analysis of the SAM. Callose is made by the GT48 (GSL/CALS) family members and transcripts were localized in either a Type 1 (GSL1, 5, 6, 10), Type 2 (GSL 8,11) or Type 4 (GSL3, 12) pattern.
Expression of GT genes during cell division
We found the mRNA of the GT2 member CSLD5 exhibits intense spots (Type 3 pattern) representing single cells against a very low background (Figure 3A). Similar intense spots were also observed for GALS2 (Figure S3, Table S3). To test whether the spotted pattern represents actively dividing cells, we carried out dual labeling of CSLD5 and GALS2 GT mRNA together with the M-phase marker, CyclinB1:1 mRNA, by using probes labeled with either cyanine5 (Cy5) or fluorescein. CSLD5 mRNA was found in most of the cells containing Cyclin B mRNA (Figure 3C, Manders coefficient M1=0.295), indicating its expression to be linked to the later portion of the cell cycle. GALS2 also largely co-expressed with CyclinB1;1 in flower primordia (Figure 3D, Manders coefficient M2=0.115). The enriched expression of CSLD5 and GALS2 in mitotic cells, together with a significant proportion of GT mRNAs showing an intense punctate labeling (i.e. Type 4 pattern), suggest that the expression of a large number of GT genes are up-regulated during cell division, probably contributing to the formation of new cross walls.
CSLD family genes function in stem cell maintenance
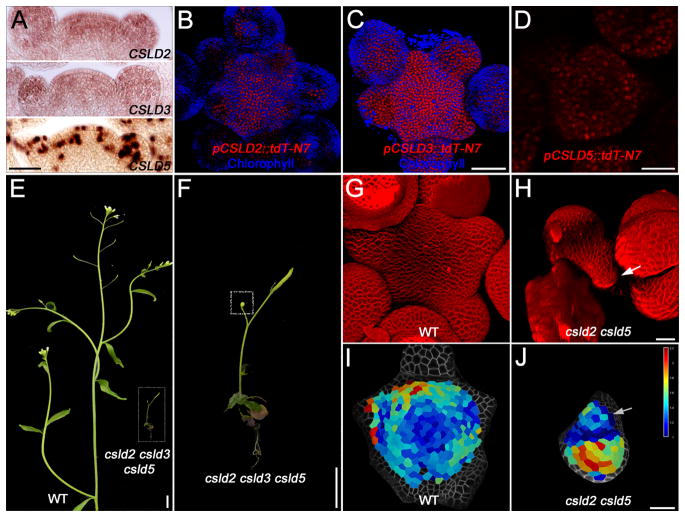
Given that CSLD5 and the other shoot apex-expressed members of the CSLD family (CSLD2, CSLD3) are found at comparatively high levels at the shoot apex (Figure 4A–D, Tabld S2), we examined SAMs from double and triple mutant combinations where whole plant phenotypes have suggested developmental defects [40]. We found severe growth retardation of the triple mutant compared to WT (Figure 4E, F). The double mutants csld2 csld5 and csld3 csld5 and the triple mutant csld2 csld3 csld5 showed highly similar phenotypes. In our growth conditions the mutant plants (double and triple mutants) produce only one to three flowers (Figure 4F), however, where flowers did form they appeared morphologically normal. For some plants no SAM was readily identifiable, especially in the triple mutant, presumably due to early termination. Confocal imaging revealed a small misshapen SAM in the csld mutants that were approximately one quarter of the diameter of WT SAMs with the flowers encompassing a large part of the meristem (compare Figures 4G and H). Over a 24-hour period individual cells were segmented and tracked to compare growth rates. For WT, cells of the CZ yield the lowest growth (blue cells in Figure 4I) with increases observed in the PZ (turquoise through yellow and red in Figure 4I). Cells that are part of flower primordia show the highest degree of growth. For a csld mutant, the SAM (Figure 4J, arrow) had no recognizable growth domains, however, the larger flower had cells achieving maximum growth rates consistent with the growth retardation being exerted at the level of the SAM, not the developing flowers. We interpret these data to mean that CSLD genes are needed for proper cell growth and proliferation in the SAM, that is, maintaining the meristematic stem cell pool, rather than being involved in specifying cell fate.
Figure 4. CSLDs Are Required for Shoot Apical Meristem Maintenance.
(A) Expression patterns of CSLD2, CSLD3 and CSLD5 as revealed by in situ hybridization. Scale bar = 50 μm.
(C–D) Confocal images showing the expression domains of CSLD2, CSLD3 and CSLD5 in SAM. CSLD2 and CSLD3 expression is active in most of the cells, while CSLD5 is enriched in dividing cells. Scale bars in (C) = 50 μm and in (D) = 25 μm.
(E and F) Whole plant phenotypes of wild-type (WT, Col-0) and the csld2 csld3 csld5 triple mutant. A close-up of the csld2 csld3 csld5 mutant is shown in (F) with the shoot apex boxed. Scale bars in (E) = 1 cm and in (F) = 0.5 mm.
(G and H) 3D renders of confocal z-stacks of wild-type and csld2 csld5 mutant SAMs. Scale bar = 20 μm.
(I and J) A growth heat map of wild-type and csld2 csld5 mutant SAMs showing relative growth increases per cell over a 24-hour period. Scale bar = 20 μm.
In the csld2 csld5 mutant, the location of the SAM is indicated by an arrow (H and J).
We also determined the cell wall composition, using linkage analysis, of SAM samples of a csld3/+ csld5 mutant in the clv3 background. The plants exhibited retarded growth compared to the clv3 single mutant and were found to contain cell walls with a marked difference in linkage composition (Figure S5A). The relative abundance of every detectable polysaccharide changed, some dramatically increasing such as heteromannan (5-fold) and others decreasing, including HG (6-fold), arabinan (2-fold) and XG (3-fold) (Figure S5B).
Discussion
The primary cell wall plays a fundamental role in plant morphogenesis via modulation of cell shape, mechanical feedback and signaling [5, 10, 12, 13, 48, 49]. Knowing what and how this wall is made, summarized in Figure 5 [50, 51], allows us to better understand plant shoot development. The SAM uses a reduced set of GTs to make its walls compared to the rest of the plant, revealed by detailed analysis of the transcriptome, and localization of the GT mRNAs revealed different categories of expression patterns are present across the SAM and the young flowers. These expression analyses, together with antibody labeling of wall components and linkage analysis have allowed us to probe the composition of cell walls in the SAM.
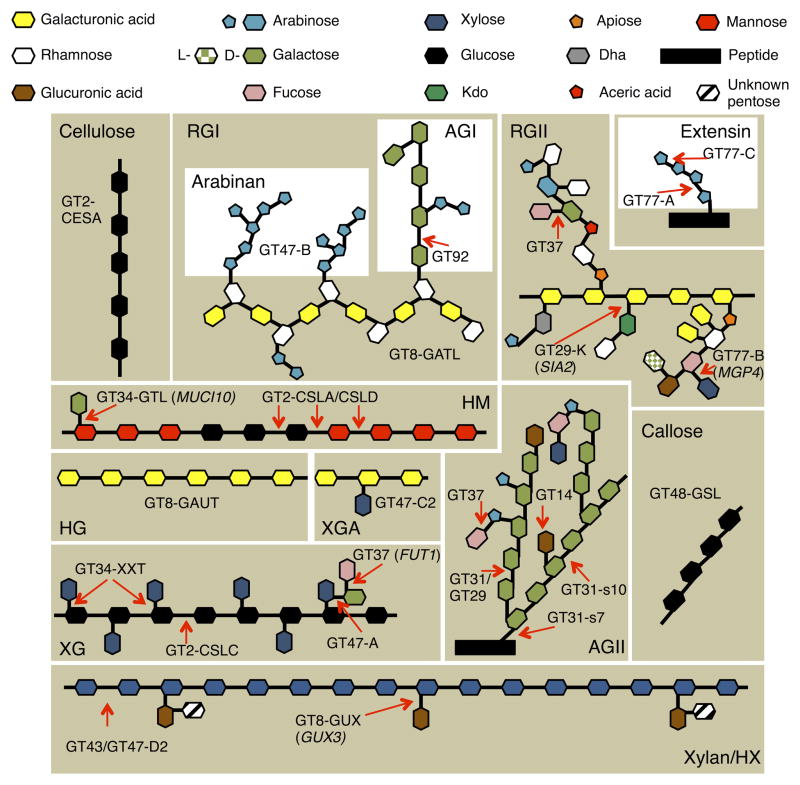
Figure 5. Graphical Representation Showing the Different Types of Polysaccharides and Their Linkages Present in the SAM Cell Wall.
Shown are GT family and subfamily assignments. Where only one candidate exists or a transcript is present >10-fold than other members of a GT family, the gene name is given in brackets. Xylan, RGI, XG and AG have diverse backbone substitution/branch patterns and the types of linkages are shown. For type II AG(P), the recently identified Arabidopsis xylosyl-terminal branch is included [50]. For xylan, the recently determined primary wall structure is shown [51]. The HM shown, as an example, is galactoglucomannan. Heteromannan (HM), rhamnogalacturanan I and II (RGI, RGII), type I and II arabinogalactan (AG), xylogalacturanan (XGA), xyloglucan (XG).
Consistent with it being the main structural component of primary cell walls, cellulose is the major polysaccharide in meristem tissues and was found at a reduced level in the SAM compared to the young flower. While the core cellulose subunits, encoded by CESA1 and CESA3, exhibit high expression in both tissues, two of the three genes encoding the third subunit are expressed predominantly in flower primordia.
Pectin is present throughout the primary walls and masks several non-cellulosic polysaccharides, as demonstrated by the requirement for pre-treatment to reveal antibody epitopes. The predominance of pectic arabinan and galactan in specific cell layers or tissues reinforce the idea that both of these polysaccharides are important as part of wall rearrangements during cellular morphogenetic events, likely attributed to their high mobility in the wall and reversible binding to cellulose [52].
For xyloglucan, the action of expansin to release the XG tethers of adjacent cellulose microfibrils has been at the heart of both cellular and tissue scale models of growth and development, especially in organ emergence from the flanks of the SAM [15]. Consistent with XG being present throughout the apical regions, we found the only confirmed gene for backbone synthesis, CSLC4, showed uniform expression. The dominance of MUR3 and several GalT genes, as well as the strong and uniform enrichment of FUT1 mRNA in the SAM suggest a high degree of galactosylation and fucosylation of XG in meristematic cells (Figure 5).
The xylan synthesis module consists predominantly of GTs encoded by IRX10L, IRX9L (but not IRX9) and IRX14/14L for backbone extension, GUX3 for (methyl)GlcA substitutions which, based on recent evidence [51], may be extended to include an unidentified pentose moiety (Figure 5). The data presented here support the IRX-LIKE genes to be associated with primary walls. Given the mostly Type 4 distribution of xylan GT transcripts, much of the deposition may occur during the de novo formation of the cross wall during cell division, and be maintained as walls mature, which might be involved in the regulation of spacing between cellulose microfibrils [51].
The localization of heteromannan in a small subset of walls that could only be detected after removal of pectin suggest that mannan is either masked by other components and modifications as cell walls mature or principally deposited early during cross wall formation and later removed. The high expression of GTL6 in the SAM, recently characterized as the GMGT MUCI10, suggests that the heteromannan in the SAM is likely galactosylated to a much higher extent than in other tissues, which might be important for proper cellulose organization [33].
Type II AG represents the glycosidic portion of AGPs that have been implicated in various aspects of plant development [53, 54]. Multiple genes from several AGP-GT families are expressed apically and their transcripts show type 1, 2 and 4 in situ patterns. The AtGALT31a mRNA was found to be particularly abundant in floral primordia and this was confirmed through live imaging of a reporter of promoter activity. The atgalt31a mutant arrests embryo development [43] and it may have an equally important role in floral organ formation but as the mutant is lethal prior to growth of the meristem, functional analysis will require targeted knockdown of the RNA late in development.
In the SAM, active cell division with intervening growth leads to the formation of leaf and flower primordia while also maintaining a pool of stem cells. Compared to animal cells that divide by forming a constriction, plants build a cell plate at the final step of cytokinesis to separate the two daughter cells. Callose and cellulose have been implicated as the major components of the cell plate, where callose decorates the nascent cross wall and is gradually replaced by cellulose as the wall matures. Callose is synthesized by members of the GSL/CALS GT48 family, exhibiting high expression in the SAM and primordia. No GSL transcripts, including GSL6 [55], were found with the Type 3 dotted pattern, suggesting no GSL enzymes are exclusive to cell plate formation. Similarly, despite the accumulation of CESA proteins on the forming cell plate [56], none of the CESA genes displayed enriched expression in dividing cells, indicating fundamental roles for callose and cellulose in both cell plate formation and wall building during post-mitotic expansion. For callose this may be limited to surrounding plasmodesmata as part of the regulatory network controlling symplastic transport [42].
Other components of the developing cell plate and their synthesis remain elusive. Antibody labelling suggests the presence of other matrix polysaccharides [57, 58], including mannan (Figure 2M), in the cell plate. Since cell plate formation is a quite transient process that usually completes in less than 60 minutes, we can speculate that a large set of GTs would be upregulated through controlled transcription and/or protein synthesis to accommodate a burst of GT enzyme activity for de novo polysaccharide synthesis during cytokinesis. Consistent with this scenario, a majority of GTs exhibited what we interpret as a cell-cycle pattern (Type 4) and suggests an important role in transcriptional reprogramming of GT genes upon cell cycle entry. The decreased expression of these GTs in non-dividing cells also indicated some transient difference in the composition and/or structure between new cross walls and mature walls, and might also imply distinct wall properties of cells within and outside of cell division. However, it is technically challenging to measure the composition of cell plates due to its transient nature and low abundance in tissues.
The expression of CSLD5 was strongly linked to cell division patterns at a time when new cross walls are laid down. CSLD5 is also found to be co-expressed with markers of the root meristem and division zone [59, 60]. Similar mitosis-enriched expression was observed for the rice homolog OsCSLD4, suggesting a conserved regulation of CSLD expression across different species [61]. The remaining shoot-expressed members, CSLD2 and CSLD3, displayed broader expression patterns across the apex. csld2 and csld3 mutants, when combined with csld5, produced plants with terminating meristems, apparently unable to maintain the size of the stem cell pool to produce more than a few flowers. It has been proposed that the CSLD gene family encodes GTs that make a type of cellulose [39] with CSLD5 making a less crystalline polysaccharide [62], however, there is also a suggestion they may make mannan [40]. A reduction in the CSLD-derived polysaccharide resulted in large changes in composition of the SAM cell wall (Figure S5) and demonstrates how changes in wall content and/or wall integrity feed back on wall biosynthesis. Based on our data, we postulate that the polysaccharide made by CSLD is important for proper cell proliferation and cell wall integrity in the SAM and is not easily compensated by the presence of other wall glycans.
In summary, we find a limited subset of GTs make the walls in the SAM. Cell wall polysaccharides, and the GTs that make them, can be uniformly distributed or focused to particular regions or cells at a given time. The phenotypes of csld mutants demonstrate a clear relationship between the cell wall and the function of the SAM for proper development. The data now allow for a targeted approach for both exploring and manipulating shoot morphogenesis.
Experimental procedures
Growth and dissection of clv3-2 tissues for RNAseq and cell wall linkage analysis
As wild type meristems were too small to obtain sufficient material for linkage analysis (see Supplemental experimental procedures), plants of the Arabidopsis thaliana clv3-2 mutant, in the Ler background [63], were grown under short day (8hr light) regimes for 4 weeks prior to transfer to long days (16 hr). Tissues (2 biological replicates of each) were collected from plants exhibiting an inflorescence stem of at least 8 cm. Enlarged SAMs were collected after careful removal of all floral organs. A razor blade was then used to remove the upper fleshy portion at the centre of the SAM (Figure 1C, upper panel) and immediately frozen in liquid N2. Early stage flowers (stage 6–7) at the periphery of the SAM were collected using fine tweezers and immediately frozen (Figure 1C, bottom panel). Tissues for the clv3-2 whole plant sample for RNA-Seq (known as the “pooled sample”) consisted of fruit (stage 17), young leaves, old leaves, roots, stem, whole inflorescence and flower (stage 15). The tissues were harvested and stored separately.
RNA-Seq sample preparation
RNA was extracted using the RNEasy plant kit (Qiagen) for each biological replicate according to the manufacturer’s protocol. For the whole plant samples, 80 ng of each sample was subsequently pooled before library making. Libraries were prepared from 500 ng total RNA using the TrueSeq Stranded Total RNA with Ribo-Zero Plant kit (Illumina).
Quantitative Reverse Transcription PCR (qRT-PCR)
For qRT-PCR analysis, total RNAs were extracted from dissected meristem, young flowers and shoot tissues using RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA (2 μg) was reverse-transcribed into cDNA using oligo(dT) primer and Transcriptor High Fidelity cDNA Synthesis Kit (Roche). The cDNA was used as templates for qRT-PCR using LightCycler 480 SYBR Green I Master (Roche) and gene specific primers (Table S4).
Sequence analysis
The six RNA-seq libraries were sent for sequencing at the Beijing Genome Institute (BGI, Hong Kong) using one full lane of Illumina HiSeq2000. The raw reads in FastQ format were obtained and analyzed in-house. We first assessed the quality of reads using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Potential adaptor contamination and low quality trailing sequences were removed using Trimmomatic [64]. Preprocessed reads were then mapped to Ler-0 reference genome [65] using Tophat [66], and possible optical duplicates from PCR during the library preparation step were then removed using the Picard tools (http://picard.sourceforge.net). The relative transcript estimation was carried out using Cufflinks [67] to obtain the fragments per kilobase of transcript per million fragments mapped (FPKM) values, and converted to Transcript Per Million (TPM) [68], and also as raw reads using HTSeq [69]. Normalization of read counts, gene ontology analysis and generation of a pairwise correlation coefficient network are described in Supplemental experimental procedures.
Polysaccharide linkage analysis
Alcohol insoluble residues (AIRs) were prepared from isolated SAMs and young flowers, and used in the determination of both neutral and acidic monosaccharide linkage composition and polysaccharide composition as previously described by Pettolino et al., (2012) [21]. For comparison with csld, crosses between clv3-2 and csld3 csld5 produced F2 plants where csld3 csld5 clv3 were not viable beyond early vegetative stage and therefore plants genotyped as CSLD3/csld3 csld5 clv3, that exhibited retarded growth, were used to generate SAM AIR for linkage analysis.
RNA in situ hybridizations of wax-embedded SAM sections
To generate gene-specific probes, cDNA fragments corresponding to each GT gene were amplified using gene-specific primers (Table S4) and ligated into the pGEM®-T Easy vector (Promega). The constructs were verified by sequencing and then used as templates for in vitro transcription using the DIG RNA Labeling Kit (Roche). Shoot apices of Arabidopsis thaliana wild type Col-0 or clv3-9 (a gift from Rüdiger Simon, Heinrich-Heine-University, Düsseldorf, Germany) were fixed in FAA (formaldehyde, acetic acid, ethanol), embedded in wax and cut into 8 μm sections. The sections were processed as described in (http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf). Briefly, after dewaxing, rehydration and dehydration, the sections were hybridized with gene-specific probes, and then incubated with anti-digoxigenin antibody (Roche). The signals were detected by the colour reaction after application of NBT/BCIP (Roche). Sense controls, yielding no hybridization with target mRNA, are shown in Figure S3.
Two-colour fluorescent in situ hybridization was used for gene co-expression analysis and the protocol is described, in detail, in Supplementary experimental procedures.
Immunohistochemistry of wall components
Wax sections generated for in situ hybridizations were used for antibody detection of wall polymers. Sections were mounted on polished slides and dewaxed using xylene followed by rehydration using an ethanol series 100% (v/v), 95%, 70%, 50%, 30%, 10%. Sections were then incubated in buffer (20 mM Tris-HCl pH8.2, 0.5mM CaCl2, 150 mM NaCl) for 20 minutes, followed by blocking in 0.5% w/v milk powder in buffer. Primary antibody, diluted in buffer, were added to sections and incubated overnight. After washes in buffer, secondary antibody incubations were carried out for 3 hours. Primary antibodies were rabbit PDM anti-mannan ([70], gift from Paul Dupree, University of Cambridge), LM28 rat anti-xylan [71] (gift from Paul Dupree), LM24 rat anti-xyloglucan [72], JIM7 rat anti-homogalacturonan [73], LM13 rat anti-arabinan [74], LM5 rat anti-galactan [75] (Plantprobes). Secondary antibodies were anti-rabbit IGG CF488A conjugate (Sigma SAB4600030), anti-rat IGG CF568 conjugate (Sigma SAB4600077) and anti-rat Alexa Fluor 647 (Life Technologies). Crystalline cellulose was detected by CBM3a [76] together with anti-his FITC secondary antibody. For enzyme treatments, sections were incubated with pectolyase (0.1% w/v, Sigma P5936) in incubation buffer (0.2M Na2HPO4, 0.1M citric acid, pH4.8) prior to primary antibody incubation. Labelled sections were mounted in ProLong Gold Antifade Mountant (Life Technologies) with a coverslip and sealed. Images were taken with a Zeiss LSM700 confocal equipped with a 20x 0.8NA dry objective.
Live cell imaging of an At1g32930 reporter and GFP-GATL6
Plants were partially dissected to remove overhanging flowers obscuring the SAM.
Live cell imaging was carried out on a Zeiss LSM700 confocal microscope equipped with 488nm and 555nm lasers and a 20x NA 1.0 water dipping objective.
Observing cell boundaries for segmentation and growth analyses in csld mutants
The csld2 csld5, csld3 csld5 and csld2 csld3 csld5 mutants were a gift from Henrik Scheller (Berkeley) and their construction have been reported by Yin et al. [40]. To view the SAM, large organs were dissected and the apex stained with the dye FM4-64 (Life Technologies, 333 ug ml-1) for 3 minutes and then carefully rinsed in water. SAMs were viewed under a Zeiss LSM700 confocal with water dipping objective where z-stacks were obtained and 3D rendered using the confocal software. For growth analyses, the acylated YFP plasma membrane marker was transformed in to heterozygous plants and double mutants identified after selection. Whole plantlets were transferred shortly after bolting to imaging boxes containing 1% w/v agar and 2.2g l−1 Murashige and Skoog (MS) medium containing Gamborg B5 vitamins (Duchefa). Confocal imaging of the YFP reporter was then carried out at two time points separated by 24hrs followed by segmentation and cell indexing using MorphographX [77] where a heat map of relative growth was selected as output upon the 3D image of the 24hr time point.
Data Availability
The sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database, accession number SRP072228.
Supplementary Material
Acknowledgments
The authors wish to thank Zachary Nimchuck for advice with reporter construction, Arun Sampathkumar for help with 3D visualization of the GALT31A reporter and Lisa Willis for help with spherical cap calculations. V.C. is in receipt of a Thailand Research Fund (TRF) grant for New Researcher (Grant Number TRG5880067), and a Research Supplement grant from Faculty of Science, Mahidol University. CB, MSD and AB acknowledge the support of the ARC Centre of Excellence in Plant Cell Walls, Australia (Grant Number CE110001007). EMM acknowledges support from the Gatsby Charitable Trust through Fellowships GAT3272/C and GAT3273-PR1 and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3406). AP acknowledges support of the EU Marie-Curie FP7 COFUND People Programme through the award of an AgreenSkills grant no. 267196. RW acknowledges support from the Leverhulme Trust (Grant RPG-2015-285).
Footnotes
Author contribution.
WY and RW generated the cell wall preparations and carried out morphological analysis of the csld mutants. WY carried out the RNA in situ hybridizations with the help of CS and RW. CS prepared the RNA-Seq libraries and GO analysis. CS and VC processed the RNA-Seq data. CTB, AB and MSD carried out the linkage analysis and calculated the polysaccharide composition. AP devised and performed the cell wall immuno-labelling protocols. RW and MSD carried out the literature searches, identified meristem glycosyltransferases and amalgamated the chemical and expression data. WY and RW prepared figures. RW, MSD, EM and AB devised the project and wrote the manuscript with contributions from the remaining authors.
References
- 1.Doblin MS, Pettolino F, Bacic A. Evans Review: Plant cell walls: the skeleton of the plant world. Funct Plant Biol. 2010;37:357. [Google Scholar]
- 2.Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–61. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 3.Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306:2206–11. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 4.Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. Arabidopsis - A powerful model system for plant cell wall research. Plant J. 2010;61:1107–1121. doi: 10.1111/j.1365-313X.2010.04161.x. [DOI] [PubMed] [Google Scholar]
- 5.Szymanski DB, Cosgrove DJ. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol. 2009;19:800–11. doi: 10.1016/j.cub.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 6.North HM, Berger A, Saez-Aguayo S, Ralet MC. Understanding polysaccharide production and properties using seed coat mutants: future perspectives for the exploitation of natural variants. Ann Bot. 2014;114:1251–63. doi: 10.1093/aob/mcu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwiatkowska D, Dumais J. Growth and morphogenesis at the vegetative shoot apex of Anagallis arvensis L. J Exp Bot. 2003;54:1585–95. doi: 10.1093/jxb/erg166. [DOI] [PubMed] [Google Scholar]
- 8.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–18. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012;22:360–7. doi: 10.1016/j.cub.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–5. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- 11.Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010;8:e1000516. doi: 10.1371/journal.pbio.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS. Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science. 2012;335:1096–9. doi: 10.1126/science.1213100. [DOI] [PubMed] [Google Scholar]
- 13.Milani P, Gholamirad M, Traas J, Arnéodo A, Boudaoud A, Argoul F, Hamant O. In vivo analysis of local wall stiffness at the shoot apical meristem in Arabidopsis using atomic force microscopy. Plant J. 2011;67:1116–23. doi: 10.1111/j.1365-313X.2011.04649.x. [DOI] [PubMed] [Google Scholar]
- 14.Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011;21:1720–6. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 15.Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of Leaf Primordia by the Cell Wall Protein Expansin. Science. 1997;276:1415–1418. [Google Scholar]
- 16.Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol. 2008;18:1943–8. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- 17.Verica JA, Medford JI. Modified MERI5 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci. 1997;125:201–210. [Google Scholar]
- 18.Priestley JH. Cell growth and cell division in the shoot of the flowering plant. New Phytol. 1929;28:54–81. [Google Scholar]
- 19.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- 20.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–67. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettolino FA, Walsh C, Fincher GB, Bacic A. Determining the polysaccharide composition of plant cell walls. Nat Protoc. 2012;7:1590–607. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- 22.Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 2013;74:423–34. doi: 10.1111/tpj.12135. [DOI] [PubMed] [Google Scholar]
- 23.Lee C, Teng Q, Zhong R, Ye ZH. Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol. 2012;53:1204–16. doi: 10.1093/pcp/pcs064. [DOI] [PubMed] [Google Scholar]
- 24.Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A. 2010;107:17409–14. doi: 10.1073/pnas.1005456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104:15572–7. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci U S A. 2006;103:5236–41. doi: 10.1073/pnas.0600120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, Orlando R, Scheller HV, Mohnen D. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci U S A. 2011;108:20225–30. doi: 10.1073/pnas.1112816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci U S A. 2007;104:8550–5. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou YH, Pogorelko G, Young ZT, Zabotina OA. Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol. 2015;56:255–67. doi: 10.1093/pcp/pcu161. [DOI] [PubMed] [Google Scholar]
- 30.Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant. 2012;5:984–92. doi: 10.1093/mp/sss032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ. Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol. 2012;195:585–95. doi: 10.1111/j.1469-8137.2012.04196.x. [DOI] [PubMed] [Google Scholar]
- 32.Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- 33.Voiniciuc C, Schmidt MH-W, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Guenl M. MUCI10 Produces Galactoglucomannan That Maintains Pectin and Cellulose Architecture in Arabidopsis Seed Mucilage. Plant Physiol. 2015:15.00851. doi: 10.1104/pp.15.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009;60:527–38. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed] [Google Scholar]
- 35.Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52:1154–68. doi: 10.1111/j.1365-313X.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- 36.Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009;57:732–46. doi: 10.1111/j.1365-313X.2008.03729.x. [DOI] [PubMed] [Google Scholar]
- 37.De Bodt S, Inzé D. A guide to CORNET for the construction of coexpression and protein-protein interaction networks. Methods Mol Biol. 2013;1011:327–43. doi: 10.1007/978-1-62703-414-2_26. [DOI] [PubMed] [Google Scholar]
- 38.Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG. Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol. 2011;155:1791–805. doi: 10.1104/pp.110.163220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol. 2011;13:973–80. doi: 10.1038/ncb2294. [DOI] [PubMed] [Google Scholar]
- 40.Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WGT, et al. The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant. 2011;4:1024–37. doi: 10.1093/mp/ssr026. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Wang L, Chen C, Xiong G, Tan XY, Yang KZ, Wang ZC, Zhou Y, Ye D, Chen LQ. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J Exp Bot. 2011;62:5161–77. doi: 10.1093/jxb/err221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21:1144–55. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T, et al. A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 2013;76:128–37. doi: 10.1111/tpj.12281. [DOI] [PubMed] [Google Scholar]
- 44.Fleming AJ, Mandel T, Roth I, Kuhlemeier C. The patterns of gene expression in the tomato shoot apical meristem. Plant Cell. 1993;5:297–309. doi: 10.1105/tpc.5.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koning AJ, Tanimoto EY, Kiehne K, Rost T, Comai L. Cell-specific expression of plant histone H2A genes. Plant Cell. 1991;3:657–65. doi: 10.1105/tpc.3.7.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV. Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012;24:5024–36. doi: 10.1105/tpc.112.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dilokpimol A, Poulsen CP, Vereb G, Kaneko S, Schulz A, Geshi N. Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol. 2014;14:90. doi: 10.1186/1471-2229-14-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jönsson H, Meyerowitz EM. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. Elife. 2014;3:e01967. doi: 10.7554/eLife.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seifert GJ, Blaukopf C. Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 2010;153:467–78. doi: 10.1104/pp.110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tryfona T, Theys TE, Wagner T, Stott K, Keegstra K, Dupree P. Characterisation of FUT4 and FUT6 α-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS One. 2014;9:e93291. doi: 10.1371/journal.pone.0093291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortimer JC, Faria-Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, Zhang Z, Stott K, Anders N, Dupree P. An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant J. 2015;83:413–426. doi: 10.1111/tpj.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin D, Lopez-Sanchez P, Gidley MJ. Interactions of pectins with cellulose during its synthesis in the absence of calcium. Food Hydrocoll. 2016;52:57–68. [Google Scholar]
- 53.Majewska-Sawka A. The Multiple Roles of Arabinogalactan Proteins in Plant Development. Plant Physiol. 2000;122:3–10. doi: 10.1104/pp.122.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson KL, Kibble NAJ, Bacic A, Schultz CJ. A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS One. 2011;6:e25154. doi: 10.1371/journal.pone.0025154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong Z, Delauney AJ, Verma DP. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell. 2001;13:755–68. doi: 10.1105/tpc.13.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miart F, Desprez T, Biot E, Morin H, Belcram K, Höfte H, Gonneau M, Vernhettes S. Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 2014;77:71–84. doi: 10.1111/tpj.12362. [DOI] [PubMed] [Google Scholar]
- 57.Moore PJ, Staehelin LA. Immunogold localization of the cell-wall-matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis inTrifolium pratense L.; implication for secretory pathways. Planta. 1988;174:433–45. doi: 10.1007/BF00634471. [DOI] [PubMed] [Google Scholar]
- 58.Northcote DH, Davey R, Lay J. Use of antisera to localize callose, xylan and arabinogalactan in the cell-plate, primary and secondary walls of plant cells. Planta. 1989;178:353–66. doi: 10.1007/BF00391863. [DOI] [PubMed] [Google Scholar]
- 59.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 60.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–63. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 61.Yoshikawa T, Eiguchi M, Hibara KI, Ito JI, Nagato Y. Rice slender leaf 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot. 2013;64:2049–61. doi: 10.1093/jxb/ert060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller HV, et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007;52:791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- 63.Bowman J. Arabidopsis: an atlas of morphology and development. New York: Springer-Verlag; 1994. [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–23. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pachter L. Models for transcript quantification from RNA-Seq. 2011 ArXiv e-prints 1104.3889. [Google Scholar]
- 69.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P. Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta. 2003;218:27–36. doi: 10.1007/s00425-003-1073-9. [DOI] [PubMed] [Google Scholar]
- 71.Cornuault V, Buffetto F, Rydahl MG, Marcus SE, Torode TA, Xue J, Crépeau MJ, Faria-Blanc N, Willats WGT, Dupree P, et al. Monoclonal antibodies indicate low-abundance links between heteroxylan and other glycans of plant cell walls. Planta. 2015;242:1321–34. doi: 10.1007/s00425-015-2375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MCF, et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem. 2012;287:39429–38. doi: 10.1074/jbc.M112.396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knox JP, Linstead PJ, King J, Cooper C, Roberts K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta. 1990;181:512–21. doi: 10.1007/BF00193004. [DOI] [PubMed] [Google Scholar]
- 74.Verhertbruggen Y, Marcus SE, Haeger A, Verhoef R, Schols HA, McCleary BV, McKee L, Gilbert HJ, Knox JP. Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 2009;59:413–25. doi: 10.1111/j.1365-313X.2009.03876.x. [DOI] [PubMed] [Google Scholar]
- 75.Jones L, Seymour GB, Knox JP. Localization of Pectic Galactan in Tomato Cell Walls Using a Monoclonal Antibody Specific to (1[->]4)-[beta]-D-Galactan. Plant Physiol. 1997;113:1405–1412. doi: 10.1104/pp.113.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281:29321–9. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- 77.de Reuille PB, Robinson S, Smith RS. Quantifying cell shape and gene expression in the shoot apical meristem using MorphoGraphX. Methods Mol Biol. 2014;1080:121–34. doi: 10.1007/978-1-62703-643-6_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database, accession number SRP072228.